Abstract
Large-scale sequencing projects have revealed an unexpected complexity in the origins, structures and functions of mammalian transcripts. Many loci are known to produce overlapping coding and non-coding RNAs with capped 5′ ends that vary in size. Methods that identify the 5′ ends of transcripts will facilitate the discovery of novel promoters and 5′ ends derived from secondary capping events. Such methods often require high input amounts of RNA not obtainable from highly refined samples such as tissue microdissections and subcellular fractions. Therefore, we have developed nanoCAGE (Cap Analysis of Gene Expression), a method that captures the 5′ ends of transcripts from as little as 10 nanograms of total RNA and CAGEscan, a mate-pair adaptation of nanoCAGE that captures the transcript 5′ ends linked to a downstream region. Both of these methods allow further annotation-agnostic studies of the complex human transcriptome.
Introduction
Analysis of the mammalian transcriptome and transcriptional network in ex vivo cells requires technologies that provide a comprehensive and unbiased view of the tissue-specific promotome (the complete set of promoters) from small amounts of RNA and the intron-exon structure of the transcripts associated with different transcription start sites (TSSs), which are marked by a cap-site in most eukaryotic RNA polymerase II-derived RNAs.
Among sequencing-based techniques to measure gene expression, tag-based methods are common. They involve reading a short sequence of a transcript that is still long enough to be mapped onto the genome. We have used Cap Analysis Gene Expression (CAGE)1–3, a cap-trapping based method which allows for systematic 5′ end profiling of capped RNAs, for the first comprehensive single-base resolution maps of TSS and promoters from human and mouse tissues4 and for deciphering transcriptional networks in the human leukemia cell line THP-15. Such large-scale characterization of TSSs showed an unprecedented complexity of the transcriptome. In contrast to classic gene models, the emerging view suggests that most genes have multiple TSSs differing by multiple bases4 and driven by various core promoters and that newly capped 5′ ends can also be created post-transcriptionaly6. Transcription can be initiated by promoters that are broad in shape, often associated with CpG islands, or by sharp promoters, which are narrow in shape and are often associated with TATA-boxes4. These promoter structures have functional implications, being associated to tissue specificity, as for example sharp promoters are, different exon usages, translation initiation sites or classes of non-coding RNAs (ncRNAs). Within the locus of a coding gene, transcription can start within and downstream of the open reading frame such as for the non-coding RNAs that can originate in genomic regions corresponding to the 3′ ends of protein coding geness4. Additionally, the capped transcriptome includes non-coding RNAs that are associated with initiation and termination of transcription6,7.
However, there are outstanding problems that could not yet be addressed with the existing technologies. CAGE requires a large quantity of starting material (~50μg of total RNA) precluding TSS transcriptome analysis of small samples, such as homogeneous cells preparation after microdissection or samples derived from cellular sub-fractionation.
Furthermore, newly identified promoters must be assigned to gene models. Although CAGE identifies new promoters, determining their connection to either downstream known gene structures or to independent novel RNAs is limited to low-throughput gene-by-gene validations. RNA shotgun sequencing approaches (RNA-seq) have been unable to distinguish multiple 5′ ends of a given gene, identifying only their most extreme boundaries at best. This constrains the functional annotation of promoters, from which accurate inference of transcriptional regulatory networks depends5 and limits the study of ncRNAs overlapping known genes. Paired-end sequencing of full-length cDNA, like the GIS (Gene Identification Signature) ditag approach8, allows for the determination of TSS and termination sites in polyadenylated mRNAs, but does not yield information on internal exons. In addition it requires large quantities of purified mRNAs.
Here we present nanoCAGE and CAGEscan technologies, which provide a genome-wide profiling of TSSs from small quantities of RNA and link them to the anatomy of transcribed RNAs. nanoCAGE was carried out with as little as 10 ng of total RNA, the equivalent of the RNA content of a thousand cells. CAGEscan provided important insights on the complexity of the promotome-transcript structure, identifying among others, RNAs that originate from a given TSS but terminate in unrelated downstream genes. Our data also provide an estimate of RNA types that populate the various cell compartments, suggesting a nuclear role for intron- and intergenic regions-derived RNAs, as well as for retrotransposon elements and antisense RNAs.
Results
Nanogram-scale RNA profiling with nanoCAGE and CAGEscan
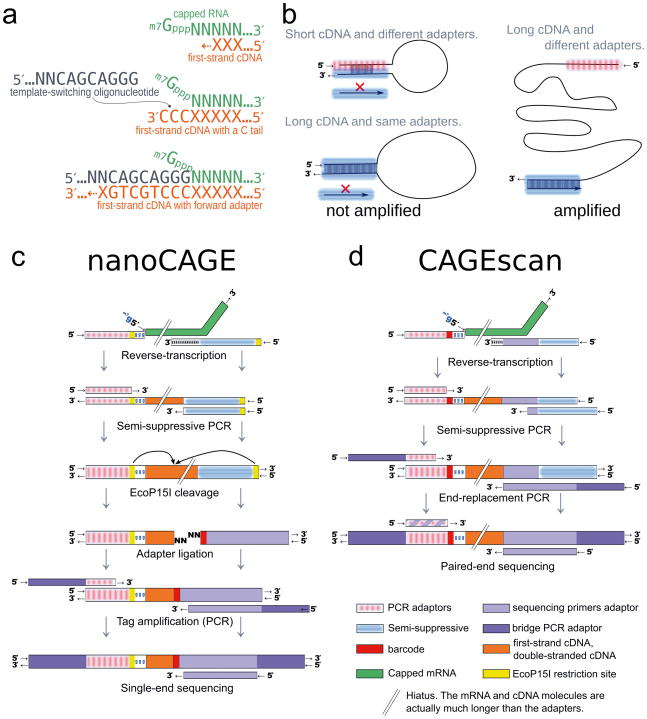
The classic CAGE protocol consists of many biochemical processing steps2,9, whereas nanoCAGE takes advantage of a peculiar property of reverse transcriptase, called “template switching”10 to select 5′ ends of capped transcripts. Template switching (TS) exploits the ability of the reverse-transcriptase to extend the cDNA using the mRNA’s cap as a template: the resulting synthesized first strand cDNA carries one to three C nucleotides that correspond to the cap structure11,12. These Cs hybridize to the ribo-G at the 3′ end of a template switching oligonucleotide (Fig. 1a). The reverse transcriptase extends cDNA polymerization using the TS oligonucleotide as template, providing extra 3′ sequence to the first-strand cDNA (Fig. 1a). which is used to prime second-strand cDNA synthesis. Although TS has been observed for blunt DNA/RNA hybrids10, we show that its efficiency on capped RNAs is far greater, therefore preferentially capturing capped, full-length transcripts (Fig. 2). TS does not require purification steps, and thus avoids loss of material.
Figure 1. Experimental outline of the nanoCAGE and CAGEscan protocols.
(a) nanoCAGE captures the 5′ ends of molecules by template switching. When polymerizing the cDNA of a capped mRNA, the reverse transcriptase adds extra cytosines that are complementary to the cap. Thus each 5′ full-length cDNAs is extended upon hybridization of the riboguanosine-tailed “template-switching” oligonucleotides to these extra cytosines. (b) In the semi-suppressive PCR, the short templates fold intramolecularly and prevent the binding of primers which precludes amplification; longer molecules are less likely to fold and are thus amplified. Templates derived from reaction artifacts form stable homo-duplexes also precluding amplification. (c) Preparation of nanoCAGE tags. After template-switching, semi-suppressive PCR and EcoP151 cleavage, 25 bp are ligated to oligonucleotide adapters that contain a sequence identifier (red box). After PCR amplification, the nanoCAGE tags are subjected to sequencing by synthesis. (d) Preparation of 5′-full-length cDNA libraries for paired-end sequencing with the CAGEscan protocol. Capped mRNAs capture is similar to a. The ends of the amplified cDNA constructs are replaced by PCR with adapters for sequencing in the Illumina Genome Analyzer, that produces paired-end reads from single cDNAs.
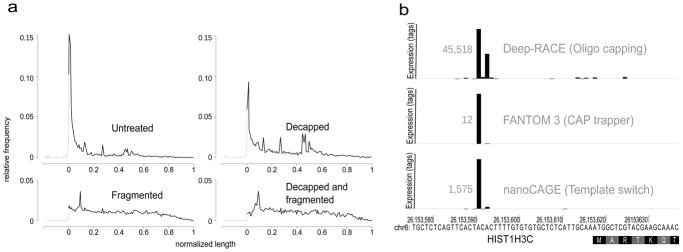
Figure 2. nanoCAGE specifically captures capped 5′ ends.
(a) nanoCAGE detects 5′ ends of capped RNA molecules. The relative frequency of CAGE tags over all RefSeq transcript models was plotted on a compound scale going from 500 bp to the start of the RefSeq (in gray) and then from 0% to 100% of the RefSeq (in black). Decapping a sample results in decreasing the prevalence of tags representing the 5′ end. Combination of decapping and fragmentation completely abolishes the detection of 5′ ends.
(b) Three independent methods of 5′ end capture, respectively based on oligo-capping, CAP trapper and template switching, detect same 5′ ends as exemplified here for the histone gene HIST1H3C, represented on a horizontal axis. The RefSeq model starts with the coding sequence at position 26,153,618 of the chromosome 6. TSSs are represented by vertical bars proportional to the number of tags they contain. The size of the highest bar is normalized for all three experiments and its expression value is written in gray at its left side.
To target non-coding, non-polyadenylated RNAs (poly-A−) and RNAs whose 3′ end has been truncated during the isolation of specific cells from the tissue of interest, we developed conditions to allow random-priming of the reverse-transcription (RT) reaction in combination with 5′ template-switching. Due to DNA-dependent polymerase activity of the reverse-transcriptase, annealing of TS oligonucleotides and RT primers to each other generates small artefactual DNA fragments that become the predominant PCR templates in subsequent steps, impairing libraries preparation (not shown). The prevalence of these artifacts has, so far, precluded the development of random primer-based cap-switch methods for whole transcriptome analysis with total RNA. To overcome this problem, we designed a “semi-suppressive PCR” method, in which the linkers at the 5′ and 3′ ends of the cDNA carry similar (but not identical) complementary sequences (Fig. 1b, Supplementary Fig. 1). Consequently, DNA templates bearing the same ending sequences (such as non-oriented cDNAs carrying two 5′ or two 3′ linkers) or small size templates (such as primer-derived artifacts) are less efficiently amplified during PCR. As a result, the majority of PCR products consist of long cDNAs properly flanked by the adapter sequences present in the TS oligonucleotide and RT primer, as they are the most efficiently amplified templates (Fig. 1b). Additionally, the removal of primer dimers by the semi-suppressive PCR enables the use of TS primer in concentrations as high as 10μM. This maximizes the efficiency of template switching since the concentration of TS oligonucleotides is the reaction-limiting factor (data not shown). The ability to use random primers considerably extends the power of this technique since (i) poly-A− transcripts constitute at least one third of the transcriptome13, (ii) long polyadenylated RNAs are often damaged by ex vivo sample preparation, such as laser capture micro-dissection of fixed tissues and (iii) PCR of oligo-dT primed cDNAs introduces strong size and representational biases regardless of potential RNA degradation.
TS oligonucleotides and RT primers used in the nanoCAGE protocol contain EcoP15I restriction sites14 to systematically generate 25 bp fragments corresponding to the 5′ end of the template-switched captured cDNAs, thus producing nanoCAGE tag libraries (Fig. 1c). Although this enzymatic cleavage might be dispensable when reading short reads with several second-generation sequencers, the standardization of tag length overcomes biases during the second round PCR and simplifies DNA molar quantification and sequencing. Additionally, EcoP15I tagging allows the introduction of a DNA sequence “barcode” at the 3′ end (Fig. 1c) and thus pooling of different libraries prior to their sequencing, resulting in dramatic cost savings15.
CAGEscan was built upon nanoCAGE, but modified to accommodate paired-end sequencing for TSS determination at 5′ ends coupled with 3′ end sequencing of cDNAs at random priming sites. Rather than cleaving the cDNAs, in CAGEscan we added adapter sequences allowing for paired-end sequencing in the Illumina Genome Analyzers16 (Fig. 1d). Thus, CAGEscan yields collections of 3′ end reads “scanning” transcripts defined by their common 5′ end, as obtained by sequencing of both the 5′ end and the 3′ end of the template-switched captured cDNAs. Yet, unlike nanoCAGE libraries that contain uniformly short sequences, CAGEscan libraries show a broader size range including fragments longer than 1 kb, which perform poorly on currently available second-generation sequencing platforms17. This problem is minimized by exclusively using highly concentrated random primers and commercial reverse transcriptases, which show little strand displacement activity. Thus, CAGEscan sequencing templates are kept relatively short, regardless the length of the original mRNA molecules.
Due to the selectivity of the template switch for capped molecules, both protocols were used on total RNA, without ribosomal RNAs (rRNA) depletion. Notably, the usage of 3′ random primer allows the detection of non-coding, non-polyadenylated RNAs13, which have been so far poorly characterized.
Reproducibility, efficiency and precision of nanoCAGE
In order to validate the reproducibility, the efficiency and the precision of nanoCAGE, we prepared libraries from serially diluted total RNA from cultured hepatocellular carcinoma cells (Hep G2) and compared them to reference TSS data.
Two duplicate sets of nanoCAGE libraries from 10, 50, 250 and 1,250 nanograms of total RNA were synthesized and sequenced with an Illumina Genome Analyzer. Extracted tags were aligned to the human genome (NCBI Build 36.1)18. We clustered TSS from all the libraries that were located less than 20 bp apart on the same genome strand4. Clusters separated by less than 400 bp were grouped in promoter regions5. We then compared expression levels, measured as number of tags per promoter region in a given library, for each pair of replicates with the same quantity of RNA. The Pearson correlation coefficients between replicas were 0.97, 0.96, 0.97 and 0.99 for, respectively, 10, 50, 250 and 1,250 ng. Replicates sequencing depth were in some cases substantially different (Supplementary Table 1) and higher correlations were observed for deeper sequenced libraries (0.99 for 1250 ng replicas). Satisfactory reproducibility was demonstrated for all RNA concentrations tested. We then pooled the tags into virtual libraries for each RNA quantity, and compared them with each other. Pearson correlation coefficients varied between 0.987 to 0.999 (Supplementary Table 2), showing similar snapshots of the transcriptome with tiny quantities of starting total RNA within a range of 10 to 1,250 ng.
A similar template-switching approach has been used with fragmented, uncapped RNA molecules19 showing that TS could also be used on uncapped 5′ ends. However, this protocol required prior depletion of ribosomal-RNA otherwise reverse-transcription of total RNA with random hexamers yielded 90–94% of ribosomal sequences20 In our hands, only 11% of Hep G2 nanoCAGE tags matched rRNA sequences, showing an 8-fold reduction in non-capped rRNA content. This demonstrates the strong preference of template-switching for capped over non-capped RNAs. To prove efficient capture of the 5′ ends of capped transcripts, we prepared nanoCAGE libraries from 100 ng of decapped, fragmented or both decapped and fragmented total RNA and analyzed the distribution of tags mapping to RefSeq transcript models21. In a library prepared with untreated RNA, 52% of the tags mapped to first exons or proximal promoters in RefSeq (defined as 500 bp to RefSeq TSS) and 31% detected potentially new promoters in intergenic regions (Fig. 2a, Supplementary Fig. 2 and Supplementary Table 3). We noted that tags mapping to internal exons and 3′ UTRs (15% in total) can also derive from genuinely capped transcripts co-localized within the boundaries of longer transcripts referenced by RefSeq4,6,9. The proportion of tags matching the 5′ end of known transcripts was halved to 23% after RNA decapping. Furthermore, this number dropped to 8.5% upon RNA fragmentation, suggesting that a large number of uncapped RNA molecules is needed to compete with capped molecules. Upon combining decapping and fragmentation, the preferential capture of 5′ ends was almost completely abolished, demonstrating that nanoCAGE distinguishes capped ends from other 5′ ends and preferentially captures the 5′ end of capped transcripts. The semi-suppressive PCR did not impair the detection of relatively short transcripts, as we detected expression for 78% of RefSeq transcripts (23,512/29,996), including 44% (271/615) of the subset shorter than 250 bp. In that respect NanoCAGE tags outperform the FANTOM3 dataset, in which only 5% (28/615) of the short RefSeq transcripts are detected (Supplementary Table 1). Furthermore, the EcoP15I cleavage did not introduce any bias as we found the CAGCAG EcoP151 restriction site in 81% of the detected transcripts for both the nanoCAGE and the FANTOM3 libraries, which were made with a different restriction enzyme, MmeI1 (see also Supplementary Fig. 3).
To confirm the precision of template-switching in detecting TSS we compared promoters identified by nanoCAGE with those found by two methods that are using different protocols for cap selection and for which Hep G2 libraries were available4,22: Deep-RACE22 (Rapid Amplification of cDNA Ends), based on oligo-capping23, and CAGE, 4. The Deep-RACE data was limited to 18 different promoters in 17 loci22. As exemplified with histone cluster 1, H3 (HIST1H3C, Fig. 2b), the main TSS was the same between nanoCAGE and Deep-RACE or FANTOM3 CAGE data in four and seven cases respectively. The HIST1H3C locus also exemplifies the ability of our random-primed approach to uncover TSSs of non-polyadenylated transcripts (HIST1H3C RefSeq model lacks any 5′ UTR information). When allowing only 4 bp discrepancy between the TSS uncovered by each methodology, the results of nanoCAGE were in agreement with Deep-RACE and FANTOM3 CAGE for 11 out of 18 promoters (Supplementary Table 4) and for 17 of the 18 Deep-RACE-validated TSS respectively. Interestingly, the two alternative promoters of PPP2R4 uncovered by Deep-RACE and CAGE were also detected by nanoCAGE and their relative differential expression levels were consistent between all three approaches (data not shown). To extend this result we compared the location of all the promoters detected by the two genome-wide libraries, nanoCAGE and CAGE. Although a large number of promoters are broad in size4, for 66% of the promoters the distance between TSS detected by both techniques was less than 5 bp (Supplementary Fig. 4, Supplementary Table 5).
Even for cells grown in culture, starting material becomes a limiting factor when cellular sub-compartments are selectively fractionated to explore specific RNA content. As part of the ENCODE project, attempts to produce CAGE libraries from nuclear RNA subfractions of the K562 myelogenous leukemia cell line (the nucleolus, the nucleoplasm, the chromatin-bound RNAs as well as from polysomal poly-A− RNA consisting mostly of rRNA) were unsuccessful due to the paucity of mRNA (not shown). Using nanoCAGE, four libraries were synthesized and between 9.5 and 13.8 million tags were sequenced for each of them. Comparing to standard poly-A− CAGE libraries, which were sequenced at the same depth, the complexity of detected 5′ ends was consistent between the two technologies for each cellular compartment (Supplementary Fig. 5). We have also found differences in TSS specificity among different compartments (not shown).
Promotome-transcriptome architecture using CAGEscan
The functional significance of novel 5′ ends is limited by the lack of information on the entire transcript. To better understand the structure of the transcripts associated with novel TSS and to better characterize the differences between nuclear and cytoplasmic transcriptomes, CAGEscan libraries were prepared in technical duplicates from four different Hep G2 cultures. In a first series of experiments, nuclear and cytoplasmic fractions were analyzed either as total RNA or as a subfraction depleted of polyadenylated transcripts (poly-A−) (Supplementary Table 1).
CAGEscan mate pair sequences were aligned against the human genome (NCBI build 36.1). The poly-A− cytoplasmic, poly-A− nuclear, total cytoplasmic and total nuclear fractions yielded together a total of 2,109,392 unique paired-end tags (Supplementary Table 6). Each of these associated a TSS to a downstream sequence in a random location. Selecting mate pairs starting within 50 bp of RefSeq transcript’s TSS and using their intron-exon structure, we estimated the length of the RNA from which mate pairs were derived. The resulting median length was 449 bp (1st and 3rd quartile: 304 and 693 bp, Supplementary Fig. 6). In comparison, the median length of RefSeq transcript models is 2,422 bp (1st and 3rd quartile: 1,509 and 3,799 bp), thus suggesting the majority of RNA Pol II transcripts are competent to produce CAGEscan mate pairs, although CAGEscan is not optimal to map the 3′ ends of transcript.
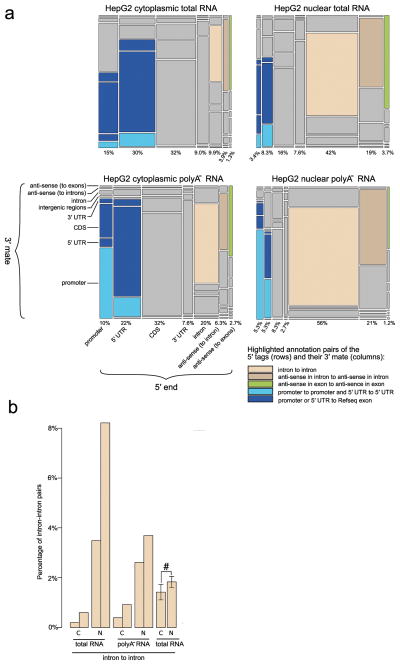
CAGEscan allows the association of TSSs detected by CAGE to otherwise orphan intergenic, intron or 3′ UTR regions. Mate pairs were then annotated with respect to RefSeq transcript models21 complemented with a proximal promoter, defined as the region comprising the 500 bp directly upstream of their 5′ end. This showed that an average of 4.24% of the transcripts were matching RefSeq transcripts, with 1.85% of them being strictly consistent with current gene models (that is starting within RefSeq promoter or 5′ UTR exon and ending within a RefSeq exon) while the rest was likely representing alternative mRNA splice forms and non-coding RNAs. These were located antisense of RefSeq, within their introns or in intergenic regions. Furthermore, the latter represented 87.5% of the total signal (Fig. 3 and Supplementary Table 7). We observed specific differences between sub-cellular fractions and between the poly-A− and total RNA (dominated by poly-A+ molecules). Paired-end tags starting and ending within RefSeq promoters or 5′ UTRs were twice more prevalent than mate pairs ending within any RefSeq exons in the poly-A− libraries than the total RNA libraries. Those may correspond in part to promoter-associated long and short RNAs (PALRs and PASRs)7. Nuclear fractions showed more paired-end tags starting and ending in intergenic and intronic regions than cytoplasmic fractions (Fig. 3a). By preparing 12 additional cytoplasmic and nuclear CAGEscan libraries from 6 HepG2 independent biological replicas, we confirmed higher abundance of intronic (Fig. 3b) and intergenic transcripts in nuclear fractions (P = 0.019 and P = 0.004 respectively, paired Student’s t-test).
Figure 3. Promotome-transcriptome analysis with CAGEscan.
(a) Annotation matrix summarizing the connections between genomic regions by CAGEscan mate pairs. We divided the genome in features that are intergenic, intron, promoter, 5′ UTR, coding sequence (CDS), 3′ UTR, antisense in introns, or antisense in exons according to RefSeq. The CAGEscan mate pairs were counted for each combination of features. For the libraries made from cytoplasmic, nuclear, cytoplasmic poly-A− and nuclear poly-A− RNAs, a matrix of 8 rows by 7 columns representing the indicated transcript features is plotted. The area of each cell is proportional to the number of pairs connecting a given combination of features. The percentages indicated below each column represent the fraction of mate pairs initiated from the same feature. The pairs starting in an intergenic feature were discarded to better visualize the differences between the other combinations. Notable combinations of features are colored. (b) The nuclear compartment contains more intron-intron pairs. Pairs of bars indicate that the libraries are technical replicates. For the experiment with six biological replicates, the percentages were averaged (error bars represent s.d., n = 6), and we observe a statistically significant difference (#) (P = 0.019. paired Student’s t-test).
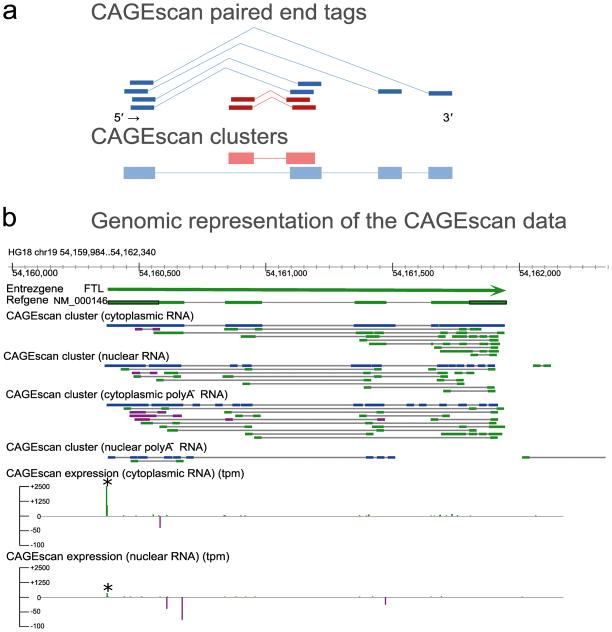
To better reconstruct transcript models, we grouped paired-end tags with overlapping 5′ ends into CAGEscan clusters (Fig. 4a). Alignment patterns of the corresponding 3′ end tags on the genome recapitulate the potential structure of the transcript resulting from a common promoter. By pooling together the poly-A− cytoplasmic, poly-A− nuclear, total cytoplasmic and total nuclear fractions libraries, we obtained 854,849 distinct CAGEscan clusters, with an average of 2.47 reads per cluster. Clustered independently, the cytoplasmic libraries produced in technical duplicates yielded between 72,666 and 34,822 clusters (with 3.5±0.6 reads per cluster) and the corresponding nuclear libraries yielded between 309,682 and 147,963 clusters (with 1.6±0.3 reads per cluster). 9% of the CAGEscan clusters (all libraries pooled) started upstream of the translation initiation site of RefSeq; of them, 76.5% reached into their 3′ UTR or into their downsteam intergenic region, associating a promoter to a 3′ UTR for 11,131 RefSeq transcripts. Comparable ratios (9.5%±2.65 and 80%±11 respectively) were obtained when considering the four libraries separately (Supplementary Table 6). The region surrounding the FTL gene, for which the complete RefSeq model is tiled with paired-end sequences, illustrates such 5′ mate pair driven clustering (Fig. 4b). We observed subcellular compartment-specific antisense expression, with most of these antisense CAGEscan clusters ending in close proximity to the promoter of FTL (Fig 4b and Supplementary Fig. 7). A total of 37,818 clusters were antisense to 9,638 RefSeqs (Supplementary Table 6). Antisense RNAs were generally more prevalent in the nuclear fractions.
Figure 4. CAGEscan connects promoters and downstream sequences.
(a) Schematic representation of CAGEscan paired-end tags clustering. Tags arising from overlapping 5′ ends are used as seed to aggregate 3′ tags into unique cluster. Depicted in blue and red are two overlapping but distinct resulting CAGEscan clusters. (b) Genomic representation of the CAGEscan data. Horizontal bars indicate annotation features (Chromosomal coordinates, Entrez Gene loci, RefSeq transcript models, CAGEscan clusters), and vertical bars represent quantitative activity of the promoters detected by the 5′ reads of the CAGEscan libraries (CAGEscan expression) in tag per million. Features and expression arising from the plus or minus strand are colored in green and purple respectively. The FTL gene (thick green bar) has a strongly active promoter (asterisk), from which originate enough CAGEscan pairs to reconstitute the gene’s intron-exon structure in a single CAGEscan cluster (blue). Antisense transcripts to FTL loci are more abundant in the nuclear libraries.
By aligning mate pairs to all exon-exon junction combinations of each transcript, we uncovered 8,462 splice junctions linked to 11,964 TSS. This also revealed the existence of 312 exon skipping events amongst 297 independent transcripts. Furthermore, clustering paired-end tags uncovered 1,569 CAGEscan clusters that initiated within the 5′ UTR of a given transcript, reached into the next downstream independent gene model in 1,198 pairs of distinct consecutive transcripts.
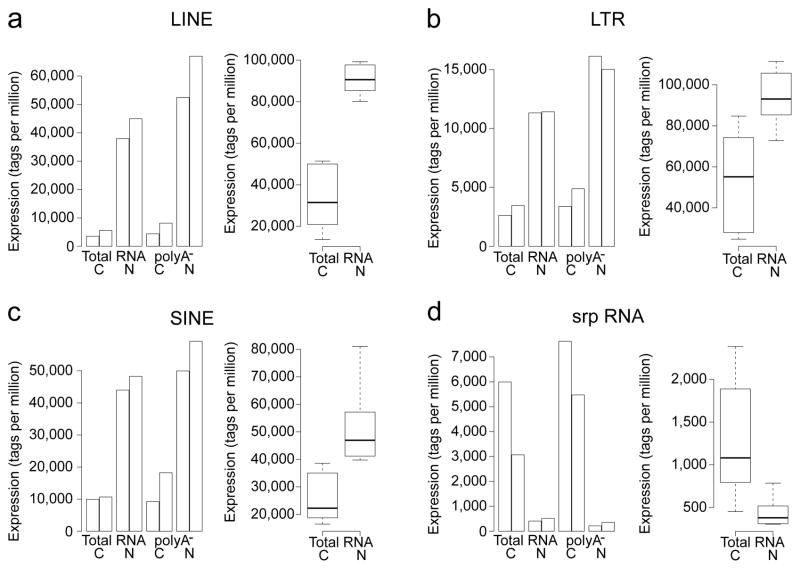
Pervasive and regulated transcription of retrotransposon elements (RE) have been observed in total RNA extracts using CAGE24. Using CAGEscan, we observed that expressed RE are more abundant in the nuclear RNA fractions than in the cytoplasmic ones. Globally, long and short interspersed nuclear elements (LINEs, SINEs) were the most highly expressed RE in HepG2, followed by long terminal repeats (LTRs). All three were strongly over-represented in the nuclear poly-A− fraction (Fig. 5a–c). As expected srpRNA (signal recognition particle) repeats were enriched in the cytoplasmic compartment (Fig. 5d).
Figure 5. Expressed repeat elements surveyed by CAGEscan.
Expression in tags per million of LINE, LTR, SINE, and srpRNA repeated elements in cytoplasmic (C) and nuclear fractions (N) from total and non-polyadenlyated RNA (poly-A−). Adjacent bars indicate technical replicates. Whisker plots summaries data from six additional biological replicates. The boxplots and whisker plots sub-panels use different scales. The nucleus appears strongly enriched for LINE, LTR and SINE transcripts, which are non-polyadenylated, while the cytoplasm appears strongly enriched in srpRNA, both in the total RNA and the non-polyadenylated fraction.
Discussion
So far, an unbiased analysis of the transcriptome and promoter usage from challenging samples such as biopsies, homogeneous population of ex vivo cell types or subcellular compartments has been hampered by the low quantity of RNA and by the use of fixatives that are detrimental to RNA integrity. The simplicity of both nanoCAGE and CAGEscan combined with decreasing sequencing cost opens the possibility for a truly high-throughput library production of pooled, multiplexed libraries followed by parallel sequencing, with applications ranging from drug screening, biopsy analysis, and whole transcriptome association studies. Therefore, we expect nanoCAGE to become the technique of choice for micro-dissected samples in experimental biology and molecular pathology. Identification of novel 5′ ends that are compartment-specific demonstrates the need and usefulness of nanoCAGE. As an added advantage, the fixed length tags generated by nanoCAGE can easily be turned into concatemers that will be advantageous when sequenced with long-read high throughput sequencers.
By linking TSSs to downstream sequences, CAGEscan provides insights into the architecture of transcripts and thus into their possible functions. Although the ability of fully scanning the 3′ end of long transcripts is currently limited by the paired-end read length range that can be simultaneously sequenced, we believe that further development of sequencing technology will overcome this limitation. CAGEscan profoundly differs from traditional inferences based on gene models such as RefSeq, which fails to grasp the complexity of the transcriptional landscape. CAGEscan analysis is data-driven and hypothesis-free, which allowed us to find non-coding RNAs, evidence of transcriptional read-through between neighboring loci, as well as novel forms of protein-coding genes. The expression level of CAGEscan promoters is indicated by the frequency of the 5′ read of the mate pairs. Furthermore CAGEscan offers a unique perspective into the relations of non-coding RNAs to the neighboring genomic/transcriptomic elements. Such novel transcription maps will be instrumental in identifying the functions of novel ncRNAs that overlap regulatory regions and are likely to regulate transcription25 or processing and recapping of RNAs26.
Supplementary Material
Acknowledgments
This work was founded by a grant of the 6th Framework of the European Union commission to the Neuro Functional Genomics consortium, by a grant of the 7th Framework to PC and SG (Dopaminet), a Grant-in-Aids for Scientific Research (A) No.20241047 for PC and a Research Grant for RIKEN Omics Science Center from MEXT to YH. Work in this project is also partially supported by the National Human Genome Research Institute grants U54 HG004557.. CP was supported by the Japanese Society for the Promotion of Science long-term fellowship number P05880. SG was funded by a career developmental award from “The Giovanni Armenise-Harvard Foundation”. We thank Alistair Forrest for critical discussions and Mylene Josserand for experimental assistance.
Footnotes
Statement of competing interest
CP, PC and RS are inventors of the Japanese patent application held by RIKEN on the moderately suppressive PCR step of the nanoCAGE protocol.
Author contributions
CP, RS and PC conceived the nanoCAGE technology. CP and PC conceived the CAGEscan technology. CP, HT, RS, MS and SO designed and performed the experiments. CP, NB, HT, TL and MV analyzed the data and interpreted the results. CP, NB, SG and PC supervised the study. DL, NH, VO, IB, HG, JD, PK, HW, CAD and TRG provided material. JS provided software. JK, YH, SG and PC provided salary support. The text and figures were drafted by PC, NB, HT, and MV, and edited by CP, NB, TL, COD and PC.
References
- 1.Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kodzius R, et al. CAGE: cap analysis of gene expression. Nat Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P. Cap-Analysis Gene Expression (CAGE): Genome-Scale Promoter Identification and Association with Expression Profile and Regulatory Networks. Pan Stanford Publishing; Singapore: 2009. [Google Scholar]
- 4.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki H, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 8.Fullwood MJ, Wei C, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valen E, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 11.Hirzmann J, Luo D, Hahnen J, Hobom G. Determination of messenger RNA 5′-ends by reverse transcription of the cap structure. Nucleic Acids Res. 1993;21:3597–3598. doi: 10.1093/nar/21.15.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohtake H, Ohtoko K, Ishimaru Y, Kato S. Determination of the capped site sequence of mRNA based on the detection of cap-dependent nucleotide addition using an anchor ligation method. DNA Res. 2004;11:305–309. doi: 10.1093/dnares/11.4.305. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 14.Meisel A, Bickle TA, Kruger DH, Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992;355:467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]
- 15.Maeda N, et al. Development of a DNA barcode tagging method for monitoring dynamic changes in gene expression by using an ultra high-throughput sequencer. BioTechniques. 2008;45:95–97. doi: 10.2144/000112814. [DOI] [PubMed] [Google Scholar]
- 16.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forrest ARR, Carninci P. Whole genome transcriptome analysis. RNA Biol. 2009;6:107–112. doi: 10.4161/rna.6.2.7931. [DOI] [PubMed] [Google Scholar]
- 18.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 19.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 20.Ozsolak F, et al. Digital transcriptome profiling from attomole-level RNA samples. Genome Res. 2010;20:519–525. doi: 10.1101/gr.102129.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivarius S, Plessy C, Carninci P. High-throughput verification of transcriptional starting sites by Deep-RACE. BioTechniques. 2009;46:130–132. doi: 10.2144/000113066. [DOI] [PubMed] [Google Scholar]
- 23.Fromont-Racine M, Bertrand E, Pictet R, Grange T. A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res. 1993;21:1683–1684. doi: 10.1093/nar/21.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cell. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 25.Carninci P. Molecular biology: The long and short of RNAs. Nature. 2009;457:974–975. doi: 10.1038/457974b. [DOI] [PubMed] [Google Scholar]
- 26.Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–21. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.