SUMMARY
Interleukin(IL)-2 and inflammation regulate effector and memory cytolytic T-lymphocyte (CTL) generation during infection. We demonstrate a complex interplay between IL-2 and inflammatory signals during CTL differentiation. IL-2 stimulation induced the transcription factor eomesodermin (Eomes), upregulated perforin (Prf1) transcription, and repressed re-expression of memory CTL markers Bcl6 and IL-7Rα. Binding of Eomes and STAT5 to Prf1 cis-regulatory regions correlated with transcriptional initiation (increased recruitment of RNA polymerase II to the Prf1 promoter). Inflammation (CpG, IL-12) enhanced expression of IL-2Rα and the transcription factor T-bet, but countered late Eomes and perforin induction while preventing IL-7Rα repression by IL-2. After infection of mice with lymphocytic choriomeningitis virus, IL-2Rα-deficient effector CD8+ T cells expressed more Bcl6 but less perforin and granzyme B, formed fewer KLRG-1+ and T-bet-expressing CTL, and killed poorly. Thus, inflammation influences both effector and memory CTL differentiation, whereas persistent IL-2 stimulation promotes effector at the expense of memory CTL development.
INTRODUCTION
Naive CD8+ T cells differentiate into effector and memory cytolytic T-lymphocytes (CTL) upon antigen stimulation in the context of infection and inflammation. During this process, the differentiating cells induce the expression of effector proteins such as the cytokine IFNγ, the pore-forming protein perforin, and a family of serine esterases known collectively as granzymes (Cruz-Guilloty et al., 2009; Harty et al., 2000). Perforin and granzymes are essential for cytolytic activity of CTL (Pipkin and Lieberman, 2007). IFNγ, perforin, and granzymes are each induced at the transcriptional level after activation, but distinct regulatory mechanisms appear to be involved—most, if not all, antigen-specific CD8+ T cells express IFNγ and granzyme B during the course of an infection, but only a fraction of these express perforin and IFNγ expression does not necessarily correlate with cytolytic activity (Harrington et al., 2008; Johnson et al., 2003; Peixoto et al., 2007; Zaiss et al., 2008). The expression of all three classes of effector genes in activated cells has been correlated with memory CTL development (Bannard et al., 2009; Harrington et al., 2008; Joshi et al., 2007; Opferman et al., 1999; Sarkar et al., 2008). However, little is known about the signals that regulate transcription of these different classes of effector genes in activated CD8+ T cells, what mechanisms are involved, and how those signals might regulate effector or memory CTL differentiation.
The factors and mechanisms that drive the differential development of effector versus memory CTL during clonal expansion are not completely understood (Badovinac and Harty, 2007; Kaech and Wherry, 2007; Williams and Bevan, 2007). A single brief T cell receptor (TCR) stimulus (signal 1) combined with costimulation (signal 2) can induce an extended period of proliferation, acquisition of effector functions, and ultimately, memory CTL formation (Kaech and Ahmed, 2001; Mercado et al., 2000; van Stipdonk et al., 2001). The duration of TCR stimulation mainly affects the magnitude of effector CD8+ T cell accumulation (Prlic et al., 2006), whereas altered TCR signaling in the context of mutant TCRs affects the balance of effector and memory CTL development (Teixeiro et al., 2009).
IL-2 signals are sometimes considered part of signal 2 (Valenzuela et al., 2002). However, the role of IL-2 signaling in CD8+ T cell differentiation has been difficult to discern in vivo because results from infection of IL-2-deficient mice have differed. This variability may reflect autoimmunity secondary to defective regulatory T cell development in IL-2-deficient mice (Bachmann and Oxenius, 2007; Malek, 2008). More recent studies that avoided these caveats have shown that IL-2 is essential for normal accumulation of effector CD8+ T cells (D’Souza et al., 2002) and for programming the ability of memory CTL to reexpand upon secondary infection in vivo (Bachmann et al., 2007; Williams et al., 2006). In addition, IL-2Rβ, an essential signaling subunit of the IL-2R complex, and STAT5, a transcription factor activated by IL-2R stimulation, are required for normal expression of perforin, granzyme B, and IFNγ in activated CD8+ T cells (Imada et al., 1998; Malek et al., 2001). Although both IL-2 and IL-15 signal through IL-2Rβ, each cytokine has different effects on CTL differentiation; stimulation of IL-2Rβ on CD8+ T cells in cell culture with IL-2, as opposed to IL-15, favors effector rather than memory CTL generation (Carrio et al., 2004; Manjunath et al., 2001), suggesting that how IL-2Rβ is activated affects gene expression.
An inflammatory signal (signal 3) provided by cytokines such as type I interferons and/or IL-12 is essential for normal effector and memory CTL generation. In different settings, signal 3 has been shown to be crucial for inducing CTL effector functions (Curtsinger et al., 2003; Mescher et al., 2006), for driving antigen-activated CD8+ T cells toward a short-lived effector cell fate (Joshi et al., 2007), and for programming contraction of the effector cell population (Badovinac et al., 2004). At the same time, type I interferons and IL-12 have also been shown to be required for memory CTL development (Xiao et al., 2009). Thus, a number of extracellular signals regulate effector and memory CTL development in vivo, but it is still unclear how these signals act individually and in combination to regulate gene expression programs in activated CD8+ T cells and control their differentiation.
In this study, we used a simple cell-culture system to investigate how the strength of IL-2R signaling regulated perforin gene (Prf1) expression after TCR activation of naive CD8+ T cells and how inflammatory signals (CpG and IL-12) influenced this process. We also investigated the requirement of IL-2 signaling for CTL responses in vivo in a setting that avoids autoimmunity; mixed bone marrow chimeric mice reconstituted with both wild-type and Il2ra-deficient cells were infected with lymphocytic choriomeningitis virus (LCMV). We found that persistent IL-2 stimulation induced Prf1 transcription and promoted effector CTL differentiation, whereas inflammatory signals had distinct effects. Our data indicate that inflammation and IL-2 signals interface in a complex way and that their relative strengths are likely to regulate the development of both effector and memory CTL.
RESULTS
IL-2R Signal Strength Regulates Perforin Expression and Cytolytic Function
To begin investigating how the “strength” of IL-2R signaling regulated the effector functions of CD8+ T cells, naive CD8+ T cells were cultured for 2 days with a strong TCR stimulus in conjunction with CD28 costimulation, then removed from stimulation and recultured in low (10 U/ml) or high (100 U/ml) IL-2 concentrations. As an indirect measure of IL-2R signal intensity, we assessed cell-surface expression of IL-2Rα, which is regulated in part through a well-characterized positive feedback loop in which STAT5 directly targets the Il2ra gene (Nakajima et al., 1997). All cells displayed uniformly high expression of IL-2Rα until day 4, as judged by flow cytometry; after day 4, cells cultured in high IL-2 maintained high IL-2Rα expression, whereas cells cultured in lower IL-2 concentrations showed decreased IL-2Rα expression (Figure S1A available online). Evaluating IL-2 signaling more quantitatively as the content of tyrosine-phosphorylated STAT5 (Figures S1B and S1C), we found that phospho-STAT5 amounts were high in cell populations cultured in either high or low IL-2 until day 5, after which they were sustained in high IL-2 but declined abruptly in cells cultured in low IL-2 (Figure S1B). At the single-cell level, flow cytometric analysis showed that 94% of the cells cultured in high IL-2, but only 15% of the cells cultured in low IL-2, maintained high amounts of phospho-STAT5 on day 6 (Figure S1C). The mean fluorescence intensities (MFIs) of phospho-STAT5 staining in each population suggested that there was an approximately 5- to 10-fold difference in IL-2 “strength” when comparing 10 versus 100 U/ml IL-2 in our cultures.
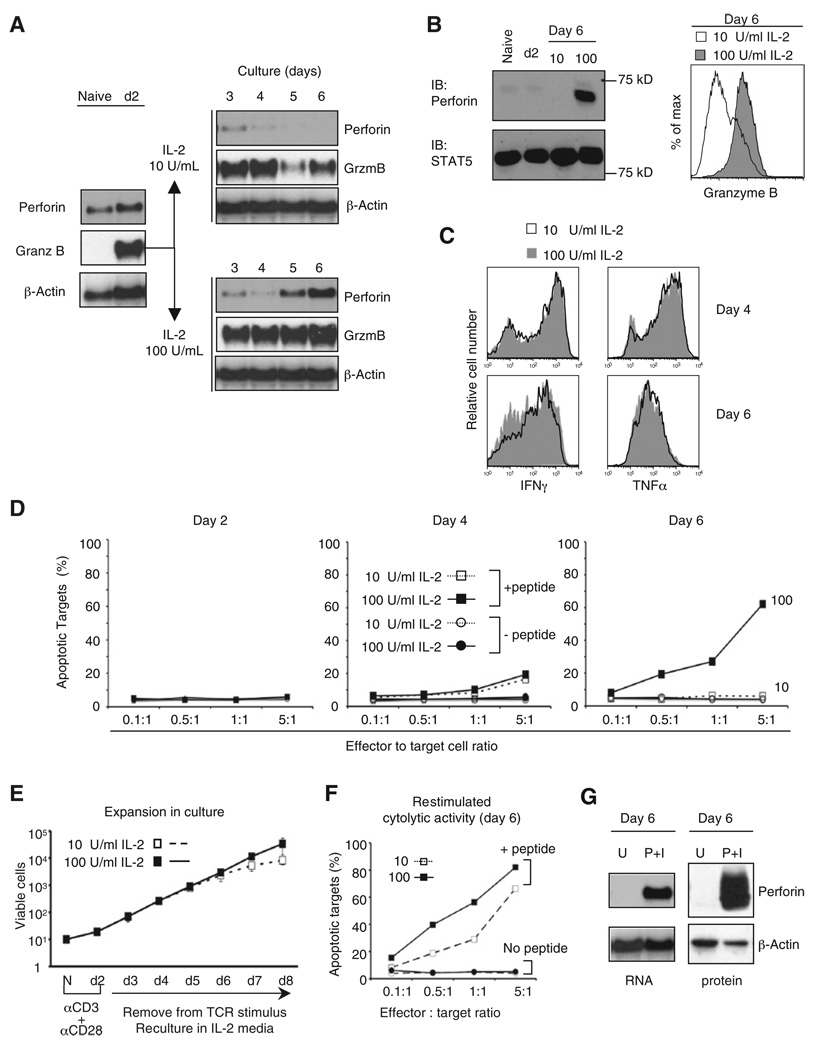
The two essential effectors of the cytolytic program, perforin and granzyme B, displayed distinct expression kinetics in activated CD8+ T cells under high or low IL-2 conditions (Figures 1A and 1B). Naive CD8+ T cells expressed perforin mRNA but very little protein, and neither was induced by 2 days of TCR stimulation. After removal from TCR stimulation, perforin mRNA expression decreased until day 4, regardless of IL-2 concentration, then continued to decrease in cells cultured in low IL-2, but increased strongly, beginning at day 5, in cells cultured with high IL-2 (Figure 1A). Similarly, perforin protein was strongly induced after day 4, when cells were cultured in high IL-2 (Figure 1B). In contrast, granzyme B mRNA was undetectable in naive T cells but was strongly induced by TCR stimulation. Although granzyme B expression was maintained in both low and high IL-2, granzyme B mRNA and protein were both more highly expressed in cells cultured in high IL-2 (Figure 1A and 1B). The concentration of IL-2 in culture did not affect the ability of CD8+ T cells to express IFNγ and TNF upon brief restimulation (Figure 1C), consistent with previous studies (Bachmann et al., 2007; Williams et al., 2006), although IL-2Rβ was likely required initially (Malek et al., 2001).
Figure 1. Different IL-2 Signal Strengths Regulate Perforin and Granzyme B Expression to Establish CTL Function.
(A) Kinetics of perforin and granzyme B mRNA expression. Purified CD8+ T cells from naive B6 mice were primed with anti-CD3 and anti-CD28, removed from the TCR stimulus after 2 days, and recultured in 10 or 100 U/ml IL-2. Total RNA was analyzed by blotting at the indicated times. Results are representative of more than three independent experiments. See also Figure S1.
(B) Perforin (left) and granzyme B (right) protein expression. Whole-cell lysates were analyzed by immunoblotting at the indicated times. Total STAT5 content was used as a control. Granzyme B expression was determined by intracellular staining and flow cytometry on day 6.
(C) Intracellular cytokine expression after restimulation. CD8+ T cells were restimulated on days 4 and 6 with 10 nM PMA + 1 µM ionomycin for 6 hr.
(D) Flow cytometry-based cytotoxicity assay for cytolytic activity. GP33-pulsed EL4 targets were coincubated with effector P14 CD8+ T cells that were differentiated as in (A). CTL activity was blocked by incubation with 5 mM EGTA (data not shown).
(E) Accumulation of CD8+ T cells after stimulation as described in (A). The data are the mean and standard deviation summarized from at least five differentiations.
(F) Cytotoxic activity after brief restimulation. P14 cells cultured until day 5 in low IL-2 were restimulated for 2 hr with PMA and Ionmycin, washed, resuspended in T cell media without IL-2, and cultured overnight before analyzing CTL activity.
(G) Perforin expression in primed CD8+ T cells cultured in 10 U/ml IL-2 upon restimulation on day 6. Lysates from cells left unstimulated (U) or restimulated with PMA (P) and ionomycin (I) as in (C) for 6 hr were analyzed by RNA blotting (left) and immunoblotting (right).
Strong IL-2 signals were necessary to induce cytolytic function. Activated CD8+ T cells cultured until day 6 in high IL-2 rapidly killed antigen-pulsed target cells in a short-term (30 min–2 hr) assay (Cruz-Guilloty et al., 2009), without prior restimulation (Figure 1D and Figure S1D), whereas those cultured in low IL-2 did not (Figure 1D). Nevertheless, cells cultured in low IL-2 were fully functional in all other respects—they accumulated exponentially for at least 6 days, with kinetics equivalent to those of cells cultured in high IL-2 (Figure 1E), and displayed efficient cytolytic activity after a brief restimulation (Figure 1F). We attribute the de novo increase in cytolytic activity to the strong induction of perforin in response to restimulation (Figure 1G).
Reciprocal Regulation of Perforin and IL-7Rα Expression
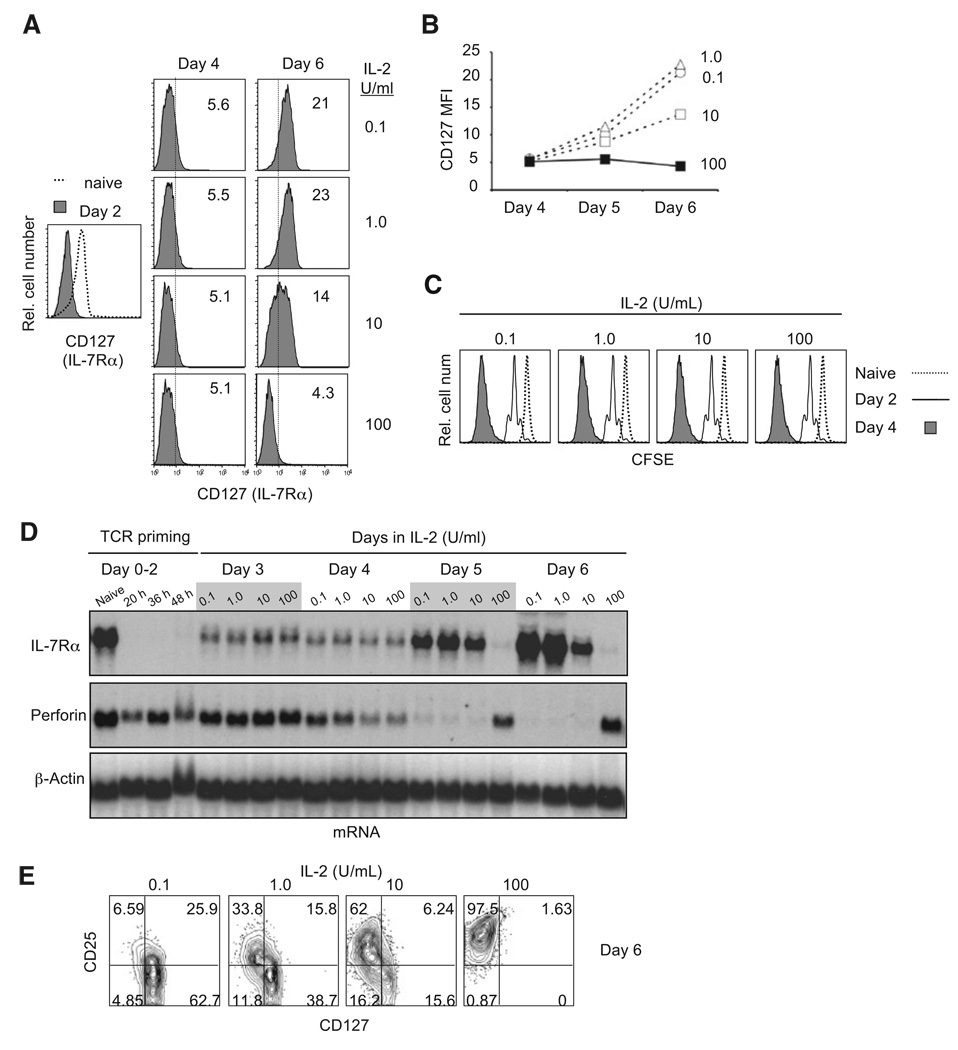
The strength of IL-2 stimulation regulated the expression of additional genes in CD8+ T cells. As expected, initial stimulation of naive P14 TCR transgenic CD8+ T cells, primed either by TCR crosslinking or by coculture with splenic APCs loaded with GP33 peptide, induced surface markers characteristic of antigen-stimulated T cells (CD44hi, CD25hi, CD69hi, CD127−). After day 4, however, cells cultured in low IL-2 quickly converted to a central memory-like phenotype (CD25lo, CD122hi, CD127 hi, CD62Lhi) and were not cytolytic, whereas the majority of those cultured in high IL-2 retained a characteristic effector phenotype (CD25 hi, CD122hi, CD127−, CD62Llo or −) and killed efficiently (Figures S2A–S2C and data not shown).
We examined IL-7Rα (CD127) regulation in more detail because effector cells that reinduce IL-7Rα at the peak of the response to some acute infections are enriched for cells that form long-lived memory CTL (Kaech et al., 2003). TCR stimulation completely downregulated surface IL-7Rα expression, and high IL-2 prevented IL-7Rα re-expression. However, IL-7Rα was reinduced after day 4 in a dose-dependent fashion if the IL-2 concentration was reduced (Figures 2A and 2B). IL-7Rα re-expression did not correlate with cell division, because all cells completely diluted CFSE (>7 divisions) by day 4 (Figure 2C) and accumulated similarly until day 5, by which time IL-7Rα had already been reinduced (Figure 2B; also see Figure 2D). At the mRNA level, IL-7Rα mRNA was repressed and perforin mRNA was reciprocally induced at the same high concentrations of IL-2 (Figure 2D). Consistent with this observation, surface expression of IL-2Rα and IL-7Rα chains was mutually exclusive: even at low IL-2 concentrations (1–10 U/ml), IL-2Rα-positive cells tended to be IL-7Rα negative (Figure 2D). Enforced expression of IL-2Rα by retroviral transduction confirmed that low IL-2 could repress IL-7Rα in cells that expressed higher IL-2Rα (Figures S2D and S2E). Thus, expression of the high-affinity IL-2Rαβγ receptor is sufficient to repress IL-7Rα re-expression, even in cells cultured in low concentrations of IL-2.
Figure 2. Reciprocal Regulation of Perforin and IL-7Rα Reexpression by IL-2.
(A) The regulation of IL-7Rα (CD127) re-expression after priming. Purified naive P14 CD8+ T cells were stained with antibodies against CD127 ex vivo (Naive), after priming for 2 days with anti-CD3 + anti-CD28 (day 2) and culture in 0.1, 1, 10 or 100 U/ml IL-2. The MFI of CD127 staining is shown in each panel. The analysis is representative of at least three separate differentiations. See also Figure S2.
(B) Graphical summary of IL-7Rα expression from the representative experiment in (A).
(C) Cell division history based on CFSE dilution after priming and culture in IL-2 as in (A).
(D) IL-7Rα and perforin mRNA expression in naive and primed P14 CD8+ T cells. Total RNA was extracted at the indicated time points from cells in (A).
(E) Flow cytometric analysis of IL-2Rα and IL-7Rα coexpression.
Strong IL-2R Signals Sustain Blimp-1 Expression and Repress Bcl-6
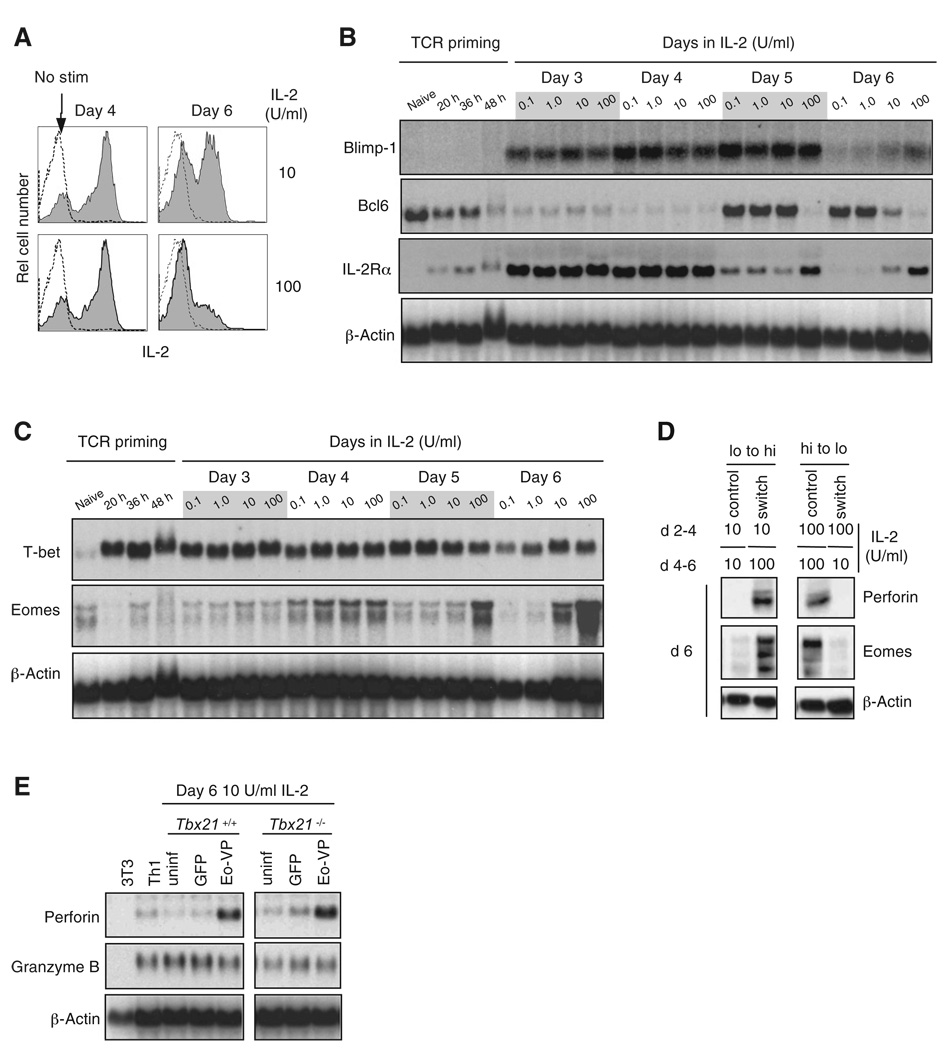
Distinct populations of effectorCD8+T cells are present early after acute viral infection that have different potential to become memory CTL, and they can be distinguished by their ability to produce IL-2 and to proliferate upon secondary stimulation ex vivo (Joshi et al., 2007; Sarkar et al., 2008). These two attributes were controlled by different strengths of IL-2 stimulation in culture. IL-2 production was not affected by different IL-2 concentrations through day 4 (Figure 3A), but by day 6, cells cultured in low IL-2 produced much more IL-2 upon restimulation than cells cultured in high IL-2 (Figure 3A); they also proliferated more strongly in response to low levels of stimulation (Figure S2F).
Figure 3. IL-2 Regulates Blimp-1, Bcl6, and Eomes Expression, and Eomes Activates the Perforin Gene.
(A) Intracellular staining of IL-2. Stainings are of unstimulated cells (outlined histogram) or cells restimulated for 4 hr with 10 nM PMA + 1 µM ionomycin (shaded histograms). Results from (A)–(E) are representative of at least two independent experiments.
(B) Blimp-1, Bcl6, and IL-2Rα mRNA expression kinetics were analyzed by RNA blotting.
(C) T-bet and Eomes mRNA expression kinetics were analyzed by RNA blotting.
(D) Eomes and perforin expression in response to IL-2 after day 4. Cultured CD8+ T cells were harvested and washed on day 4 and switched from low IL-2 to high IL-2 and vice versa (switch) or returned to their original IL-2 media (control) for an additional 2 days. Whole-cell lysates were generated and analyzed by immunoblotting on day 6.
(E) Perforin and granzyme B (GrzmB) mRNA expression upon Eomes-VP16 transduction. On day 6 of culture in 10 U/ml IL-2, total RNA was analyzed by RNA blotting; uninfected (uninf), Eomes-VP16 (Eo-VP), or empty cassette (GFP) transduced; Th1 and NIH 3T3 cells are shown as controls. Transduction efficiency in each culture was equivalent between constructs. See also Figure S3.
Studies of gene-disrupted mice have shown that Blimp-1 and Bcl6 are required in vivo for the development of effector and memory CTL, respectively (Ichii et al., 2002; Rutishauser et al., 2009). We therefore examined whether IL-2 regulated expression of these two transcription factors. Blimp-1 mRNA was not expressed in naive cells and was only induced upon removal from the TCR stimulus and culture in IL-2 (Figure 3B). By day 6, Blimp-1 mRNA expression was not maintained in low IL-2, but Blimp-1 remained expressed in high IL-2. Conversely, Bcl6 mRNA was expressed in naive CD8+ T cells, downregulated during TCR stimulation, and re-expressed after day 4 in low, but not high, IL-2, and this correlated inversely with expression of IL-2Rα mRNA (Figure 3B). Thus, the inverse expression of Blimp-1 and Bcl6 typical of effector cells in vivo was regulated by the degree of IL-2 stimulation.
IL-2 Induces Perforin Gene Transcription through STAT5 and Eomes, but not T-bet
The T-box transcription factors T-bet and Eomes are both required for normal CTL differentiation (Intlekofer et al., 2008; Intlekofer et al., 2005; Joshi et al., 2007). We asked whether IL-2 stimulation regulated expression of T-bet and Eomes upon CD8+ T cell activation. Expression of T-bet and Eomes mRNA and protein was low or undetectable in naive CD8+ T cells. Two days of TCR stimulation strongly induced T-bet (Figure 3C), and its expression was maintained in an IL-2-independent manner through day 6 (Figure 3C). In contrast, Eomes was induced by day 4 and was upregulated through day 6 in a manner strongly dependent on IL-2 concentration in culture (Figure 3C). By switching cells from low to high IL-2 and vice versa on day 4 of culture, we confirmed that Eomes and perforin expression were both regulated by IL-2, whereas T-bet expression was not (Figure 3D and data not shown).
The kinetics and IL-2 dependence of Eomes expression was closely paralleled by perforin expression (Figures 3C and 2D), leading us to ask whether Eomes could induce perforin without strong IL-2 stimulation. Under these conditions, endogenous Eomes was not highly expressed and STAT5 phosphorylation was low (Figures 3C and 3D and Figures S1B and S1C). We activated purified naive CD8+ T cells from wild-type and T-bet-deficient B6 mice for 2 days, transduced them with a retrovirus expressing a hyperactive form of Eomes (Eomes-VP16) linked to IRES-GFP, then cultured the cells in low IL-2. Cells infected with Eomes-VP16, but not the control GFP retrovirus, strongly expressed perforin mRNA on day 6 (Figure 3E). Eomes-VP16 had no effect on the expression of granzyme B mRNA (Figure 3C). Control experiments showed that the effect of Eomes could not be attributed to a large increase in IL-2Rα or IL-2Rβ expression (Figure S3). Thus, Eomes most likely acts directly at Prf1 after induction by IL-2.
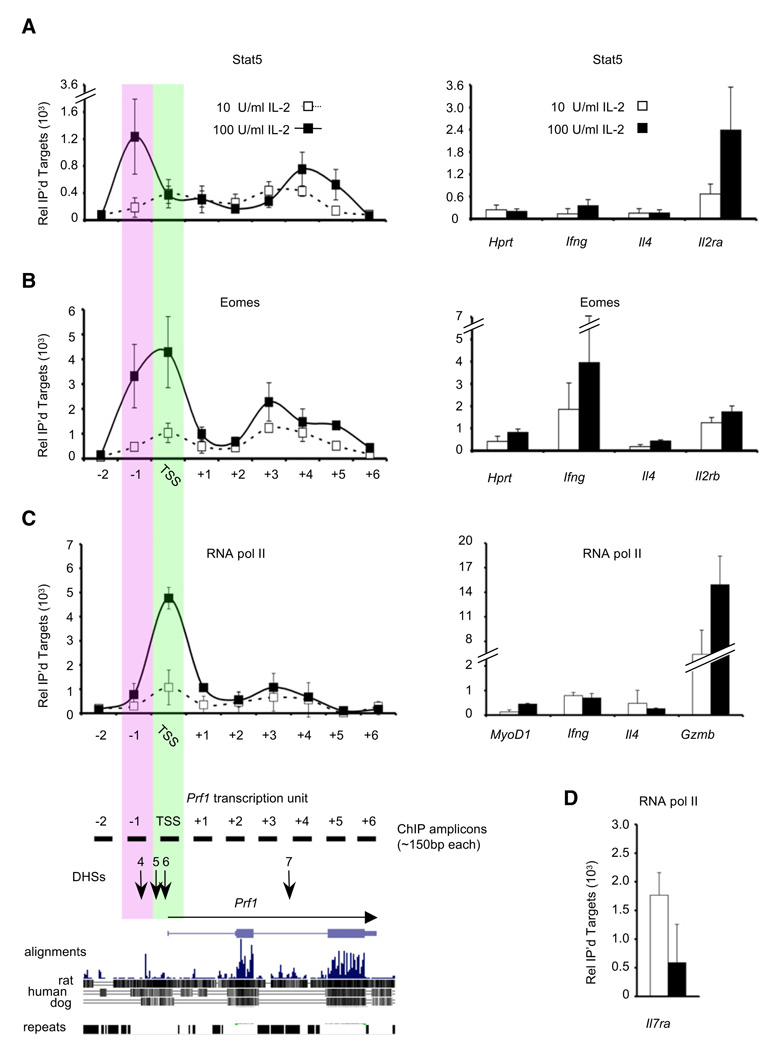
We tested this hypothesis by chromatin immunoprecipitation (ChIP) (Figure 4). Endogenous Eomes and STAT5 proteins both bound to Prf1 on day 6 in cells cultured in high IL-2, although their binding patterns were distinct. STAT5 primarily bound DNase I hypersensitive (DHS) site 4 at –1 kb and within intron 3 downstream of DHS7, comparable to its binding to the TSS of Il2ra, a known STAT5 target gene (Nakajima et al., 1997) (Figure 4B). Eomes bound to the Prf1 transcription start site (TSS) and to a lesser degree DHS4 (Cruz-Guilloty et al., 2009), comparable to its binding to the TSSs of Il2rb and Ifng, genes that are direct targets of Eomes (Intlekofer et al., 2005). Eomes also bound modestly near DHS7 (Figure 4B). Compared to cells cultured in high IL-2, cells cultured in low IL-2 showed considerably less binding of STAT5 and Eomes to Prf1, consistent with their lower phospho-STAT5 content and lower Eomes expression (Figures 4A and 4B). However, the amount of binding was still somewhat greater than that observed at the TSS of ll4 and Hprt, genes that are not expressed in CD8+ T cells or that are expressed, but not regulated, by IL-2, respectively. Therefore, even low binding of phospho-STAT5 and Eomes to the Prf1 gene may be relevant.
Figure 4. IL-2 Regulates RNA Polymerase II Recruitment and the Binding of STAT5 and Eomes to Prf1.
(A) ChIP analysis of endogenous STAT5. Chromatin was isolated from cells on day 6. The efficiency of recovery relative to input at the −1 kb region of Prf1 was 0.39% in high-IL-2 conditions for STAT5. The data in panels (A) and (B) show the mean and standard deviation of duplicate measurements, pooled from at least two immunoprecipitations of chromatin prepared from two independent CD8+ T cell differentiations. See also Figure S4.
(B) ChIP analysis of endogenous Eomes. The efficiency of recovery relative to input for the −1 kb region of Prf1 was 0.97% in high-IL-2 conditions for Eomes.
(C) ChIP analysis of RNA pol II. Data show the mean and standard deviation of duplicate measurements from separate immunoprecipitations from at least two independent differentiations; data from each differentiation was normalized based upon binding of RNA pol II to the Hprt TSS. The efficiency of recovery of the Hprt TSS was ~0.6% and was approximately six times greater than from the Ifng TSS.
(D) ChIP analysis of RNA pol II at the Il7ra TSS from the chromatin analyzed in (C).
Prf1 Transcription Is Regulated by Recruitment of RNA Polymerase II
TCR stimulation of naive CD4 T cells leads them to differentiate into distinct subsets with respect to expression of cytokine genes, a process that involves differential chromatin remodeling of specific cytokine gene loci (Ansel et al., 2006; Avni et al., 2002; Fields et al., 2002; Grogan et al., 2001). Because activated CD8+ T cells differentiated in high IL-2 express at least 20 times more perforin mRNA and exhibit a differentially remodeled DNase I hypersensitivity (DHS) site pattern in the Prf1 locus compared to differentiated Th1 cells (Pipkin et al., 2007), we asked whether differential perforin expression by CD8+ T cells in response to low versus high IL-2 was accompanied by differences in the DHS site pattern surrounding Prf1 (Figure S4) (Pipkin and Lichtenheld, 2006). Unexpectedly, we found that there was no difference in the pattern of DHS sites across ~200 kb of the Prf1 locus regardless of whether the cells had been differentiated in low or high IL-2 (Figure S4 and data not shown). Thus, IL-2 signals did not appear to “open” IL-2 specific cis-regulatory regions in the Prf1 gene but, rather, acted on a previously opened locus.
We asked whether IL-2 stimulation regulated recruitment of RNA polymerase II (Pol II) (Figure 4C). Relative to cells differentiated in low IL-2, cells differentiated in high IL-2 exhibited dramatically increased recruitment of Pol II to the Prf1 TSS at day 6, along with a small increase in Pol II density across the gene body (Figure 4C). Conversely, Pol II was recruited efficiently to the Il7ra TSS in CD8+ T cells cultured in low IL-2, but not in cells cultured in high IL-2 (Figure 4D). In both high and low IL-2, the level of Pol II binding at the Prf1 TSS was greater than that observed at the TSS of the muscle-specific gene MyoD1, which is not expressed in T cells (Figure 4C). Pol II binding at the TSS of the housekeeping gene Hprt (not regulated by IL-2) was comparable to the level of Pol II binding at the Prf1 TSS in high IL-2 (data not shown). Thus, the strength of IL-2 stimulation determined whether Pol II was recruited to the Prf1 and Il7ra genes in activated CD8+ T cells.
IL-2Rα Deficiency Impairs the Differentiation of Effector CD8+ T Cells into CTL In Vivo
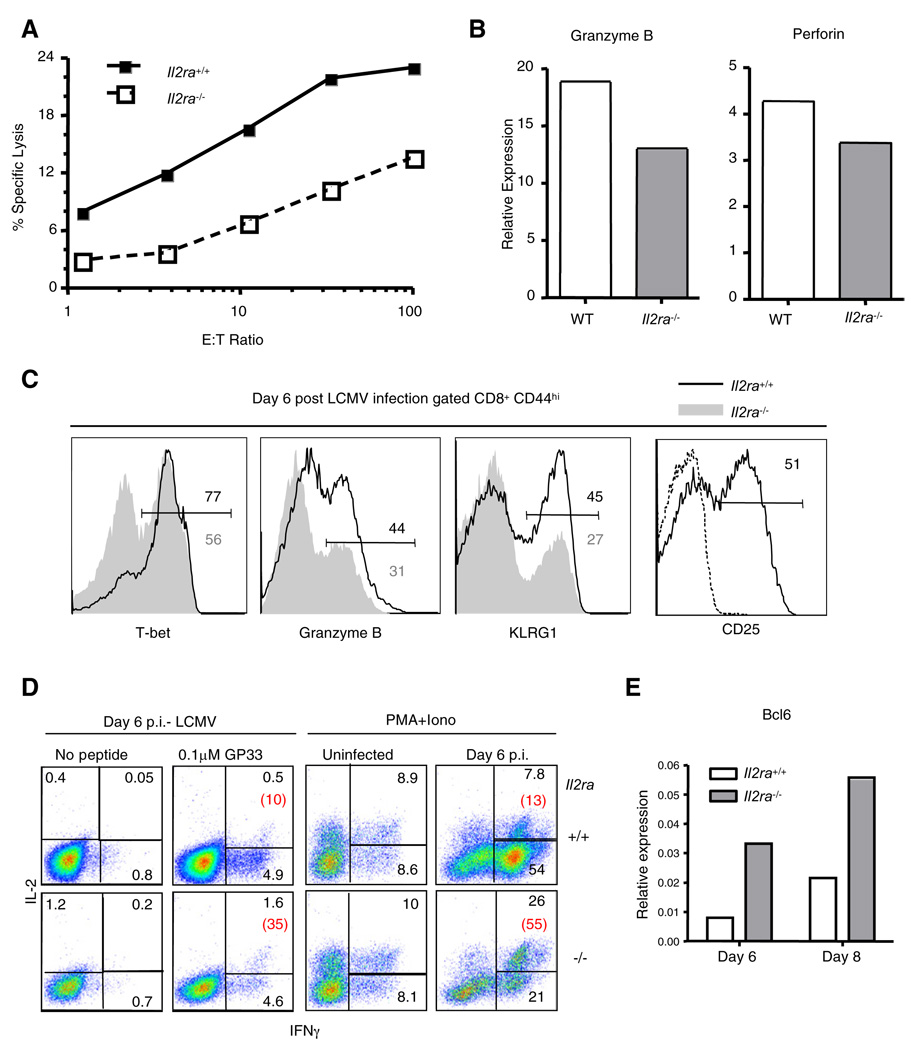
To examine the effect of IL-2 upon effector CD8+ T cell differentiation, we generated mixed bone marrow chimeras by transferring a mixture of congenically distinct wild-type and IL-2Rα-deficient bone marrow cells into lethally irradiated B6 mice and infecting them with LCMV (Bachmann et al., 2007; Williams et al., 2006). Eight days after infection, CD44hi, antigen-experienced Il2ra+/+, and Il2ra−/− CD8+ T cells were sorted and their cytotoxic activity was analyzed in a standard chromium release assay using GP33 peptide-loaded targets. Despite equal representation of GP33-specific cells among wild-type and Il2ra−/− effectors based on tetramer staining (data not shown), Il2ra−/− CD8+ T cells were severely defective in their capacity to lyse targets: 10-fold more effectors were needed for comparable target cell lysis (Figure 5A). This impairment correlated with decreased expression of perforin and granzyme B mRNA in sorted Il2ra−/− CD44hi cells compared to wild-type cells (Figure 5B). In addition, on day 6 after infection, when responding wild-type CD8+ T cells expressed maximal IL-2Rα, fewer total Il2ra−/− CD8+ T cells expressed granzyme B, T-bet, or KLRG-1, compared to wild-type cells, whereas proportionally more knockout cells expressed IL-7Rα and CD62L (Figure 5C and data not shown) (Williams et al., 2006). Furthermore, Il2ra−/− CD8+ T cells contained a much larger proportion of effector cells that coproduced IFNγ and IL-2 upon restimulation and expressed much more Bcl6 mRNA (Figures 5D and 5E). Thus, CD8+ T cells that could not signal via high affinity IL-2Rs differentiated inefficiently into effector CTL, even in an environment containing normal cells and inflammatory signals.
Figure 5. IL-2Rα-Deficient CD8+ T Cells Are Impaired in CTL Differentiation upon LCMV Infection.
(A) Il2ra+/+ and Il2ra−/− mixed bone marrow chimeric (BMC) mice were infected with LCMV. Eight days postinfection, Il2ra+/+ and Il2ra −/− CD44hi CD8+ effector cells were sorted and their ability to kill GP33 peptide-pulsed target cells was assayed.
(B) Quantitative RT-PCR analysis of cDNA prepared from sorted CD44hi Il2ra+l+ and Il2ra−/− CD8+ T cells. Values were normalized based on amplification of Hprt mRNA.
(C) Percent of total Il2ra+/+ (upper number) and Il2ra−/− (lower, gray number) CD8+ T cells from mixed BMC mice that express the indicated proteins was determined 6 days post-LCMV infection. CD25 expression by the Il2ra+/+ cells is depicted. Dashed line shows staining in uninfected BMC mice.
(D) Intracellular cytokine production of IL-2 and IFNγ by Il2ra+/+ and Il2ra−/− effector CD8+ T cells. Splenocytes from uninfected BMC mice and BMC mice on day 6 postinfection were stimulated for 4 hr with GP33 peptide, or PMA (50 ng/ml) and Ionomycin (500 ng/ml). Red numbers indicate the percentage of IL-2+ events among IFNγ+ cells.
(E) Quantitative RT-PCR analysis of cDNA prepared from sorted CD44hi Il2ra+/+ and Il2ra−/− CD8+ T cells. Values were normalized based on amplification of Hprt mRNA.
Panels (A)–(E) are representative results from at least three independent experiments with at least two mice per time point.
IL-2 Signaling Reduces Memory CTL Formation, but not Secondary Re-expansion
To determine the in vivo fate of in vitro-activated CD8+ T cells, we transferred primed P14 TCR-transgenic CD8+ T cells that had been cultured in high or low IL-2 to naive B6 mice (mixed at a 1:1 ratio, Figure 6). Engraftment of cells cultured in high and low IL-2 18 hr after transfer was equivalent in both the spleen (Figure 6A) and the lung (data not shown), and this equal representation was maintained when the mice were bled 10 days later (data not shown). When the same mice were analyzed 35 days after transfer (41 days after initial TCR activation in vitro), the P14 cells that had been cultured in low IL-2 maintained their frequencies, whereas those that had been cultured in high IL-2 had decreased in frequency and absolute number in all tissues analyzed (Figures 6A and 6B). Although cells from the high IL-2 cultures maintained some CD62Llo cells both 10 and 35 days after transfer, cells from the low IL-2 cultures were essentially all CD62Lhi (Figure 6C). Thus, the differential phenotypic programming observed in vitro was preserved upon in vivo transfer, indicating that T cells that had received higher and more prolonged IL-2 signals were impaired for memory differentiation.
Figure 6. Strong IL-2 Signals Negatively Impact CD8+ T Cell Memory Formation without Impairing Secondary Expansion.
(A) 2.5 × 105 congenically marked P14 cells that had been differentiated in high (100 U/ml) or low (10 U/ml) concentrations of IL-2 were transferred in a 1:1 mix to the same recipient. Representative mice were analyzed for the presence of the transferred cells in the spleen or lung (data not shown) 18 hr and 35 days after transfer.
(B) Thirty-five days after transfer, the presence of the transferred P14 cells was determined (pLNs, peripheral lymph nodes). FACS plots are gated on total CD8+ T cells. High (H) IL-2 (Thy1.2+); Low (L) IL-2 (Thy1.2−).
(C) CD62L expression on transferred P14 cells in the blood.
(D) Thirty-five days after P14 transfer, recipient mice were infected with LCMV. Five days later, the frequency of the transferred P14s of total CD8+ T cells was determined in the liver and mesenteric lymph nodes (mLNs).
(E) Absolute number of P14 T cells was determined in the spleens of representative mice before (day 35 after transfer) and after (day 5) LCMV infection. Data depicts average numbers ± SEM of at least two mice per time point per group.
Panels (A)–(E) are representative results from two independent experiments.
As a functional read-out for the fitness of the memory P14 cells derived from culture in varying concentrations of IL-2,we infected recipient mice with LCMV 35 days after transfer. Five days after infection, both populations of P14 T cells robustly expanded in the spleen, lymph nodes ,and liver; despite the fact that the P14 cells differentiated in low IL-2 were represented at an increased frequency prior to rechallenge, they did not exhibit an advantage upon re-expansion (Figures 6D and 6E). Thus, with CD8+ T cells from this in vitro system, alterations in IL-2 exposure altered their differentiation to memory but did not subsequently influence their ability to respond to secondary stimulation.
Inflammatory Stimuli and IL-2R Signals Have Cooperative and Opposing Effects
Given that both IL-2 and inflammation regulate CTL development, we examined how inflammatory stimuli interfaced with IL-2 signals to control gene expression in activated CD8+ T cells. We primed naive P14 CD8+ T cells with APCs and GP33 peptide, with or without inflammation (CpG), followed by culture in high or low IL-2 with or without IL-12 (Figure 7). The kinetics of perforin and granzyme B mRNA expression in cells primed with peptide and APC were similar to those previously observed after priming with plate-bound αCD3 and αCD28 (Figure 7A, compare with Figure 1A). However, compared to cells stimulated with GP33 peptide alone, cells stimulated in the presence of CpG followed by culture in IL-12 and low IL-2 upregulated T-bet and IL-2Rα mRNA, as well as CD25 surface expression (Figures 7A and 7B). IL-2Rα upregulation occurred prior to the initial cell division (Figure 7C) in a manner that was largely resistant to treatment with blocking antibodies to IL-2 (Figure 7D; Figure S5), suggesting that IL-2Rα expression was induced prior to, and independently of, feedback through IL-2/STAT5. In addition, inflammation increased the peak amount of IL-2Rα mRNA and surface protein expression in cells cultured in low IL-2 and sustained CD25 expression beyond the precipitous drop otherwise seen at day 3 in the absence of inflammation (Figures 7A and 7B). Although inflammation itself enhanced IL-2Rα expression, high IL-2 still upregulated IL-2Rα expression further in cells primed with inflammation (Figure 7A). Together, these data showed that by increasing the early expression of IL-2Rα, inflammation enhanced the duration and responsiveness of activated CD8+ T cells to IL-2 signals.
Figure 7. Inflammatory Signals and IL-2 Signal Strength Induce Distinct Transcriptional Regimes.
(A) The kinetics of mRNA expression in the presence and absence of inflammation. Naive P14 CD8+ T cells were stimulated by coculture with GP33 peptide (blue) and APCs, with or without CpG (red), for 2 days (d2) and then were recultured, with or without IL-12 (CpG/IL-12), in low (10 U/ml) or high (100 U/ml) IL-2. See also Figure S5.
(B) The kinetics of surface IL-2Rα expression. P14 CD8+ T cells were primed with GP33 and APCs and cultured under the indicated conditions. IL-2Rα expression was determined by flow cytometry.
(C) IL-2Rα expression prior to the first cell division. Naive P14 cells were loaded with CFSE and primed as shown in (B). After 36 hr, cells that had not diluted CFSE were gated, and IL-2Rα expression was determined by flow cytometry and is shown relative to naive cells stained prior to stimulation.
(D) IL-2Rα expression in the presence of blocking αIL-2 antibody during priming. Cells were cultured as in (B).
Nevertheless, some of the effects of high IL-2, namely upregulation of perforin and Eomes, were different when inflammatory signals were present (Figure 7A). Inflammatory signals during priming impaired the ability of high IL-2 to induce Eomes and perforin at later times (Figure 7A; Figure S5). In addition, they impaired the ability of high IL-2 to repress IL-7Rα mRNA expression. Thus, inflammatory stimuli prolonged IL-2 responsiveness but, paradoxically, also interfered with the ability of IL-2 to regulate certain genes.
DISCUSSION
Here, we tested the hypothesis that different “strengths” of IL-2 stimulation induce different transcriptional responses that alter CD8+ T cell differentiation. We showed that increasing IL-2R signal strength promoted effector CTL differentiation in a simple cell-culture system and that IL-2R signals were required for normal gene expression and accumulation of effector CTL during viral infection. Moreover, we showed that inflammatory stimuli (CpG/IL-12) potentiated early IL-2 responsiveness but simultaneously altered the transcriptional programs induced by IL-2. Our findings are consistent with those of an accompanying study (Kalia et al., 2010) reporting that viral infection induces some effector cells to express more IL-2Rα for a longer duration than others and that these cells are both more responsive to IL-2 and more prone to differentiate into effector rather than memory CTL.
CD8+ T cell differentiation upon infection is complex and integrates multiple signals including strength of TCR stimulus and costimulation, IL-2R signals, and inflammation (Williams and Bevan, 2007). We found that IL-2 regulated perforin and granzyme B expression independently of inflammation (CpG and IL-12), a result that seems to contradict previous studies showing that inflammatory signals were obligatory for inducing cytolytic function (Curtsinger et al., 2003). The difference could lie in the strength of initial TCR signals: Curstsinger et al. primed naive cells with MHC I-peptide and B7-coated microspheres to mimic APC, and under these conditions, initial IL-2Rα induction was low and transient and strongly dependent on inflammation for sustained expression in the presence of low IL-2 concentrations (Curtsinger et al., 2003; Valenzuela et al., 2002). In contrast, we used plate-bound anti-CD3 and anti-CD28 or live APC with high concentrations of peptide, conditions that maintained IL-2Rα expression without a requirement for inflammatory signals. Our studies revealed that IL-2 enhanced perforin transcription, whereas CpG and IL-12 did not. We hypothesize that in conditions of weak TCR stimulation, inflammation enables the IL-2 responsiveness that is necessary for expression of perforin and granzyme B; however, inflammation is less critical under conditions of strong TCR stimulation.
Increased inflammation promotes “short-lived” effector CTL development, programs clonal contraction, and induces CTL effector functions (Badovinac et al., 2004; Curtsinger et al., 2003; Joshi et al., 2007). Somewhat counterintuitively, inflammatory signals also appear to be required for memory CTL development (Xiao et al., 2009). In the simplified cell-culture setting, our results showed that inflammation could regulate both effector and memory CTL development in the context of different strengths of IL-2 signals. On the one hand, inflammatory signals increased T-bet and IL-2Rα expression even in low IL-2 conditions, whereas without inflammation, strong IL-2R signals increased Eomes, perforin, and granzyme B expression. Thus, both inflammation and IL-2 promoted aspects of effector CTL differentiation. On the other hand, however, as best seen in our high IL-2 conditions, inflammatory signals attenuated the late expression of perforin and Eomes and increased IL-7Rα expression, counteracting certain effects of persistent IL-2 signals. Thus, the generation of effector and memory CTL during infection in vivo is likely to be determined by the relative balance of TCR, costimulatory, inflammatory, IL-2, and IL-15 signals encountered by individual responding cells and possibly additional unidentified signals as well.
Analysis of CD8+ T cell differentiation after activation in vitro and the CD8+ T cell response to LCMV in the absence of IL-2R signals in vivo supports two conclusions regarding the roles of IL-2. First, IL-2Rα-deficient CD8+ T cells in vivo resembled cells cultured in low concentrations of IL-2: both types of cells showed reduced perforin and granzyme B expression, as well as premature re-expression of IL-7Rα, CD62L, and Bcl6. Thus, strength of IL-2 stimulation directly regulated expression of genes characteristic of both effector and memory CTL. Second, IL-2Rα-deficient CD8+ T cells developed proportionally fewer KLRG-1-and T-bet-expressing effector cells relative to wild-type CD8+ T cells near the peak of the LCMV response. This was unlikely to be due to a direct effect of IL-2 upon T-bet expression because IL-2 did not regulate T-bet in CD8+ T cells primed in culture. Therefore, IL-2 appeared to drive selectively the accumulation of primed effector CD8+ T cells that had already induced T-bet. These two conclusions are consistent with those of an accompanying study, demonstrating that CD25hi effector CD8+ T cells (i.e., more IL-2-responsive) generated during LCMV infection exhibited a more effector-like gene expression profile, proliferated more extensively at the tail-end of the effector phase, and contributed inefficiently to the formation of memory CTL relative to CD25lo effector cells (Kalia et al., 2010).
Our results help to clarify the regulation of T-bet and Eomes during CD8+ T cell activation. As expected, T-bet was induced upon TCR stimulation; however, we found that Eomes was induced in response to IL-2 several days after removal from the TCR stimulus. The induction of T-bet by TCR signals, and Eomes by IL-2 stimulation, might partially explain the sequential upregulation of T-bet and Eomes during infection (Intlekofer et al., 2005). In addition, we found that the presence of CpG at priming was sufficient to repress later Eomes induction by IL-2 (data not shown). Previous studies have shown that T-bet and Eomes are inversely regulated in activated CD8+ T cells by IL-12 (Takemoto et al., 2006). However, CpG stimuli did not enhance T-bet expression, unless exogenous IL-12 was also provided (Joshi et al., 2007; Szabo et al., 2002). Thus, T-bet-independent pathways are likely to prevent Eomes upregulation by effector cells.
Both T-bet and Eomes can positively regulate IL-2Rβ expression in CTL (Intlekofer et al., 2008; Intlekofer et al., 2005; Joshi et al., 2007). Here, we showed that IL-2 induced Eomes. In conjunction with the previous studies, our results are consistent with the hypothesis that T-bet and Eomes operate in feedback loops. For example, T-bet could initiate elevated IL-2Rα expression to enable later IL-2-induced Eomes upregulation that feeds back to increase Il2rb even further; Eomes together with phospho-STAT5 would then feed forward to activate late effector genes such as Prf1. Our data suggest that inflammation could be important to initiate this process by enhancing T-bet and IL-2Rα expression. However, we also showed that strong inflammation inhibited Eomes upregulation by IL-2 and, thus, could result in effector cells that fail to upregulate Eomes and IL-2Rβ expression efficiently. Thus, the paradoxical positive and negative effects of inflammation on the development of both effector and memory CTL in vivo could relate in part to positive and negative effects of inflammation on IL-2-regulated gene expression.
Finally, our data confirm previous studies implicating IL-2 as an important regulator of Prf1 transcription (Zhang et al., 1999) and provide additional mechanistic insights as to how IL-2 mediates Prf1 gene activation. Under the conditions we examined, IL-2 did not induce global chromatin remodeling of previously reported DHS sites (Pipkin et al., 2007) but acted on a previously “opened” locus. Our data suggest that the main mechanism of gene activation by IL-2 is increased transcription initiation via increased RNA Pol II recruitment to the TSS, as opposed to stimulated elongation of preinitiated Pol II complexes. In addition, RNA Pol II loading at the promoter was coordinated with binding of endogenous STAT5 at an enhancer located at −1 kb, previously shown to be controlled by STAT5 in transient reporter assays (Zhang et al., 1999). Moreover, Eomes bound to the −1 kb enhancer and the TSS concurrently with STAT5 and RNA Pol II, suggesting that STAT5 and Eomes might participate in recruiting RNA Pol II. At the same time, RNA Pol II was recruited away from the Il7ra transcription start site under persistent IL-2 signaling, suggesting that IL-2-mediated regulation of RNA Pol II recruitment to and away from accessible genes might be a general mechanism for controlling the differentiation of activated CD8+ T cells.
EXPERIMENTAL PROCEDURES
Mice, Chimeras, and Infections
CD8+ T cells were isolated from 4- to 12-week-old Tcrα −/− × P14 TCR transgenic (Taconic), C57BL6/J or Tbx21−/− mice (Jackson Laboratory). P14 CD8+ T cells were uniformly naive based on staining with antibodies recognizing CD25, CD44, CD62L, and CD122. All mice were maintained in specific pathogen-free barrier facilities and used according to protocols approved by the Immune Disease Institute and Harvard Medical School animal care and use committees. For bone marrow chimeric mice and LCMV infection, 6- to 8-week-old B6.SJL-PtprcaPep3b/BoyJ (CD45.1) mice were purchased from the Jackson Laboratory. B6.SJL × B6 (CD45.1/CD45.2, heterozygote) and B6.129S4-Il2ratm1Dw/J (Il2ra−/−, CD45.2) mice were bred in our facility at the University of Washington (Seattle, WA) under specific pathogen-free conditions. Bone marrow preparations from the femur and tibia of donor mice were incubated with anti-CD3-, CD4- and CD8-biotin, followed by anti-biotin magnetic beads, and applied to LS columns for depletion of T cells (Miltenyi). 2–5 × 106 total T cell-depleted bone marrow cells, containing a roughly 1:1 mixture of wild-type and IL-2Rα-deficient bone marrow, were injected intravenously into lethally irradiated B6.SJL hosts (1000 Rad). Mice were infected with 2 × 105 PFU of LCMV-Armstrong intraperitoneally 10–12 weeks posttransplant.
Isolation, Culture, and Retroviral Transduction of Primary CD8+ T Cells
CD8+ T cells were purified (>95% purity) by negative selection (Invitrogen) from RBC-lysed single-cell suspensions from pooled spleen and lymph node cells. For stimulation, purified CD8+ T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum, 2 mM L-glutamine, penicillin-streptomycin, nonessential amino acids, sodium pyruvate, vitamins, 10 mM HEPES, and 50 µM 2-mercaptoethanol as 4 × 105 cells/cm2 in T25 flasks coated with anti-CD3 (clone 2C11) and anti-CD28 (clone 37.51) (1 µg/ml) by precoating with 300 µg/ml goat anti-hamster IgG. After 48 hr, cells were removed from the TCR signal and recultured at a concentration of 5 × 105 cells/ml in media supplemented with the indicated concentration of recombinant human IL-2 (rhIL-2). For activation of P14 CD8+ T cells with APCs, combined spleen and lymph nodes were used to generate single-cell suspensions after RBC lysis. Washed cells resuspended in complete in T cell media as 4 × 106 cells/ml and GP33 peptide was added to 1 µM final concentration and were untreated or supplemented with 3 ng/ml CpG (ODN1826, Invivogen, Inc.). After 48 hr, CD8+ T cells were purified by negative selection for direct analysis, or cocultures were counted and recultured as 5 × 105 cells/ml in rhIL-2-containing media either with or without a single dose of IL-12 to 5 ng/ml. Purified naive P14 CD8+ T cells were also activated with mitomycin C-treated APCs from B6 Tcra−/− spleens and gave similar results (data not shown). Every 24 hr, cells were counted and readjusted to 5 × 105 cells/ml with fresh media containing 10 or 100 U/ml rhIL-2. Viral supernatants were generated by transfection of Phoenix packaging cells and concentration by overnight centrifugation at 6000 × g. At ~42 hr after TCR priming of 106 CD8+ T cells in 1 ml per well in 12-well plates, the culture media was replaced with complete media supplemented with 8 µg/ml polybrene containing concentrated virus. The plates were centrifuged at 700 × g for 1 hr at room temperature and then incubated at 37°C for 5 hr. Retroviral constructs for Eomes-VP16 and the MIG control empty vector were a gift from Dr. Steve L. Reiner (Intlekofer et al., 2005).
Cytotoxicity Assays
For flow cytometric killing analysis, EL-4 thymoma target cells were loaded with 0 or 1 µM GP33 peptide for 2 hr before a 2 hr coincubation with P14 CD8+ T cells in 96-well round-bottom plates (Cruz-Guilloty et al., 2009). After coincubation, cells were stained with AnnexinV-FITC and anti-CD8+-APC. For standard killing assays, bulk, polyclonal CD44hi wild-type, and Il2ra−/− CD8+ effector T cells from spleens of mixed BMC mice, infected 8 days prior with LCMV, were FACS-sorted based on congenic marker expression. EL-4 cells were labeled with 51Cr (Perkin Elmer) for 1 hr at 37°C, washed extensively, and then incubated with effector cells (adjusted for proportion of antigen-specific cells based on staining an aliquot of each population with Db-GP33 tetramer) with either 10 µM GP33 peptide or no peptide for 4 to 5 hr at 37°C. Percent specific lysis from duplicate or triplicate wells was determined relative to the spontaneous (targets alone) and total release (2% Triton X-100 detergent) controls, as described previously (Tyznik et al., 2004).
Chromatin Immunoprecipitation and Real-Time PCR Analysis
Chromatin immunoprecipitation (ChIP) was performed as described (Cruz-Guilloty et al., 2009). Briefly, formaldehyde-fixed chromatin was isolated from 2–4 × 107 CD8+ T cells for each immunoprecipitation and was sonciated to yield 0.5–1 kb chromatin fragments. Immunoprecipitation was performed by adding optimized antibody amounts (see Supplemental Experimental Procedures), followed by overnight incubation at 4°C; protein A-sepharose beads were added for the last 3 hr of the incubation. After bead washes, chromatin was treated with RNase A for 1 hr at 37°C, followed by addition of Proteinase K and overnight incubation at 65°C to reverse crosslinking, and DNA was purified with QIAquick columns (QIAGEN). For real-time PCR detection of immunoprecipitated targets using the SYBR Green PCR kit, a standard curve was generated for each sample based on amplification of serial dilutions of input DNA. ChIP DNA PCR reactions were performed in duplicates. Agarose gel analysis and melt curves were analyzed to ensure amplification of specific target sequences.
Additional Experimental Procedures
Additional Experimental Procedures can be found in Supplemental Experimental Procedures available with this article online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. S. Reiner for generously providing the Eomes-VP16 retroviral construct. This work was funded by NIH grants AI44432 and AI707088 (to A.R.) and AI19335 (to M.J.B.); University of Miami Developmental Center for AIDS Research 5P30AI073961 (to M.G.L.); and the National Cancer Institute F32CA126247 (to M.E.P). F.C.G. was a predoctoral fellow of the Ryan Foundation and was supported by a Ford Foundation Predoctoral Fellowship.
Footnotes
SUPPLEMENTAL INFORMATION
The Supplemental Information include five figures, Supplemental Experimental Procedures, and one table and can be found with this article online at doi:10.1016/j.immuni.2009.11.012.
REFERENCES
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- Bachmann MF, Oxenius A. Interleukin 2: from immunostimulation to immunoregulation and back again. EMBO Rep. 2007;8:1142–1148. doi: 10.1038/sj.embor.7401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann MF, Wolint P, Walton S, Schwarz K, Oxenius A. Differential role of IL-2R signaling for CD8+ T cell responses in acute and chronic viral infections. Eur. J. Immunol. 2007;37:1502–1512. doi: 10.1002/eji.200637023. [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Harty JT. Manipulating the rate of memory CD8+ T cell generation after acute infection. J. Immunol. 2007;179:53–63. doi: 10.4049/jimmunol.179.1.53. [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Porter BB, Harty JT. CD8+ T cell contraction is controlled by early inflammation. Nat. Immunol. 2004;5:809–817. doi: 10.1038/ni1098. [DOI] [PubMed] [Google Scholar]
- Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323:505–509. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrio R, Bathe OF, Malek TR. Initial antigen encounter programs CD8+ T cells competent to develop into memory cells that are activated in an antigen-free, IL-7- and IL-15-rich environment. J. Immunol. 2004;172:7315–7323. doi: 10.4049/jimmunol.172.12.7315. [DOI] [PubMed] [Google Scholar]
- Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger JM, Lins DC, Mescher MF. Signal 3 determines tolerance versus full activation of naive CD8 T cells: dissociating proliferation and development of effector function. J. Exp. Med. 2003;197:1141–1151. doi: 10.1084/jem.20021910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza WN, Schluns KS, Masopust D, Lefrancois L. Essential role for IL-2 in the regulation of antiviral extralymphoid CD8 T cell responses. J. Immunol. 2002;168:5566–5572. doi: 10.4049/jimmunol.168.11.5566. [DOI] [PubMed] [Google Scholar]
- Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J. Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452:356–360. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]
- Harty JT, Tvinnereim AR, White DW. CD8+ T cell effector mechanisms in resistance to infection. Annu. Rev. Immunol. 2000;18:275–308. doi: 10.1146/annurev.immunol.18.1.275. [DOI] [PubMed] [Google Scholar]
- Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, Arima M, Kuroda Y, Tokuhisa T. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat. Immunol. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW, Leonard WJ. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J. Exp. Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BJ, Costelloe EO, Fitzpatrick DR, Haanen JB, Schumacher TN, Brown LE, Kelso A. Single-cell perforin and granzyme expression reveals the anatomical localization of effector CD8+ T cells in influenza virus-infected mice. Proc. Natl. Acad. Sci. USA. 2003;100:2657–2662. doi: 10.1073/pnas.0538056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat. Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Wherry EJ. Heterogeneity and Cell-Fate Decisions in Effector and Memory CD8(+) T Cell Differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. 2Rα expression on virus-specific CD8+ T cells directs terminal effector differentiation in vivo. Immunity. 2010;32(this issue):91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Malek TR. The biology of interleukin-2. Annu. Rev. Immunol. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- Malek TR, Yu A, Scibelli P, Lichtenheld MG, Codias EK. Broad programming by IL-2 receptor signaling for extended growth to multiple cytokines and functional maturation of antigen-activated T cells. J. Immunol. 2001;166:1675–1683. doi: 10.4049/jimmunol.166.3.1675. [DOI] [PubMed] [Google Scholar]
- Manjunath N, Shankar P, Wan J, Weninger W, Crowley MA, Hieshima K, Springer TA, Fan X, Shen H, Lieberman J, von Andrian UH. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J. Clin. Invest. 2001;108:871–878. doi: 10.1172/JCI13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado R, Vijh S, Allen SE, Kerksiek K, Pilip IM, Pamer EG. Early programming of T cell populations responding to bacterial infection. J. Immunol. 2000;165:6833–6839. doi: 10.4049/jimmunol.165.12.6833. [DOI] [PubMed] [Google Scholar]
- Mescher MF, Curtsinger JM, Agarwal P, Casey KA, Gerner M, Hammerbeck CD, Popescu F, Xiao Z. Signals required for programming effector and memory development by CD8+ T cells. Immunol. Rev. 2006;211:81–92. doi: 10.1111/j.0105-2896.2006.00382.x. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Liu XW, Wynshaw-Boris A, Rosenthal LA, Imada K, Finbloom DS, Hennighausen L, Leonard WJ. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity. 1997;7:691–701. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- Opferman JT, Ober BT, Ashton-Rickardt PG. Linear differentiation of cytotoxic effectors into memory T lymphocytes. Science. 1999;283:1745–1748. doi: 10.1126/science.283.5408.1745. [DOI] [PubMed] [Google Scholar]
- Peixoto A, Evaristo C, Munitic I, Monteiro M, Charbit A, Rocha B, Veiga-Fernandes H. CD8 single-cell gene coexpression reveals three different effector types present at distinct phases of the immune response. J. Exp. Med. 2007;204:1193–1205. doi: 10.1084/jem.20062349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Lichtenheld MG. A reliable method to display authentic DNase I hypersensitive sites at long-ranges in single-copy genes from large genomes. Nucleic Acids Res. 2006;34:e34. doi: 10.1093/nar/gkl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Lieberman J. Delivering the kiss of death: progress on understanding how perforin works. Curr. Opin. Immunol. 2007;19:301–308. doi: 10.1016/j.coi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Pipkin ME, Ljutic B, Cruz-Guilloty F, Nouzova M, Rao A, Zuniga-Pflucker JC, Lichtenheld MG. Chromosome transfer activates and delineates a locus control region for perforin. Immunity. 2007;26:29–41. doi: 10.1016/j.immuni.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Prlic M, Hernandez-Hoyos G, Bevan MJ. Duration of the initial TCR stimulus controls the magnitude but not functionality of the CD8+ T cell response. J. Exp. Med. 2006;203:2135–2143. doi: 10.1084/jem.20060928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 2006;177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- Teixeiro E, Daniels MA, Hamilton SE, Schrum AG, Bragado R, Jameson SC, Palmer E. Different T cell receptor signals determine CD8+ memory versus effector development. Science. 2009;323:502–505. doi: 10.1126/science.1163612. [DOI] [PubMed] [Google Scholar]
- Tyznik AJ, Sun JC, Bevan MJ. The CD8 population in CD4-deficient mice is heavily contaminated with MHC class II-restricted T cells. J. Exp. Med. 2004;199:559–565. doi: 10.1084/jem.20031961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela J, Schmidt C, Mescher M. The roles of IL-12 in providing a third signal for clonal expansion of naive CD8 T cells. J. Immunol. 2002;169:6842–6849. doi: 10.4049/jimmunol.169.12.6842. [DOI] [PubMed] [Google Scholar]
- van Stipdonk MJ, Lemmens EE, Schoenberger SP. Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat. Immunol. 2001;2:423–429. doi: 10.1038/87730. [DOI] [PubMed] [Google Scholar]
- Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu. Rev. Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- Williams MA, Tyznik AJ, Bevan MJ. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441:890–893. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Casey KA, Jameson SC, Curtsinger JM, Mescher MF. Programming for CD8 T cell memory development requires IL-12 or type I IFN. J. Immunol. 2009;182:2786–2794. doi: 10.4049/jimmunol.0803484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss DM, Sijts AJ, Mosmann TR. Enumeration of cytotoxic CD8 T cells ex vivo during the response to Listeria monocytogenes infection. Infect. Immun. 2008;76:4609–4614. doi: 10.1128/IAI.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Scordi I, Smyth MJ, Lichtenheld MG. Interleukin 2 receptor signaling regulates the perforin gene through signal transducer and activator of transcription (Stat)5 activation of two enhancers. J. Exp. Med. 1999;190:1297–1308. doi: 10.1084/jem.190.9.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.