Abstract
PURPOSE
To define the molecular signature of limbal SP cells and identify signaling pathways associated with the phenotype of these putative stem cells.
METHODS
Primary cultures of pig limbal epithelial cells stained with Hoechst 33342 were sorted by flow cytometry into SP and non-SP cells, and purified RNA was processed for microarray analysis with an oligonucleotide spotted array. Expressed transcripts for which SP and non-SP expressions differed by more that 1.5-fold in each paired set and by twofold overall were considered to be differentially expressed. Differential expression was validated by quantitative PCR and immunostaining. Data-mining methods were used to identify cellular processes that are either salient or depressed in the SP cells.
RESULTS
The microarray identified approximately 9000 distinct, expressed, and identifiable genes. Of those, 382 and 296 were either over- or underexpressed in the SP cells, respectively. Overrepresentation analysis indicated that SP cells are in a low metabolic and biosynthetic state. In addition, a pattern of elevated MXD1, MAXI2, DUSP5, p27/KIP1, and p57/KIP2 and decreased Cyclin D and CDK genes can be expected to slow intrinsic and mitogen-induced G1-to-S cell cycle transition. SP cells were also rich in genes associated with stem cell phenotype and genes providing protection against oxidative and/or xenobiotic damage.
CONCLUSIONS
Microarray analysis of pig limbal SP cells yielded a molecular signature underscoring a phenotype characterized by slow cycling and low metabolic activity. The results provide valuable insights for the preservation and/or replication of epithelial stem cells.
Stem cells are critical for the function of the ocular surface. In the limbocorneal system, stem cells are concentrated in the limbus.1–5 Limbal damage due to chemical or thermal injury, microbial infection, or autoimmune reactions results in limbal stem cell deficiencies reflected in corneal opacities, neovascularization, and/or generalized inflammation.6–8 Advances in the experimental research and clinical application of limbal transplantation for the treatment of limbal stem deficiencies have been rapid, progressing from the straightforward transplantation of contralateral biopsies to pre-expansion of the donor material in culture.9–13
The isolation and characterization of limbal stem cells should facilitate optimal enhancement of precursor cell proliferation during epithelial cell expansion in culture, thereby increasing the reconstructive capacity of expanded cell patches and reducing the size of the initial donor cell pool that needs to be excised from a healthy contralateral eye for successful transplantation.
During the past decade, putative stem cells have been isolated from multiple organs by using an established correlation between stemness and the ability to efflux large aromatic compounds, in particular Hoechst 33342 by the ABCG2/BCRP transporter.14–18 Hoechst 33342-transporting cells are easily recognized and sorted by flow cytometry on the basis of their fluorescent emission characteristics and are presently referred as side population (SP) cells. Several laboratories, including ours, have isolated SP cells from mammalian CNJ and limbal epithelia and subjected these to stemness tests.19–25 Using in vivo BrdU labeling and long-term (>2 months) chasing in young rabbits, we showed that in the epithelia (E) of both limbus and conjunctiva (CNJ), SP cells were highly enriched in cells that have been slow cycling in vivo and displayed other features associated with stem cells in vitro.21 In most organs examined, SP cells are quite rare; they typically amount to between 0.05% and 0.5% of the total cells in each studied tissue or organ. These low numbers are consistent with the experiments in bone marrow indicating that SP cells are between the most primitive, or basic, stem cell.26
We have recently completed a microarray-based study of differentially expressed genes, or molecular signature, of SP cells isolated from the human CNJE.27 The rarity of these cells poses unique challenges and their investigation, such as in differential gene expression studies, has been problematic. Preparations of cadaveric donor corneas typically yielded 250,000 to 500,000 limbal cells and fewer than 1,000 SP cells. Hence, with a view to delineating a molecular signature of limbal SP cells, we opted in this study to employ specimens obtained from the pig, a species from which we could simultaneously obtain a large number of fresh corneas from young animals and for which microarrays are commercially available.
Many of the genes differentially expressed in the limbal SP cells may underpin functional features that underscore the SP cell elsewhere and/or universal features of stem cells, such as in vivo slow cycling. Hence, to probe for the possible general relevance of genes that are differentially expressed genes in the limbal SP cells, we performed similar microarray measurements for the pig CNJE, a tissue that shares with the corneal epithelium a common environment, developmental origin, and PAX6 expression28,29 and used these results and recently obtained microarray data for the human CNJE27 for comparative assessments.
METHODS
Tissue Procurement, Cell Isolation, and Culture
Fresh pig eyes with intact eyelids enucleated from killed 3-month-old pigs were delivered within 30 hours of excision by Pel Freeze (Rogers, AR). A human cornea from an unidentifiable adult Caucasian male was obtained from the National Disease Research Interchange (NDRI, Philadelphia, PA).
Pig corneas were excised and, after careful removal of all perilimbal conjunctival tissue, limbal strips were microdissected into quarter sections. The limbal strips and quartered conjunctivas were incubated for 16 to 20 hours at 4°C in 5 mg/mL Dispase (Roche, Nutley, NJ) dissolved in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-buffered (hb), penicillin-streptomycin–complemented Dulbecco’s modified Eagle’s medium and Ham’s F12 1:1 mix (D/F-12), with a side-to-side tilting motion. Most of the limbal epithelial sheets spontaneously fully or partially separated from the underlying stroma and are easily scooped out of the enzymatic solution. Conjunctival sheets were separated from the subepithelial matrix by gentle mechanical prodding. Epithelial sheets were trypsinized for 20 to 25 minutes at 37°C with orbital agitation. The dissociated cells were sieved through 100- and 40-µm filters, pelleted, resuspended in bicarbonate-buffered D/F12 complemented with 5% FBS, 10 ng/mL cholera toxin, 10 ng/mL epidermal growth factor (EGF), insulin, transferrin, selenium, and 0.5% dimethyl sulfoxide (SHEM), plated in 75-cm2 flasks at a density of 80,000 to 100,000 cells/cm2 and cultured for 16 hours at 37°C in a 5% CO2 incubator. The three limbal experiments performed used 30 corneas each and the duplicate conjunctival experiments were based on pools of four tissues. Unless stated otherwise, all reagents were purchased from Sigma (St. Louis, MO).
Flow Cytometry
After overnight culture, the medium was refreshed with prewarmed solution to remove floating cells (60%–70% of the total plated) and complemented with 5 µg/mL Hoechst 33342 for 1.5 hours. The treated cells were then released by quick trypsinization, spun down, and resuspended in ice-cold phenol red-free D/F12 complemented with 4% serum and 1 µg/mL propidium iodide (PI). Cell sorting was performed with a flow cytometer (INFLUX; Cytopeia, Seattle, WA). The source tube was maintained at 4°C and SP and non-SP (nSP) cells were sorted directly into 750 µL RNA isolation solution (Tri-Reagent LS; Molecular Research Center [MRC] Cincinnati, OH). We used a specific range of forward (FSC) and side (SSC) light-scattering levels to exclude the great majority of lymphocytes (cells with very low SSC) and to limit the amount of other complex nonepithelial cells present within the epithelial strata (cells with very high SSC such as melanocytes and dendritic cells).27 Fumitremorgin C (FTC; a generous gift from Susan Bates, National Cancer Institute, Bethesda, MD) was used to determine the involvement of ABCG2 in the observed SP cells.22
Microarray Processing
RNA isolation solutions containing the collected SP and nSP cells were adjusted to 1.0 mL with the addition of H2O and 1 µL poly acryl carrier (MRC). RNA was isolated according to the manufacturer’s instructions and dissolved in 5-µL water. RNA yields, determined using a quantitation reagent (RiboGreen; Molecular Probes, Eugene, OR), ranged from 2.0 to 2.4 pg RNA/cell. RNA integrity was evaluated with microarrays (2100 Bioanalyzer 6000 nanochips; Agilent Technologies, Santa Clara, CA). cDNA was synthesized with reverse transcriptase (SuperScript Choice; Invitrogen, Rockville, MD) and a modified oligo dT primer (Affymetrix, Santa Clara, CA). The initial cDNA was subjected to two cycles of in vitro transcription (IVT; ENZO BioArray HighYield Kit; Affymetrix) to generate amplified biotin-labeled cRNA. After electro-phoretic assessment of the suitability of this cRNA product for microarray processing, appropriately fragmented (restricted) cRNA was hybridized to the gene microarray (GeneChip Porcine Genome Array; probe sets interrogate approximately 23,256 transcripts; Affymetrix). Hybridized microarrays were stained with a streptavidin-phycoerythrin reagent and fluorescence images were captured with a laser scanner (G2500A; Agilent).
Data Analyses
Normalized signal intensities (SIs; relative measures of gene expression) and signal quality properties in the form of gene present (P) or not-present calls were extracted from the fluorescent images (Microarray Suite, ver. 5.0 [MAS 5.0]; Affymetrix) and collected in spreadsheet format (lists) for downstream processing. To identify transcripts that are differentially expressed for the SP/nSP comparison, we first derived an expressed transcript spreadsheet incorporating only those transcripts that received the MAS5 present rating in all three experiments in at least one cell type: SP or nSP. The Hierarchical Clustering Explorer (http://www.cs.umd.edu/hcil/hce/ provided in the public domain by the Human Computer Interaction Lab, University of Maryland, College Park, MD) were used to generate a heat map for these transcripts. From the list of expressed genes we derived two primary differential lists, one incorporating only those transcripts for which the SP/nSP expression (SI) ratio (R) is greater than 1.5 in each of the three SP/nSP comparisons conducted and a second one limited to those transcripts for which the same ratios were less than 1:1.5. Transcripts included in these primary lists were considered over- or underexpressed if the overall SP/nSP expression ratio was greater than 2.0 or smaller than 0.5, respectively. The combination of these two latter lists is taken to represent a SP molecular signature. The duplicate experiments from pig conjunctiva and four replicates of human conjunctival epithelium27 were processed in an identical manner.
Pig microarray transcript-to-gene conversions were based on multispecies gene homology annotations generously provided by Christopher K. Tuggle (Iowa State University, Ames, IA; Couture et al., manuscript in preparation). A shortened description of the annotation methodology is provided in Wang et al.30 The list was further enriched by adding annotations (from the NetAffx Analysis Center; Affymetrix; www.affymetrix.com/analysis/index.affx) that were not included in the data from Iowa State University.
Annotations indicating moderate to strong homology to known genes from other species were used to derive HGNC (HUGO Gene Nomenclature Committee) gene symbols for the primary differential lists. The HGNC symbols lists were submitted to The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/ provided in the public domain by the National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD) to identify overrepresented biological or molecular processes within the differentially expressed transcripts. DAVID analysis probes each gene list against a relevant population list, and calculates scores as probabilities and/or false-discovery rates (FDRs) reporting on the likelihood of overrepresentation within the ontological systems derived from the Gene Ontology (GO) consortia and other public genomic resources.
Real-Time PCR
Total RNA was prepared from 23,500 sorted pig limbal SP and nSP cells. Three quarters of this RNA was reverse transcribed (Omniscript enzyme; RT+ product; Qiagen). The remaining RNA was processed in parallel under identical conditions but with omission of the enzyme, to generate an RT− product. Real-time PCR (40 synthesis cycles at 60°C) of the RT+ (triplicates, each using one twelfth of the total RT reaction) and RT− (single tube using one fourth of the reaction) products were performed in a sequence-detection system (Prism 7900HT; Applied Biosystems, Foster City, CA; using the RT2 SYBR Green/ROX PCR Master Mix; SuperArrays, Frederick, MD), with PCR forward and reverse primers for pig β-actin (aagtactccgtgtggatcgg and cagtccgccta-gaagcattt; yields 131-bp amplicon), CXCR4 (ctgagaagcatgacggacaa and tcccaaagtaccagtttgcc; 113-bp amplicon), GPX2 (aacagcctcaagtacgtccg and gtcgtcataagggtagggca; 133-bp amplicon), and DUSP5 (ggttgaagcag-caagatggt and gagaggtgcaacgagaaagg; 113 bp amplicon). SP/nSP message ratios in the amplified material were calculated from cycle thresholds (Cts) according to the ΔΔCt method, by using β-actin Cts for normalization.27
Immunostaining
Cultures in the printed glass slides were stained with FITC-conjugated anti-keratin14 (clone LL002; Abcam, Cambridge, MA). Cryosections of pig and human limbocorneal sections were stained using an antibody against aquaporin 3 (a generous gift from Dennis Brown, Harvard Medical School, Cambridge, MA).
RESULTS
SPs in Briefly Cultured Pig Limbal and Conjunctival Cells
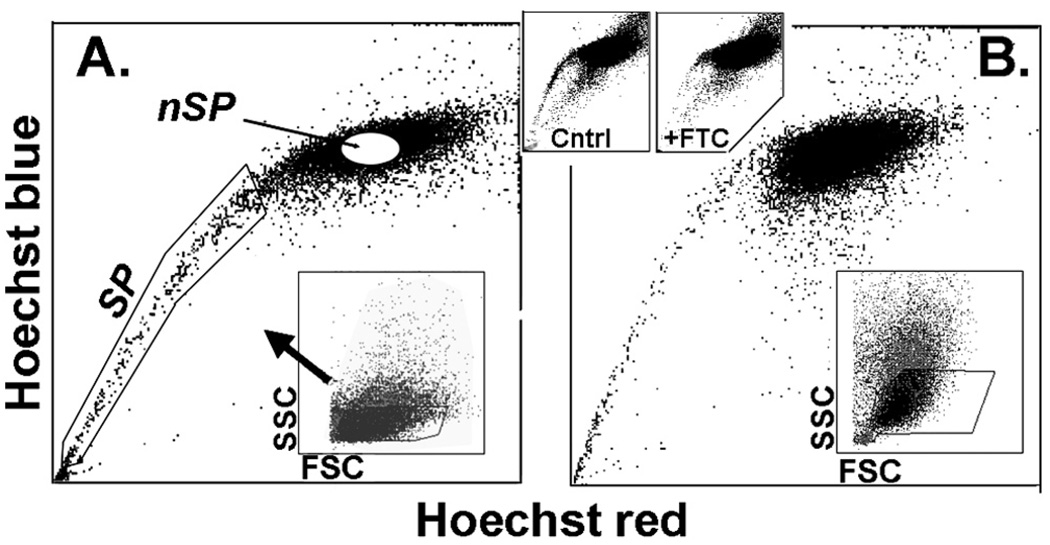
After an overnight culture in SHEM between 20% and 25% of pig limbal cells harvested by trypsinization firmly adhered to the plastic substratum. Incubation of the attached cells with Hoechst 33342 led to well-defined side populations for both limbal and conjunctival cells (Fig. 1). Those side populations extended from the main Hoechst saturated G0/G1 group down to cells that fully excluded the dye. This degree of Hoechst exclusion is higher than we observed in earlier studies with cells incubated in suspension soon after trypsinization.22 This improvement may reflect a better metabolic status of the cells, on which Hoechst efflux is dependent, after recovery in culture. Limbal SP cells accounted for 0.89% (n = 5) of the live gated cells. As previously shown for human or rabbit ocular surface cells, the SP feature was largely abolished when the incubation included the ABCG2-specific inhibitor FTC.
FIGURE 1.
Side populations in pig ocular surface cells. (A) Limbus. (B) Conjunctiva. SP and nSP cell regions are indicated in (A). Bottom insets: images indicate that cells were collected only from limited side (SSC)- and forward (FSC)-scatter ranges. Top insets: data show that the SP was greatly reduced by inclusion of the ABCG2-specific inhibitor FTC.
RNA and Microarray Quality Controls
The need to trypsinize, centrifuge, resuspend, and maintain the epithelial cells in cold storage for 20 to 40 minutes before sorting and lysis resulted in moderate DNA damage as assessed by biochip (2100 Bioanalyzer; Agilent) (data not shown). Although the damage was within acceptable limits, careful retrospective analysis of data integrity was warranted. In addition to RNA damage before amplification, normal early termination of the processivity of the reverse transcription and IVT enzymes invariably contribute to a gradual decrease in the concentration of available probe as a function of the probed region distance from the priming (polyA) 3′ end. If these factors occur unevenly across the samples, the SI ratio could display a dependence on probe location relative to that priming end.
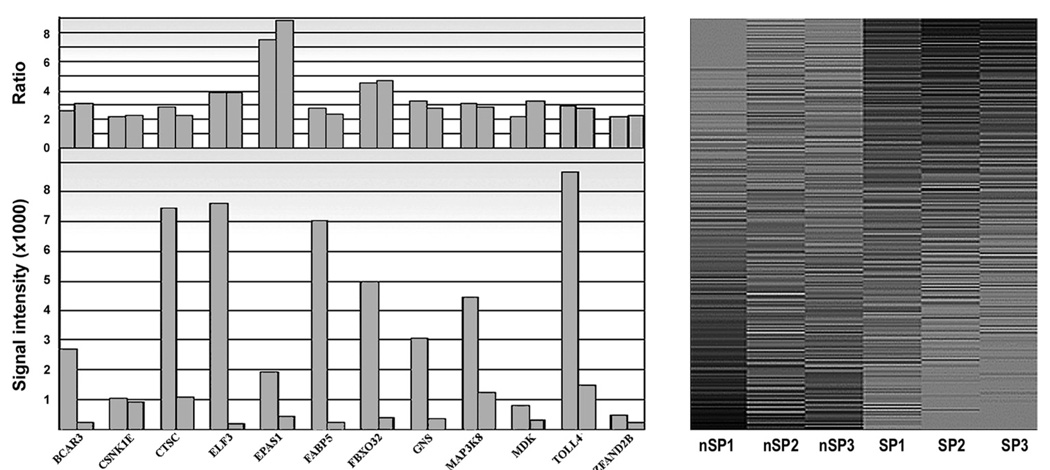
The results for three actin transcripts, a housekeeping gene, one near the poly(A) end, one in the middle of the gene and one near the 5′ end of the ∼1000-bp gene, showed that, whereas SI values decreased along the 3′ to 5′ direction, the SP/nSP expression ratio remained essentially unchanged, whether we used them as the ratio between the SP and nSP averages, or as the average of SP/nSP ratios for each individual paired experiment (Table 1). Ratios for transcripts of differentially expressed genes were also not demonstrably affected by the SI values of different representative transcripts, even when these values differed by as much as 50-fold (Fig. 2A). Finally, heat maps of expressed transcripts (Fig. 2B) showcased the similarity between nSP or SP lists and the difference between the two cell types.
TABLE 1.
Relationship between Probe Locations Respective to the PolyA Site and Signal Intensities and SP/nSP SI Ratios for the β-Acting Gene
| Aver- SP |
Aver- nSP |
R | R- Exp1 |
R- Exp2 |
R- Exp3 |
Aver- Rs |
|
|---|---|---|---|---|---|---|---|
| Actin-5′ | 13,016 | 13,874 | 0.94 | 0.59 | 1.43 | 0.95 | 0.99 |
| Actin-M | 10,267 | 10,807 | 0.95 | 0.83 | 0.92 | 1.11 | 0.95 |
| Actin-3′ | 1,375 | 1,424 | 0.97 | 0.98 | 0.80 | 1.07 | 0.95 |
FIGURE 2.
Quality controls for microarray processing. Right: SP/nSP SI ratios calculated from different transcripts of selected overexpressed genes. Gene names are indicated on the x-axis. Note that calculated expression ratios are not dependent on the signal intensity for a given transcript. Left: Gray-scale heat map of the three nSP and three SP lists of expressed transcripts arranged in descending SP/nSP SI ratio. The lists was limited to transcript with an average SI value greater than 1% of the highest SI value measured. Darkness of the shading correlates with SI value.
Differential Expression
The total transcript-present list included 11,300 probes of which, 518 and 447 were SP over- and underexpressed, respectively. By removing transcripts that presently cannot be linked to a known gene and based on a redundancy gene representation (i.e., more than one transcript probe per gene) of approximately 20%, we calculated that the total gene-present list, when fully curated, would include approximately 9000 distinct genes. Likewise, limiting the differentially expressed lists to transcripts that can be ascribed to a known (named) gene produced over- and underexpressed gene lists containing 382 and 296 genes, respectively, for a total of 678 genes or 7.5% of the total identified. This large percentile underscores the unique phenotype of limbal epithelial SP cells, as previously observed for the human conjunctival epithelium.27 The 40 genes with the greatest over- or underexpression ratios and average SI greater than 100 (we assumed that most low SI genes are less likely to be relevant for cell phenotype and function), are displayed in Table 2A and Table 2B and the complete differentially expressed gene list is provided in the Appendix, online at http://www.iovs.org/cgi/content/full/50/12/5630/DC1.
TABLE 2.
SP Genes Expressed in the Pig Limbal Epithelium
| A. SP Overexpressed Genes and Correlation with nSP/SP SI Ratios for Pig and Human Conjunctival Epithelia | ||||||
|---|---|---|---|---|---|---|
| Affy ID | Symbol | R | Gene Name/Annotation | R-pCNJ | R-hCNJ | |
| 1 | Ssc.7176.1.A1_at | CXCR4 | 14.01 | Chemokine (C-X-C motif) receptor 4 | 24.31 | 14.51 |
| 2 | Ssc.22100.1.S1_at | SH3BGRL3 | 12.70 | SH3-binding domain glutamic acid-rich protein | A | 1.09* |
| 3 | Ssc.3901.1.A1_at | MXD1 | 9.02 | Max dimerization protein 1 | 9.53 | 4.07 |
| 4 | Ssc.18918.1.A1_at | GPX2 | 8.74 | Glutathione peroxidase 2 | 4.96 | 2.17 |
| 5 | Ssc.3549.1.S1_at | EPAS1 | 7.55 | Endothelial PAS domain protein 1 | 2.37 | 0.53* |
| 6 | Ssc.1013.1.A1_at | MGST1 | 6.69 | Microsomal glutathione S-transferase 1 | 3.51 | 1.88 |
| 7 | Ssc.8261.1.A1_at | CYP2C19 | 6.42 | Cytochrome P450 | 1.17* | 2.04 |
| 8 | Ssc.24193.1.S1_at | GRTP1 | 6.10 | Growth hormone regulated TBC protein 1 | 3.55 | 4.22 |
| 9 | Ssc.9522.1.A1_at | RNF144B | 5.96 | Ring finger protein 144b | 2.04 | A |
| 10 | Ssc.10618.1.A1_at | NET1 | 5.72 | Neuroepithelial cell trans gene 1 | 0.97* | 1.74* |
| 11 | Ssc.23022.1.S1_at | RHCG | 5.70 | Rhesus type C glycoprotein | 5.07 | 2.04 |
| 12 | Ssc.5978.3.S1_a_at | RHBDD2 | 5.54 | Rhomboid, veinlet-like 7 isoform 1 | 3.27 | 1.31* |
| 13 | Ssc.6738.1.S1_at | LNX1 | 5.54 | Ligand of numb-protein X 1 | 2.62 | 8.43 |
| 14 | Ssc.29575.1.A1_at | VNN2 | 5.44 | Vanin 2 isoform 1 | 2.82 | 5.42 |
| 15 | Ssc.28055.1.A1_at | ZAR1 | 5.36 | Zygote arrest 1-like protein | A | A |
| 16 | Ssc.773.2.S1_at | HIST1H2BD | 5.34 | Histone cluster 1, h2bd | 3.74 | 4.42 |
| 17 | Ssc.3832.1.S1_at | AQP3 | 5.01 | Aquaporin 3 | 3.35 | 1.19 |
| 18 | Ssc.4026.1.S1_at | GPN2 | 4.92 | GPN-loop gtpase 2 | 1.45* | NL |
| 19 | Ssc.5314.1.S1_at | LGALS4 | 4.83 | Lectin, galactoside-binding, 4 (galectin 4) | 7.07 | 0.57* |
| 20 | Ssc.7256.1.A1_at | RPS20 | 4.69 | Ribosomal protein S20 | 0.66* | 0.81* |
| 22 | Ssc.14531.1.S1_at | ACADS | 4.60 | Acyl-Coenzyme A dehydrogen. Short chain | 2.19 | A |
| 23 | Ssc.19335.1.S1_at | MYOM1 | 4.60 | Myomesin 1 isoform 2 | A | 0.52* |
| 24 | Ssc.13553.1.A1_at | GNA14 | 4.54 | Guanine nucleotide binding protein alpha14 | 2.72 | 4.04 |
| 25 | Ssc.13913.1.S1_at | ZMYND8 | 4.50 | Zinc finger, MYND-type containing 8 | 4.01 | NL |
| 26 | Ssc.4368.1.S1_at | FBXO32 | 4.49 | F-box protein 32 | 4.53 | 8.16 |
| 27 | Ssc.23465.1.S1_at | TJP2 | 4.37 | Tight junction protein 2 isoform 1 | 5.03 | 0.83* |
| 28 | Ssc.19823.1.A1_at | ABLIM1 | 4.37 | Actin-binding LIM protein 1 isoform s isoform 4 | 2.30 | 1.81 |
| 29 | Ssc.17990.1.A1_at | EEA1 | 4.34 | Early endosome antigen 1 | A | A |
| 30 | Ssc.10025.3.S1_at | CEBPD | 4.29 | CCAAT/enhancer binding protein delta | 9.76 | 1.69* |
| 31 | Ssc.8594.1.A1_at | BLNK | 4.28 | B-cell linker isoform 1 | 2.89 | 2.47 |
| 32 | Ssc.28513.1.S1_at | NRARP | 4.23 | NOTCH-regulated ankyrin repeat protein | 2.29 | NL |
| 33 | Ssc.3198.1.S1_at | CGN | 4.21 | Cingulin | 2.57 | 3.60 |
| 34 | Ssc.17970.1.A1_at | TMM54 | 4.13 | Transmembrane protein 54 | 3.72 | NL |
| 35 | Ssc.10931.1.S1_at | CRYAB | 4.10 | Crystallin, alpha B | 2.40 | 2.13 |
| 36 | Ssc.13069.1.A1_at | MPN | 4.06 | Marapsin | 2.89 | NL |
| 37 | Ssc.3476.1.A1_at | PIR | 4.04 | Pirin | 3.17 | 2.08 |
| 38 | Ssc.114.1.S1_at | HSP70 | 3.99 | Heat shock protein 70 | 4.97 | NL |
| 39 | Ssc.8871.2.A1_at | CDKN1C | 3.98 | Cyclin-dependent kinase inhib. 1C (p57/kip2) | 1.66* | 2.43 |
| 40 | Ssc.779.1.S1_at | ELF3 | 3.86 | E74-like factor 3 | 5.63 | 7.47 |
| B. SP Underexpressed Genes | ||||
|---|---|---|---|---|
| Affy ID | Symbol | R | Gene Name/Annotation | |
| 1 | Ssc.28492.1.S1_at | DNAJC2 | 0.22 | Zuotin related factor 1 |
| 2 | Ssc.30861.1.A1_at | FLRT3 | 0.22 | Fibronectin leucine rich transmembrane |
| 3 | Ssc.28424.1.A1_at | RRP15 | 0.22 | Ribosomal Rna processing 15 homolog |
| 4 | Ssc.16209.1.S1_at | TNC | 0.23 | Tenascin c |
| 5 | Ssc.8555.1.A1_at | INHBA | 0.24 | Inhibin a |
| 6 | Ssc.12822.1.A1_at | IL24 | 0.25 | Interleukin 24 |
| 7 | Ssc.6093.2.A1_at | MAP1B | 0.26 | Microtubule-associated protein 1b |
| 8 | Ssc.6662.1.S1_at | SEMA6D | 0.27 | Semaphorin 6d |
| 9 | Ssc.7528.1.A1_at | MBNL3 | 0.27 | Muscleblind-like 3 isoform g |
| 10 | Ssc.16743.1.S1_at | FN1 | 0.27 | Fibronectin |
| 11 | Ssc.12695.1.S1_at | ACSL4 | 0.28 | Acyl-coa synthetase long-chain family member |
| 12 | Ssc.29550.1.A1_at | ZNF239 | 0.29 | Zinc finger protein 239 |
| 13 | Ssc.27410.1.S1_at | MYCN | 0.29 | V-myc myelocytomatosis viral related |
| 14 | Ssc.15629.1.S1_at | WDR3 | 0.29 | Wd repeat-containing protein 3 |
| 15 | Ssc.24731.1.A1_at | MEX3B | 0.29 | Mex-3 homolog b |
| 16 | Ssc.19744.1.S1_at | NNP73 | 0.30 | Nucleolar protein 73 |
| 17 | Ssc.29120.1.A1_at | AHCTF1 | 0.30 | At hook containing transcription factor 1 |
| 18 | Ssc.30843.1.A1_at | ADAMDEC | 0.30 | Disintegrin and metalloproteinase domain-like |
| 19 | Ssc.14318.2.A1_at | BBS7 | 0.30 | Bardel-biedl syndrome 7 |
| 20 | Ssc.24741.2.S1_at | DNTTIP2 | 0.30 | Deoxynucleotidyltransferase, interacting |
| 21 | Ssc.4814.1.A1_at | PAN2 | 0.30 | Poly(a) specific ribonuclease subunit homolog |
| 22 | Ssc.10709.1.A1_at | CNBP | 0.31 | Zinc finger protein 2 isoform 2 |
| 23 | Ssc.7062.1.A1_at | ASNSD1 | 0.31 | Asparagine synthetase domain containing 1 |
| 24 | Ssc.19236.1.S1_at | SPRED1 | 0.31 | Sprouty-related, evh1 domain containing 1 |
| 25 | Ssc.31124.1.S1_at | COL14A1 | 0.31 | Collagen, type xiv, alpha 1 |
| 26 | Ssc.30737.1.A1_s_a | PNPT1 | 0.32 | Polyribonucleotide nucleotidyltransferase 1 |
| 27 | Ssc.6670.1.A1_at | SLC39A6 | 0.32 | Solute carrier family 39 (zinc transporter) |
| 28 | Ssc.30853.1.S1_at | FGF2 | 0.32 | Fibroblasr growth factor 2 |
| 28 | Ssc.25265.1.S1_a_ | PRP22 | 0.32 | Death (asp-glu-ala-his) box polypeptide 29 |
| 29 | Ssc.4597.1.A1_at | MASP2 | 0.32 | Mannan-binding lectin serine peptidase 2 |
| 30 | Ssc.27518.1.A1_at | PVRL3 | 0.32 | Poliovirus receptor-related 3 |
| 31 | Ssc.22032.1.A1_at | LARP4 | 0.32 | La ribonucleoprotein domain family, member 4 |
| 32 | Ssc.17313.2.S1_at | CALD1 | 0.32 | Caldesmon 1 isoform 5 |
| 33 | Ssc.27693.1.A1_a_ | ADCY3 | 0.32 | Adenylate cyclase 3 |
| 34 | Ssc.19406.1.S1_at | RBM34 | 0.32 | Rna binding motif protein 34 |
| 35 | Ssc.2227.1.S1_at | GLT25D1 | 0.33 | Glycosyltransferase 25 domain containing 1 |
| 36 | Ssc.13548.1.S1_at | NMD3 | 0.33 | Nmd3 homolog |
| 37 | Ssc.21447.1.S1_at | EPRS | 0.33 | Glutamyl-prolyl-trna synthetase |
| 39 | Ssc.29187.1.A1_at | GJA1 | 0.33 | Gap junction protein, alpha 1 |
| 40 | Ssc.24285.1.S1_at | RPP40 | 0.33 | Ribonuclease P 40kda subunit, isoform 1 |
NL, not listed n the array; A, absent.
Does not comply with the definition for overexpression.
The limbal data included in Table 2A were compared with data obtained in a similar fashion from two replicated SP/nSP microarray analyses of pig conjunctiva epithelium and with the data earlier data from human conjunctiva.27 The great majority of the 40 genes included in Table 2A were also overexpressed in SP cells of pig CNJ; only 8 did not comply with the overexpression criteria (Table 2A, RpCNJ column). Extending the correlation to human CNJ (Table 2A, RhCNJ column) provided a convenient means of reinforcing the identification of genes that may have an intrinsic relationship to either stem cells in general, epithelial stem cells at large, or to stem cells of the PAX-6 positive epithelial of the ocular surface. From Table 2 A it is possible to identify genes that may be overexpressed in stem cells of the ocular surface in either a species-independent (e.g., CXCR4, MXD1, GPX2, EPAS1, MGST1, GRTP1, LNX1, VNN2, and FBXO32) or species-dependent (e.g., RHBDD2, AQP3) manner, and others that may be unique to the corneal epithelium (e.g., ZAR1, EEA1).
Overall, gene underexpression was less pronounced than overexpression. No gene was strongly underexpressed, and whereas there were 104 genes with an SP/nSP ratio greater than 3, only 20 underexpressed genes possessed nSP/SP ratios greater than this substantial threshold.
Validations
Due to limited amounts of SP RNA available per experiment, validation by real-time PCR was performed only for three markedly overexpressed genes. The PCR-derived SP/nSP ratios for CXCR4, GPX2, and DUSP5 were 24-, 12- and 3.7- fold, respectively, higher in all three cases than the averages measured in the microarray experiment (Table 2A, lines 1, 4; and Table 3, respectively). Agarose gel electrophoresis of the real-time PCR reaction products confirmed that for all four genes used, a single product of the expected size was observed (see the Methods section).
TABLE 3.
Selected Categories of Genes Differentially Expressed in the Pig Limbal SP Cells
| Category | Symbol | R | Gene Name//Annotation |
|---|---|---|---|
| Associated with stem cell phenotype |
CXCR4 | 14.51 | Cytokine receptor 4* |
| ALDH1A1 | 3.59 | Aldehyde dehydrogenase (cytosolic)* | |
| KRT19 | 3.06 | Keratin 19* | |
| KRT15 | 2.64 | Keratin 15* | |
| GJA1 | 0.33 | Connexin43* | |
| Basal cell markers | KRT5 | 1.07 | Keratin 5 |
| KRT14 | 1.12 | Keratin 14 | |
| Control of MAPK phosphorylation |
DUSP5 | 2.77 | Dual specificity phosphatase 5* |
| DUSP1 | 2.13 | Dual specificity phosphatase 1* | |
| DUSP14 | 2.14 | Dual specificity phosphatase 14* | |
| DUSP16 | 2.07 | Dual specificity phosphatase 16 | |
| Modulation of G1/S transition by MYC |
MXD1 | 9.02 | Max dimerization protein 1//MAD1* |
| MXI1 | 3.45 | Myc associated protein??MAD2* | |
| MYC | 0.86 | C-myc* | |
| G1/S transition cyclins | CCND2 | 0.56 | Cyclin D2* |
| CDK6 | 0.56 | Cyclin-dependent kinase 6* | |
| Stem cell replication | HES1 | 2.58 | Hairy and enhancer of split 1* |
| ID2 | 2.47 | Inhibitor of DNA/differentiation 2* | |
| ID1 | 2.19 | Inhibitor of DNA/differentiation 1* | |
| CNDJ2 | 0.22 | MIDA1\\Inhib. ID1 induction of prolifer. | |
| Protection against radical, oxidative and xenobiotic damage |
SH3BGRL3 | 12.70 | Glutaredoxin activity* |
| GPX2 | 8.74 | Glutathione peroxidase 2* | |
| MGST1 | 6.69 | Microsomal glutathione S-transf. 1 | |
| MGST2 | 2.34 | Microsomal glutathione S-transf. 2 | |
| GSR | 2.25 | Glutathione reductase* | |
| HAGH | 2.07 | Hydroxyacyl glutathione hydrolase | |
| CYP2C19 | 6.42 | Cyt. P450, xenobiotic processing | |
| CYP1A1 | 3.52 | Cyt. P450, xenobiotic processing* | |
| SQRDL | 2.87 | Sulfide:quinone oxidoreductase | |
| DHRS3 | 2.68 | Dioxin-induciblereductase m. 3* | |
| NQO1 | 2.56 | Oxidoreductase 1* | |
| RRM2 | 2.46 | Ribonucleotide reductase M2 | |
| DHRS8 | 2.34 | Dehydrogenase m.8 |
Similarly over-, under-, or indistinctly expressed, respectively, in both pig and human conjunctival SP cells.
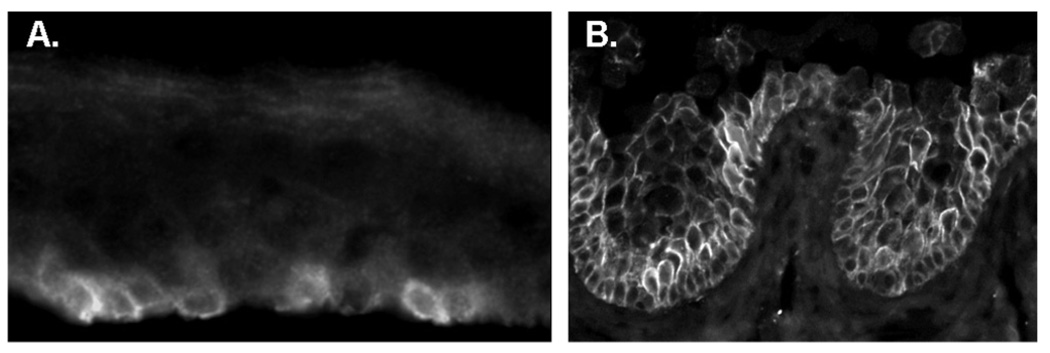
The scarcity of antibodies known to recognize pig protein homologues is a serious impediment for validation at the polypeptide level. Aquaporin 3 is highly overexpressed in the pig but not in the human (Table 2A). We identified an antibody that recognizes the pig and human proteins. Consistent with what would be expected for a stem cell biomarker, in the pig the antibody generated intense staining in isolated basal cells (Fig. 3A). In stark contrast, in human samples, the protein was strongly expressed in all basal and early suprabasal limbal (Fig. 3B) and conjunctival (not shown) cells.
FIGURE 3.
Expression of aquaporin 3 in the limbus, In pig, strong expression occurs only in isolated basal cells. In human the expression occurs throughout the basal and early suprabasal cells.
DAVID Cluster and Gene Categories Analysis
Differentially expressed genes with SI averages greater than 100 were subjected to DAVID’s functional annotation clustering using DAVID’s human gene set as background. For the overexpressed genes, the largest annotation clusters were those associated with regulation of cellular and biological developmental processes (59 and 52 genes of the 215 recognized; enrichment scores of 2.68 and 2.33, respectively). The GO_TERMS with the most significant overrepresentation with FDRs under 2% are shown in Table 4. It is noteworthy that the most overrepresented terms are related to regulatory inhibition of cellular biological and metabolic activity. It also identifies transcription from the RNA polymerase II promoter (genes, EPAS1, MAFB, CHD4, SAP18, EGR1, LMCD1, ID1, KLF7, ID2, TNFRSF1A, CDKN1C, HES1, and MYD88). Conversely, DAVID’s overrepresentation analysis of the underexpressed gene set essentially showed that the SP cells were underactive in terms of the synthesis of ribosomes and cellular organelles, a feature consistent with an undifferentiated, generally quiescent cell.
TABLE 4.
Over- and Underrepresented GO Terms within the Differentially Expressed Genes
| Term | n | % | FDR |
|---|---|---|---|
| Overrepresented | |||
| GO:0048519≈negative regulation of biological process | 29 | 13.49 | 0.313 |
| GO:0048523≈negative regulation of cellular process | 28 | 13.02 | 0.377 |
| GO:0009892≈negative regulation of metabolic process | 15 | 6.98 | 0.908 |
| GO:0006366≈transcription from RNA pol. II promoter | 19 | 8.84 | 1.210 |
| GO:0048869≈cellular developmental process | 37 | 17.21 | 1.387 |
| Underrepresented | |||
| GO:0042254≈ribosome biogenesis and assembly | 15 | 5.81 | < 10−3 |
| GO:0022613≈ribonucleoprot. biogenesis & assembly | 20 | 7.75 | < 10−3 |
| GO:0005730≈nucleolus | 17 | 6.59 | < 10−3 |
| GO:0003723≈RNA binding | 34 | 13.18 | < 10−3 |
| GO:0006396≈RNA processing | 23 | 8.91 | < 10−3 |
| GO:0043233≈organelle lumen | 35 | 13.57 | < 10−3 |
| GO:0006364≈rRNA processing | 9 | 3.49 | 0.003 |
| GO:0043227≈membrane-bound organelle | 132 | 51.16 | 0.009 |
| GO:0044428≈nuclear part | 34 | 13.18 | 0.012 |
| GO:0006996≈organelle organization and biogenesis | 35 | 13.57 | 0.020 |
| GO:0005634≈nucleus | 93 | 36.05 | 0.0442 |
| GO:0048522≈positive regulation of cellular process | 27 | 10.47 | 0.954 |
| GO:0045182≈translation regulator activity | 9 | 3.49 | 1.158 |
| GO:0044238≈primary metabolic process | 131 | 50.78 | 1.349 |
| GO:0044446≈intracellular organelle part | 69 | 26.74 | 1.397 |
| GO:0005578≈proteinaceous extracellular matrix | 14 | 5.43 | 1.425 |
| GO:0019538≈protein metabolic process | 69 | 26.74 | 1.430 |
| GO:0031012≈extracellular matrix | 14 | 5.43 | 1.685 |
| GO:0044237≈cellular metabolic process | 130 | 50.39 | 1.932 |
Columns describe the number of genes included (n), percent of total considered (%) and false-discovery rate (%).
Topical Gene Analysis
Even though the differentially expressed gene list is quite diverse and includes many genes whose functions remain poorly defined, several features of expression were compatible with the concept that these SP cells form part of the limbal stem cell population (Table 3). The SP cells overexpresses CXCR4, a pleiotropic cytokine receptor which plays a variety of roles, such as being a critical component of hematopoietic stem cells homing in bone marrow,31,32 the homing of mesen-chymal stem cells to bone33 and epithelial cell proliferation.34 ALDH1A1 is a cytosolic enzyme that has been shown to be generally associated with stem cells in multiple organs or tissues, and its overexpression may increase overall oxidative protection.35 Overexpression of keratin-15 and -19 and reduced CJA1/connexin43 is a well-documented feature of hair follicle stem cells36 and limbal stem cells,37 respectively. The differential expression of several categories of genes involved in the control of MAPK phosphorylation, cell cycling, stem cell replication and survival mechanisms (Table 3) need to be considered.
DISCUSSION
SP cell populations are quite small, typically comprising between 0.05% and 0.5% of the total organ/tissue cell count. Sound microarray studies require, at minimum, nanogram quantities of RNA, amounts that are difficult to derive from either human or rodent sources. Given this context, to delineate the molecular signature of limbal SP cells we resorted to the pig, a species possessing appropriate ocular dimensions and abundantly accessible tissue for which commercial microarrays are available. Although sequencing of the pig genome is far from complete and the available microarray has a limited gene representation span (e.g., neither p63, a gene commonly included in limbal stem cell studies or ABCG2, the prospective SP-generating transporter is represented), our studies yielded a robust molecular signature that incorporates nearly 700 genes of the ∼9000 recognized as limbal-expressed by the microarray. These results clearly show the unique nature of the SP cells.
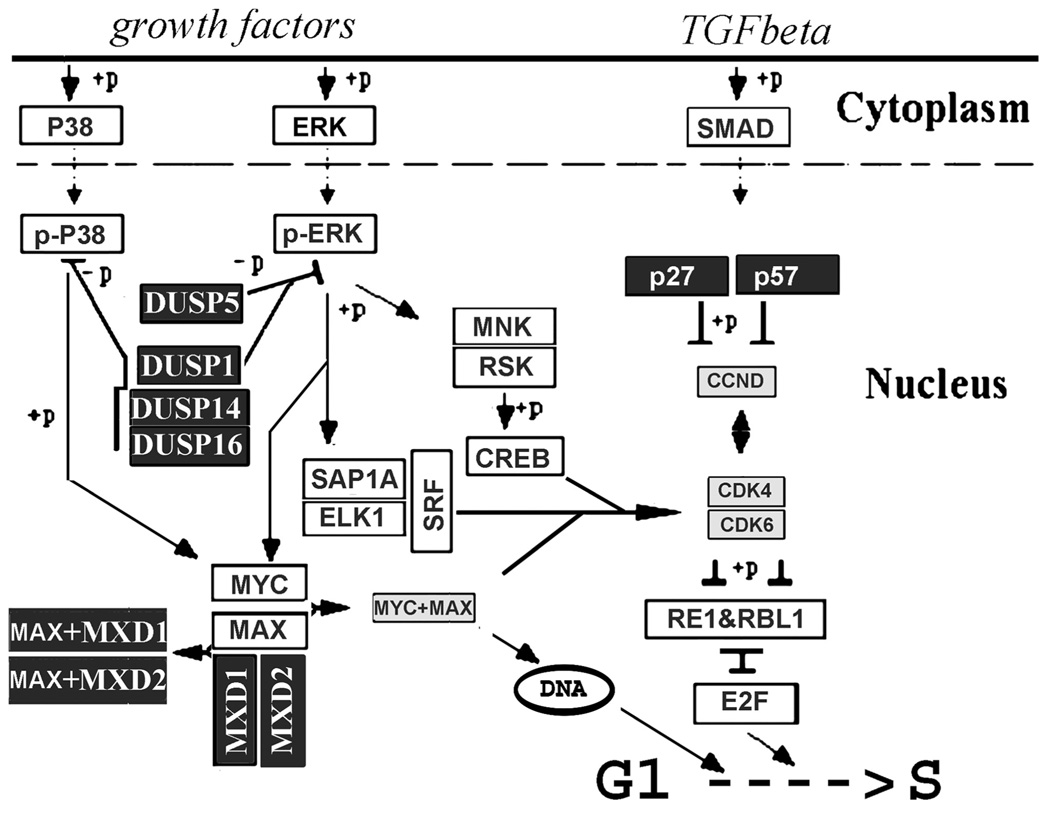
Expression patterns that are consistent with known stem cell features can be gleaned from the overall differentially expressed gene list. Slow cycling and the implicit low responsiveness to growth stimulation under steady state conditions represent the most recognized features of epithelial stem cells. The differentially expressed gene list incorporates several expression patterns that will confer or may contribute to slow cycling or quiescence (Fig. 4). Multiple dual specificity phosphatases (DUSPs), the primary inhibitors of ERK, p38, andJNK, the terminal mitogen-activated protein kinases38 are overex-pressed (Table 3), and none is underexpressed (Appendix). Since ERK1/2, mediate growth factor-initiated activation of nuclear transcription factors that control the G1-to-S restriction (R) point, overexpression of DUSPs, in particular DUSP5, with its capacity for ERK dephosphorylation and nuclear sequestra-tion,39 can be predicted to have an inhibitory effect on the stimulation of proliferation by circulating mitogens. We have recently confirmed proliferative inhibition by this enzyme in a corneal cell line (Wolosin et al., unpublished data, 2009). DUSP14 has been directly shown to slow pancreatic β-cell proliferation.40
FIGURE 4.
Schematic description of nuclear protein gene patterns that may affect proliferation of SP cells. Overexpression of multiple nuclear dual specificity phosphatases may simultaneously decrease proliferative and migratory responses to growth factors in the SP cells. Large overexpression of MXD1 will add to the blockade of growth factors responses by sequestering MAX, the cofactor for MYC. Overexpression of the cyclin D kinase inhibitors and simultaneous under-expression of cyclin D and its dependent kinases could also have similar effects.
The overexpression of the MAD proteins MXD1 and MXI1, through its effect on MYC, can also have a negative impact on proliferation (Table 3, Fig. 4). MAPK-activated MYC promotes transcriptional activities that facilitate the G1-to-S transition and affect multiple cell cycle stages.41 MYC transcriptional effects require its association with MYC associated factor X (MAX).42 MAX availability, however, is negatively controlled by its alternative association with MAD proteins. Hence, markedly augmented expression of MXD1/MAD1 and MXI1/MAD2 levels in the SP cells is likely to further dull the responsiveness of these cells to mitogen stimuli.
The third notable expression pattern with the potential to inhibit the rate of G1 to S transition in a mitogen-independent manner involves cyclin D, the protein that integrates signals controlling G1-to-S transition via retinoblastoma. SP cells overexpress the two main kinase inhibitors of this cyclin, p27/CIP1, and p57/KIP2, and conversely moderately underexpress cyclin D2 and CDK kinases that activate cell cycle progression after association with nonphosphorylated cyclin D isoforms (Table 3, Fig. 4).
HES1 and ID are dominant negative blockers for several differentiation-inducing helix-loop-helix transcription factors. Recently Nakamura et al.43 have shown that HES1 is a critical survival factor for the mouse limbal stem cell, in similarity to the effect of both HES1 and ID1 and in the hematopoietic system.44,45
In addition to slow cycling, geno- and cytoprotection are two functions that seem naturally associated with long-lived stem cells. The overexpressed gene list includes a large number of genes that have been shown to be involved in defense against free radicals, oxidative stress, and removal of aromatic DNA damage inducers or xenobiotic compounds (Table 3).
The potential significance of the results to human biology is supported by the results displayed in Table 2. More than 70% of the genes overexpressed in the limbal SP cells are also overexpressed in the conjunctival SP cells, and likewise most of the genes overexpressed in these two systems are also overexpressed in the human conjunctiva SP cells. However, as suggested by the case of aquaporin 3, whose overexpression appears to be a pig-specific phenomenon (Table 2, Fig. 3), such extrapolations necessitate a degree of caution and/or specific reconfirmation.
From a practical clinical perspective, with the proviso that further study solidifies the notion that SP cells are part of the limbal stem cell population, many of the features indicated by the gene expression patterns observed in these cells may prevent prompt expansion of stem and early precursor cells within total cultured populations for use in surgical limbal reconstructive procedures. It is the transient amplifying cells that are more likely to undergo rapid gains in these cultures. Modification of ex vivo cell behavior through either pharmacologic means or reversible ectopic gene or antisense expression may be useful to improve the fraction of stem/precursor cells within expanded limbal populations.
In summary, notwithstanding the present limitations imposed by incomplete pig genome sequence data, the differential global gene expression analysis outlined in this report demonstrated the feasibility and value of the microarray studies used in advancing limbal stem cell research.
Acknowledgments
The authors thank the Mount Sinai School of Medicine for the use of the Flow Cytometry, Microarray, and Real Time PCR shared facilities, without which the study could not have been completed.
Supported by National Institutes of Health Grants EY 014878, EY 015132, and EY01867 (Core Center Grant) and an Unrestricted grant from Research to Prevent Blindness, Inc.
Footnotes
Disclosure: M.A.M. Akinci, None; H. Turner, None; M. Taveras, None; J.M. Wolosin, None
References
- 1.Davanger M, Evensen A. Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature. 1971;229:560–561. doi: 10.1038/229560a0. [DOI] [PubMed] [Google Scholar]
- 2.Schermer A, Galvin S, Sun TT. Differentiation-related expression of a major 64 K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62. doi: 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 4.Tseng SC. Concept and application of limbal stem cells. Eye. 1989;3:141–157. doi: 10.1038/eye.1989.22. [DOI] [PubMed] [Google Scholar]
- 5.Wolosin JM, Xiong X, Schutte M, Stegman Z, Tieng A. Stem cells and differentiation stages in the limbo-corneal epithelium. Prog Retin Eye Res. 2005;19:223–255. doi: 10.1016/s1350-9462(99)00005-1. [DOI] [PubMed] [Google Scholar]
- 6.Chen JJ, Tseng SC. Corneal epithelial wound healing in partial limbal deficiency. Invest Ophthalmol Vis Sci. 1990;31:1301–1314. [PubMed] [Google Scholar]
- 7.Puangsricharern V, Tseng SC. Cytologic evidence of corneal diseases with limbal stem cell deficiency. Ophthalmology. 1995;102:1476–1485. doi: 10.1016/s0161-6420(95)30842-1. [DOI] [PubMed] [Google Scholar]
- 8.Huang AJ, Tseng SC. Corneal epithelial wound healing in the absence of limbal epithelium. Invest Ophthalmol Vis Sci. 1991;32:96–105. [PubMed] [Google Scholar]
- 9.Kenyon KR, Tseng SC. Limbal autograft transplantation for ocular surface disorders. Ophthalmology. 1989;96:709–722. doi: 10.1016/s0161-6420(89)32833-8. [DOI] [PubMed] [Google Scholar]
- 10.Tsai RJ, Tseng SC. Human allograft limbal transplantation for corneal surface reconstruction. Cornea. 1994;13:389–400. doi: 10.1097/00003226-199409000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Tsubota K, Satake Y, Ohyama M, et al. Surgical reconstruction of the ocular surface in advanced ocular cicatricial pemphigoid and Stevens-Johnson syndrome. Am J Ophthalmol. 1996;122:38–52. doi: 10.1016/s0002-9394(14)71962-2. [DOI] [PubMed] [Google Scholar]
- 12.Pellegrini G, Traverso CE, Franzi AT, Zingirian M, Cancedda R, De Luca M. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet. 1996;349:990–993. doi: 10.1016/S0140-6736(96)11188-0. [DOI] [PubMed] [Google Scholar]
- 13.Hayashida Y, Nishida K, Yamato M, et al. Ocular surface reconstruction using autologous rabbit oral mucosal epithelial sheets fabricated ex vivo on a temperature-responsive culture surface. Invest Ophthalmol Vis Sci. 2005;46:1632–1639. doi: 10.1167/iovs.04-0813. [DOI] [PubMed] [Google Scholar]
- 14.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou S, Schuetz JD, Bunting KD, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 20012;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 16.Zhou S, Morris JJ, Barnes Y, Lan L, Schuetz JD, Sorrentino BP. Bcrp1 gene expression is required for normal numbers of side population stem cells in mice, and confers relative protection to mitoxantrone in hematopoietic cells in vivo. Proc Natl Acad Sci U S A. 2002;99:12339–12344. doi: 10.1073/pnas.192276999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono M, Maruyama T, Masuda H, et al. Side population in human uterine myometrium displays phenotypic and functional characteristics of myometrial stem cells. Proc Natl Acad Sci U S A. 2007;104:18700–18705. doi: 10.1073/pnas.0704472104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshi N, Kusakabe T, Taylor BJ, Kimura S. Side population cells in the mouse thyroid exhibit stem/progenitor cell-like characteristics. Endocrinology. 2007;148:4251–4158. doi: 10.1210/en.2006-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolosin JM, Tseng SCG, Budak MT, et al. The limbo-corneal epithelium and its stem cells: principles and applications. In: Greer VE, editor. Focus in Stem Cell Research. New York: Nova Science Publishers; 2004. pp. 59–92. [Google Scholar]
- 20.Paiva CS, Chen Z, Corrales RM, Pflugfelder SC, Li DQ. ABCG2 transporter identifies a population of clonogenic human limbal epithelial cells. Stem Cells. 2005;23:63–73. doi: 10.1634/stemcells.2004-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budak MT, Alpdogan OS, Zhou M, Lavker RM, Akinci MA, Wolosin JM. Ocular surface epithelia contain ABCG2-dependent side population cells exhibiting features associated with stem cells. J Cell Sci. 2005;118:1715–1724. doi: 10.1242/jcs.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epstein SP, Wolosin JM, Asbell PA. P63 expression levels in side population and low light scattering ocular surface epithelial cells. Trans Am Ophthalmol Soc. 2005;103:187–199. [PMC free article] [PubMed] [Google Scholar]
- 23.Wolosin JM. Cell markers and the side population phenotype in ocular surface epithelial stem cell characterization and isolation. Ocul Surf. 2006;4:10–23. doi: 10.1016/s1542-0124(12)70261-6. [DOI] [PubMed] [Google Scholar]
- 24.Umemoto T, Yamato M, Nishida K, Yang J, Tano Y, Okano T. Limbal epithelial side-population cells have stem cell-like properties, including quiescent state. Stem Cells. 2006;24:86–94. doi: 10.1634/stemcells.2005-0064. [DOI] [PubMed] [Google Scholar]
- 25.Park KS, Lim CH, Min BM, et al. The side population cells in the rabbit limbus sensitively increased in response to the central cornea wounding. Invest Ophthalmol Vis Sci. 2006;47:892–900. doi: 10.1167/iovs.05-1006. [DOI] [PubMed] [Google Scholar]
- 26.Goodell MA, Rosenzweig M, Kim H, et al. Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat Med. 1997;3:1337–1345. doi: 10.1038/nm1297-1337. [DOI] [PubMed] [Google Scholar]
- 27.Akinci MAM, Turner H, Taveras M, et al. Molecular profiling of the of conjunctival epithelial stem side population cells identifies atypical cell surface markers and sources of a slow cycling phenotype. Invest Ophthalmol Vis Sci. doi: 10.1167/iovs.08-2861. Published online March 25, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koroma BM, Yang JM, Sundin OH. The Pax-6 homeobox gene is expressed throughout the corneal and conjunctival epithelia. Invest Ophthalmol Vis Sci. 1997;38:108–120. [PubMed] [Google Scholar]
- 30.Wang Y, Couture OP, Qu L, et al. Analysis of porcine transcriptional response to Salmonella enterica serovar Choleraesuis suggests novel targets of NFkappaB are activated in the mesenteric lymph node. BMC Genomics. 2008;9:437. doi: 10.1186/1471-2164-9-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratajczak MZ, Zuba-Surma E, Kucia M, Reca R, Wojakowski W, Ratajczak J. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20:1915–1924. doi: 10.1038/sj.leu.2404357. [DOI] [PubMed] [Google Scholar]
- 32.Chute JP. Stem cell homing. Curr Opin Hematol. 2006;13:399–406. doi: 10.1097/01.moh.0000245698.62511.3d. [DOI] [PubMed] [Google Scholar]
- 33.Kitaori T, Ito H, Schwarz EM, et al. Stromal cell-derived factor 1/CXCR4 signaling is critical for the recruitment of mesenchymal stem cells to the fracture site during skeletal repair in a mouse model. Arthritis Rheum. 2009;60:813–823. doi: 10.1002/art.24330. [DOI] [PubMed] [Google Scholar]
- 34.Florin L, Maas-Szabowski N, Werner S, Szabowski A, Angel P. Increased keratinocyte proliferation by JUN-dependent expression of PTN and SDF-1 in fibroblasts. J Cell Sci. 2005;118:1981–1989. doi: 10.1242/jcs.02303. [DOI] [PubMed] [Google Scholar]
- 35.Moreb JS. Aldehyde dehydrogenase as a marker for stem cells. Curr Stem Cell Res Ther. 2008;3:237–246. doi: 10.2174/157488808786734006. [DOI] [PubMed] [Google Scholar]
- 36.Kloepper JE, Tiede S, Brinckmann J, et al. Immunophenotyping of the human bulge region: the quest to define useful in situ markers for human epithelial hair follicle stem cells and their niche. Exp Dermatol. 2008;7:592–609. doi: 10.1111/j.1600-0625.2008.00720.x. [DOI] [PubMed] [Google Scholar]
- 37.Matic M, Petrov IN, Chen S, Wang C, Dimitrijevich SD, Wolosin JM. Stem cells of the corneal epithelium lack connexins and metabolite transfer capacity. Differentiation. 1997;61:251–260. doi: 10.1046/j.1432-0436.1997.6140251.x. [DOI] [PubMed] [Google Scholar]
- 38.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 39.Mandl M, Slack DN, Keyse SM. Specific inactivation and nuclear anchoring of extracellular signal-regulated kinase 2 by the inducible dual-specificity protein phosphatase DUSP5. Mol Cell Biol. 2005;25:1830–1845. doi: 10.1128/MCB.25.5.1830-1845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klinger S, Poussin C, Debril MB, Dolci W, Halban PA, Thorens B. Increasing GLP-1-induced beta-cell proliferation by silencing the negative regulators of signaling cAMP response element modulator-alpha and DUSP14. Diabetes. 2008;57:584–593. doi: 10.2337/db07-1414. [DOI] [PubMed] [Google Scholar]
- 41.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates Cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura T, Ohtsuka T, Sekiyama E, et al. Hes1 regulates corneal development and the function of corneal epithelial stem/progenitor cells. Stem Cells. 2008;26:1265–1274. doi: 10.1634/stemcells.2007-1067. [DOI] [PubMed] [Google Scholar]
- 44.Kunisato A, Chiba S, Nakagami-Yamaguchi E, et al. HES-1 preserves purified hematopoietic stem cells ex vivo and accumulates side population cells in vivo. Blood. 2003;101:1777–1783. doi: 10.1182/blood-2002-07-2051. [DOI] [PubMed] [Google Scholar]
- 45.Perry SS, Zhao Y, Nie L, Cochrane SW, Huang Z, Sun XH. Id1, but not Id3, directs long-term repopulating hematopoietic stem-cell maintenance. Blood. 2007;110:2351–2360. doi: 10.1182/blood-2007-01-069914. [DOI] [PMC free article] [PubMed] [Google Scholar]