Abstract
During eukaryotic chemotaxis, external chemical gradients guide the crawling motion of cells. This process plays an important role in a large variety of biological systems and has wide ranging medical implications. New experimental techniques including confocal microscopy and microfluidics have advanced our understanding of chemotaxis while numerical modeling efforts are beginning to offer critical insights. In this short review, we survey the current experimental status of the field by dividing chemotaxis into three distinct “modules”: directional sensing, polarity and motility. For each module, we attempt to point out potential new directions of research and discuss how modeling studies interact with experimental investigations.
Keywords: actin, cAMP, motility, pseudopod, modeling, Dictyostelium , neutrophils
Chemotaxis is defined as directed movement of cells up or down a chemical gradient. Here we focus on chemotaxis in eukaryotic cells where the gradient sensing results in directed cell motion in the form of crawling. Eukaryotic chemotaxis is thought to be a key component in a multitude of biological processes. In some processes, in vitro experiments have unambiguously shown that cells respond to ligand concentration gradients set up by pipettes or gradient chambers. Examples include neuronal patterning where nerve growth factors determine the direction of axonal growth [1], wound healing where the platelet-derived growth factor guides fibroblast to the injury area [2] and cancer metastasis where tumor cells respond chemotactically to gradients of epidermal growth factor [3]. For other processes, including germ cell migration [4] and hemocyte migration [5], the evidence for chemotaxis is less direct and typically relies on showing that ectopic expressions of a ligand can alter the migration of the motile cell.
Much of our present understanding of eukaryotic chemotaxis has come from studies in the social amoeba, Dictyostelium discoideum, and cells of the mammalian immune system. Amoebae respond to the chemoattractant cAMP by crawling towards the high end of a concentration gradient. The resulting motion is much faster than found in the examples mentioned above: typical speeds of 10µm/min are observed, equivalent to roughly one body length per minute and comparable to that of the other “champion” of chemotaxis, neutrophils. Studies on these rapid responders have shown that many of the molecular pathways are common to Dictyostelium and mammalian cells [6, 7]. Furthermore, several of the Dictyostelium experiments have been performed in neutrophils as well [8]. The powerful forward and reverse microbial style genetics of Dictyostelium has been able to pin-point critical genes and proteins and has shown how they work. The use of proteins fused to the Green Fluorescent Protein, GFP, combined with biochemical examination of changes in enzymatic activities, has lead to quantitative real-time measurements of the dynamics of subcellular localization of these molecules. In addition, the wealth of experimental data has encouraged a large number of theoretical studies. In fact, it is fair to say that the information flow between experiments and modeling is no longer unidirectional and that theory now often drives experiments.
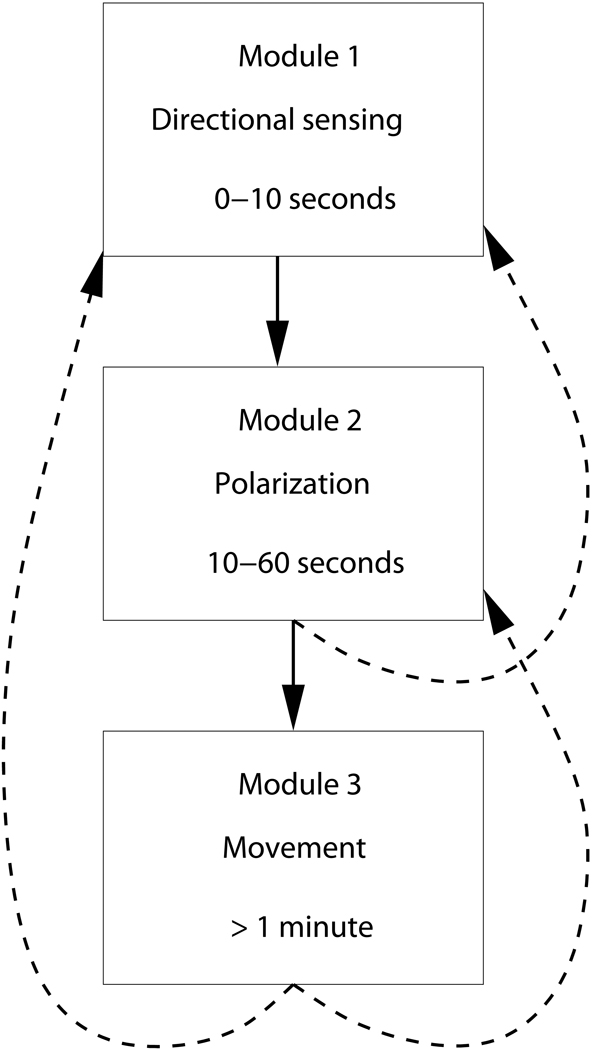
We divide eukaryotic chemotaxis into three distinct modules, each representing a different temporal stage of chemotaxis:
directional sensing; processes that occur on a time scale of 0–10 s after a gradient change and that are characterized by subcellular localization of several key signaling components but do not involve the reorganization of the cytoskeleton.
formation of a stable leading edge and cell polarity; processes that occur on longer timescales (10–45 s) and that lead to cell regions that can clearly be identified as front, back and sides. Polarization can occur in response to a gradient, in which case it is coupled to the gradient sensing process, or can occur spontaneously.
motility; processes that occur after the polarity is established and which include the actual movement of the cell (>1 minute).
Of course, the boundaries between these stages, schematically shown in Fig. 1, are not absolute and overlap exists. Nevertheless, this subdivision of the chemotaxis response is becoming commonly accepted [9]. Our review of these modules can only be cursory and the interested reader is encouraged to consult the reference articles for further details. Our goal here is to give a brief overview of the current status and to indicate our personal ideas where experimental and theoretical progress can be made.
Fig. 1.
Modules in chemotaxis. The arrows represent interactions between the modules which can be forward or reverse.
Directional sensing
The first stage of the cellular response involves the establishment of a leading edge of the cell and occurs during the first 0–10 sec after binding of the ligand to the receptors. A number of critical components in the directional sensing pathway have been identified greatly aided by the readily available genetic mutants in Dictyostelium [10]. Furthermore, through the use of GFP reporters, it has become clear that a number of these display a distinct subcellular re-organization during the initial events after chemoattractant stimulation [11–13].
A general picture of the initial events after chemoattractant stimulation has emerged that is conserved between Dictyostelium and neutrophils and that can be summarized as follows: chemoattractant binds to G protein-coupled receptors (GPCRs) that are uniformly distributed on the cell surface, leading to the dissociation of the Gα and Gβγ subunits of the coupled heterotrimeric G protein that activates downstream effectors. The earliest known downstream event is the localized activation of Ras at the leading edge, which was visualized by using a GFP fusion of a Ras binding domain (GFP-RBD) that only interacts with GTP bound Ras [14]. This leads to the activation of PI3K and the production of PI(3,4)P2/PI(3,4,5)P3, producing binding sites for a subclass of PH (Pleckstrin Homology) domain-containing proteins including CRAC [15], Akt/PKB [15, 16] and PhdA [17].
Recruitment of PI3K forms a feedback loop, mediated through localized F-actin polymerization, that further increases PIP3 at the front [14, 18]. This ultimately leads to F-actin protrusions and cell movement. The phosphoinositide 3 lipid phosphatase PTEN shows a reciprocal pattern of localization: it delocalizes from the activated leading edge but remains associated with the lateral sides and posterior of the cell [19, 20]. We have listed the currently known molecules that are spatial re-organized, along with the typical timescales for localization in Table 1.
Table 1.
Spatial reorganization and timescales of various signaling components following a stimulus from a pipette
| Molecule | Activation time scale | Asymmetry |
|---|---|---|
| G protein | <1 s | None |
| Ras | 3 s | Front/back |
| PH-domain proteins | 5 s | Front/back |
| PI3K | 15 s | Front/back |
| PTEN | 10 s | Back/front |
| Actin | 5 s | Front/back |
| PAKa | 30 s | Back/front |
| MyoII | 30 s | Back/front |
Recently, it has become clear that the PI3K pathway might not be as critically important as previously thought. Evidence for this comes from experiments in which all five PI3Ks and PTEN were genetically removed, thus eliminating the PIP3 pathway [21]. In steep gradients, these cells were found to be chemotactically responsive. However, it was found that inhibiting either the PIP3 pathway or a phospholipase A2 (PLA2) dependent pathway using pharmacological interventions eliminates chemotaxis in shallow gradients [22]. These results suggest that multiple redundant pathways are responsible for proper chemotactic behavior [23, 24]. Precisely how these parallel pathways interact with each other is currently unclear and is a challenge for the future although it is interesting to note that both pathways deplete local PIP2.
The picture that has emerged from these experiments is that the directional sensing machinery must rely on some form of communication between the back and the front of the cell and is not a purely local phenomenon. The latter can be ruled out by realizing that, typically, the stimulation in these experiments is presented to the cell in the form of a pipette containing the chemoattractant cAMP. Due to the fast diffusion of cAMP around the cell, the back of the cell (i.e. the side farthest away from the pipette) experiences a rapid increase in cAMP concentration. Thus, the stimulus levels at the front and the back rise rapidly and approach comparable levels within less than a second. A purely local mechanism would then lead to near-equal response levels at the back and the front. The experiments, however, show a large response at the front and an almost completely suppressed response at the back [14, 25]. Of course, it is possible that the stimulation level at the back is below a certain threshold. However, the existence of such a threshold can be ruled out by the fact that subcellular re-organization takes place over a wide range of stimulus strengths. We should also note here that a temporal mechanism, in which rapid protrusions sense their local concentrations, is worth considering.
The precise nature of the global intracellular communication and the origin of the switch-like behavior is the subject of intense research, and a number of different model mechanisms have been proposed [26–34]. Most of these models invoke the presence of a diffusible inhibitor which plays the role of the communicator. However, despite several decades of research efforts this inhibitor remains elusive.
The unraveling of these mechanisms requires precise experimental control of the external conditions and has lead to the application of new techniques including microfluidic devices, photo-uncaging of caged cAMP and modern pipette techniques [35–38]. These techniques allow for careful control of the cAMP stimulus, both spatially and temporally. For example, it is now possible to independently stimulate two sides of the cell and directly address the validity of some of the proposed mechanisms. Furthermore, the microfluidic devices are able to produce precisely controlled gradients of any shape. This can be used to investigate how the detection threshold depends on the steepness of the gradient and on the average background concentration. Clearly, future experiments will see an increase in the use of these quantitative experimental techniques and will undoubtedly play a crucial role in the further elucidation of directional sensing.
Polarization
As we already indicated in Fig. 1, the directional sensing module feeds directly into the polarity module. By polarity, we mean the symmetry-broken configuration of the cell’s internal components, including the cytoskeleton, allowing for the determination of regions that can be identified as front, back and sides. A number of essential components of polarity formation have been identified [13, 39–41]. It is important to realize, however, that polarization does not require a gradient stimulus: spontaneous polarization is observed in the absence of a gradient [42]. Thus, the polarity module needs to be able to work even when it does not receive direct input from the directional sensing module.
Our understanding of the events leading to cytoskeletal reorganization has dramatically improved over the past 10 years. This is mainly due to novel experimental techniques that allow for direct visualization of intracellular pathway components. Through these experiments, it has become clear that the cAMP chemoattractant receptors and their associated heterotrimeric G proteins are distributed uniformly around the cell perimeter and are activated in proportion to the concentration of cAMP [43]. Yet, the leading edge and posterior of the cells display highly polarized distributions of specific signaling components. PI3K, PtdIns(3,4,5)P3 (PIP3), and the PH domain-containing proteins that bind PIP3 (Akt, CRAC, and PhdA) preferentially localize to the edge of the cell that is closest to the chemoattractant source [15, 16, 25, 44]. Molecules that are associated with F-actin polymerization, including WASP, SCAR, and the GTP-bound forms of their activators (Rac GEFs), are found in the front and, to a lesser degree, at the posterior [45, 46]. PTEN, a negative regulator of the PI3K pathway, is excluded from the leading edge and accumulates at the lateral sides and posterior, hydrolyzing PIP3 and thereby restricting recruitment of PH proteins to these sites and simultaneously suppressing lateral pseudopod formation [19, 20]. Myosin II (myoII) is found along the lateral sides and at the posterior, mediates cortical tension, restricts pseudopod extension, and promotes contraction of the posterior [47]. MyoII in the rear is controlled, in part, by PAKa, a p21-activated protein kinase, which preferentially localizes to the cell’s posterior [48]. The disassembly of MyoII at the leading edge is mediated by MHCKA (Myosin Heavy Chain Kinase A), which is recruited to the leading edge and activated by F-actin [49, 50]. Myosin I and PAKc preferentially localize to the leading edge and are essential for proper chemotaxis [51]. Activated ERK1, a mitogen-activated protein kinase, and its upstream kinase MEK1 also localize to the cell’s anterior [52].
Our understanding of the role of various pathway components that are required for the formation of cell polarity has been obtained through the analysis of mutant strains. For example, unstimulated pten null cells display a marked increase in the formation of spontaneous protrusions while pten null cells stimulated with chemoattractant show an elevated and extended actin polymerization response. Compared to wild-type cells, PIP3 levels remain elevated in pten null cells for a prolonged time after stimulation and chemotaxis is severely disrupted [19, 20, 53]. These results suggest a crucial role for PIP3 in polarity formation. Deleting the major two forms of Class I PI3Ks in Dictyostelium or the addition of chemical PI3K inhibitors (LY294002 and wortmannin) impair cell polarity [19, 54].
Polarity is maintained and further stabilized through myoII assembly in the back of the cell. Cells that lack guanylyl cyclases (GCs) are defective in myoII assembly and the ability to suppress lateral pseudopod formation [40]. Also, these cells exhibit an expanded shape for anterior pseudopods. By contrast, cells carrying mutations in the cGMP-phosphodiesterases accumulate high levels of cGMP and become hyperpolarized, displaying a narrow dominant pseudopod at the leading edge [55, 56]. An important aspect of cGMP is that it is able to rapidly diffuse throughout the cell and thereby allow different local regions to coordinate their behavior; as already mentioned, this type of coordination is critical for the polarity system.
As is the case for the directional sensing pathways, a number of mechanistic questions pertaining to polarity remain unanswered. For example, it is not clear how directional sensing leads to polarity. Also, recent experiments suggest that cells can become polarized when stimulated globally or even in the absence of a stimulus [42]. One hypothesis is that this pre-existing polarity can interfere with directional sensing accuracy. This might explain the significant number of cells with “wrong” subcellular organization following a gradient stimulation [38]. Finally, it is not well understood how the polarized state is globally established in the cell and how stable this state is.
Again, progress towards answering these questions demands experimental techniques with a greater quantitative control than is available using standard pipette assays. Particularly promising are experiments in which the gradient can be rapidly reversed. These can be used to start addressing how cells are able to redirect themselves upon a change in chemoattractant gradient. Gradients in microfluidics are stable, controllable and rapidly reversible and offer the possibility to study re-establishment of polarity following a reversal of the cAMP gradient in a quantitative fashion.
Motility
A large body of literature has built up on almost all aspects of actin-based motility [57, 58]. Almost all crawling cells use the polymerization dynamics of actin [58–60], coupled to adhesive contacts with the substrate as well as active contraction (due to myosin motors), to propel themselves forward. The growth of an actin network, due at least in part to the action of Arp2/3, pushes against the cell membrane at the forward part of the cell, causing protrusion. Further behind, the network is disassembled, thereby providing a source of actin monomers that are transported by diffusion and (possibly) cytoplasmic flow back to the polymerizing front. Not all such protrusions are effective in actually moving the cell forward. Finally, myosin motors interact with actin near the rear of the cell, generating contraction forces that bring up the rear [39].
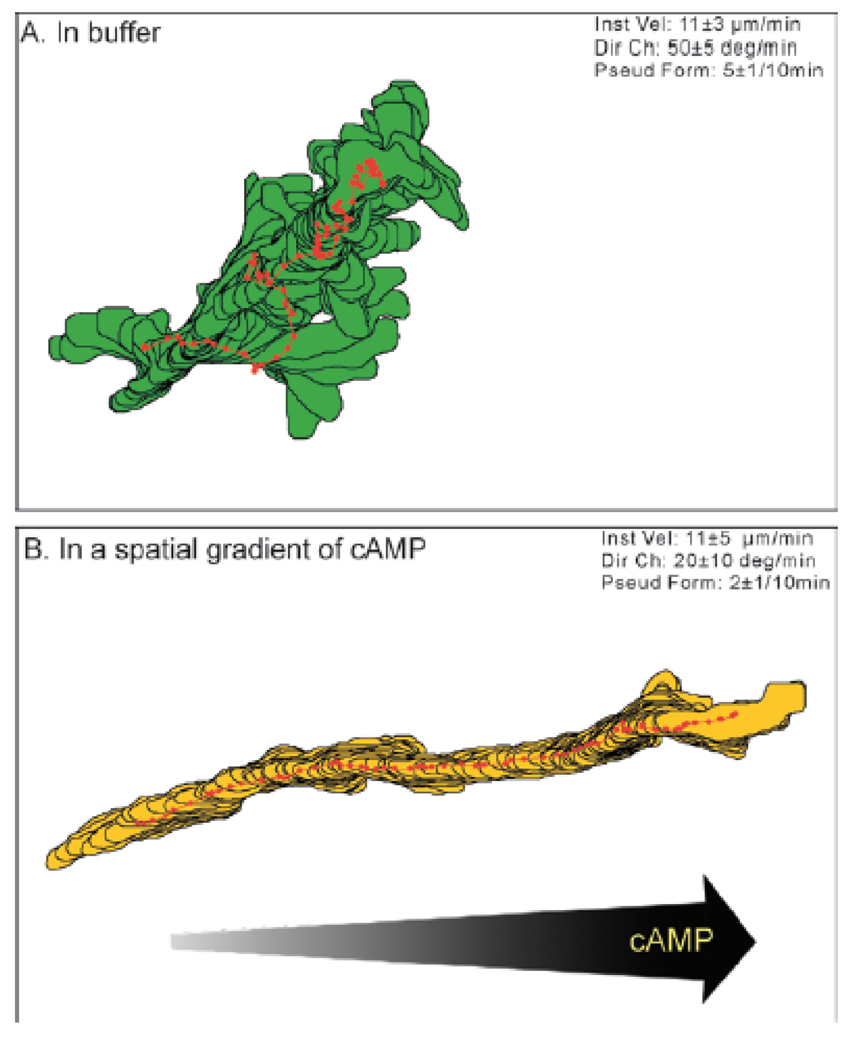
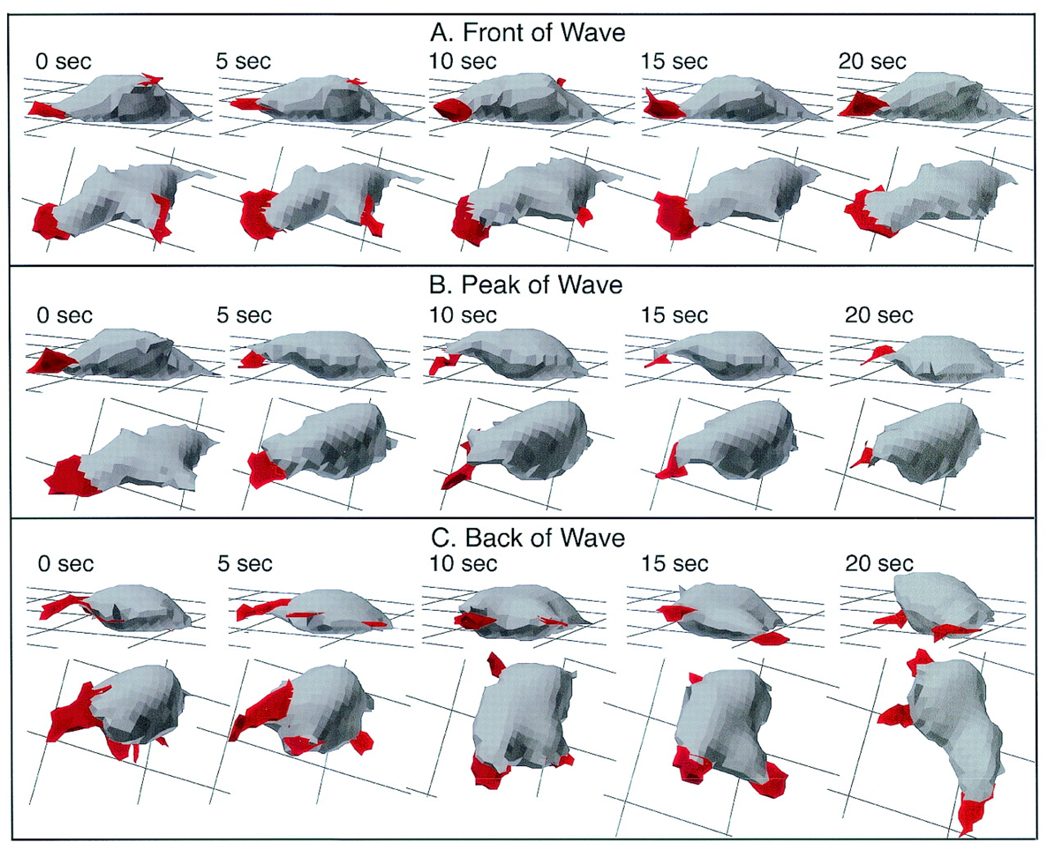
Much insight regarding motility has been obtained in recent years using computer assisted motion analysis [61, 62]. This analysis, mostly carried out in two dimensions (2D DIAS) but also possible in three dimensions (3D DIAS), traces the outline of the cell using boundary detection software. This outline, obtained from video frames of the experiment, is then used to determine various motility quantities including instantaneous velocity, directional change and directional persistence. A typical example of the output of 2D DIAS is shown in Fig. 2, where the outlines of a cell in buffer and in a gradient are plotted every 4 s. The position of the centroid is marked by a red dot. Recently, this analysis has been extended to perform a three dimensional reconstruction of cells obtained using optical sectioning. An example of a time-sequence of a moving cell reconstructed using 3D DIAS is shown in Fig. 3.
Fig. 2.
Computer generated cell outlines and centroids (represented by red dots) of Dictyostelium cells in. A: basic motility of cells in buffers showing random, non-directed motion. B: Cells subjected to a spatial gradient exhibit directed motion in the direction of the gradient (from [62]).
Fig. 3.
3D reconstructions of a representative Ax4 cell in the front (A), at the peak (B), and in the back (C) of a simulated temporal wave of cAMP generated in a perfusion chamber. Nonparticulate pseudopodial zones are demarcated in red. The cell is viewed at each time point at angles of 15° and 60° from the surface. Note that the Ax4 cell is elongate along the substratum in the front of the wave, rounds up and retracts the dominant pseudopod at the peak of the wave, and resumes pseudopod formation but in all directions and without cell elongation in the back of the wave. The behavior of this cell is representative of that of nine additional Ax4 cells reconstructed in 3D in a similar fashion (from [71]).
A distinction needs to be made between motion in the absence of external signals, basic motility, and motion in a temporally or spatially varying signal, directed motion. When placed in buffer, cells are not stationary. They continuously extend pseudopods and move about, as can be seen in Fig. 2. Some basic properties of this process, obtained using 2D DIAS, are summarized in Table 2. On average, a new pseudopod is formed every two minutes and, for the most part, only one pseudopod is present at any given time. Furthermore, the directional change, defined as change in direction during a time interval, is high. Thus, the location of the new pseudopod is not correlated to the old position and pseudopod formation is random in the absence of a chemotactic gradient. It should be noted that the motility properties of Dictyostelium cells are not independent of the developmental state of the cell. For example, the typical velocity of individual cells is low at the beginning of development, peaking roughly at the onset of aggregation and then decreases once again [63]. The numbers presented in the table are representative for the peak motility rate.
Table 2.
Basic quantities for cells in buffer (basic motility) and in a spatial gradient (chemotaxis)
| Experimental condition |
Instantaneous velocity |
Frequency of pseudopod formation |
Directional change |
|---|---|---|---|
| Basic motility | 11±3 µm/min | 5±1/10min | 50°±5°/min |
| Chemotaxis | 11±3 µm/min | 2±1/10min | 20°±10°/min |
The basic properties of cells moving in a spatial gradient are also summarized in Table 2. When comparing these properties to the ones representing basic motility, it is striking that the instantaneous velocities are identical. What changes drastically, however, are the pseudopod formation rate and the directional change, both of which show a significant reduction. This reflects the directed motion of the cell: instead of randomly located, the new pseudopod is highly correlated to the position of the old one. As a result, the cells are moving in a direction that is guided by the gradient (see Fig. 2B).
The formation of pseudopods has also been characterized in Dictyostelium and neutrophil cells that were forced to crawl under agarose and so were flattened to essentially two dimensions [64]. It was found that in a shallow gradient of chemoattractant new pseudopods usually bifurcated off existing ones rather than being formed de novo and that pseudopod generation appears to be independent of chemotactic signaling. Furthermore, the addition of a PI3K inhibitor did not change the accuracy of new pseudopods but did alter the frequency of pseudopod generation, suggesting that the PIP3 pathway is not essential for chemotaxis under these conditions. Future experiments that combine this experimental technique with techniques that can determine the subcellular distribution of proteins and that can precisely control the gradient have the promise to start unraveling the pathways underlying motility.
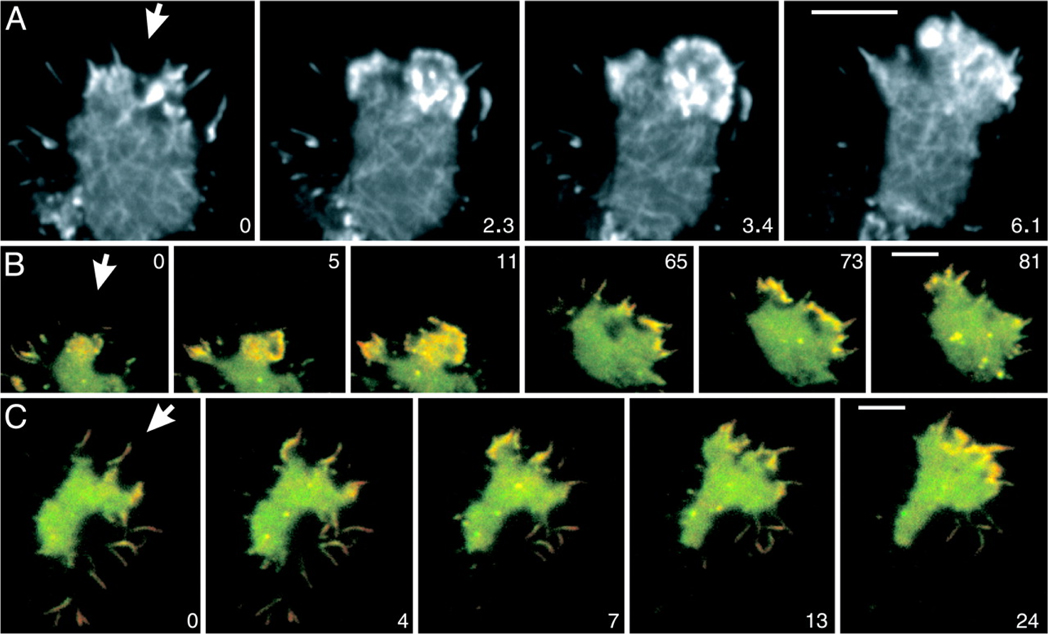
Several groups have attempted to formulate computational models of cells. The early models of motion [65, 66] focused on simple 1-D models with the "cell" running from the front to the back along a line. While these have some predictive power, they obviously cannot properly address the dynamics of cell shape. More recent work [67] has attempted to deal with a two-dimensional representation of the actin-myosin dynamics and its coupling to the cell shape. To date, these models have been applied to keratocytes and fibroblasts, but not to Dictyostelium or neutrophils. However, recent data have demonstrated quite dramatically that motility in all these cells, especially that which arises via the response to a chemotactic signal, operates using the same principles. In particular, the role of Arp2/3 in nucleating actin network linkage at the leading edge of the cell, as a part of the overall chemotaxis response, can be directly visualized (see Fig. 4, [68]).
Fig. 4.
Actin and Arp2/3 dynamics in chemotaxis. The cells shown respond to cAMP applied through a micropipet that is placed outside the frames shown. The direction of the diffusion gradients is indicated by arrows. (A) A cell labeled with full-length LimE-GFP recorded at a frame rate of 100 msec. (B and C) Cells labeled with GFP-Arp3 (green) and mRFP-LimEcoil (red). Yellow regions indicate merge of the two labels. The pattern of dense actin assemblies in B resembles the pattern in A, showing that these structures are enriched in the Arp2/3 complex. At subsequent stages, Arp2/3-rich actin structures are distributed along the region of the cell border that is exposed to higher cyclic-AMP concentrations. (C) Arp2/3-rich actin structures are preferentially accumulated at the base of filopods that point toward the source of the gradient. (In B and C, network structures are not visible, because the resolution of dual-emission fluorescence recordings is limited.) Time is indicated in seconds. (Scale bar: 5 µm.). From [68].
This modeling research focuses exclusively on basal motility and does not attempt to incorporate the signaling steps that connect from the sensory apparatus that detects external signals, to the control of actin-based protrusion. A full chemotaxis model would undoubtedly involve modeling the two-way coupling between the ligand receptor and its downstream effectors and the actin-myosin cytoskeletal dynamics, a truly challenging task. Not surprisingly, previous work has focused on purely phenomenological models that directly relate rules of motion to external chemical gradients. However, we believe that the time is ripe to attempt a more involved treatment which couples a directional sensing module to a polarity module and a motility module, much like we sketched in Fig. 1. Again, data from new experiments will prove essential in this endeavor and one new technique is particularly promising. This technique uses semi-flexible substrates to measure the forces exerted by crawling cells onto their substrate [69, 70]. Through the coupling of the observed deformations of the substrate with the known material parameters it is now possible to derive traction force fields in a rapid and systematic fashion. This offers the possibility to systematically study the effects of mutations on the motility of cells. These forces are clearly an important input parameter in any motility model and provide yet another example where combined modeling and experiment can lead to substantial progress.
Acknowledgements
We thank Herbert Levine and Richard A. Firtel for useful discussions. This work was supported by the NIH (PO1 GM078586).
References
- 1.Schnorrer F, Dickson BJ. Axon guidance: morphogens show the way. Curr Biol. 2004;vol. 14:19–21. doi: 10.1016/j.cub.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 2.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. 1999;vol. 341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 3.Condeelis J, Singer R, Segall JE. The Great Escape: When Cancer Cells Hijack the Genes for Chemotaxis and Motility. Annu Rev Cell Dev Biol. 2005 doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 4.Raz E, Reichman-Fried M. Attraction rules: germ cell migration in zebrafish. Curr Opin Genet Dev. 2006;vol. 16:355–359. doi: 10.1016/j.gde.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Cho NK, Keyes L, Johnson E, Heller J, Ryner L, Karim F, Krasnow MA. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell. 2002;vol. 108:865–876. doi: 10.1016/s0092-8674(02)00676-1. [DOI] [PubMed] [Google Scholar]
- 6.van Es S, Devreotes PN. Molecular basis of localized responses during chemotaxis in amoebae and leukocytes. Cell. Mol. Life Sci. 1999;vol. 55:1341–1351. doi: 10.1007/s000180050374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: A focus on leukocytes and Dictyostelium. Annu. Rev. Cell Biol. 1988;vol. 4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- 8.Bourne HR, Weiner O. A chemical compass. Nature. 2002;vol. 419:21. doi: 10.1038/419021a. [DOI] [PubMed] [Google Scholar]
- 9.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem. 2003;vol. 278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 10.Manahan CL, Iglesias PA, Long Y, Devreotes PN. Chemoattractant signaling in dictyostelium discoideum. Annu Rev Cell Dev Biol. 2004;vol. 20:223–253. doi: 10.1146/annurev.cellbio.20.011303.132633. [DOI] [PubMed] [Google Scholar]
- 11.van Haastert PJM, Devreotes PN. Chemotaxis: signalling the way forward. Nature Rev. Mol. Cell Biol. 2004;vol. 5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 12.Merlot S, Firtel RA. Leading the way: directional sensing through phosphatidylinositol 3-kinase and other signaling pathways. J. Cell Sci. 2003;vol. 116:3471–3478. doi: 10.1242/jcs.00703. [DOI] [PubMed] [Google Scholar]
- 13.Williams HP, Harwood AJ. Cell polarity and Dictyostelium development. Curr. Opin. Microbiol. 2003;vol. 6:621–627. doi: 10.1016/j.mib.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;vol. 167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;vol. 95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 16.Meili R, Ellsworth C, Lee S, Reddy TBK, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;vol. 18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funamoto S, Milan K, Meili R, Firtel RA. Role of phosphatidylinositol 3' kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 2001;vol. 153:795–809. doi: 10.1083/jcb.153.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charest PG, Firtel RA. Feedback signaling controls leading-edge formation during chemotaxis. Curr Opin Genet Dev. 2006;vol. 16:339–347. doi: 10.1016/j.gde.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;vol. 109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 20.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;vol. 109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 21.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;vol. 17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 22.van Haastert PJ, Keizer-Gunnink I, Kortholt A. Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J Cell Biol. 2007;vol. 177:809–816. doi: 10.1083/jcb.200701134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Iijima M, Tang M, Landree MA, Huang YE, Xiong Y, Iglesias PA, Devreotes PN. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev Cell. 2007;vol. 12:603–614. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veltman DM, Keizer-Gunnik I, Van Haastert PJ. Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J Cell Biol. 2008;vol. 180:747–753. doi: 10.1083/jcb.200709180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 2004;vol. 101:8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narang A, Subramanian KK, Lauffenburger DA. A mathematical model for chemoattractant gradient sensing based on receptor-regulated membrane phospholipid signaling dynamics. Ann Biomed Eng. 2001;vol. 29:677–691. doi: 10.1114/1.1385805. [DOI] [PubMed] [Google Scholar]
- 27.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys. J. 2002;vol. 82:50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J. Cell Sci. 1999;vol. 112:2867–2874. doi: 10.1242/jcs.112.17.2867. [DOI] [PubMed] [Google Scholar]
- 29.Rappel WJ, Thomas PJ, Levine H, Loomis WF. Establishing direction during chemotaxis in eukaryotic cells. Biophys. J. 2002;vol. 83:1361–1367. doi: 10.1016/S0006-3495(02)73906-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine H, Kessler DA, Rappel WJ. Directional sensing in eukaryotic chemotaxis: a balanced inactivation model. Proc Natl Acad Sci U S A. 2006;vol. 103:9761–9766. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gamba A, de Candia A, Di Talia S, Coniglio A, Bussolino F, Serini G. Diffusion-limited phase separation in eukaryotic chemotaxis. Proc Natl Acad Sci U S A. 2005;vol. 102:16927–16932. doi: 10.1073/pnas.0503974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skupsky R, Losert W, Nossal RJ. Distinguishing modes of eukaryotic gradient sensing. Biophys J. 2005;vol. 89:2806–2823. doi: 10.1529/biophysj.105.061564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier-Schellersheim M, Xu X, Angermann B, Kunkel EJ, Jin T, Germain RN. Key role of local regulation in chemosensing revealed by a new molecular interaction-based modeling method. PLoS Comput Biol. 2006;vol. 2:e82. doi: 10.1371/journal.pcbi.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iglesias PA, Devreotes PN. Navigating through models of chemotaxis. Curr Opin Cell Biol. 2008 doi: 10.1016/j.ceb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Xu X, Meier-Schellersheim M, Jiao X, Nelson LE, Jin T. Quantitative imaging of single live cells reveals spatiotemporal dynamics of multistep signaling events of chemoattractant gradient sensing in Dictyostelium. Mol Biol Cell. 2005;vol. 16:676–688. doi: 10.1091/mbc.E04-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song L, Nadkarni SM, Bodeker HU, Beta C, Bae A, Franck C, Rappel WJ, Loomis WF, Bodenschatz E. Dictyostelium discoideum chemotaxis: Threshold for directed motion. Eur J Cell Biol. 2006;vol. 85:981–989. doi: 10.1016/j.ejcb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Beta C, Wyatt D, Rappel WJ, Bodenschatz E. Flow photolysis for spatiotemporal stimulation of single cells. Anal Chem. 2007;vol. 79:3940–3944. doi: 10.1021/ac070033y. [DOI] [PubMed] [Google Scholar]
- 38.Samadani A, Mettetal J, van Oudenaarden A. Cellular asymmetry and individuality in directional sensing. Proc Natl Acad Sci U S A. 2006;vol. 103:11549–11554. doi: 10.1073/pnas.0601909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Affolter M, Weijer CJ. Signaling to cytoskeletal dynamics during chemotaxis. Dev Cell. 2005;vol. 9:19–34. doi: 10.1016/j.devcel.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Postma M, Bosgraaf L, Loovers HM, van Haastert PJM. Chemotaxis: signalling modules join hands at front and tail. EMBO Rep. 2004;vol. 5:35–40. doi: 10.1038/sj.embor.7400051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dormann D, Weijer CJ. Chemotactic cell movement during development. Curr. Opin. Genet. Devel. 2003;vol. 13:358–364. doi: 10.1016/s0959-437x(03)00087-x. [DOI] [PubMed] [Google Scholar]
- 42.Postma M, Roelofs J, Goedhart J, Gadella TWJ, Visser AJWG, van Haastert PJM. Uniform cAMP stimulation of Dictyostelium cells induces localized patches of signal transduction and pseudopodia. Mol. Biol. Cell. 2003;vol. 14:5019–5027. doi: 10.1091/mbc.E03-08-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao Z, Zhang N, Murphy DB, Devreotes PN. Dynamic distribution of chemoattractant receptors in living cells during chemotaxis and persistent stimulation. J. Cell Biol. 1997;vol. 139:365–374. doi: 10.1083/jcb.139.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang YE, Iijima M, Parent CA, Funamoto S, Firtel RA, Devreotes P. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P3 to the leading edge of chemotaxing cells. Mol. Biol. Cell. 2003;vol. 14:1913–1922. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myers SA, Han JW, Lee Y, Firtel RA, Chung CY. A Dictyostelium Homologue of WASP Is Required for Polarized F-Actin Assembly during Chemotaxis. Mol Biol Cell. 2005;vol. 16:2191–2206. doi: 10.1091/mbc.E04-09-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park KC, Rivero F, Meili R, Lee S, Apone F, Firtel RA. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. Embo J. 2004;vol. 23:4177–4189. doi: 10.1038/sj.emboj.7600368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yumura S, Uyeda TQP. Myosins and cell dynamics in cellular slime molds. Int. Rev. Cytol. 2003;vol. 224:173–225. doi: 10.1016/s0074-7696(05)24005-6. [DOI] [PubMed] [Google Scholar]
- 48.Chung CY, Firtel RA. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 1999;vol. 147:559–575. doi: 10.1083/jcb.147.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steimle PA, Licate L, Cote GP, Egelhoff TT. Lamellipodial localization of Dictyostelium myosin heavy chain kinase A is mediated via F-actin binding by the coiled-coil domain. FEBS Lett. 2002;vol. 516:58–62. doi: 10.1016/s0014-5793(02)02494-8. [DOI] [PubMed] [Google Scholar]
- 50.Rubin H, Ravid S. Polarization of myosin II heavy chain-protein kinase C in chemotaxing Dictyostelium cells. J. Biol. Chem. 2002;vol. 277:36005–36008. doi: 10.1074/jbc.M205986200. [DOI] [PubMed] [Google Scholar]
- 51.Lee S, Rivero F, Park KC, Huang E, Funamoto S, Firtel RA. Dictyostelium PAKc is required for proper chemotaxis. Mol Biol Cell. 2004;vol. 15:5456–5469. doi: 10.1091/mbc.E04-04-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sobko A, Ma H, Firtel RA. Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev. Cell. 2002;vol. 2:745–756. doi: 10.1016/s1534-5807(02)00186-7. [DOI] [PubMed] [Google Scholar]
- 53.Comer FI, Parent CA. PI 3-kinases and PTEN: how opposites chemoattract. Cell. 2002;vol. 109:541–544. doi: 10.1016/s0092-8674(02)00765-1. [DOI] [PubMed] [Google Scholar]
- 54.Chen LF, Janetopoulos C, Huang YE, Iijima M, Borleis J, Devreotes PN. Two phases of actin polymerization display different dependencies on PI(3,4,5)P-3 accumulation and have unique roles during chemotaxis. Mol. Biol. Cell. 2003;vol. 14:5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bosgraaf L, Russcher H, Smith JL, Wessels D, Soll DR, van Haastert PJM. A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 2002;vol. 21:4560–4570. doi: 10.1093/emboj/cdf438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimmel AR, Firtel RA. Breaking symmetries: regulation of Dictyostelium development through chemoattractant and morphogen signal-repsonse. Curr. Opin. Genet. Devel. 2004;vol. 14:540–549. doi: 10.1016/j.gde.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Bray D. Cell movements: from molecules to motility. New York: Garland Pub.; 2001. [Google Scholar]
- 58.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;vol. 112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 59.Rafelski SM, Theriot JA. Crawling toward a unified model of cell mobility: spatial and temporal regulation of actin dynamics. Annu Rev Biochem. 2004;vol. 73:209–239. doi: 10.1146/annurev.biochem.73.011303.073844. [DOI] [PubMed] [Google Scholar]
- 60.Mogilner A, Oster G. Polymer motors: pushing out the front and pulling up the back. Curr Biol. 2003;vol. 13:R721–R733. doi: 10.1016/j.cub.2003.08.050. [DOI] [PubMed] [Google Scholar]
- 61.Wessels D, Voss E, Von Bergen N, Burns R, Stites J, Soll DR. A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil. Cytoskel. 1998;vol. 41:225–246. doi: 10.1002/(SICI)1097-0169(1998)41:3<225::AID-CM4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 62.Soll DR, Wessels D, Heid PJ, Zhang H. A contextual framework for characterizing motility and chemotaxis mutants in Dictyostelium discoideum. J. Muscle Res. Cell Motil. 2002;vol. 23:659–672. doi: 10.1023/a:1024459124427. [DOI] [PubMed] [Google Scholar]
- 63.Varnum B, Edwards KB, Soll DR. Dictyostelium amebae alter motility differently in response to increasing versus decreasing temporal gradients of cAMP. J. Cell Biol. 1985;vol. 101:1–5. doi: 10.1083/jcb.101.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;vol. 9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 65.Gracheva ME, Othmer HG. A continuum model of motility in ameboid cells. Bull Math Biol. 2004;vol. 66:167–193. doi: 10.1016/j.bulm.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 66.DiMilla PA, Barbee K, Lauffenburger DA. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys J. 1991;vol. 60:15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mogilner A, Rubinstein B. The physics of filopodial protrusion. Biophys J. 2005;vol. 89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diez S, Gerisch G, Anderson K, Muller-Taubenberger A, Bretschneider T. Subsecond reorganization of the actin network in cell motility and chemotaxis. Proc Natl Acad Sci U S A. 2005;vol. 102:7601–7606. doi: 10.1073/pnas.0408546102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Del Alamo JC, Meili R, Alonso-Latorre B, Rodriguez-Rodriguez J, Aliseda A, Firtel RA, Lasheras JC. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc Natl Acad Sci U S A. 2007;vol. 104:13343–13348. doi: 10.1073/pnas.0705815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lombardi ML, Knecht DA, Dembo M, Lee J. Traction force microscopy in Dictyostelium reveals distinct roles for myosin II motor and actin-crosslinking activity in polarized cell movement. J Cell Sci. 2007;vol. 120:1624–1634. doi: 10.1242/jcs.002527. [DOI] [PubMed] [Google Scholar]
- 71.Zhang H, Heid PJ, Wessels D, Daniels KJ, Pham T, Loomis WF, Soll DR. Constitutively active protein kinase A disrupts motility and chemotaxis in Dictyostelium discoideum. Euk. Cell. 2003;vol. 2:62–75. doi: 10.1128/EC.2.1.62-75.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]