Abstract
Persistent hepatitis C virus infection is associated with progressive hepatic fibrosis and liver cancer. Acute infection evokes several distinct innate immune responses, but these are partially or completely countered by the virus. Hepatitis C virus proteins serve dual functions in replication and immune evasion, acting to disrupt cellular signaling pathways leading to interferon synthesis, subvert Jak-STAT signaling to limit expression of interferon-stimulated genes, and block antiviral activities of interferon-stimulated genes. The net effect is a multilayered evasion of innate immunity, which negatively influences the subsequent development of antigen-specific adaptive immunity, thereby contributing to virus persistence and resistance to therapy.
Keywords: Inflammation, Innate Immunity, Interferon, Liver Injury, Signal Transduction, Toll-like Receptors (TLR), Viral Protease, Virus, RIG-I-like Helicases, Immune Evasion
Introduction
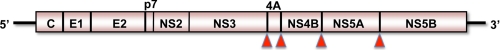
Hepatitis C virus (HCV)2 is a small, hepatotropic, positive-strand RNA virus classified within a unique genus of the Flaviviridae family (1). Its genome is <10 kb in length and encodes only 10 different proteins (Fig. 1), yet HCV is able to survive the immune response in most acutely infected persons and establish lifelong persistent infection. Such chronic infections are associated with a significant risk of progressive liver fibrosis and hepatocellular carcinoma. Worldwide, >130 million people are persistently infected with HCV, resulting in an estimated 366,000 deaths due to cirrhosis and cancer annually (2). HCV is thus a significant public health threat. It is also difficult and expensive to treat. Current standard-of-care therapies combine pegylated interferon-α (Peg-IFN-α) and ribavirin but cure <50% of patients with genotype 1 virus infection. Direct-acting specific antiviral therapies are on the near horizon but will be limited by rapid selection of resistant virus unless administered as part of a multidrug mixture (3). Peg-IFN-α is thus likely to remain a mainstay of therapy for years to come.
FIGURE 1.
Organization of the positive-sense RNA genome of HCV. Genomic RNA contains a single large open reading frame flanked by 5′- and 3′-untranslated RNA segments. The open reading frame encodes a polyprotein of >3000 amino acids that undergoes processing by cellular and viral proteases to produce 10 mature proteins. The core (C) and envelope proteins E1 and E2 are structural components of the infectious virus particle, whereas the remaining proteins are nonstructural (NS) and required for RNA replication (NS3–NS5B) or particle assembly and egress from the cell (p7 and NS2). Major viral protease activities include NS2, which cleaves in cis at the NS2-NS3 junction, and NS3/4A, a noncovalent complex of the N-terminal domain of NS3 and an accessory peptide sequence from NS4A that directs cleavage at the sites indicated by the red arrowheads and also targets the cellular signaling proteins MAVS and TRIF for proteolysis.
Antigen-specific adaptive T cell immunity is key to determining the outcome of acute HCV infection, with virus-specific CD4+ T cell help essential for effective T cell control (reviewed in Ref. 4). However, earlier innate immune responses are also critical. They are induced upon host recognition of common molecular patterns expressed by HCV and other viruses, and they provide relatively immediate protection against infection. IFNs play a central role in the innate immune response to viruses and comprise multiple classes of soluble cytokines, each with a distinct receptor system. Those of known relevance to HCV include type I IFNs (IFN-β and over a dozen distinct IFN-α subtypes, encoded by genes clustering on human chromosome 9), type II IFN (IFN-γ, a single IFN type encoded by a gene on chromosome 12), and type III IFN (IFN-λ, three subtypes known otherwise as interleukin (IL)-29, IL-28A, and IL-28B) (5, 6). Overlapping signaling pathways induce the expression of type I and III IFNs, and this occurs in many types of cells infected with viruses. In turn, these IFNs induce similar if not identical antiviral states in cells, albeit through different receptors with distinct cell-type distributions. IFN-γ is markedly different. Its production is restricted to certain types of immune cells, such as natural killer cells and cytotoxic T cells, and its actions are primarily immunomodulatory.
Sequence polymorphisms in human genes encoding IFN-λ3 (IL-28B) (7), IFN-γ (8), and the natural killer cell receptor KIR2DL3 and its human leukocyte antigen C group 1 ligand (9) influence the outcome of HCV infection. The association between outcome and IFN-λ3 genotype is particularly strong and also evident in the response to Peg-IFN-α therapy (10). These genome-wide association studies provide clinical evidence of the close linkage that exists between the innate and adaptive immune responses. Type I IFN regulation of components of the class I antigen processing pathway, including PA28 (proteasome activator 28) subunits and endoplasmic reticulum aminopeptidases (11), is one example of such linkage. IFN-λ3 regulation of regulatory T cells is another (12). There are undoubtedly many others (13).
IFN Signaling in the Liver
IFN-stimulated genes (ISGs) are expressed at high levels in the liver in many patients with chronic hepatitis C and indeed are induced within 48 h of acute infection in chimpanzees (14–16). However, other chronically infected patients show little or no evidence of ISG expression in the liver (16), despite similar levels and duration of virus infection. At present, there is no good understanding of this or the activation status of the innate immune signaling pathways and transcription factors controlling IFN synthesis within infected and uninfected cells in the HCV-infected liver.
IRF3 (IFN regulatory factor 3), a highly regulated transcription factor constitutively expressed in the cytoplasm in a latent inactive form, plays a key role in regulating the synthesis of IFN-β (17). Virus infections typically induce specific C-terminal phosphorylation of IRF3, resulting in its dimerization and transport to the nucleus. In coordination with NF-κB and ATF2/c-Jun, IRF3 forms an enhanceosome complex on the IFN-β promoter, leading to transcriptional activation and synthesis of IFN-β (18). Secreted IFN-β binds the type I IFN-α/β receptor (IFNAR), resulting in autocrine/paracrine activation of the Jak (Janus kinase)-STAT (signal transducer and activator of transcription) signaling pathway, which leads in turn to the expression of many dozens of ISGs controlled by promoters containing IFN-stimulated response elements. IRF7 synthesis is similarly induced and is capable of forming heterodimers with IRF3; its role as an amplifier is critical to the induction of most IFN-α genes (19). Activated IRF3 is also capable of directly inducing the transcription of a subset of ISGs, some of which (like ISG56) have direct antiviral activity against HCV (20, 21). IRF3 also controls transcription of IFN-λ (22).
IRF3 is activated by cell receptors that sense “pathogen-associated molecular patterns” (PAMPs) residing in viral proteins and RNAs. Defining HCV-specific PAMPs and the receptors they bind in the liver has been a daunting task, as available experimental systems are generally poor mimics of conditions in vivo. Wild-type virus does not replicate well in cell culture, and there are no good small animal models of hepatitis C. Nonetheless, studies in cell culture indicate that two distinct PAMP receptors, retinoic acid-inducible gene I (RIG-I) (23, 24) and TLR3 (Toll-like receptor 3) (25), sense HCV infection, induce IFN-β promoter activity, and partially restrict viral replication. Both of these PAMP receptors recognize viral RNAs. TLR2 may also sense infection, inducing inflammatory signals upon binding HCV proteins (26). A fourth potentially important sensor is TLR7. Little is known specifically about its role in the induction of IFN responses in the HCV-infected liver, but it is a likely player. Although they act to some extent redundantly, these receptors differ in terms of the PAMPs they sense, where they are expressed within the cell, and whether they are likely to be expressed in hepatocytes infected with HCV. They also differ in their utilization of signaling adaptor proteins, two of which are targeted for degradation by HCV.
RIG-I Signaling in HCV Infection
RIG-I (DDX58) is a DExD box helicase expressed ubiquitously within the cytoplasm of most cell types. It senses short non-self double-stranded RNAs (dsRNAs) with free 5′-triphosphates (Fig. 2, left) (27, 28). Ligand recognition is dependent upon an ATP-driven translocase activity (29) and results in the recruitment of RIG-I to the mitochondrial surface, where it interacts with MAVS (mitochondrial antiviral signaling protein; also known as IPS-1, VISA, and Cardif) resident on the outer mitochondrial membrane (30–32). This occurs through shared caspase recruitment domains (CARDs) in RIG-I and MAVS. Tandem CARDs near the N terminus of RIG-I become accessible upon RNA binding, inducing a conformational change relieving the effect of a C-terminal regulatory domain (33). This promotes RIG-I self-association as well as interactions with the MAVS adaptor, leading to assembly of a signaling complex on the mitochondrial surface that activates downstream non-canonical kinases of the IκB kinase complex, TBK-1 (TANK-binding kinase 1) and IκB kinase-ϵ, that phosphorylate IRF3 (34).
FIGURE 2.
RIG-I and TLR3 sensing of viral RNAs induces IFN-β synthesis. Two distinct signaling pathways, one initiated by the recognition of 5′-triphosphate-containing cytosolic viral RNAs by the DExD box helicase RIG-I (left) and the other by TLR3 recognition of viral dsRNA >40–50 bp within the lumen of vesicles in an early endosomal compartment (right), lead to phosphorylation of IRF3, activation of NF-κB, and induction of IFN-β transcription. Both pathways are disrupted by expression of the viral NS3/4A protease, which cleaves the adaptor proteins MAVS and TRIF, respectively. See text for details. In addition to those proteins shown, roles have been suggested for MEKK1, FADD, and RIP in RIG-I signaling leading to IFN-β synthesis. TM, transmembrane domain; PI3K, phosphatidylinositol 3-kinase; IKK, IκB kinase; MITA, mediator of IRF3 activation.
A complete discussion of RIG-I signaling is well beyond the scope of this minireview, but a considerable number of host proteins are essential for optimal induction of IFN-β synthesis via this pathway (Fig. 2, left). The RING finger domain-containing E3 ubiquitin ligase TRIM25 (tripartite motif-containing protein 25) mediates ubiquitylation of RIG-I at Lys-63, and this is essential for efficient MAVS binding and downstream induction of IFN synthesis (35). TRAF3 (tumor necrosis factor receptor-associated factor 3) is also required for optimal IFN induction, interacting with MAVS and helping to link it to the downstream kinases responsible for IRF3 phosphorylation (36, 37). TRAF6 is also recruited to the MAVS complex and, along with MEKK1, a member of the MAP3K family, is required for optimal activation of NF-κB, an essential component of the IFN-β enhanceosome (18, 38). TRADD, a tumor necrosis factor receptor adaptor protein, is also recruited to MAVS and has been implicated in IRF3 and NF-κB activation (39). NF-κB activation involves an interaction with CARD9 (CARD family member 9) and Bcl-10 (B cell CLL/lymphoma 10) protein that is MAVS-dependent but not required for IRF3 activation (Fig. 2, left) (40). On the other hand, NLRX1, a conserved nucleotide-binding domain and leucine-rich repeat-containing family member, is localized to the mitochondrial outer membrane, where it interacts with MAVS and may negatively regulate signaling (41).
Upstream of MAVS, RIG-I interacts with ASC (apoptosis-associated speck-like protein containing CARD; PYCARD) (Fig. 2, left), linking RIG-I sensing of viral RNA to MAVS-independent activation of a caspase-1-dependent inflammasome (40). Inflammasome-mediated production of mature IL-1β is influenced by signaling through MAVS, as it is dependent upon NF-κB-mediated stimulation of pro-IL-1β expression. Although IL-1β expression occurs in the context of viral hepatitis, it is not known if hepatocytes (the cell type within which HCV replicates) are capable of mounting an inflammasome response.
RIG-I senses infection by many RNA virus types, including those with positive- and negative-strand as well as double-stranded genomes (42). Picornaviruses are an exception, as their RNAs lack a free 5′-triphosphate and are sensed by a related CARD-containing helicase, MDA5 (melanoma differentiation-associated gene 5) (42). RIG-I is functionally expressed within hepatocytes and most hepatocyte-derived cell lines (43, 44). It senses HCV infection in Huh-7 hepatoma cells, leading to transient nuclear translocation of IRF3 and activation of the IFN-β promoter (23). Hydrodynamic transfection of synthetic HCV RNAs has shown that a conserved poly(U/C) sequence from the 3′-untranslated RNA segment of the HCV genome stimulates ISG expression in mouse liver, where it is recognized by RIG-I, presumably in conjunction with 5′-triphosphate, as an HCV PAMP (43).
Prior to the identification of RIG-I, the major protease expressed by HCV, NS3/4A, was found to inhibit Sendai virus-induced activation of the IFN-β promoter in cells containing self-amplifying HCV replicon RNAs (45). This was shown subsequently to be due to NS3/4A-mediated cleavage of the RIG-I adaptor protein MAVS (32, 46). Scission occurs between Cys-508 and His-509, close to the C terminus of MAVS, releasing MAVS from the mitochondrial membrane and eliminating its ability to function in signaling (Fig. 2, left). Oligomerization of MAVS is essential to its ability to signal; this occurs through its mitochondrial transmembrane domain and is blocked by NS3/4A cleavage (47). Although RIG-I has been shown to recognize HCV infection in Huh-7 cells, signaling is shut down as infection progresses, NS3/4A accumulates, and MAVS is degraded (23). This can be reversed by ketoamide and macrocyclic peptidomimetic inhibitors of NS3/4A now being developed as potential therapies for hepatitis C but only at concentrations significantly greater than the antiviral EC50 (48). Why higher drug concentrations are required for restoration of signaling is unknown. It is possible that HCV infection impacts signaling at another step in the pathway (49). The NS3 protein, a component of NS3/4A, interacts directly with TBK-1, and this may add to the disruption of IRF3 activation (50). Overexpression studies suggest that another HCV nonstructural protein, NS4B, may also interfere with RIG-I signaling (51), but the reliability of this experimental approach is questionable.
There is little doubt that HCV functionally disrupts RIG-I signaling in vivo. Although infection is limited to a minority of hepatocytes (52), immunoblots show MAVS to be cleaved in liver biopsies from some patients with chronic hepatitis C (23). This suggests that the intrahepatic IFN responses observed in many patients (16) either are induced through alternative signaling pathways or possibly originate in newly infected cells that have yet to accumulate sufficient NS3/4A to ablate signaling. The cleavage of MAVS by NS3/4A is likely to contribute to HCV pathogenesis, but it is not unique or a proximate cause of virus persistence. The protease of a closely related flavivirus, GB virus B, also cleaves MAVS, yet this virus typically causes self-limited infection in its natural hosts (53). In addition, the 3ABC protease precursor expressed by hepatitis A virus, a picornavirus associated only with acute hepatitis in humans, is targeted to the mitochondrial membrane, where it also cleaves MAVS with similar effects on signal transduction (54).
TLR3 Signaling in Hepatitis C
TLR3 senses dsRNA formed during the replication of positive-strand RNA viruses as well as some DNA viruses (55). A membrane-bound protein, it is expressed predominantly in an early endosomal compartment, where it senses extracellular ligand (56). Structural models suggest that dsRNA complexes >40–50 bp in length bind sites at opposite ends of a horseshoe-shaped ectodomain containing leucine-rich repeats (57). This drives dimerization of TLR3, which initiates signaling through its cytosolic TIR (Toll/IL-1 receptor homology) domain. TLR3 dimerization results in recruitment of an adaptor protein, TRIF (TIR domain-containing adaptor-inducing IFN-β; also known as TICAM-1), that interacts with TLR3 through homotypic TIR domains (Fig. 2, right) (58, 59). This interaction is transient, after which TRIF relocates to distinct punctate cytoplasmic bodies, possibly as an oligomer (56). At some point in this process, TRAF6, TRAF3, and TBK-1 are recruited to TRIF, leading to phosphorylation of IRF3 (37, 58). Endosomal acidification is required for recognition of dsRNA by TLR3 (60), and phosphorylation of TLR3 by phosphatidylinositol 3-kinase is essential for full activation of IRF3 and induction of IFN-β synthesis (61). TLR3 ligation of dsRNA also strongly induces NF-κB activity and pro-inflammatory cytokine synthesis via interactions of TRIF with RIP-1 (receptor-interacting protein 1) kinase (58, 62).
TLR3 is expressed in a variety of cell types in the liver, including stellate cells, resident macrophages (Kupffer cells), myeloid dendritic cells, and biliary epithelial cells (63). It is expressed at low levels in normal hepatocytes but can be demonstrated directly in uninfected human liver by two-photon microscopy (25, 64). Primary cultures of human hepatocytes strongly up-regulate expression of ISGs, including TLR3 itself, when stimulated by extracellular poly(I:C), a dsRNA surrogate (25, 65). The impact of HCV on TLR3 signaling has been difficult to study because Huh-7 hepatoma cells that are commonly used for propagation of HCV do not express TLR3 (44). This may be because TLR3 expression is regulated by p53, which is mutated in Huh-7 and many other hepatocellular carcinoma cell lines.
However, TLR3-mediated poly(I:C) stimulation of IRF3- and NF-κB-responsive promoters is inhibited in HeLa cells containing a replicating subgenomic HCV replicon RNA, as well as in HCV-infected Huh-7.5 cells in which TLR3 expression was reconstituted by retroviral gene transduction (Huh7.5-TLR3 cells) (25, 66). TLR3 signaling was also disrupted in osteosarcoma cells conditionally expressing NS3/4A but could be partially restored following treatment with a specific NS3/4A protease inhibitor (66). TRIF is cleaved in vitro by NS3/4A (Fig. 2, right), and its expression is significantly reduced in HeLa cells containing HCV replicons or in Huh-7.5 cells infected with HCV (25, 66). Brief treatment of infected cells with an NS3/4A inhibitor partially restores TRIF expression (25). In some cell types, however, NS3/4A overexpression has been reported not to reduce TRIF abundance (65, 67). This may reflect cell type-specific differences in TRIF or NS3/4A localization or technical difficulties assessing TRIF abundance, as it is normally expressed at very low levels.
NS3/4A-mediated cleavage of TRIF occurs at a unique site in the molecule bearing remarkable sequence homology to the NS4B/5A cleavage site in the viral polyprotein (66, 68). Whereas sites of NS3/4A scission in the polyprotein have an acidic residue at the P6 position, the P6 position in TRIF marks the end of an extended polyproline tract that likely assumes a left-handed polyproline II helical conformation (68). A peptide substrate containing this polyproline sequence is nonetheless processed efficiently by the protease (66, 68). NMR studies suggest that the polyproline II helix in TRIF may associate with a 310 helix near the NS3 catalytic triad, anchoring the substrate and promoting its cleavage (68).
TLR3 senses HCV infection and induces ISG expression in Huh7.5-TLR3 cells that are deficient in RIG-I signaling (25). This partially restricts HCV replication when cells are infected at low multiplicity, but the protective effect is overwhelmed by a high multiplicity of infection. Unlike RIG-I, TLR3 can potentially sense viral dsRNA released by other cells into the extracellular milieu. Because NS3/4A is expressed only within infected cells, TLR3-mediated responses in uninfected hepatocytes and other cell types may contribute to IFN responses observed in some patients.
HCV Infection and Other TLRs
TLR2 senses bacterial and fungal cell wall components or viral structural proteins and induces expression of pro-inflammatory cytokines when bound by ligand (69). Expressed on the plasma membrane of monocytes and other cell types, including hepatocytes (63), it acts cooperatively with TLR1 or TLR6 to signal via a TRIF- and MAVS-independent pathway involving sequential recruitment of the adaptor proteins TIRAP (TIR domain-containing adaptor protein) and MyD88 (myeloid differentiation factor 88). Downstream signaling involves IL-1 receptor-associated kinases (IRAKs) and TRAF6, resulting in activation of NF-κB. TLR2 signaling is stimulated in primary human monocytes and macrophages by recombinant HCV core and NS3 protein, inducing synthesis of tumor necrosis factor-α and IL-10 (26). Whether this occurs in vivo is unknown, but TLR2 signaling could contribute to inflammatory changes in the HCV-infected liver. TLR2 also signals from an endosomal location in inflammatory monocytes (a discrete population of Ly6C+CD11b+CD11c− cells in bone marrow and spleen), inducing IRF3 activation and type I IFN synthesis in response to viral ligands (70). Whether such cells exist within the liver or would sense HCV infection is not known. Overexpression studies suggest that the viral NS5A protein may interact with MyD88, preventing the recruitment of IRAK and inhibiting TLR signaling (71), but the biological relevance of this is uncertain, as evidence that HCV infects monocytes or macrophages is lacking.
TLR7 is expressed by plasmacytoid dendritic cells (pDCs), which can produce 200–1000-fold more type I IFN than any other type of cell in the blood (72). TLR7 senses single-stranded RNA within an endosomal compartment, signaling via MyD88, TRAF6, and IRAK-4 to activate IRF7, which is constitutively expressed at high abundance in pDCs. BDCA3+ pDCs are abundant within some HCV-infected livers (73), and it seems likely that TLR7 signaling in such cells may be a source of IFN in patients with strong ISG responses. Consistent with this, pDCs are triggered to produce type I IFN through a TLR7-dependent pathway when co-cultured with Huh-7 cells containing replicating HCV RNA (74). Data concerning the influence of HCV infection on pDC function in vivo are conflicting (75, 76).
TLR7 is also expressed at low level in hepatocytes (77). Consistent with this, a potent TLR7 agonist induced an antiviral response in Huh-7 cells containing replicating HCV RNA (77). A G/U-rich single-stranded 20-nucleotide sequence from the polyprotein-coding region of the HCV genome, as well as a poly(U) sequence from the 3′-untranslated RNA, stimulated production of IFN-α when transfected into pDCs and also induced NF-κB activation in Huh-7 cells (78). These segments of the HCV genome appear to function as PAMPs, but it is not certain that the response observed in Huh-7 cells was mediated by TLR7. HCV infection did not induce NF-κB activation in Huh-7 cells in this study (78), but Huh-7 cells typically display high basal NF-κB activity. It is not known whether HCV specifically disrupts TLR7 signaling.
IFN-induced Intracellular Signaling
Type I IFN-α and IFN-β mediate gene transcription by signaling in a paracrine and autocrine fashion after binding the heterodimeric IFNAR on the plasma membrane (Fig. 3). Overexpression of the HCV core protein interferes with IFN signaling downstream of the IFNAR, most likely because of a direct interaction with STAT1, leading to reduced phospho-STAT1 (Fig. 3) (79, 80). However, although HCV protein expression impaired downstream IFN signaling in transgenic mice, tyrosine phosphorylation of STAT proteins by Jak was not affected (81). PP2A (protein phosphatase 2A) was up-regulated in these animals, perhaps as a result of endoplasmic reticulum stress (82, 83). This was associated with reduced methylation of STAT1, presumably due to direct inhibition of PRMT1 (protein arginine methyltransferase 1) by PP2A (82, 84). Hypomethylation of STAT1 promotes its association with PIAS1 (protein inhibitor of activated STAT1), a negative regulator of STAT1-mediated gene transcription (Fig. 3). The finding of reduced arginine methylation of STAT1 and increased STAT1-PIAS1 association in HCV-infected human liver tissues provides support for this mechanism in vivo (82, 84).
FIGURE 3.
Suppression of IFN-induced Jak-STAT signaling in hepatitis C. Type I (IFN-α/β) and III (IFN-λ) IFNs initiate signaling by binding to distinct heterodimeric receptors on the plasma membrane but then signal through a common pathway involving Tyk-2 and Jak-1 phosphorylation of STAT1 and STAT2 and subsequent recruitment of IRF9 to form the transcription factor ISGF3, which stimulates transcription of ISGs under the control of IFN-stimulated response elements. The HCV core protein may confound these responses by interacting directly with STAT1, whereas endoplasmic reticulum stress within infected cells may induce PP2A activity that acts indirectly through PRMT1 and PIAS to suppress signaling. Type I IFN signaling may also be impeded by induction of USP18 that interacts directly with the IFNAR2 subunit of the IFNAR. See text for details. ISRE, IFN-stimulated response element.
Clinical data indicate that enhanced expression of SOCS3 (suppressor of cytokine signaling 3) within the HCV-infected liver is associated with poor treatment outcome and thus may impede Jak-STAT signaling (80, 85). On the other hand, recent studies implicate USP18 (ubiquitin-specific peptidase 18; or UBP43) rather than SOCS1 or SOCS3 in long-term refractoriness to IFN in mice dosed repeatedly with IFN-α (86). This situation may mimic that in many untreated hepatitis C patients, in which there may be persistent stimulation of type I IFN expression. In addition to its ISG15-deconjugating activity, USP18 interacts with the IFNAR2 subunit of the IFNAR and may thus act to suppress Jak-STAT signaling induced by type I IFNs (87). RNA interference-mediated silencing of USP18 potentiates the antiviral activity of IFN-α against HCV in cell culture (88). Importantly, USP18 gene expression is increased in the livers of many HCV-infected patients and is associated with poor response to Peg-IFN-α therapy (15, 16). In summary, Jak-STAT signaling initiated by IFN-α/β appears likely to be suppressed in HCV infection, possibly through multiple mechanisms.
Type III IFNs bind a distinct heterodimeric membrane receptor composed of IL-10R2 and IFNLR1 subunits but utilize the same Jak-STAT signaling pathway downstream involving Jak-1 and Tyk-2 to activate gene transcription (Fig. 3) (6). IFN-λ-induced signaling is thus subject to the same potential suppression by PP2A and SOCS3 as IFN-α/β but would not be affected by the interaction of USP18 with IFNAR2 (87).
Interference with IFN Effector Mechanisms
In addition to interfering with the induction of IFN synthesis and IFN-induced intracellular signaling, HCV may specifically target IFN-induced effector mechanisms. NS5A (and possibly also the envelope protein E2) binds to and antagonizes the dsRNA-activated kinase protein kinase R (PKR) (89–91). An inducible ISG, PKR regulates cellular translation through dsRNA-stimulated autophosphorylation and subsequent phosphorylation of the translation initiation factor eIF2α. Although the viral internal ribosome entry site controlling translation of the HCV polyprotein is relatively insensitive to phospho-PKR and phospho-eIF2α (92), PKR nonetheless negatively regulates HCV replication non-cytolytically in cultured cells (93, 94). The activation of PKR by HCV impairs ISG protein expression in cell culture, as it causes a general repression of translation, and it has been suggested that this may paradoxically impede the antiviral actions of IFN (95). It is not clear, however, that such a mechanism would be operative in vivo or consistent with continued survival of infected hepatocytes.
NS5A also interacts with 2′,5′-oligoadenylate synthetase, potentially interfering with its activity (96). Data indicating an association between sequence polymorphisms in NS5A and treatment outcome support a role for NS5A in IFN antagonism in vivo (97). Transgenic mice expressing NS5A also show defective PKR, IFN-β, and 2′,5′-oligoadenylate synthetase responses to virus infection and are slow in clearing adenovirus from the liver due to impaired IFN-γ expression (98, 99). It is possible that enhanced intrahepatic expression of USP18 (15, 16) could also blunt IFN responses through its ISG15-deconjugating activity (100).
Conclusions
HCV infection evokes a number of innate immune responses, many of which are partially or completely countered by the virus. Reflecting successful adaptation to its human host, HCV has evolved mechanisms that disrupt signaling pathways involved in the induction of IFN synthesis, subvert Jak-STAT signaling to limit the expression of ISGs, or directly block the antiviral activities of some ISGs. The net effect is a multilayered evasion of innate immune responses, which is likely to exert a profound negative influence on the subsequent development of adaptive immunity to HCV, contributing to virus persistence and resistance to therapy.
Supplementary Material
Acknowledgments
I thank Kui Li and Zongdi Feng for critical review of the manuscript.
This work was supported in part by National Institutes of Health Grants U19-AI40035-14 and R21-AI081058-1 from NIAID. This is the third article in the Thematic Minireview Series on Hepatitis C Virus. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- HCV
- hepatitis C virus
- Peg-IFN-α
- pegylated interferon-α
- IL
- interleukin
- ISG
- IFN-stimulated gene
- IFNAR
- IFN-α/β receptor
- PAMP
- pathogen-associated molecular pattern
- RIG-I
- retinoic acid-inducible gene I
- TLR
- Toll-like receptor
- dsRNA
- double-stranded RNA
- CARD
- caspase recruitment domain
- IRAK
- IL-1 receptor-associated kinase
- pDC
- plasmacytoid dendritic cell
- PKR
- protein kinase R.
REFERENCES
- 1.Knipe D., Howley P., Griffin D. E., Martin M. A., Lamb R. A., Roizman B., Straus S. E. (2007) Fields Virology, 5th Ed., pp. 1253–1304, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 2.Perz J. F., Armstrong G. L., Farrington L. A., Hutin Y. J., Bell B. P. (2006) J. Hepatol. 45, 529–538 [DOI] [PubMed] [Google Scholar]
- 3.Shimakami T., Lanford R. E., Lemon S. M. (2009) Curr. Opin. Pharmacol. 9, 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehermann B. (2009) J. Clin. Invest. 119, 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pestka S. (2007) J. Biol. Chem. 282, 20047–20051 [DOI] [PubMed] [Google Scholar]
- 6.Uzé G., Monneron D. (2007) Biochimie 89, 729–734 [DOI] [PubMed] [Google Scholar]
- 7.Thomas D. L., Thio C. L., Martin M. P., Qi Y., Ge D., O'Huigin C., Kidd J., Kidd K., Khakoo S. I., Alexander G., Goedert J. J., Kirk G. D., Donfield S. M., Rosen H. R., Tobler L. H., Busch M. P., McHutchison J. G., Goldstein D. B., Carrington M. (2009) Nature 461, 798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y., Yang H., Borg B. B., Su X., Rhodes S. L., Yang K., Tong X., Tang G., Howell C. D., Rosen H. R., Thio C. L., Thomas D. L., Alter H. J., Sapp R. K., Liang T. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khakoo S. I., Thio C. L., Martin M. P., Brooks C. R., Gao X., Astemborski J., Cheng J., Goedert J. J., Vlahov D., Hilgartner M., Cox S., Little A. M., Alexander G. J., Cramp M. E., O'Brien S. J., Rosenberg W. M., Thomas D. L., Carrington M. (2004) Science 305, 872–874 [DOI] [PubMed] [Google Scholar]
- 10.Ge D., Fellay J., Thompson A. J., Simon J. S., Shianna K. V., Urban T. J., Heinzen E. L., Qiu P., Bertelsen A. H., Muir A. J., Sulkowski M., McHutchison J. G., Goldstein D. B. (2009) Nature 461, 399–401 [DOI] [PubMed] [Google Scholar]
- 11.Shin E. C., Seifert U., Urban S., Truong K. T., Feinstone S. M., Rice C. M., Kloetzel P. M., Rehermann B. (2007) J. Interferon Cytokine Res. 27, 985–990 [DOI] [PubMed] [Google Scholar]
- 12.Morrow M. P., Pankhong P., Laddy D. J., Schoenly K. A., Yan J., Cisper N., Weiner D. B. (2009) Blood 113, 5868–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabelitz D., Medzhitov R. (2007) Curr. Opin. Immunol. 19, 1–3 [DOI] [PubMed] [Google Scholar]
- 14.Bigger C. B., Brasky K. M., Lanford R. E. (2001) J. Virol. 75, 7059–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L., Borozan I., Feld J., Sun J., Tannis L. L., Coltescu C., Heathcote J., Edwards A. M., McGilvray I. D. (2005) Gastroenterology 128, 1437–1444 [DOI] [PubMed] [Google Scholar]
- 16.Sarasin-Filipowicz M., Oakeley E. J., Duong F. H., Christen V., Terracciano L., Filipowicz W., Heim M. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7034–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiscott J. (2007) J. Biol. Chem. 282, 15325–15329 [DOI] [PubMed] [Google Scholar]
- 18.Panne D., Maniatis T., Harrison S. C. (2004) EMBO J. 23, 4384–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. (2000) Immunity 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 20.Grandvaux N., Servant M. J., tenOever B., Sen G. C., Balachandran S., Barber G. N., Lin R., Hiscott J. (2002) J. Virol. 76, 5532–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C., Pflugheber J., Sumpter R., Jr., Sodora D. L., Hui D., Sen G. C., Gale M., Jr. (2003) J. Virol. 77, 3898–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onoguchi K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., Fujita T. (2007) J. Biol. Chem. 282, 7576–7581 [DOI] [PubMed] [Google Scholar]
- 23.Loo Y. M., Owen D. M., Li K., Erickson A. K., Johnson C. L., Fish P. M., Carney D. S., Wang T., Ishida H., Yoneyama M., Fujita T., Saito T., Lee W. M., Hagedorn C. H., Lau D. T., Weinman S. A., Lemon S. M., Gale M., Jr. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumpter R., Jr., Loo Y. M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S. M., Gale M., Jr. (2005) J. Virol. 79, 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang N., Liang Y., Devaraj S., Wang J., Lemon S. M., Li K. (2009) J. Virol. 83, 9824–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang S., Dolganiuc A., Szabo G. (2007) J. Leukocyte Biol. 82, 479–487 [DOI] [PubMed] [Google Scholar]
- 27.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 28.Schlee M., Roth A., Hornung V., Hagmann C. A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K. A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. (2009) Immunity 31, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myong S., Cui S., Cornish P. V., Kirchhofer A., Gack M. U., Jung J. U., Hopfner K. P., Ha T. (2009) Science 323, 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 31.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 32.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 33.Saito T., Hirai R., Loo Y. M., Owen D., Johnson C. L., Sinha S. C., Akira S., Fujita T., Gale M., Jr. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 35.Gack M. U., Shin Y. C., Joo C. H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J. U. (2007) Nature 446, 916–920 [DOI] [PubMed] [Google Scholar]
- 36.Saha S. K., Pietras E. M., He J. Q., Kang J. R., Liu S. Y., Oganesyan G., Shahangian A., Zarnegar B., Shiba T. L., Wang Y., Cheng G. (2006) EMBO J. 25, 3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. (2006) Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 38.Yoshida R., Takaesu G., Yoshida H., Okamoto F., Yoshioka T., Choi Y., Akira S., Kawai T., Yoshimura A., Kobayashi T. (2008) J. Biol. Chem. 283, 36211–36220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michallet M. C., Meylan E., Ermolaeva M. A., Vazquez J., Rebsamen M., Curran J., Poeck H., Bscheider M., Hartmann G., König M., Kalinke U., Pasparakis M., Tschopp J. (2008) Immunity 28, 651–661 [DOI] [PubMed] [Google Scholar]
- 40.Poeck H., Bscheider M., Gross O., Finger K., Roth S., Rebsamen M., Hannesschläger N., Schlee M., Rothenfusser S., Barchet W., Kato H., Akira S., Inoue S., Endres S., Peschel C., Hartmann G., Hornung V., Ruland J. (2010) Nat. Immunol. 11, 63–69 [DOI] [PubMed] [Google Scholar]
- 41.Moore C. B., Bergstralh D. T., Duncan J. A., Lei Y., Morrison T. E., Zimmermann A. G., Accavitti-Loper M. A., Madden V. J., Sun L., Ye Z., Lich J. D., Heise M. T., Chen Z., Ting J. P. (2008) Nature 451, 573–577 [DOI] [PubMed] [Google Scholar]
- 42.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 43.Saito T., Owen D. M., Jiang F., Marcotrigiano J., Gale M., Jr. (2008) Nature 454, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li K., Chen Z., Kato N., Gale M., Jr., Lemon S. M. (2005) J. Biol. Chem. 280, 16739–16747 [DOI] [PubMed] [Google Scholar]
- 45.Foy E., Li K., Wang C., Sumpter R., Jr., Ikeda M., Lemon S. M., Gale M., Jr. (2003) Science 300, 1145–1148 [DOI] [PubMed] [Google Scholar]
- 46.Li X. D., Sun L., Seth R. B., Pineda G., Chen Z. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baril M., Racine M. E., Penin F., Lamarre D. (2009) J. Virol. 83, 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Y., Ishida H., Lenz O., Lin T. I., Nyanguile O., Simmen K., Pyles R. B., Bourne N., Yi M., Li K., Lemon S. M. (2008) Gastroenterology 135, 1710–1718.e2 [DOI] [PubMed] [Google Scholar]
- 49.Cheng G., Zhong J., Chisari F. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8499–8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otsuka M., Kato N., Moriyama M., Taniguchi H., Wang Y., Dharel N., Kawabe T., Omata M. (2005) Hepatology 41, 1004–1012 [DOI] [PubMed] [Google Scholar]
- 51.Tasaka M., Sakamoto N., Itakura Y., Nakagawa M., Itsui Y., Sekine-Osajima Y., Nishimura-Sakurai Y., Chen C. H., Yoneyama M., Fujita T., Wakita T., Maekawa S., Enomoto N., Watanabe M. (2007) J. Gen. Virol. 88, 3323–3333 [DOI] [PubMed] [Google Scholar]
- 52.Liang Y., Shilagard T., Xiao S. Y., Snyder N., Lau D., Cicalese L., Weiss H., Vargas G., Lemon S. M. (2009) Gastroenterology 137, 1448–1458 [DOI] [PubMed] [Google Scholar]
- 53.Chen Z., Benureau Y., Rijnbrand R., Yi J., Wang T., Warter L., Lanford R. E., Weinman S. A., Lemon S. M., Martin A., Li K. (2007) J. Virol. 81, 964–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y., Liang Y., Qu L., Chen Z., Yi M., Li K., Lemon S. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7253–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 56.Funami K., Sasai M., Ohba Y., Oshiumi H., Seya T., Matsumoto M. (2007) J. Immunol. 179, 6867–6872 [DOI] [PubMed] [Google Scholar]
- 57.Liu L., Botos I., Wang Y., Leonard J. N., Shiloach J., Segal D. M., Davies D. R. (2008) Science 320, 379–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato S., Sugiyama M., Yamamoto M., Watanabe Y., Kawai T., Takeda K., Akira S. (2003) J. Immunol. 171, 4304–4310 [DOI] [PubMed] [Google Scholar]
- 59.Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. (2003) Nat. Immunol. 4, 161–167 [DOI] [PubMed] [Google Scholar]
- 60.Matsumoto M., Funami K., Tanabe M., Oshiumi H., Shingai M., Seto Y., Yamamoto A., Seya T. (2003) J. Immunol. 171, 3154–3162 [DOI] [PubMed] [Google Scholar]
- 61.Sarkar S. N., Peters K. L., Elco C. P., Sakamoto S., Pal S., Sen G. C. (2004) Nat. Struct. Mol. Biol. 11, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 62.Meylan E., Burns K., Hofmann K., Blancheteau V., Martinon F., Kelliher M., Tschopp J. (2004) Nat. Immunol. 5, 503–507 [DOI] [PubMed] [Google Scholar]
- 63.Seki E., Brenner D. A. (2008) Hepatology 48, 322–335 [DOI] [PubMed] [Google Scholar]
- 64.Nakamura M., Funami K., Komori A., Yokoyama T., Aiba Y., Araki A., Takii Y., Ito M., Matsuyama M., Koyabu M., Migita K., Taniguchi K., Fujioka H., Yatsuhashi H., Matsumoto M., Ishibashi H., Seya T. (2008) Hepatol. Int. 2, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jouan L., Melancon P., Rodrigue-Gervais I., Raymond V., Selliah S., Boucher G., Bilodeau M., Grandvaux N., Lamarre D. (2010) J. Hepatol. 52, 167–175 [DOI] [PubMed] [Google Scholar]
- 66.Li K., Foy E., Ferreon J. C., Nakamura M., Ferreon A. C., Ikeda M., Ray S. C., Gale M., Jr., Lemon S. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dansako H., Ikeda M., Ariumi Y., Wakita T., Kato N. (2009) Arch. Virol. 154, 801–810 [DOI] [PubMed] [Google Scholar]
- 68.Ferreon J. C., Ferreon A. C., Li K., Lemon S. M. (2005) J. Biol. Chem. 280, 20483–20492 [DOI] [PubMed] [Google Scholar]
- 69.Takeuchi O., Akira S. (2009) Immunol. Rev. 227, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbalat R., Lau L., Locksley R. M., Barton G. M. (2009) Nat. Immunol. 10, 1200–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abe T., Kaname Y., Hamamoto I., Tsuda Y., Wen X., Taguwa S., Moriishi K., Takeuchi O., Kawai T., Kanto T., Hayashi N., Akira S., Matsuura Y. (2007) J. Virol. 81, 8953–8966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilliet M., Cao W., Liu Y. J. (2008) Nat. Rev. Immunol. 8, 594–606 [DOI] [PubMed] [Google Scholar]
- 73.Lau D. T., Fish P. M., Sinha M., Owen D. M., Lemon S. M., Gale M., Jr. (2008) Hepatology 47, 799–809 [DOI] [PubMed] [Google Scholar]
- 74.Takahashi K., Asabe S., Wieland S., Garaigorta U., Gastaminza P., Isogawa M., Chisari F. V. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7431–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Decalf J., Fernandes S., Longman R., Ahloulay M., Audat F., Lefrerre F., Rice C. M., Pol S., Albert M. L. (2007) J. Exp. Med. 204, 2423–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanto T., Hayashi N. (2007) Hepatol. Res. 37, Suppl. 3, S319–S326 [DOI] [PubMed] [Google Scholar]
- 77.Lee J., Wu C. C., Lee K. J., Chuang T. H., Katakura K., Liu Y. T., Chan M., Tawatao R., Chung M., Shen C., Cottam H. B., Lai M. M., Raz E., Carson D. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1828–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y. L., Guo Y. J., Bin L., Sun S. H. (2009) J. Hepatol. 51, 29–38 [DOI] [PubMed] [Google Scholar]
- 79.Lin W., Kim S. S., Yeung E., Kamegaya Y., Blackard J. T., Kim K. A., Holtzman M. J., Chung R. T. (2006) J. Virol. 80, 9226–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bode J. G., Ludwig S., Ehrhardt C., Albrecht U., Erhardt A., Schaper F., Heinrich P. C., Häussinger D. (2003) FASEB J. 17, 488–490 [DOI] [PubMed] [Google Scholar]
- 81.Blindenbacher A., Duong F. H., Hunziker L., Stutvoet S. T., Wang X., Terracciano L., Moradpour D., Blum H. E., Alonzi T., Tripodi M., La Monica N., Heim M. H. (2003) Gastroenterology 124, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 82.Duong F. H., Filipowicz M., Tripodi M., La Monica N., Heim M. H. (2004) Gastroenterology 126, 263–277 [DOI] [PubMed] [Google Scholar]
- 83.Christen V., Treves S., Duong F. H., Heim M. H. (2007) Hepatology 46, 558–565 [DOI] [PubMed] [Google Scholar]
- 84.Duong F. H., Christen V., Filipowicz M., Heim M. H. (2006) Hepatology 43, 796–806 [DOI] [PubMed] [Google Scholar]
- 85.Kim K. A., Lin W., Tai A. W., Shao R. X., Weinberg E., De Sa Borges C. B., Bhan A. K., Zheng H., Kamegaya Y., Chung R. T. (2009) J. Hepatol. 50, 705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sarasin-Filipowicz M., Wang X., Yan M., Duong F. H., Poli V., Hilton D. J., Zhang D. E., Heim M. H. (2009) Mol. Cell. Biol. 29, 4841–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Malakhova O. A., Kim K. I., Luo J. K., Zou W., Kumar K. G., Fuchs S. Y., Shuai K., Zhang D. E. (2006) EMBO J. 25, 2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Randall G., Chen L., Panis M., Fischer A. K., Lindenbach B. D., Sun J., Heathcote J., Rice C. M., Edwards A. M., McGilvray I. D. (2006) Gastroenterology 131, 1584–1591 [DOI] [PubMed] [Google Scholar]
- 89.Gale M. J., Jr., Korth M. J., Tang N. M., Tan S. L., Hopkins D. A., Dever T. E., Polyak S. J., Gretch D. R., Katze M. G. (1997) Virology 230, 217–227 [DOI] [PubMed] [Google Scholar]
- 90.Taylor D. R., Tian B., Romano P. R., Hinnebusch A. G., Lai M. M., Mathews M. B. (2001) J. Virol. 75, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pflugheber J., Fredericksen B., Sumpter R., Jr., Wang C., Ware F., Sodora D. L., Gale M., Jr. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4650–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shimoike T., McKenna S. A., Lindhout D. A., Puglisi J. D. (2009) Antiviral Res. 83, 228–237 [DOI] [PubMed] [Google Scholar]
- 93.Jiang D., Guo H., Xu C., Chang J., Gu B., Wang L., Block T. M., Guo J. T. (2008) J. Virol. 82, 1665–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang J. H., Kato N., Muroyama R., Taniguchi H., Guleng B., Dharel N., Shao R. X., Tateishi K., Jazag A., Kawabe T., Omata M. (2010) Liver Int. 30, 311–318 [DOI] [PubMed] [Google Scholar]
- 95.Garaigorta U., Chisari F. V. (2009) Cell Host Microbe 6, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taguchi T., Nagano-Fujii M., Akutsu M., Kadoya H., Ohgimoto S., Ishido S., Hotta H. (2004) J. Gen. Virol. 85, 959–969 [DOI] [PubMed] [Google Scholar]
- 97.Enomoto N., Sakuma I., Asahina Y., Kurosaki M., Murakami T., Yamamoto C., Ogura Y., Izumi N., Marumo F., Sato C. (1996) N. Engl. J. Med. 334, 77–81 [DOI] [PubMed] [Google Scholar]
- 98.Kriegs M., Bürckstümmer T., Himmelsbach K., Bruns M., Frelin L., Ahlén G., Sällberg M., Hildt E. (2009) J. Biol. Chem. 284, 28343–28351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kanda T., Steele R., Ray R., Ray R. B. (2009) J. Virol. 83, 8463–8469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ritchie K. J., Hahn C. S., Kim K. I., Yan M., Rosario D., Li L., de la Torre J. C., Zhang D. E. (2004) Nat. Med. 10, 1374–1378 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.