Abstract
The fungal iterative nonreducing polyketide synthases (NRPKSs) synthesize aromatic polyketides, many of which have important biological activities. The product template domains (PT) embedded in the multidomain NRPKSs mediate the regioselective cyclization of the highly reactive polyketide backbones and dictate the final structures of the products. Understanding the sequence-activity relationships of different PT domains is therefore an important step toward the prediction of polyketide structures from NRPKS sequences and can enable the genome mining of hundreds of cryptic NRPKSs uncovered via genome sequencing. In this work, we first performed phylogenetic analysis of PT domains from NRPKSs of known functions and showed that the PT domains can be classified into five groups, with each group corresponding to a unique product size or cyclization regioselectivity. Group V contains the formerly unverified PT domains that were identified as C6-C11 aldol cyclases. The regioselectivity of PTs from this group were verified by product-based assays using the PT domain excised from the asperthecin AptA NRPKS. When combined with dissociated PKS4 minimal PKS, or replaced the endogenous PKS4 C2-C7 PT domain in a hybrid NRPKS, AptA-PT directed the C6-C11 cyclization of the nonaketide backbone to yield a tetracyclic pyranoanthraquinone 4. Extensive NMR analysis verified that the backbone of 4 was indeed cyclized with the expected regioselectivity. The PT phylogenetic analysis was then expanded to include ∼100 PT sequences from unverified NRPKSs. Using the assays developed for AptA-PT, the regioselectivities of additional PT domains were investigated and matched to those predicted by the phylogenetic classifications.
Keywords: Antibiotics, Aspergillus, Enzyme Catalysis, Fungi, NMR, Cyclase, Genome Mining, Megasynthase
Introduction
Natural products such as polyketides and nonribosomal peptides isolated from filamentous fungi have played indispensable roles in human health care (1). During the past decade, advances in rapid DNA sequencing techniques have enabled the complete genome sequencing of many fungi species (2–6). One important outcome of the sequencing efforts is the revelation of large numbers of natural product biosynthetic pathways encoded in these organisms. Among them, putative polyketide synthases (PKSs)2 and nonribosomal peptide synthetases (NRPSs) are particularly abundant. Many of the encoded pathways are silent under laboratory culturing conditions, and as a result, the metabolites associated with these pathways have not been observed or isolated (7). Mining these uncharacterized pathways is therefore an important objective toward discovery of new bioactive molecules and novel enzymatic machineries (8).
One method of predicting the structures of polyketides encoded in cryptic pathways is by analyzing the sequence of the PKSs. This method has been successfully used in the prediction of bacterial polyketides by examining the linear arrangement of catalytic sites along the modular type I PKSs (9, 10). However, this method is not readily applicable to fungal polyketides because of the differences in enzyme architectures and more importantly, the lack of the colinearity rule. Fungal PKSs are large megasynthases that assemble single copies of catalytic domains in one polypeptide. The domains function in an iterative fashion and use built-in programming rules to dictate chain length, cyclization, reductive tailoring, and other structural features (11). Therefore, understanding the relationships between protein sequence and catalytic activity, as well as substrate specificity, of each domain is essential to unlocking the biosynthetic potential of fungal PKSs.
Among the different families of fungal iterative PKSs, the nonreducing PKSs (NRPKSs) are responsible for the synthesis of a number of important metabolites, including the well-known mycotoxin aflatoxin (12). Other representative products of NRPKSs include pigment precursors (1,3,6,8-tetrahydroxylnaphthalene, THN), compounds with antibiotic (viridicatumtoxin) and anticancer (bikaverin) properties. Polyketides produced from NRPKSs are aromatic compounds with sizes ranging from monocyclic aromatics to those containing multiple fused rings. Starting from the N terminus of the megasynthase, the following domains are typically found in an NRPKS: a starter unit:ACP transacylase (SAT) that selects the starter unit (13); a ketosynthase (KS) that catalyzes repeated decarboxylative condensation to elongate the polyketide backbone; a malonyl-CoA:ACP transacylase (MAT) that selects and transfers the extender unit malonyl-CoA; a product template (PT) domain that controls the immediate cyclization regioselectivity of the reactive polyketide backbone (14); an acyl-carrier protein (ACP) that serves as the tether of the growing and completed polyketide via its phosphopantetheinyl arm (15); and a thioesterase/Claisen-like cyclase (TE/CLC) domain that cyclizes and releases the product (16). Other domains such as methyltransferase (MT) and reductase (R) can be found in subclades of the NRPKSs (6). Establishment of domain boundaries using computation tools such as the Udwary-Merski algorithm (UMA) (17), and dissection of domains (12, 18–20), have been used to verify the function of individual domains. Expression of intact megasynthases in heterologous hosts such as Aspergillus oryzae (21, 22) and Escherichia coli (23) has been used to reconstitute the activities of the entire megasynthase through structural determination of the polyketide products.
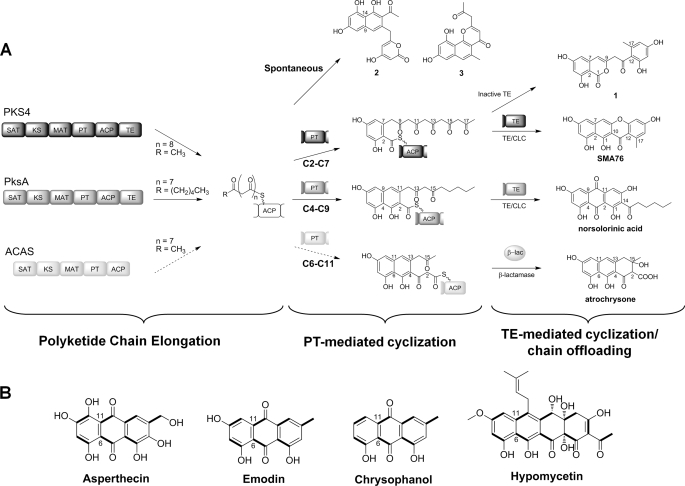
Because the poly-β-ketone backbone of a newly synthesized polyketide is extremely reactive, the NRPKSs must suppress spontaneous cyclization and control the regioselectivity of the first cyclization step to afford the F-mode folding of the fungal aromatic polyketides (24). In this mode, the first aromatic ring contains two intact acetate units, whereas the two bridging carbons are derived from two different acetate units. As shown in Fig. 1, the three most commonly observed F-mode folding patterns are C2-C7, C4-C9, and C6-C11. Recently, the PT domain has been shown to be responsible for this important first step in tailoring the completed polyketide backbone. Using dissected domains, Townsend and co-workers demonstrated that the PT from PksA (from Aspergillus parasiticus) can regioselectively cyclize a hexanoyl-primed octaketide via C4-C9 and C2-C11 aldol condensations en route to norsolorinic acid, the key intermediate of aflatoxin biosynthesis (12) (Fig. 1A). The structural basis of PksA-PT has also been elucidated to confirm its catalytic role as an aldol cyclase (14). Ma et al. (23) have reconstituted the intact bikaverin synthase PKS4 (from Gibberella fujikuroi) in E. coli to synthesize the C2-C7 cyclized nonaketide SMA76a. Systematic removal of individual domains from the intact PKS4 was then conducted to verify the functional roles of the TE and PT domains (18, 19). Removal of the TE/CLC domain (named PKS4–99) produced the C2-C7 cyclized SMA93 1 and confirmed the role of the TE/CLC in catalyzing the C1-C10 Claisen reaction (18) (Fig. 1A). Further excision of the PT domain from PKS4–99 abolished the production of 1 and led to the biosynthesis of aberrantly cyclized products 2 and 3, confirming the role of the PT domain in catalyzing the C2-C7 aldol condensation (19).
FIGURE 1.
A, biosynthesis of fungal aromatic polyketides. C2-C7, C4-C9/C2-C11, and C6-C11/C4-C13 are commonly observed F-mode folding patterns catalyzed by fungal NRPKSs. Product template domains that mediate C2-C7 and C4-C9/C2-C11 folding modes have been characterized in the study of PKS4 and PksA, respectively. TE/CLC domains catalyze the cyclization of the last ring and off-loading. In the absence of PT-mediated cyclization, the PKS4 minimal PKS produces spontaneously cyclized products 2 and 3. B, C6-C11/C4-C13 cyclization regioselectivity has been observed in various fungal aromatic polyketide such as asperthecin, emodin, chrysophanol, and hypomycetin.
The commonly observed C6-C11 F-mode cyclization is exemplified by the model product emodin and related compounds (25–28) (Fig. 1B). NRPKSs that synthesize C6-C11 cyclized aromatic polyketides include AptA and AN0150 involved in the synthesis of asperthecin and emodin in Aspergillus nidulans, respectively (25, 26), as well as ACAS that synthesizes atrochrysone in Aspergillus terreus (Fig. 1A) (29). A common feature observed among these PKSs is the lack of a fused TE/CLC domain. For ACAS, a discrete β-lactamase-type TE (ACTE) is employed to release the product from ACAS (29). Unlike the PT domains from PksA and PKS4, the activities of the PT domains from C6-C11 regiospecific NRPKSs have not been verified.
Because of the important roles played by PT domains in controlling the polyketide structures, being able to precisely predict the cyclization regioselectivity of PT domains is an important step toward suggesting the potential products of NRPKS. Toward this end, we sought to 1) establish a sequence-activity relationship through bioinformatics analysis; 2) establish a simple experimental platform to assay the activities of PT domains; and 3) use the experimental platform to confirm the predicted regioselectivity of PT domains from cryptic NRPKSs.
EXPERIMENTAL PROCEDURES
Materials
[1,2,3-13C]Malonate was purchased from Cambridge Isotope Laboratories. CoA was purchased from Sigma Aldrich. A. nidulans FGSC A4 and A. niger ATCC 1015 were obtained from NRRL. The E. coli strain XL-1 Blue utilized for DNA manipulation and BL21(DE3) used for protein expression were purchased from Stratagene. The protein expression strain BAP1 described by Pfeifer et al. (30) was from Prof. Chaitan Khosla.
General Technique for DNA Manipulation
The A. nidulans and A. niger genomic DNA were prepared using the ZYMO (Orange, CA) ZR fungal/bacterial DNA kit according to supplied protocols. PCR reactions were performed with the Phusion high-fidelity DNA polymerase (NEB) and Platinum Pfx DNA polymerase (Invitrogen). The PCR products are cloned into the pCR-blunt vector obtained from Invitrogen. Restriction enzymes (NEB) and T4 ligase (Invitrogen) were used to digest and ligate the DNA fragments. The primers used to amplify the genes were synthesized by IDT and Operon.
Phylogenetic Analysis of PT Domains
The 128 PT sequences were obtained from National Center for Biotechnology Information (NCBI) through blasting the PT domains and KS domains of PKS4, ACAS and PksA. The sequence for VrtA from Penicillium aethiopicum was provided by Y. H. Chooi. All of these 129 NRPKSs belong to subclades I and II as defined in Ref. (6). PT domains of each NRPKS were carefully extracted according to the boundary of PksA-PT (12). The PT sequences were aligned with CLUSTALX 2.0 (31), and phylogenetic analyses were conducted using MEGA version 4 (32) by means of the boot strap minimum evolution method. The evolutionary history was inferred using the Minimum Evolution method (33). The evolutionary distances were computed using the Poisson correction method (34) and are in the units of the number of amino acid substitutions per site.
Cloning of the Stand-alone PT Domains
Primers used to amplify AptA-PT, which contains one intron, are listed in supplemental Table S1. The first exon was amplified by PCR from A. nidulans genomic DNA with the forward primer AptA-PT-f and reverse primer AptA-PT-i-r; and the second exon was amplified using the primer AptA-PT-i-f and the reverse primer AptA-PT-r. Splice by overlapping extension PCR (SOE-PCR) was used to fuse the two pieces to synthesize a DNA fragment encoding AptA-PT flanked with NdeI and NotI. The DNA fragment of AptA-PT was inserted into pET24 vector digested with NdeI and NotI to yield pYR94. The PT domains of AN0150 and VrtA were cloned using similar methods, and the primers are listed in supplemental Table S1. Following amplification, the gene encoding AN0150-PT was inserted into pET24 vector digested with NdeI and NotI to yield pYR160, and the gene encoding VrtA-PT was inserted into pET28 vector digested with NdeI and EcoRI to yield pYR149.
Construction of PKSH1
Primers used to amplify AptA-PT and PKS4-ACP are listed in supplemental Table S1. AptA-PT and PKS4-ACP were amplified and fused together by SOE-PCR to synthesize a DNA fragment encoding AptA-PT-PKS4-ACP flanked with NotI. pWJ279 is derived from pET28 and encodes the N-terminal hexahistidine tagged PKS4-SAT-KS-MAT between NdeI and NotI. The DNA fragment of AptA-PT-PKS4-ACP flanked by NotI was inserted into the NotI-digested pWJ279 to yield pYR123.
Expression and Purification of Proteins
Expression of stand-alone PT domains and full megasynthases follow identical procedures. The example of AptA-PT is detailed here. pYR94 was used to transform the BL21(DE3) for protein expression. The cells were cultured at 37 °C and 250 rpm in 500 ml LB medium supplemented with 35 μg/ml kanamycin to a final OD600 between 0.4 and 0.6. The culture was then incubated on ice for 10 min before addition of 0.1 m isopropylthio-β-d-galactoside (IPTG) for protein expression. The cells were further cultured at 18 °C for 12–16 h. The cells were then harvested by centrifugation (3,500 rpm, 15 min, 4 °C), resuspended in ∼25 ml lysis buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl, 10 mm imidazole) and lysed by sonication on ice. Cellular debris was removed by centrifugation (15,000 rpm, 30 min, 4 °C), and Ni-NTA agarose resin was then added to the supernatant (1–2 ml/liter of culture). The suspension was swirled at 4 °C for 2 h and loaded into a gravity column. The protein-bound resin was washed with one column volume of 10 mm imidazole in buffer A (50 mm Tris-HCl, pH 7.9, 2 mm EDTA, 2 mm DTT), followed by 0.5 column volume of 20 mm imidazole in buffer A. The protein was then eluted with 250 mm imidazole in buffer A. Purified AptA-PT was concentrated and exchanged into buffer A+10% glycerol with the centriprep filter devices (Amicon), aliquoted and flash frozen. Protein concentration was determined with the Bradford assay using bovine serum albumin as a standard.
In Vitro Reconstitution of PKSH1
The reactions were performed at 100 μl scale in 100 mm phosphate buffer (pH 7.4) in the presence of 100 mm sodium malonate, 2 mm DTT, 7 mm MgCl2, 20 mm ATP, 5 mm coenzyme A, 20 μm MatB, and 10 μm PKSH1. The reactions were terminated with 1 ml of 99% ethyl acetate (EA)/1% acetic acid (AcOH). The organic phase was separated, evaporated to dryness, redissolved in 20 μl of DMSO and analyzed with a Shimadzu 2010 EV Liquid Chromatography Mass Spectrometer by using both positive and negative electrospray ionization and a phenomenex Luna 5 μm, 2.0 × 100 mm C18 reverse-phase column. Compounds were separated on a linear gradient of 5% acetonitrile (CH3CN, v/v) in water (0.1% formic acid) to 95% CH3CN in water (0.1% formic acid) over 30 min with a flow rate of 0.1 ml/min.
In Vitro Characterization of the Stand-alone PT Domains
The reactions were performed at 100 μl scale in 100 mm phosphate buffer (pH 7.4) in the presence of 100 mm sodium malonate, 2 mm DTT, 7 mm MgCl2, 20 mm ATP, 5 mm coenzyme A, 20 μm MatB, 10 μm PKS4 KS-MAT didomain, 50 μm PKS4-ACP, and 50 μm PT domain. The reactions were terminated, extracted, and analyzed with the same procedure as in the in vitro reconstitution of PKSH1.
Large Scale Synthesis of 4 and NMR Characterization
To obtain a sufficient quantity of 4, the 100 μl in vitro assay was scaled up to 100 ml. The reaction mixture was shaken gently at room temperature and the reaction progress was monitored by HPLC. After ∼48 h when the product level had reached a plateau, the reaction mixture was extracted three times with equal volume of EA (1% AcOH). The resultant organic extracts were combined and evaporated to dryness, washed with methanol, redissolved in DMSO, and purified by reverse-phase HPLC (Alltech Apollo 5 μm, 250 mm × 4.6 mm) on a linear gradient of 45 to 95% CH3CN (v/v) over 15 min and 95% CH3CN (v/v) for 15 min in water (0.1% trifluoroacetic acid) at a flow rate of 1 ml/min. The heteronuclear multiple quantum coherence (HMQC) and heteronuclear multiple bond correlation (HMBC) spectra of 4 were performed on the Bruker DRX-600 spectrometer using DMSO-d6 as the solvent, the 1H spectrum was performed on a Bruker DRX-500 spectrometer, and 13C spectrum was performed on a Bruker ARX-500 spectrometer.
Construction, Purification, and Reconstitution of the PKSH2
Primers used to amplify An03g05440-PT and PKS4-ACP are listed in supplemental Table S1. An03g05440-PT was first fused to PKS4-ACP through SOE-PCR to yield the DNA fragments encoding PT-ACP flanked with NotI. With the same procedure as in constructing PKSH1, the DNA fragment of An03g05440-PT-PKS4-ACP flanked by NotI was inserted into the NotI-digested pWJ279 to yield pYR243. pYR243 was expressed and purified with the same procedure as PKSH1. Small scale in vitro assays of 100 μl were performed at room temperature and analyzed by LC-MS.
RESULTS
Phylogenetic Analysis of PTs
To establish a sequence-activity relationship, we performed a phylogenetic analysis of PT domains from NRPKSs that have been associated with known fungal aromatic polyketides (supplemental Table S2). A previous phylogenetic analysis of the KS sequences classified the NRPKSs into subclades I and II that consist of PKS4 and PksA; and a subclade III of which the PKSs all contain a MT domain embedded between ACP and TE/CLC domains (6). Because of this architectural difference and phylogenetic distances to subclades I and II, subclade III NRPKSs were not included in this study. The subclades I and II PT sequences were aligned and phylogenetically analyzed by means of the boot strap minimum evolution method (32). The resulting phylogenetic tree shown in Fig. 2 clearly divides these NRPKSs into five major groups.
FIGURE 2.
Phylogenetic analysis of PT domains from 30 NRPKSs that have been related to known aromatic polyketides. The NRPKSs have been classified into five major groups: group I, C2-C7 monocyclic PKSs; group II, THN synthases (C2-C7 bicyclic PKSs); group III, C2-C7 multicyclic PKSs; group IV, C4-C9 PKSs; and group V, C6-C11 PKSs. The evolutionary distances are in the units of the number of amino acid substitutions per site. The sum of branch length is 10.38. The tree is drawn to scale (see scale bar), with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Group I includes the NRPKSs that synthesize compounds containing a single aromatic ring, exemplified by the recently discovered orsellinic acid synthase AN7909 from A. nidulans (26, 35). Group I also consists of the NRPKSs that are involved in the synthesis of the aromatic portions of the resorcylic acid lactones, including the zearalenone PKS13 (36, 37), the hypothemycin Hpm3 (38, 39), and the radicicol RDC1/RADS2 (38, 40). Primed by different starter units, Group I NRPKSs synthesize a tetraketide backbone and the PT domains perform the regioselective C2-C7 cyclization to yield the aromatic ring. Group II contains most of the known THN synthases (THNSs). The PT domain of this group catalyzes the cyclization of pentaketide backbones via C2-C7 aldol condensation, followed by TE/CLC catalyzed cyclization of the second ring and product release (41).
The remaining three groups contain NRPKSs that synthesize longer polyketide chains and multiply fused-ring structures. Group III contains NRPKSs that cyclize first ring via C2-C7 regioselectivity, such as the nonaketide synthase PKS4 (23, 42) and the aurofusarin heptaketide synthase PKS12 from G. zeae (43). An interesting member of the phylogenetic tree is the WA synthase that synthesizes THN from A. nidulans. This NRPKS is a member of Group III instead of Group II. This classification is consistent with previous biochemical analysis, which showed that synthesis of THN by WA is a result of chain shortening of the heptaketide product YWA1 (44). The well-studied PksA and other NRPKSs that are involved in the synthesis of C4-C9/C2-C11 cyclized aromatic polyketides are clearly separated into Group IV. This group contains other known aflatoxin synthases that are analogues of PksA (45, 46), as well as the heptaketide synthase cercosporin synthase CTB1 (47). Within this group, NRPKSs primed with different starter units are clearly divergent in sequence, as illustrated by the phylogenetic distance between the acetate-primed CTB1 and the hexanoate-primed aflatoxin synthases. Lastly, Group V consists of AptA, AN0150, and ACAS, which putatively cyclize the nascent polyketide via C6-C11/C4-C13 regioselectivity. Because all members of this group lack the C terminus TE/CLC domain, the Group V PT domains appear to be responsible for all the enzyme-catalyzed polyketide cyclization reactions.
Validating the Group V PT Domain Activities
Our analysis with a limited number of PT domains indicated that cyclization specificities can be readily correlated and classified based on protein sequence. To validate this phylogenetic classification of PT functions, we attempted to confirm the predicted but unverified C6-C11/C4-C13 cyclization regioselectivity of Group V PT domains. To do so, we hypothesized that PT domains can be extracted from the native megasynthase and functionally combined with a well-characterized heterologous minimal PKS. By analyzing the structure of the product synthesized by the hybrid components, the regiospecificity of the unknown PT domain can be deciphered. This approach may be especially useful for NRPKSs of which the intact megasynthase or the endogenous minimal PKS domains cannot be solubly expressed or functionally reconstituted.
The G. fujikuroi PKS4 is well-suited to serve as the template for PT domain swapping because (i) the megasynthases can be expressed at high levels and the minimal PKS (KS-MAT, ACP) can produce ample amounts of the nonaketide products in vitro, which facilitates product structure elucidation; (ii) the minimal PKS domain does not significantly influence the cyclization regioselectivity of the nascent polyketide (19). We therefore anticipated that a heterologous PT could dictate the cyclization outcome; and (iii) other built-in cyclization domain, such as the TE/CLC domain can be removed from the assembly line without affecting the product elongation and turnover. Hence, we can attribute all cyclization steps to that of the transplanted PT domain.
We chose to study the PT domain from the asperthecin synthase AptA (AptA-PT) (25) of Group V. The gene encoding AptA-PT was amplified from aptA in accordance with the boundaries of the crystallized PksA PT domain (12) (supplemental Fig. S1). AptA-PT was then expressed from E. coli strain BL21(DE3) and purified to a final yield of 13 mg/liter. To assay the regioselectivity, AptA-PT was combined with the PKS4 minimal PKS. We used the malonyl-CoA synthase MatB (48) from Rhizobium trifolii to generate the malonyl-CoA extender units from malonate, ATP and CoA. The reaction was allowed to proceed for 12 h, after which the organic products were extracted, dried, and analyzed by LC-MS (Fig. 3B). As expected, the minimal PKS alone produced polyketides that are not cyclized in accordance with the F-modes, including 2 and 3 (supplemental Fig. S2). In contrast, in the presence of AptA-PT, the spontaneous cyclization modes were suppressed and a predominant product that corresponds to a new compound 4 (retention time, RT = 25.5 min) was produced (Fig. 3B). Compound 4 has a parent ion peak [M-H]− at m/z 337, which is consistent with a nonaketide backbone that has undergone three dehydrations and one oxidation. The nonaketide backbone of 4 was further confirmed when an increase of 9 mu was observed for 4 upon using [2-13C]malonate as the substrate of MatB (supplemental Fig. S3). The UV absorbance of 4 displays λmax at 247, 315, 474 nm, which suggests that this compound contains a highly conjugated chromophore (supplemental Fig. S3).
FIGURE 3.
A, construction and SDS-PAGE analysis of PKSH1. PKSH1 (196 kDa) is constructed through domain shuffling of PT domain between PKS4 and AptA using PKS4–99 as a template. For SDS-PAGE: lane M, bench mark ladder; lane E, Ni-NTA-purified PKSH1. B, LC-MS (325 nm) analysis of the polyketides synthesized by PKSH1 in vitro. Trace i, PKS4 minimal PKS alone synthesizes nonaketides 2 and 3, which are derived from spontaneous cyclization of the backbone; trace ii, PKS4 minimal PKS domains and AptA-PT interact efficiently to afford a new nonaketide 4 as the predominant product; trace iii, parent megasynthase PKS4–99 synthesizes 1 as a major product; and trace iv, PKSH1 synthesizes 4 as the predominant product.
We next tested whether the AptA-PT domain can functionally replace the PKS4 PT domain in PKS4 to create a hybrid megasynthase. Using the TE-less PKS4 (PKS4–99) as the template and SOE PCR, we constructed PKSH1 in which the original PT domain was replaced with the AptA-PT (Fig. 3A). PKSH1 was expressed in E. coli strain BAP1, which contains a chromosomally encoded copy of the broadly specific phosphopantetheinyl transferase, Sfp (30). This hybrid megasynthase was soluble-expressed in BAP1, and was purified with nickel affinity chromatography to near homogeneity (∼2 mg/liter) (Fig. 3A). Interestingly, when PKSH1 was assayed for product formation in vitro, a similar product profile was observed compared with the reaction mixture containing the dissociated enzymatic components. Whereas PKS4–99 produced the expected C2-C7 cyclized 1 as the main product, PKSH1 synthesized 4 as the predominant product (Fig. 3B). This result indicates that while the hybrid megasynthases remain functional to produce polyketides, the regioselectivity of the cyclization steps is completely altered upon replacement of the PT domain. The robust turnover of products by PKSH1 further confirms the dexterity of PKS4 toward domain engineering experiments.
Elucidation of the Structure of 4
To determine the structure and cyclization pattern of 4, we scaled up the in vitro assay with PKSH1 with [12C]malonate that is enriched with 10% of 1,2,3-13C-malonate. The 13C-enriched 4 was purified from the crude extract through reverse-phase HPLC and thoroughly characterized by NMR spectroscopy. Seven signals were observed in the 1H NMR spectrum including one methyl (δH = 2.29 ppm), four aromatic (δH = 6.65, 6.81, 7.16, 7.69 ppm), and two phenolic protons (δH = 12.11, 13.74 ppm). A spin system of CH (δH = 6.65 ppm) -CH (δH = 7.16 ppm) was observed with a coupling constant of JHH = 2.4 Hz. The 13C NMR spectrum (Table 1, supplemental Fig. S4) showed 18 signals, each appearing as doublets due to isotopic enrichment. The signals include those that are associated with a quinone (δC = 181.4 and δC = 188.4 ppm). An additional carbonyl peak (δC = 158.1 ppm) was observed and is indicative of a pyrone moiety. 1H-13C HMQC (supplemental Fig. S4) correlations enabled the assignment of the four hydrogen atoms to the directly bonded carbon atoms (8-C/H, 10-C/H, 14-C/H, and 16-C/H). The coupling constants calculated from the 13C spectrum allowed establishment of nine sets of neighboring carbons. Together with the remaining one-dimensional and two-dimensional NMR data, including 1H-13C HMBC (supplemental Fig. S4) correlations, the structure of 4 was elucidated as an α-pyranoanthraquinone as shown in Fig. 4.
TABLE 1.
NMR spectral data for 4 in DMSO-d6
The 1H and 13C NMR spectrum were recorded at 500 MHz, the 1H-13C HMBC and HMQC spectra were collected at 600 MHz.
| No. | 13C δ | JCC | 1H δ | 1H-13C HMBC |
|---|---|---|---|---|
| ppm | Hz | ppm (m, area, JHH (Hz)) | ||
| 1 | 158.1 | 75.2 | ||
| 2 | 110.3a | 75.7 | ||
| 3 | 164.8 | 64.8 | 13.74 (s, 1H) | 2, 4 |
| 4 | 113.7 | 63.9 | ||
| 5 | 188.4 | 59.6 | ||
| 6 | 109.3 | 59.6 | ||
| 7 | 164.5 | 69.8 | 12.11 (s, 1H) | 6, 7, 8 |
| 8 | 108.5 | 69.3 | 6.65 (d, 1H, 2.4) | 6, 10 |
| 9 | 165.5 | 61.7 | ||
| 10 | 108.8 | 62.0 | 7.16 (d, 1H, 2.4) | 6, 8, 12 |
| 11 | 134.9 | 54.4 | ||
| 12 | 181.1a | 54.6 | ||
| 13 | 137.1 | 62.6 | ||
| 14 | 115.0 | 63.4 | 7.69 (s, 1H) | 2, 4, 12, 16 |
| 15 | 145.7 | 53.4 | ||
| 16 | 104.2 | 53.3 | 6.81 (s, 1H) | 2, 14, 17 |
| 17 | 160.0 | 51.8 | ||
| 18 | 19.4 | 51.1 | 2.29 (s, 3H) | 16, 17 |
a Weak signals and were observable in the 1H-13C HMBC.
FIGURE 4.
The biosynthesis of 4 and the 1H-13C HMBC correlations of 4. 4 is synthesized from a nonaketide backbone synthesized by the PKS4 minimal PKS domains, followed by the AptA-PT-mediated C6-C11 and C4-C13 cyclizations as shown in this study. Cyclization of the third ring via C2-C15 cyclization is proposed to occur spontaneously. The proposed, spontaneous C1-O17 esterification forms the forth ring and releases the product, followed by second ring oxidation to afford 4.
Based on the structure and the [1,2-13C]acetate incorporation pattern of 4, the cyclizations steps catalyzed by PKSH1 are shown in Fig. 4. Importantly, this verified that AptA-PT indeed catalyzes the cyclization of polyketide backbone via C6-C11 aldol condensation. Cyclization of the second ring via C4-C13 is most likely to be also facilitated by this PT domain. Closure of the third ring via C2-C15 is likely spontaneous, which was proposed for the ACAS (29) and is commonly observed for most bacterial anthraquinone polyketides (49). Finally, a spontaneous C1-O17 esterification reaction takes place to form the α-pyrone fourth ring and concomitant product release, followed by second ring oxidation to yield 4 (Fig. 4). To generalize this finding, we further dissected the PT domain of A. nidulans AN0150 (26) as a second example of Group V PTs (supplemental Fig. S1). When combined, PKS4 minimal PKS and AN0150-PT also synthesized 4 as the predominant product (supplemental Fig. S5).
Therefore, we have functionally verified the regioselectivity of Group V PT domains through heterologous recombination with PKS4 minimal PKS, both as dissociated enzymes and as a hybrid megasynthase. Together, the C2-C7, C4-C19, and the C6-C11 PT domains represent the three known fungal “cyclases” reconstituted so far. It is interesting to note that the AptA PT domain functions efficiently on a nonaketide backbone, while asperthecin was predicted to derive from an octaketide backbone (25). Thus the PT domains may also be functional when paired with minimal PKSs that are of different chain length specificities.
Prediction of PT Regioselectivity from NRPKS of Unknown Function
Given that all five groups of PT functions have been functionally verified, the phylogenetic tree may therefore serve as a predictive tool of product structures synthesized from NRPKSs. To do so, we included 99 additional PT sequences extracted from NRPKSs of unknown functions (supplemental Table S3) into the alignment. The inclusion of these sequences did not significantly influence the phylogenetic classification and most of the sequences fall into these five groups discussed above (supplemental Fig. S6).
To test whether the PT regioselectivity agrees with the phylogenetic classification, we chose to functionally test a PT domain extracted from an unknown NRPKSs. The PT domain chosen was that of An03g05440 from A. niger (assigned as e_gw1_14.257 in A. niger ATCC 1015). The An03g05440-PT is located within group III, yet belongs to a branch that does not contain any functionally verified NRPKS. The gene fragment encoding the An03g05440-PT was amplified from the A. niger genomic DNA and replaced pks4 PT in the PKS4–99 template to afford the hybrid megasynthase PKSH2. PKSH2 was similarly expressed in BAP1, and purified to a final yield of ∼1.5 mg/liter (Fig. 5A). PKSH2 was assayed in vitro and the products were analyzed with LC-MS (Fig. 5B). Similar to PKS4–99, PKSH2 synthesized 1 as the major product with similar yields. This confirms An03g05440-PT indeed directs the cyclization of the nonaketide backbone via C2-C7 regioselectivity and suggests that the parent NRPKS is responsible for the synthesis of a product that contains this structural feature. In accordance with the recent review on the secondary metabolites synthesized by A. niger (50), the C2-C7 An03g05440 may be involved in the synthesis of the YWA1 analogues such as fonsecin, rubrofusarin, or the dimeric aurasperone B.
FIGURE 5.
A, construction and SDS-PAGE analysis of PKSH2. PKSH2 (193 kDa) is constructed through domain shuffling of PT domain between PKS4 and An03g05440 using PKS4–99 as a template. For SDS-PAGE: lane M, bench mark ladder; lane E,, Ni-NTA-purified PKSH2. B, LC-MS (325 nm) analysis of the polyketides synthesized by PKSH2 in vitro. Trace i, the parent megasynthase PKS4–99 synthesizes 1 as a major product; and trace ii, PKSH2 also synthesizes 1 as the major product, confirming the phylogenetic prediction of An03g05440-PT as a C2-C7 aldol cyclase.
Viridicatumtoxin is a tetracycline-like fungal aromatic polyketide (supplemental Fig. S7). Recently, the gene cluster has been located in P. aethiopicum and the NRPKS involved was shown to be vrtA (51). Previous 13C enrichment study of viridicatumotoxin led to two possible cyclization pathways (52), which can originate from either C6-C11 or C8-C13 first ring cyclization (supplemental Fig. S7). Our phylogenetic analysis finds VrtA PT domain is located in Group V (supplemental Fig. S6). To clarify the regioselectivity of VrtA, we dissected VrtA-PT (supplemental Fig. S1) and assayed it with PKS4 minimal PKS domains in trans. As expected from phylogenetic prediction, 4 was synthesized as a major product (supplemental Fig. S7). This result indicates that the formation of the viridicatumotoxin aglycon initiates with the C6-C11 regioselective cyclization.
DISCUSSION
With an increasing number of sequenced genomes from filamentous fungi, it has become apparent that these organisms are “underachievers” as natural product producers. A majority of sequenced biosynthetic gene clusters are silent under laboratory conditions and as a result, the metabolites associated with these pathways have remained cryptic. Therefore, being able to predict structures or partial structures of compounds based on sequence analysis of pathways is an important goal toward mining the fungal biosynthetic potential (53). In this work, we used phylogenic analysis of the PT domains to classify fungal NRPKSs into five groups and showed a correlation between protein sequence and cyclization regioselectivity. The five groups cover the known cyclizations modes of C2-C7, C4-C9, and C6-C11. The PT domains that control the C2-C7 aldol cyclizations were further grouped separately based on the ring sizes of the final products. Through verification of the regioselectivities of the Group V PT domains, we established a straightforward experimental approach to assay the function of PT domains. The assay is based on heterologous combinations of target PT domains with an NRPKS minimal PKS. Combining the bioinformatic and the simple biochemical approaches, we were able to predict and confirm the activities of two NRPKS PT domains uncovered from genome sequencing. Similarly, fundamental understanding of sequence-activity relationships of domains that control chain length, such as the KS, can provide additional insights into the structures of polyketides produced by cryptic NRPKSs.
The assay developed here is highly robust and the dominant polyketide produced reflects the regioselectivity of the target PT domains. The heterologous PT domains were able to interact compatibly with the PKS4 ACP domain, illustrating this key protein-protein interaction is not specific between endogenous partners. The mix and match of minimal PKS components with different PT domains are reminiscent of the combinatorial biosynthesis experiments performed with the bacterial type II PKSs and cyclases (49, 54). In these studies, the cyclases were found to be broadly specific and can be combined with different type II minimal PKSs to generate a number of regioselectively controlled unnatural aromatic polyketides. Our work here indicates that this approach can also be extended to fungal iterative type I PKSs to synthesize different compounds cyclized via the F-modes.
The construction of functional, hybrid NRPKSs demonstrates modularity of the domains in fungal PKSs, in that key domains can be readily swapped without major compromises to megasynthase structure, activity, and processivity. Such modularity provides further evidence that NRPKS domains may be evolutionarily derived from the juxtaposition of dissociated catalytic domains. The dimeric PT domains likely play a key role in maintaining the overall structural integrity of the megasynthases. Our results therefore suggest the overall folds of the different PT domains from different groups may be similar, thereby maintaining the endogenous communication and protein-protein interactions between the upstream KS-AT didomain and the downstream ACP domain. We compared the sequences of Group V PT domains to those from the other groups and found the sequential similarities between different groups are moderate. Group V PT domains show no more than 30% identities and 40% similarities to members of the other groups. For example, AptA-PT and PksA-PT share only 19% identity and 33% similarity, even though the final products are both anthraquinone polyketides. Hence, the sequence divergence must contribute to major differences in the sizes and geometries of the cyclization chamber (14), leading to different orientation of the polyketide backbone in the PT domains. Structural analysis of C2-C7 and C6-C11 PT domains, and comparison to that of the C4-C9 PksA PT domain will provide insights into these differences.
Whereas using dissociated catalytic domains can provide a rapid method of assaying PT activities, using hybrid megasynthases such as PKSH1 and PKSH2 has several advantages for combinatorial biosynthesis applications. First, the simplicity of cloning and expressing a single gene is an advantage over having to manipulate multiple genes. This was demonstrated in this work, where we used a single PKSH1 instead of three separate proteins (KS-MAT, PT, and ACP) to produce sufficient amounts of 4 for structure elucidation. Second, the in cis interactions between the different domains will lead to increased catalytic efficiency and product turnover, especially under in vivo conditions. Indeed, while compound 4 can be recovered from the cell pellets of E. coli expressing PKSH1, no product can be detected from E. coli expressing different domains as stand-alone proteins. This was similarly demonstrated using PKS4 minimal PKS in E. coli, where artificially linking the KS-MAT with ACP led to significant improvement in product turnover in vivo (19).
Supplementary Material
Acknowledgment
We thank Yit Heng Chooi and Wenjun Zhang for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01GM085128 and a David and Lucile Packard Fellowship (to Y. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1–S3.
- PKS
- polyketide synthase
- DTT
- dithiothreitol
- NRPKS
- nonreducing polyketide synthase
- PT
- product template
- NRPS
- nonribosomal peptide synthetases
- THN
- 1,3,6,8-tetrahydroxylnaphthalene
- HMBC
- heteronuclear multiple bond correlation
- HMQC
- heteronuclear multiple quantum coherence.
REFERENCES
- 1.Hoffmeister D., Keller N. P. (2007) Nat. Prod. Rep. 24, 393–416 [DOI] [PubMed] [Google Scholar]
- 2.Yoder O. C., Turgeon B. G. (2001) Curr. Opin. Plant Biol. 4, 315–321 [DOI] [PubMed] [Google Scholar]
- 3.Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R., Batzoglou S., Lee S. I., Bastürkmen M., Spevak C. C., Clutterbuck J., Kapitonov V., Jurka J., Scazzocchio C., Farman M., Butler J., Purcell S., Harris S., Braus G. H., Draht O., Busch S., D'Enfert C., Bouchier C., Goldman G. H., Bell-Pedersen D., Griffiths-Jones S., Doonan J. H., Yu J., Vienken K., Pain A., Freitag M., Selker E. U., Archer D. B., Peñalva M. A., Oakley B. R., Momany M., Tanaka T., Kumagai T., Asai K., Machida M., Nierman W. C., Denning D. W., Caddick M., Hynes M., Paoletti M., Fischer R., Miller B., Dyer P., Sachs M. S., Osmani S. A., Birren B. W. (2005) Nature 438, 1105–1115 [DOI] [PubMed] [Google Scholar]
- 4.Machida M., Asai K., Sano M., Tanaka T., Kumagai T., Terai G., Kusumoto K., Arima T., Akita O., Kashiwagi Y., Abe K., Gomi K., Horiuchi H., Kitamoto K., Kobayashi T., Takeuchi M., Denning D. W., Galagan J. E., Nierman W. C., Yu J., Archer D. B., Bennett J. W., Bhatnagar D., Cleveland T. E., Fedorova N. D., Gotoh O., Horikawa H., Hosoyama A., Ichinomiya M., Igarashi R., Iwashita K., Juvvadi P. R., Kato M., Kato Y., Kin T., Kokubun A., Maeda H., Maeyama N., Maruyama J., Nagasaki H., Nakajima T., Oda K., Okada K., Paulsen I., Sakamoto K., Sawano T., Takahashi M., Takase K., Terabayashi Y., Wortman J. R., Yamada O., Yamagata Y., Anazawa H., Hata Y., Koide Y., Komori T., Koyama Y., Minetoki T., Suharnan S., Tanaka A., Isono K., Kuhara S., Ogasawara N., Kikuchi H. (2005) Nature 438, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 5.Nierman W. C., Pain A., Anderson M. J., Wortman J. R., Kim H. S., Arroyo J., Berriman M., Abe K., Archer D. B., Bermejo C., Bennett J., Bowyer P., Chen D., Collins M., Coulsen R., Davies R., Dyer P. S., Farman M., Fedorova N., Feldblyum T. V., Fischer R., Fosker N., Fraser A., García J. L., García M. J., Goble A., Goldman G. H., Gomi K., Griffith-Jones S., Gwilliam R., Haas B., Haas H., Harris D., Horiuchi H., Huang J., Humphray S., Jiménez J., Keller N., Khouri H., Kitamoto K., Kobayashi T., Konzack S., Kulkarni R., Kumagai T., Lafon A., Latgé J. P., Li W., Lord A., Lu C., Majoros W. H., May G. S., Miller B. L., Mohamoud Y., Molina M., Monod M., Mouyna I., Mulligan S., Murphy L., O'Neil S., Paulsen I., Peñalva M. A., Pertea M., Price C., Pritchard B. L., Quail M. A., Rabbinowitsch E., Rawlins N., Rajandream M. A., Reichard U., Renauld H., Robson G. D., Rodriguez de Córdoba S., Rodriguez-Peña J. M., Ronning C. M., Rutter S., Salzberg S. L., Sanchez M., Sánchez-Ferrero J. C., Saunders D., Seeger K., Squares R., Squares S., Takeuchi M., Tekaia F., Turner G., Vazquez de Aldana C. R., Weidman J., White O., Woodward J., Yu J. H., Fraser C., Galagan J. E., Asai K., Machida M., Hall N., Barrell B., Denning D. W. (2005) Nature 438, 1151–1156 [DOI] [PubMed] [Google Scholar]
- 6.Kroken S., Glass N. L., Taylor J. W., Yoder O. C., Turgeon B. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brakhage A. A., Schuemann J., Bergmann S., Scherlach K., Schroeckh V., Hertweck C. (2008) Prog. Drug. Res. 66, 3–12 [DOI] [PubMed] [Google Scholar]
- 8.Corre C., Challis G. L. (2009) Nat. Prod. Rep. 26, 977–986 [DOI] [PubMed] [Google Scholar]
- 9.Khosla C., Tang Y., Chen A. Y., Schnarr N. A., Cane D. E. (2007) Annu. Rev. Biochem. 76, 195–221 [DOI] [PubMed] [Google Scholar]
- 10.Udwary D. W., Zeigler L., Asolkar R. N., Singan V., Lapidus A., Fenical W., Jensen P. R., Moore B. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10376–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox R. J. (2007) Org. Biomol. Chem. 5, 2010–2026 [DOI] [PubMed] [Google Scholar]
- 12.Crawford J. M., Thomas P. M., Scheerer J. R., Vagstad A. L., Kelleher N. L., Townsend C. A. (2008) Science 320, 243–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford J. M., Dancy B. C., Hill E. A., Udwary D. W., Townsend C. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16728–16733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford J. M., Korman T. P., Labonte J. W., Vagstad A. L., Hill E. A., Kamari-Bidkorpeh O., Tsai S. C., Townsend C. A. (2009) Nature 461, 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wattana-amorn P., Williams C., Ploskoń E., Cox R. J., Simpson T. J., Crosby J., Crump M. P. (2010) Biochemistry 49, 2186–2193 [DOI] [PubMed] [Google Scholar]
- 16.Fujii I., Watanabe A., Sankawa U., Ebizuka Y. (2001) Chem. Biol. 8, 189–197 [DOI] [PubMed] [Google Scholar]
- 17.Udwary D. W., Merski M., Townsend C. A. (2002) J. Mol. Biol. 323, 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma S. M., Zhan J., Xie X., Watanabe K., Tang Y., Zhang W. (2008) J. Am. Chem. Soc. 130, 38–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W., Li Y., Tang Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20683–20688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y., Smith L. H., Cox R. J., Beltran-Alvarez P., Arthur C. J., Simpson F. R. (2006) ChemBioChem 7, 1951–1958 [DOI] [PubMed] [Google Scholar]
- 21.Tsai H. F., Fujii I., Watanabe A., Wheeler M. H., Chang Y. C., Yasuoka Y., Ebizuka Y., Kwon-Chung K. J. (2001) J. Biol. Chem. 276, 29292–29298 [DOI] [PubMed] [Google Scholar]
- 22.Bailey A. M., Cox R. J., Harley K., Lazarus C. M., Simpson T. J., Skellam E. (2007) Chem. Commun. 4053–4055 [DOI] [PubMed] [Google Scholar]
- 23.Ma S. M., Zhan J., Watanabe K., Xie X., Zhang W., Wang C. C., Tang Y. (2007) J. Am. Chem. Soc. 129, 10642–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas R. (2001) ChemBioChem 2, 612–627 [DOI] [PubMed] [Google Scholar]
- 25.Szewczyk E., Chiang Y. M., Oakley C. E., Davidson A. D., Wang C. C., Oakley B. R. (2008) Appl. Environ. Microbiol. 74, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bok J. W., Chiang Y. M., Szewczyk E., Reyes-Domingez Y., Davidson A. D., Sanchez J. F., Lo H. C., Watanabe K., Strauss J., Oakley B. R., Wang C. C., Keller N. P. (2009) Nat. Chem. Biol. 5, 462–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bringmann G., Noll T. F., Gulder T. A., Grüne M., Dreyer M., Wilde C., Pankewitz F., Hilker M., Payne G. D., Jones A. L., Goodfellow M., Fiedler H. P. (2006) Nat. Chem. Biol. 2, 429–433 [DOI] [PubMed] [Google Scholar]
- 28.Breinholt J., Jensen G. W., Kjaer A., Olsen C. E., Rosendahl C. N. (1997) Acta Chem. Scand. 51, 855–860 [Google Scholar]
- 29.Awakawa T., Yokota K., Funa N., Doi F., Mori N., Watanabe H., Horinouchi S. (2009) Chem. Biol. 16, 613–623 [DOI] [PubMed] [Google Scholar]
- 30.Pfeifer B. A., Admiraal S. J., Gramajo H., Cane D. E., Khosla C. (2001) Science 291, 1790–1792 [DOI] [PubMed] [Google Scholar]
- 31.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 32.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 33.Rzhetsky A., Nei M. (1992) Mol. Biol. Evol. 9, 945–967 [Google Scholar]
- 34.Zuckerkandl E., Pauling L. (1965) in Evolving Genes and Proteins: A Symposium Held at the Institute of Microbiology of Rutgers, pp 97–166, The State University, NY [Google Scholar]
- 35.Schroeckh V., Scherlach K., Nützmann H. W., Shelest E., Schmidt-Heck W., Schuemann J., Martin K., Hertweck C., Brakhage A. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14558–14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H., Zhan J., Watanabe K., Xie X., Tang Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6249–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y. T., Lee Y. R., Jin J., Han K. H., Kim H., Kim J. C., Lee T., Yun S. H., Lee Y. W. (2005) Mol. Microbiol. 58, 1102–1113 [DOI] [PubMed] [Google Scholar]
- 38.Reeves C. D., Hu Z., Reid R., Kealey J. T. (2008) Appl. Environ. Microbiol. 74, 5121–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H., Qiao K., Gao Z., Meehan M. J., Li J. W. H., Zhao X., Dorrestein P. C., Vederas J. C., Tang Y. (2010) J. Am. Chem. Soc., 4530–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S., Xu Y., Maine E. A., Wijeratne E. M., Espinosa-Artiles P., Gunatilaka A. A., Molnár I. (2008) Chem. Biol. 15, 1328–1338 [DOI] [PubMed] [Google Scholar]
- 41.Fujii I., Mori Y., Watanabe A., Kubo Y., Tsuji G., Ebizuka Y. (2000) Biochemistry 39, 8853–8858 [DOI] [PubMed] [Google Scholar]
- 42.Linnemannstöns P., Schulte J., del Mar Prado M., Proctor R. H., Avalos J., Tudzynski B. (2002) Fungal Genet. Biol. 37, 134–148 [DOI] [PubMed] [Google Scholar]
- 43.Kim J. E., Han K. H., Jin J., Kim H., Kim J. C., Yun S. H., Lee Y. W. (2005) Appl. Environ. Microbiol. 71, 1701–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujii I., Yasuoka Y., Tsai H. F., Chang Y. C., Kwon-Chung K. J., Ebizuka Y. (2004) J. Biol. Chem. 279, 44613–44620 [DOI] [PubMed] [Google Scholar]
- 45.Brown D. W., Yu J. H., Kelkar H. S., Fernandes M., Nesbitt T. C., Keller N. P., Adams T. H., Leonard T. J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1418–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tominaga M., Lee Y. H., Hayashi R., Suzuki Y., Yamada O., Sakamoto K., Gotoh K., Akita O. (2006) Appl. Environ. Microbiol. 72, 484–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H., Lee M. H., Daub M. E., Chung K. R. (2007) Mol. Microbiol. 64, 755–770 [DOI] [PubMed] [Google Scholar]
- 48.An J. H., Kim Y. S. (1998) Eur. J. Biochem. 257, 395–402 [DOI] [PubMed] [Google Scholar]
- 49.McDaniel R., Ebert-Khosla S., Hopwood D. A., Khosla C. (1995) Nature 375, 549–554 [DOI] [PubMed] [Google Scholar]
- 50.Nielsen K. F., Mogensen J. M., Johansen M., Larsen T. O., Frisvad J. C. (2009) Anal. Bioanal. Chem., 1225–1242 [DOI] [PubMed] [Google Scholar]
- 51.Chooi Y.-H., Cacho R., Tang Y. (2010) Chem. Biol. 17, 483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jesus A., Hull W., Steyn P., Heerden F., Vleggaar R. (1982) J. Chem. Soc., Chem. Commun. 902–904 [Google Scholar]
- 53.Schümann J., Hertweck C. (2006) J. Biotechnol. 124, 690–703 [DOI] [PubMed] [Google Scholar]
- 54.Rawlings B. J. (1999) Nat. Prod. Rep. 16, 425–484 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.