Abstract
Biological sulfate reduction is a process with high environmental significance due to its major contribution to the carbon and sulfur cycles in anaerobic environments. However, the respiratory chain of sulfate-reducing bacteria is still poorly understood. Here we describe a new respiratory complex that was isolated as a major protein present in the membranes of Desulfovibrio vulgaris Hildenborough. The complex, which was named Qrc, is the first representative of a new family of redox complexes. It has three subunits related to the complex iron-sulfur molybdoenzyme family and a multiheme cytochrome c and binds six hemes c, one [3Fe-4S]+1/0 cluster, and several interacting [4Fe-4S]2+/1+ clusters but no molybdenum. Qrc is related to the alternative complex III, and we show that it has the reverse catalytic activity, acting as a Type I cytochrome c3:menaquinone oxidoreductase. The qrc genes are found in the genomes of deltaproteobacterial sulfate reducers, which have periplasmic hydrogenases and formate dehydrogenases that lack a membrane subunit for reduction of the quinone pool. In these organisms, Qrc acts as a menaquinone reductase with electrons from periplasmic hydrogen or formate oxidation. Binding of a menaquinone analogue affects the EPR spectrum of the [3Fe-4S]+1/0 cluster, indicating the presence of a quinone-binding site close to the periplasmic subunits. Qrc is the first respiratory complex from sulfate reducers to have its physiological function clearly elucidated.
Keywords: Bacterial Metabolism, Cytochromes, Energy Metabolism, Iron-Sulfur protein, Membrane Proteins, Quinones, Respiratory Chain, Hydrogen Metabolism, Hydrogenase, Sulfate Reduction
Introduction
Respiratory chains rely on membrane-bound protein complexes to perform a stepwise transduction of the free energy of redox reactions into an electrochemical potential difference across the membrane that drives the synthesis of ATP. In aerobic eukaryotic organisms, a fixed and mostly linear respiratory chain is present, in contrast to bacteria and archaea whose flexible and usually highly branched respiratory chains reflect their ability to use an array of different electrons donors and acceptors (1). This flexibility enables prokaryotes to explore virtually every possible energy source and oxidant pairs that can provide energy to sustain life and, thus, colonize multiple habitats on earth, including the most extreme environments. In addition, the recycling of elements provided by bacterial energy metabolism is a crucial component for the maintenance of life in our planet (2). The study of bacterial respiratory complexes (BRCs)3 has highlighted the diversity of mechanisms by which energy conservation can be achieved (3, 4). BRCs usually have simpler compositions than their eukaryotic counterparts and are, therefore, useful models to understand the molecular mechanisms involved in those processes. Furthermore, bacteria contain many unique BRCs whose architectures have highly modular characters, with different arrangements of modules giving rise to different proteins and physiological functions (5, 6).
An example of a novel BRC has recently been described that is a functional substitute for the bc1 complex, or complex III (7–9). This complex, named alternative complex III (ACIII), is found in many groups of bacteria whose genome lacks a complex III but includes heme-copper oxygen reductases that receive electrons from soluble periplasmic proteins (thus requiring the presence of a protein to transfer electrons from the quinone pool to these proteins). The function of ACIII as a quinol:periplasmic electron acceptor oxidoreductase has recently been demonstrated with the complex from Chloroflexus aurantiacus (10), whereas in Rhodothermus marinus it was shown to transfer electrons directly to an oxygen reductase (11). The ACIII complex includes subunits with homology to those of the complex iron-sulfur molybdoenzyme (CISM) family (6), which are widespread bacterial oxidoreductases such as formate dehydrogenase or nitrate, dimethyl sulfoxide, thiosulfate, polysulfide and tetrathionate reductases. Many enzymes in this family are part of anaerobic respiratory chains and have a typical three-subunit composition consisting of a catalytic subunit with a molybdopterin cofactor, an electron transferring subunit with four [FeS] centers, and an integral membrane subunit that anchors the protein complex to the membrane and is involved in electron exchange with the quinone pool. The subunit composition of ACIII is more complex and includes four integral membrane proteins, two cytochromes c, and a large periplasmic protein with two domains, homologous to the [FeS] and catalytic subunits of the CISM enzymes. However, no molybdenum cofactor is present in the so far described ACIII complexes of R. marinus and C. aurantiacus, in agreement with its function as an electron transfer complex (8, 9).
A distinct BRC related to ACIII but with a simpler subunit composition was described to be present in the sulfate-reducing bacteria Desulfovibrio vulgaris and Desulfovibrio desulfuricans (8). Sulfate reduction is a widespread process in anaerobic habitats and has a very high environmental significance, being a major contributor to the sulfur and carbon cycles (12). Yet, it is one of the last few major biological processes that remains to be elucidated at a molecular level. The respiratory chain of sulfate-reducing bacteria (SRB) is very interesting and unusual due to the presence of several unique components, namely several BRCs that are distinct from those found in other organisms (13). The SRB complexes described to date are either involved in oxidation of the menaquinol pool or direct transmembrane electron transfer. No complex has yet been described in SRB with the function of reducing the quinone pool. Here, we describe the isolation and characterization of a new complex from D. vulgaris Hildenborough and show that it functions as a Type I cytochrome c3:menaquinone oxidoreductase. We propose to name this complex as quinone-reductase complex (Qrc), encoded by the operon qrcABCD. The D. vulgaris QrcABCD complex is the first described example of a new family of BRC that is related to the ACIII complex but performs the reverse function of this by reducing the menaquinone pool with electrons transferred from a periplasmic cytochrome c. We show also that Qrc is efficiently reduced by periplasmic hydrogenase and formate dehydrogenase via the cytochrome c3 and, thus, acts as the linker to reduce the quinone pool during growth on H2 or formate. An essential role of the qrcB gene (previously designated as mopB) for growth of D. desulfuricans G20 in hydrogen or formate with sulfate has recently been described (14). The involvement of the Qrc complex in sulfate respiration is discussed within the context of genome data available for SRB.
EXPERIMENTAL PROCEDURES
Bacterial Growth and Protein Purification
D. vulgaris Hildenborough (DSM 644) growth and preparation of solubilized membrane proteins were conducted as previously described (15). All chromatographic steps were performed at 4 °C. The purifications were followed using UV-visible spectroscopy and BN- and SDS-PAGE. The solubilized proteins were applied to a fast flow Q-Sepharose column equilibrated with 20 mm Tris-HCl buffer with 10% glycerol, 0.1% n-dodecyl-β-d-maltoside (Buffer A), and a quarter of a CompleteTM tablet per liter. A stepwise gradient of increasing NaCl concentration was performed, the fractions eluting at ∼250 mm NaCl were concentrated, and their ionic strength was lowered. This sample was subjected to a second passage on a Q-Sepharose HP using the same protocol. The heme-containing fractions were concentrated and loaded in an IMAC-Sepharose HP column, saturated with Ni2+, and equilibrated with Buffer A with 0.4 m NaCl. A stepwise gradient was performed with imidazole to elute a fraction containing the Qrc complex at 50 mm. The [FeFe] and [NiFeSe] hydrogenases and Type I cytochrome c3 (TpIc3) were isolated from D. vulgaris by established procedures. The FdhAB formate dehydrogenase from this organism was purified inside the anaerobic chamber monitoring its catalytic activity.
Biochemical Analysis
Protein concentration, BN- and Tricine-SDS-PAGE, N-terminal protein sequence analysis, and covalent and non-covalent heme content determination was performed as described (15). Molybdenum and tungsten were analyzed by inductively coupled plasma. The pterin cofactor extraction and fluorescence analysis was performed according to Sebban et al. (16). Determination of the molecular mass of the complex was performed by gel filtration in a Superdex 200 column and of the subunits in a matrix-assisted laser desorption ionization time-of-flight spectrometer.
Spectroscopic Techniques
Room temperature UV-visible spectra were obtained in a Shimadzu UV-1603 or UV-1203 spectrophotometer inside the anaerobic chamber. Fluorescence measurements were recorded on a Cary Varian Eclipse instrument. EPR spectra were recorded on a Bruker EMX spectrometer equipped with an ESR-900 continuous flow helium cryostat.
Redox Titration
Potentiometric titrations were performed as described in Pires et al. (15) in 50 mm Tris-HCl and 0.1% n-dodecyl-β-d-maltoside, pH 7.6, as buffer, with 1 μm concentration of purified protein and 2 μm concentration of the reported mixture of redox mediators including also antraquinone-2-sulfonate.
Quinone Interaction Experiments
All experiments were performed in an anaerobic chamber (95% Ar, 5% H2). The reduction of the Qrc complex with quinols was measured monitoring the heme reduction at the Soret band. The buffer used in all cases was 20 mm Tris-HCl, pH 7.6, plus 5 mm EDTA and 0.1% n-dodecyl-β-d-maltoside. 2,3-Dimethyl-1,4-naphthoquinol and menaquinol-4 (vitamin K2) were prepared from the respective quinones in ethanol by reduction with metallic zinc in 5 m HCl. Menadiol was reduced from menadione according to Pires et al. (15) and also solubilized in ethanol. For the reverse reaction the complex was reduced with a substoichiometric amount of dithionite, and an ethanolic solution of 2,3-dimethyl-1,4-naphthoquinone, menadione, or menaquinone-4 was added as the electron acceptor. As a control experiment, the reduced protein was treated with an equal volume of ethanol. To test the effect of HQNO by EPR, a concentrated solution of HQNO in ethanol was added to a final concentration of 0.5 mm. In a control sample the same volume of ethanol was added.
In Vitro Electron Transfer Assays
The assays were performed inside the anaerobic chamber. It was tested if the purified [FeFe] and [NiFeSe] hydrogenases as well as FdhAB formate dehydrogenase were able to reduce the Qrc. Hydrogenases were first activated under H2 overpressure and when applicable incubated with TpIc3 at room temperature for 10 min before being added to the Qrc in the cuvette. Fdh was activated by incubation with 20 mm sodium formate and 1 mm dithiothreitol for 5 min. In every case the enzyme activity was first tested with methyl viologen. The assay mixture contained 50 mm Tris-HCl, pH 7.6, which was H2-saturated in the case of hydrogenase assays and contained 1 mm dithiothreitol for Fdh assays. Qrc was present at 0.6 μm, and the enzymes and cytochromes were added in catalytic amounts (5 nm). Qrc reduction was measured by following the increase in absorption at 419 nm, and sodium dithionite was added in the end of each assay to verify the complete reduction of Qrc.
Cytochrome TpIc3:Menaquinone-4 Oxidoreductase Assay
The electron transfer activities were measured at 25 °C inside an anaerobic glove box after the oxidation of TpIc3 at 552 nm (absorption coefficient, 116 mm−1 cm−1) in 50 mm phosphate buffer, pH 7.3, 0.01% n-dodecyl-β-d-maltoside. TpIc3 was first reduced stepwise but not completely (to prevent excess of reductant) with a diluted dithionite solution (∼1 mm). The reaction was started by the addition of Qrc (100 nm) and menaquinone-4 to the reduced TpIc3 in the cuvette. The unspecific oxidation of TpIc3 by menaquinone-4 was subtracted to give the final activity. Reaction rates were determined from the initial linear oxidation rate. Kinetic parameters (Km and Vmax) were determined by nonlinear curve-fitting using Hyper32 software (University of Liverpool).
Sequence Analysis Tools
Blast searches were performed at the NCBI website (www.ncbi.nlm.nih.gov). Genome sequence data were retrieved and analyzed at the Integrated Microbial Genomes website, the Comprehensive Microbial Resource website, and VIMSS Comparative Genomics website. Multiple alignments were performed using ClustalX. Sequence analysis and transmembrane helix predictions were done using programs available at ExPASy.
RESULTS
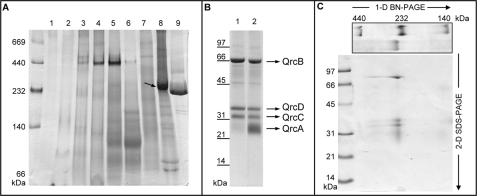
The only BRC that has been characterized so far from D. vulgaris is the Tmc complex (17). To further investigate the respiratory chain of this model organism, membrane protein complexes were analyzed by BN-PAGE after separation on ion-exchange chromatography. A major band was observed at ∼230 kDa, which stained positive for hemes c (Fig. 1A). This band is not present in a similar analysis of membrane extracts from D. desulfuricans ATCC 27774, but its high abundance indicates it is one of the major proteins in the membranes of D. vulgaris. After optimization of a purification protocol, the new complex could be obtained in a pure form. Tricine-SDS-PAGE revealed the presence of four subunits with apparent molecular masses of 70, 38, 33, and 29 kDa (Fig. 1B). The 29-kDa band stains poorly with Coomassie Blue but is well visible with heme-staining. A two-dimensional gel confirmed that the four subunits are present in the BN-PAGE ∼230-kDa band (Fig. 1C). To identify the new complex in the D. vulgaris genome, N-terminal sequences were obtained for the four subunits. The 70- and 33-kDa bands gave clear results (ALDRR and SSFKEF) that enabled their assignment as the products of genes DVU0694 and DVU0693 that are part of a predicted 3-gene operon in D. vulgaris (DVU0692-0694). The N-terminal sequences of the 38- and 29-kDa bands gave very weak signals, suggesting they were blocked. However, several amino acid residues of the N terminus of the membrane protein DVU0692 could be identified. Because the isolated complex contains four subunits, we reanalyzed the DNA sequence corresponding to the two neighboring genes DVU0695 and DVU0696, which are annotated as hypothetical proteins. This analysis showed that a gene (which we named 0695*) encoding for a 23-kDa multiheme cytochrome is found upstream from DVU0694. Several of the predicted N-terminal amino acids of DVU0695* are present in the N-terminal sequence analysis of the 29-kDa band (ME_RQL), confirming this assignment and showing that the gene cluster (Fig. 2A) encodes the four subunits present in the isolated complex. The molecular mass of the complex determined by gel filtration is 240 ± 25 kDa. This agrees with a predicted stoichiometry of 1:1:1:1 for the QrcABCD subunits (theoretical molecular mass, 180 kDa), based on analogy with the CISM family, with the difference accounted for by the detergent micelle.
FIGURE 1.
Gel electrophoresis of the Qrc complex. A, shown is BN-PAGE (5–15%) of fractions 1–9 from the first purification step, stained for hemes over the Coomassie Blue. The arrow points to the Qrc band. B, Tricine-SDS-PAGE of the purified Qrc (20 μg) is shown. 1, stained with Coomassie Blue; 2, as 1, but also stained for hemes c. C, shown is a two-dimensional (2-D) BN-Tricine-SDS-PAGE gel of Qrc. 1-D, one-dimensional.
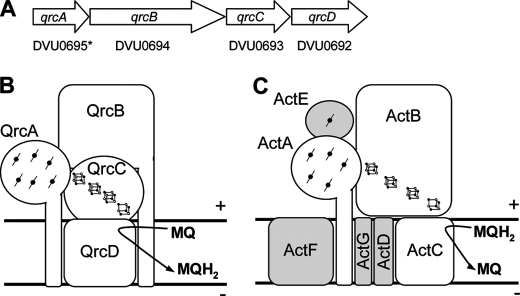
FIGURE 2.
The Qrc and ACIII complexes. A, shown is the qrc gene cluster in D. vulgaris (DVU0692-0695*). A schematic representation of the D. vulgaris Qrc complex (B) and the related ACIII (C) are shown. Similar subunits in the two complexes are in white (ActB is a fusion of QrcB- and QrcC-like proteins). MQ, menaquinone; MQH2, menaquinol.
Sequence Analysis of the D. vulgaris qrcABCD Genes
The D. vulgaris gene cluster DVU0692–0695* was previously identified by Yanyushin et al. (8) as encoding a new class of bacterial oxidoreductase that is related to the ACIII. A number of factors, discussed below, clearly distinguish the ACIII complex from the one described here. Given these differences and the fact that they perform distinct physiological functions, we opted to name the new complex as quinone-reductase complex (Qrc) and the genes as qrcABCD. The QrcABCD complex is present in the genomes of several organisms, which strikingly are all sulfate-reducing Deltaproteobacteria (with one exception, Solibacter usitatus, which likely acquired it through lateral gene transfer). A schematic representation of the QrcABCD complex, based on sequence analysis and the characterization described below, is shown in Fig. 2B.
The qrcA gene (DVU0695*) encodes a 23-kDa cytochrome c with six heme binding motifs. The N-terminal analysis of this protein shows that the Sec-translocating signal peptide is not cleaved and forms a transmembrane helix that probably serves as a membrane anchor. The closest homologues of qrcA are the actA genes that are part of the ACIII complexes (8, 9). Most QrcA and ActA proteins include only five heme binding sites, and there are five strictly conserved residues (four His and one Met) that are the probable distal ligands of the five hemes. Binding of six hemes to D. vulgaris QrcA was confirmed by mass spectrometry (see below). Sequence analysis indicates that both QrcA and ActA proteins form a separate group when compared with other multiheme c cytochromes such as NrfA, NrfB, NapC/NrfH, cytochromes c3, hydroxylamine oxidoreductase, cytochrome c554, or octaheme cytochromes (data not shown).
The genes qrcBCD are related to the genes of the three subunit CISM molybdoenzyme family (6). The qrcB gene (DVU0694) encodes a 72-kDa protein similar to the molybdo-bis(pyranopterin guanidine dinucleotide) cofactor-containing catalytic subunit of CISM and includes a twin-arginine signal peptide indicating it is periplasmic. Similarly to QrcA, the signal peptide is still present in the mature protein and likely serves as a membrane anchor. The qrcC gene (DVU0693) encodes a 29-kDa iron-sulfur protein that belongs to the family of four cluster proteins. These are electron-transferring subunits found in many respiratory enzymes such as CISM and contain four cubane iron-sulfur clusters (FS1-FS4). The clusters form a pathway for electron transfer between the membrane quinone pool and the catalytic center, as described in the structures of Fdh-N formate dehydrogenase, Nar nitrate reductase, and Psr polysulfide reductase (18–20). Sixteen Cys residues involved in binding of four iron-sulfur clusters are conserved in QrcC. Although QrcC has no signal peptide, it should be co-translocated with QrcB as happens with CISM enzymes. The qrcD gene (DVU0692) encodes a 48-kDa integral membrane protein with 10 transmembrane helices that belongs to the family of CISM proteins that interact with the quinone pool without the involvement of hemes b, such as NrfD or PsrC (21).
The QrcB and QrcC proteins are most closely related to the two domains of the ActB subunit of ACIII that probably resulted from a gene fusion event, and QrcD is most closely related to the two ActC and ActF subunits of ACIII, which are very similar and resulted from a gene duplication event. However, a dendogram of the known sequences of QrcB, domain 1 of ActB, and other proteins in the family clearly shows that QrcB forms a separate cluster from ActB (supplemental Figs. S1) even though the two proteins have a common origin as previously proposed (8). This is also the case for QrcC, domain 2 of ActB, and related proteins (data not shown). Thus, sequence analysis indicates unambiguously that QrcB, QrcC, and QrcD constitute novel groups within their respective families. In Geobacter metallireducens and Thermus thermophilus the corresponding operons encode ACIII and not Qrc complexes as previously suggested (8). Interestingly, in G. metallireducens, Geobacter uraniumreducens, and Mariprofundus ferrooxydans the two domains of ActB are encoded by two separate genes, as happens in the Qrc complex, and these are the ACIII sequences most closely related to Qrc subunits.
Several of the catalytic subunits of CISM molybdoenzymes include a [4Fe-4S]2+/1+ center named FS0 that is involved in electron transfer between the molybdopterin cofactor and the FeS centers of the four-cluster protein. Curiously, the four Cys that bind the FS0 cluster are only present in some QrcB proteins (but not in D. vulgaris) and in some ActB proteins (e.g. G. metallireducens) The loss of the FS0 center in several QrcB and ActB proteins agrees with the fact that a molybdopterin cofactor is not present in these proteins (see below).
Characterization of the Qrc Redox Centers
We first tested whether a molybdenum (or tungsten) pterin cofactor is present in the Qrc complex, as this is absent in ACIII complexes (8, 9). No molybdenum or tungsten was detected in the Qrc complex by inductively coupled plasma, and no molybdenum or tungsten signal was observed by EPR spectroscopy (see below). Attempted extraction of a pterin group followed by fluorescence analysis was also negative. This shows that the QrcB protein does not bind a molybdo-bis(pyranopterin guanidine dinucleotide) cofactor. Besides QrcB and ActB proteins, there is a third example of a protein belonging to the family of molybdopterin oxidoreductases, that also does not contain the Mo cofactor; that is, the Nqo3/NuoG subunit of the respiratory complex I (NADH:quinone oxidoreductase), which includes a molybdopterin oxidoreductase-like domain of unknown function (22). Using sequence analysis of QrcB, ActB, and Nqo3 versus other molybdopterin oxidoreductases for which the structure is known, we could not identify sequence features that could explain the absence of the pterin, due to a low level of sequence identity between the different families.
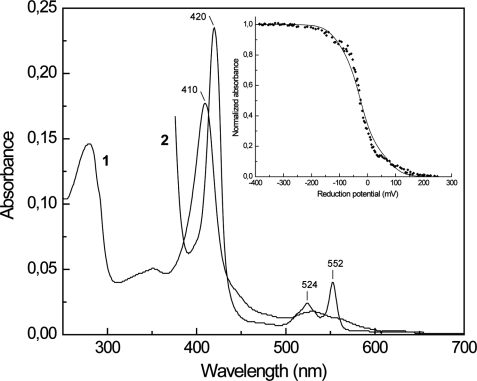
The UV-visible spectrum of the Qrc complex indicates the presence of only hemes c (Fig. 3). The absence of hemes b was confirmed by the pyridine hemochrome assay and HPLC analysis of extractable hemes. Mass spectrometry of the Qrc complex subunits gave a mass of 27,450 Da for the QrcA protein, which confirms that six hemes are covalently bound. The macroscopic redox potentials of the hemes were determined through a redox titration monitored by visible spectroscopy following the Soret and α bands at pH 7.6 (Fig. 3, inset). The hemes started to be reduced at ∼+150 mV and were fully reduced at ∼−250 mV. This broad potential range reflects the presence of multiple heme centers with overlapping redox potentials, and the contribution of individual hemes cannot be resolved. The experimental points were simulated with the sum of six Nernst equations with mid-point redox potentials of +80, −25, and −110 mV (±10 mV) in a 1:4:1 ratio, without taking into account possible redox interactions. These redox potentials give an indication of the macroscopic redox behavior of the system and are not individual heme potentials. The first heme to be reduced is likely to correspond to the His/Met bound heme and the others to the bis-His hemes, which typically have lower redox potentials.
FIGURE 3.
UV-visible spectra of the Qrc complex. Oxidized (1) and reduced (2) forms are shown. Inset, redox titration to the Qrc hemes followed by UV-visible spectroscopy at 420 nm; the solid line corresponds to a theoretical simulation with the sum of six Nernst equations with redox potentials of +80, −25, and −110 mV in a ratio of 1:4:1.
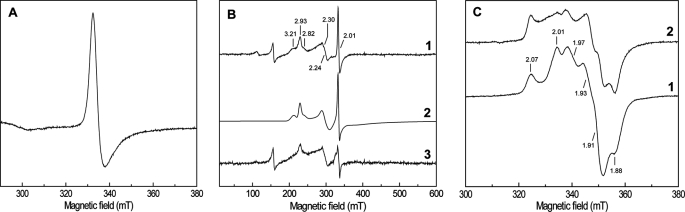
The Qrc cofactors were also studied by EPR spectroscopy. The spectrum of the complex in the oxidized state shows a strong signal at g ∼ 2.01 typical of [3Fe-4S]1+ clusters (Fig. 4A) as observed in NarGHI (23, 24) or FrdABCD fumarate reductase (25), both of which contain a [3Fe-4S]1+/0 cluster. A signal corresponding to a [3Fe-4S]1+/0 cluster is also present in the R. marinus ACIII complex (7). In addition, there are several overlapping signals for low-spin ferric hemes with gmax values of 3.21, 2.93, and 2.82 and a gmed of ∼2.24 (Fig. 4B). These three signals can be well simulated with a stoichiometry of 3:2:1. The intensity of the g ∼ 2.01 signal relative to the heme signals was constant in several preparations and was also estimated by spectral simulation giving a relative proportion of one [3Fe-4S]1+/0 center for six low-spin hemes, in agreement with the sequence-based predictions. At high temperatures, no signals attributable to molybdenum or tungsten species were detected. Reduction with ascorbate leads to almost complete reduction of the [3Fe-4S]1+/0 signal and partial reduction of the low-spin hemes (Fig. 4B). Upon dithionite reduction a complex signal appears with features at 2.07, 2.01, 1.97, 1.93, 1.91, and 1.88 (Fig. 4C), in agreement with the presence of several reduced [4Fe-4S]2+/1+ clusters in QrcC. The shape of the signal is indicative of magnetic interactions between the several centers, as has been described for this protein family, where the FeS clusters are within close distance of each other (23, 24, 26). Overall, the spectral characterization indicates that all the cofactors predicted from the Qrc gene sequences (six hemes c and four cubane clusters) are present in the isolated complex, with one of these clusters being a [3Fe-4S]1+/0 one. Furthermore, no hemes b are present in the QrcD membrane subunit.
FIGURE 4.
EPR spectra of Qrc complex. A, shown is the [3Fe-4S]+1 signal present in the oxidized sample. B, shown are low-spin heme signals present in the oxidized sample (1), theoretical simulation obtained by the sum of four components (three low-spin hemes and one [3Fe-4S]+1 center) in a 3:2:1:1 ratio (hemes: ga = 3.16, 1.9, 1.3; gb = 2.93, 2.21, 1.5; gc = 2.825, 2.28, 1.6; [3Fe-4S]+1 center, g = 2.01, 2.01, 1.2) (2), and after reduction with ascorbic acid 0.2 m (3). C, shown is the spectrum after dithionite reduction with some indicative g values shown for reference (1) and after reduction with 0.5 μm [NiFeSe] hydrogenase/0.5 μm TpIc3 in the presence of H2 (2). Spectra were recorded under the following conditions: microwave frequency, 9.38 GHz; microwave power, 20.1 milliwatts (A) and 2.4 milliwatts (B and C); modulation frequency, 100 kHz; modulation amplitude, 1 millitesla; temperature, 10 K.
Physiological Function of the Qrc Complex
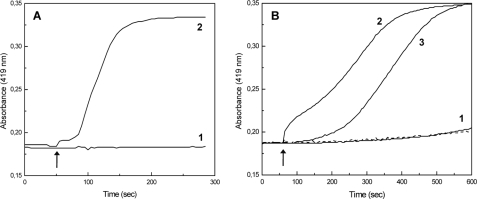
Given the similarity of Qrc to the ACIII complex, it was first tested whether it could have a similar function of oxidizing menaquinol and reducing a periplasmic protein (10). Initially, the reduction of Qrc with two menaquinol analogues (menadiol, 2,3-dimethyl-1,4-naphthoquinol) and with menaquinol-4 (vitamin K2) was tested in anaerobic conditions, but no reduction of the hemes c was observed even with large excess of the quinols. However, the opposite reaction i.e. oxidation of reduced Qrc complex by the menaquinones was extremely fast, suggesting that Qrc could function preferably with the reverse activity of ACIII by acting as a reducer of the menaquinone pool. This agrees with the fact that the redox potentials displayed by the hemes in D. vulgaris Qrc are significantly lower than those reported for R. marinus ACIII (+235 mV, 2 hemes; +80 mV, 1 heme; −45 mV, 2 hemes) (7). Furthermore, no complex in SRB has been positively identified to act as a menaquinone reductase, whose presence is essential when SRB grow on hydrogen, as this is oxidized by periplasmic hydrogenases that in many organisms, namely Desulfovibrio spp., do not contain a cytochrome b subunit for transferring electrons to the quinone pool but, rather, reduce the periplasmic TpIc3 (for review, see Refs. 27 and 28). A similar situation is observed for the periplasmic formate dehydrogenases. Thus, the Qrc complex could act to transfer electrons from the reduced TpIc3 to the menaquinone pool. To test this hypothesis we first checked whether the Qrc complex could be reduced by a periplasmic hydrogenase and formate dehydrogenase of D. vulgaris Hildenborough directly or via the TpIc3. With the [FeFe] hydrogenase, direct reduction of the Qrc hemes in the presence of H2 was negligible but increased dramatically when catalytic amounts of TpIc3 were present (Fig. 5A). A similar result was observed with the [NiFeSe] hydrogenase. With the FdhAB formate dehydrogenase and formate, direct reduction of Qrc was also quite slow, and a fast rate was observed in the presence of TpIc3 (Fig. 5B). Using the monohemic cytochrome c553 (a proposed electron acceptor of formate dehydrogenase in Desulfovibrio (16)) no catalytic effect was detected, and with the two cytochromes together the rate of reduction was equivalent to that obtained for the TpIc3 alone. Reduction of the Qrc FeS clusters by hydrogenase/TpIc3/H2 was also checked by EPR (Fig. 4C), which showed that all the hemes, the [3Fe-4S]1+/0 cluster, and at least two of the [4Fe-4S]2+/1+ clusters of the Qrc complex are reduced. These results show that reduction of the Qrc complex by the TpIc3 indeed occurs and that this complex can receive electrons from oxidation of either periplasmic H2 or formate.
FIGURE 5.
Reduction of Qrc with periplasmic proteins. A, reduction of Qrc with [FeFe] hydrogenase under H2 in the absence (1) or presence of TpIc3 (2) is shown. B, reduction of Qrc by FdhAB and formate in the absence of any cytochrome (1, dashed line) or in the presence of cytochrome c553 (1, solid line), TpIc3 (2), or both cytochromes (3) is shown. The arrow indicates the time of enzyme/cytochrome addition. Reduction was monitored by the increased absorption at 419 nm.
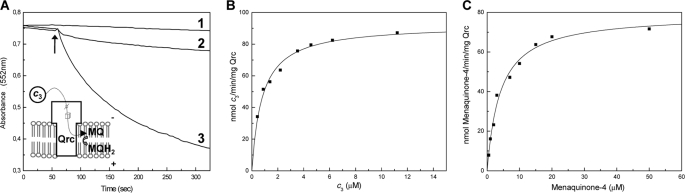
Based on these experiments, we next tested whether Qrc could catalyze the reduction of menaquinone from TpIc3. For this activity we used menaquinone-4 as substrate and followed the oxidation of reduced TpIc3 in strictly anaerobic conditions. The direct oxidation of reduced cytochrome c3 by menaquinone-4 was very slow, but the Qrc complex did indeed catalyze this reaction (Fig. 6A). A kinetic analysis of the TpIc3:menaquinone activity of Qrc showed that it followed Michaelis-Menten kinetics with a maximal activity of 92 nmol of TpIc3/min/mg (16.6 min−1, assuming a molecular mass of 180 kDa) and a Km of 0.8 μm for the cytochrome and 4.0 μm for menaquinone-4 (Fig. 6, B and C). Overall, these results confirm that the Qrc complex is a menaquinone reductase and provide the first functional characterization of this novel family.
FIGURE 6.
Catalytic activity of the Qrc complex. A, shown is oxidation of TpIc3 (7 μm) after the addition of menaquinone-4 (20 μm) (1), Qrc (2), or both (3). The arrow indicates the time of menaquinone-4 and/or Qrc addition. Reaction rates are shown of Qrc TpIc3:menaquinone-4 oxidoreductase activity by varying the cytochrome concentration with 20 μm menaquinone-4 (B) or the quinone concentration with 3.5 μm TpIc3 (C). Each point corresponds to the initial linear rate, and the solid line is the Michaelis-Menten-fitted curve obtained with nonlinear regression analysis. MQ, menaquinone; MQH2, menaquinol.
The Qrc Complex Quinone-binding Site
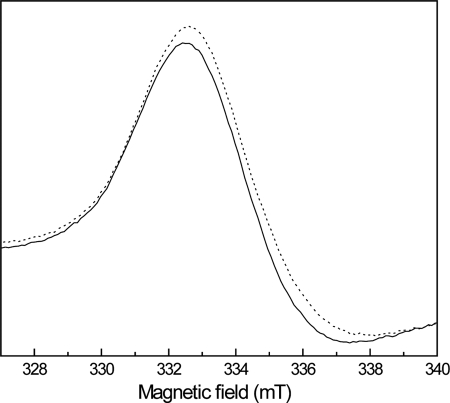
The location of the quinone-binding site relative to the membrane is a crucial aspect of quinone-interacting proteins, as the site of proton release or proton uptake upon quinone oxidation or reduction may be crucial to determine whether the reaction will be electrogenic or not (3, 4). Because there are no redox cofactors bound to the membrane QrcD subunit, it seems most plausible that the quinone-binding site of Qrc will be situated on the periplasmic side of the membrane, within suitable distance for electron transfer from the Qrc redox centers. In Escherichia coli fumarate reductase and in a mutant of DMSO reductase (where the [4Fe-4S] FS4 cluster has been turned into a [3Fe-4S] one), a close proximity between the quinone-binding site in the membrane subunit and the FS4 cluster has been observed through the effect of HQNO, a menasemiquinone analogue, on the EPR signal of the [3Fe-4S]+1 center (26, 29, 30). We performed a similar experiment and incubated the Qrc complex with HQNO before analysis by EPR. Relative to a control to which only ethanol was added, we observed no difference in the heme signals of the HQNO-treated sample, but an effect was detected in the g value of the [3Fe-4S]+1 signal, which shifted from 2.010 to 2.024 (Fig. 7). This effect is of the same order of magnitude as that observed for the mutated DMSO reductase (26, 29) and indicates that the menaquinone binding site of the Qrc complex is within close distance of the [3Fe-4S]+1 center of the QrcC subunit and, thus, located on the periplasmic side of the membrane.
FIGURE 7.
Effect of HQNO on the [3Fe-4S]1+ center of Qrc. EPR spectra of the Qrc [3Fe-4S]1+ center in the absence (solid line) and in the presence of 0.5 mm HQNO (dotted line) is shown. In a control sample to which only ethanol was added, no effect was observed. Spectra were recorded under the following conditions: microwave frequency, 9.38 GHz; microwave power, 20.1 milliwatts; modulation frequency, 100 kHz; modulation amplitude, 1 millitesla; temperature, 10 K.
DISCUSSION
We report here the isolation and detailed characterization of a new membrane complex from D. vulgaris that constitutes the first example of a novel family of BRC belonging to the superfamily of CISM proteins (6). This complex, named QrcABCD, is a major component among membrane proteins in D. vulgaris. The QrcABCD complex is composed of four subunits: a multiheme cytochrome c and three subunits related to the subunits of CISM family, including a protein related to the molybdopterin-containing subunit (but which has no molybdenum cofactor), an electron-transfer four cluster protein, and an integral membrane protein. These subunits show highest similarity to some of the subunits of ACIII, a recently described family of BRC (8–10). ACIII is widespread in bacteria that lack a bc1 complex, but only two examples have been characterized so far, from R. marinus (7, 9) and C. aurantiacus (8, 10). Besides the different composition, there are several factors which indicate that the Qrc complex should have a different physiological function from the ACIII complex; (i) in most organisms the ACIII genes are found next to genes coding for heme copper cytochrome oxidases (that will oxidize the cytochrome c or another periplasmic protein, reduced by ACIII), whereas this is never observed for Qrc genes; (ii) several organisms (e.g. Desulfohalobium retbaense or Syntrophobacter fumaroxidans) have a Qrc complex but lack genes coding for a cytochrome oxidase; (iii) at least one organism, Desulfobacterium autotrophicum HRM2, has genes coding for both Qrc and ACIII complexes, with only the latter being next to genes coding for an oxidase. In addition, sequence analysis clearly indicates that the Qrc proteins form a separate group from the related ACIII proteins. Yanyushin et al. (8) proposed that the ACIII complex arose from Qrc by the acquisition of additional subunits. In this respect it is interesting to note that in the ACIII complex of the Deltaproteobacteria Geobacter spp., the molybdopterin-like subunit, and the FeS subunit are not fused as in other ACIII but are encoded by separate genes as in Qrc, supporting the evolutionary link between the two complexes. The Qrc complex may in turn have evolved from a CISM enzyme by acquisition of a cytochrome c subunit and loss of the molybdopterin cofactor. It is, thus, a most interesting evolutionary cross-point in the CISM family in which one function has been lost while another is evolving.
Our results show that the Qrc complex is a cytochrome c3:menaquinone oxidoreductase, whereas ACIII oxidizes menaquinol. These contrasting functions agree with the lower redox potentials observed for the hemes of QrcA relative to R. marinus ActA (7), as in Qrc the electrons are transferred from the low-redox potential hemes of TpIc3 (−325 to −170 mV) to menaquinone (−70 mV), whereas in ACIII they go from menaquinol to a high potential cytochrome c. This study provides the first spectroscopic characterization of the redox centers from a Qrc complex, which is an important contribution as only a few CISM proteins have been studied in detail (6) and only partial characterization of the redox centers is available for one ACIII (7, 9). We obtained evidence for the presence of six hemes c, one [3Fe-4S]1+/0, and three [4Fe-4S]2+/1+, but no molybdopterin cofactor, in the D. vulgaris Qrc complex. Based on the crystal structures of the CISM enzymes Fdh-N, Nar, and Psr (18–20), it is likely that the cubane clusters of QrcC form a wire that electrically links the hemes c of QrcA to a menaquinone-binding site in QrcD situated close to the [3Fe-4S]1+/0 center. In the absence of a catalytic site, the function of the large QrcB subunit is unknown and may be to provide a structural scaffold for the other subunits.
Sulfate respiration is an ancient mode of energy metabolism (31) that is ubiquitous in anaerobic habitats and has very important environmental and economical consequences (12). There is still a major gap in our understanding of bioenergetics in SRB, as the sites of energy conservation have not been clearly established even though a quinone-based chemiosmotic mechanism is likely present (13, 32). Most SRB lack important respiratory complexes such as complex I or the bc1 complex, but two unique BRCs are conserved, QmoABC (33) and DsrMKJOP (15). These are possible sites of energy conservation, although this could not yet be experimentally verified. The subunit composition of Qmo indicates that it transfers electrons from the quinone pool to the cytoplasm, whereas this is less clear for the Dsr complex, which is probably involved in direct transmembrane electron transfer. One of the major energy sources for SRB in their natural habitats is hydrogen, which is oxidized by periplasmic hydrogenases that are very abundant in SRB (27). In most bacteria the periplasmic uptake hydrogenases include a membrane cytochrome b subunit that transfers electrons directly to the quinone pool. Strikingly, most SRB are very unusual in that their periplasmic hydrogenases, and formate dehydrogenases lack the cytochrome b subunit and instead transfer electrons to a pool of soluble cytochromes c, of which the characteristic TpIc3 is the most abundant (28, 35). Thus, a membrane protein is required to transfer electrons from TpIc3 to the quinone pool or directly to the cytoplasm for reduction of sulfate. This last function may be performed by the Hmc (36) and Tmc complexes (17), which are reduced by the TpIc3 (37, 38), in contrast to Dsr. No complex had yet been described in SRB that could reduce the quinone pool. We show here that the Qrc complex is a TpIc3:menaquinone oxidoreductase and is efficiently reduced by either hydrogenases or formate dehydrogenase through this cytochrome. All organisms that contain the Qrc complex are deltaproteobacterial SRB (with a single exception), indicating that its function is specifically associated with sulfate respiration. Analysis of available SRB genome sequences show that the presence of qrc genes correlates with the presence of periplasmic hydrogenases and formate dehydrogenases lacking membrane subunits for quinone interaction (with a single exception of a Fdh in Desulfatibacillum alkenivorans). Conversely, in the few deltaproteobacterial SRB that lack the Qrc complex, either the hydrogenases and formate dehydrogenases include membrane subunits for quinone reduction (e.g. Desulfotalea psychrophila or D. desulfuricans ATCC27774), or a complex that can perform a similar function is present, such as the nine-heme cytochrome complex (Nhc) (13) (e.g. Desulfovibrio piger).
A recent study in D. desulfuricans G20 provided important evidence to support this physiological function of the Qrc complex (14). In this study a mutant library of D. desulfuricans G20 was screened to identify genes involved in H2 metabolism based on a deficiency to grow syntrophically with Methanospirillum hungatei on lactate. Three mutants were identified, with transposon insertions in genes coding for the [FeFe] hydrogenase small subunit (hydB, Dde_0082), TpIc3 (cycA, Dde_3182), and gene Dde_2933, which is annotated as a putative molybdopterin oxidoreductase molybdopterin-binding subunit (and, thus, named mopB), and which corresponds to qrcB. The mopB/qrcB mutant is unable to grow with H2 or formate as electron donors but grows normally with lactate. The three mutants also accumulate higher levels of hydrogen and formate during growth on lactate/sulfate. These results further support the presence of a metabolic pathway involving periplasmic hydrogenases or formate dehydrogenases, TpIc3, and the Qrc complex and show an essential role of the Qrc complex during growth on H2 or formate. In several transcriptome studies (39–42) the expression of the qrc genes follows the same behavior as the genes for TpIc3, [FeFe] hydrogenase, and genes related to sulfate respiration (dsrAB, aprAB, qmoABC, dsrMKJOP and genes for ATP synthase), suggesting a co-regulation of the qrc and sulfate reduction genes. A similar effect was observed in a proteomic study (43).
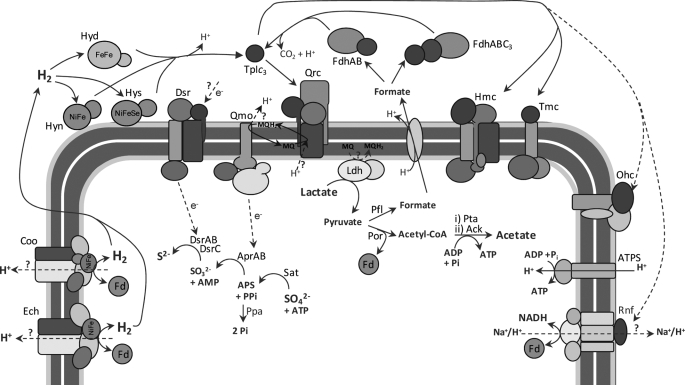
A model for the electron transfer chain of sulfate respiration in D. vulgaris is shown in Fig. 8. Periplasmic oxidation of hydrogen or formate during growth on these substrates or when they are formed as intermediaries in the metabolism of carbon compounds, such as lactate, results in reduction of the TpIc3. This transfers electrons to the Qrc complex, which reduces menaquinone at the periplasmic side of the membrane close to the QrcC [3Fe-4S]+1/0 center. If the protons required for quinone reduction are taken up from the periplasm, then no energy conservation will occur during this process. However, based on what has been proposed for Psr (20), which includes a membrane subunit of the same family as QrcD, it is possible that there are channels in QrcD for proton uptake from the cytoplasm, in which case quinone reduction by the Qrc complex would be electrogenic. Further experiments have to be carried out to elucidate this point. The reduced menaquinone pool will be oxidized by the Qmo complex, which should transfer electrons to the cytoplasmic adenosine phosphosulfate reductase. It is most likely that oxidation of menaquinol by Qmo occurs at the periplasmic side of the membrane, releasing protons to the periplasm, as this complex includes two hemes b in its membrane subunit with suitable redox potentials to perform transmembrane electron transfer for the cytoplasmic subunits (33). Thus, the Qrc and Qmo complexes likely constitute the two arms of an energy-conserving redox loop (3, 4), contributing to proton motive force generation during sulfate reduction. But why do most SRB prefer to have a pathway involving the soluble TpIc3 and the Qrc complex rather than using the more usual mechanism of direct quinone reduction through a membrane subunit of hydrogenases and formate dehydrogenases? One possible explanation is that the involvement of TpIc3 increases the metabolic flexibility of the organism, as electrons may be shuttled through several alternative pathways, as indicated by the presence of other membrane redox complexes that are also associated with a cytochrome c subunit such as Tmc, Hmc, Ohc, and Rnf (28), of which Hmc and Tmc have already been shown to act as electrons acceptors of TpIc3 (17, 37, 38). Indeed, sulfate reducers such as D. psychrophila or Archaeoglobus fulgidus that lack the TpIc3 also do not contain these various complexes and probably have a less plastic metabolism. Both Tmc and Hmc have a cytoplasmic subunit that is homologous to DsrK and so may be part of alternative pathways for sulfite reduction involving DsrAB (34). The Rnf complex present in several other organisms may provide a link to the ferredoxin and NAD(H)/NADP(H) pools (44).
FIGURE 8.
Model for the respiratory chain of D. vulgaris during growth on hydrogen, or lactate, and sulfate. The model is based on previous observations (for review, see Refs. 13 and 28) and the present results as discussed in the text under (“Discussion”). Ldh, lactate dehydrogenase; Por, pyruvate:ferredoxin oxidoreductase; Pta, phosphate acetyltransferase; Ack, acetate kinase; Fd, ferredoxin; Ech and Coo, membrane-bound cytoplasmic-facing hydrogenases; Hyn1, [NiFe] hydrogenase isozyme 1; Hyd, [FeFe] hydrogenase; Hys, Pfl, pyruvate-formate lyase; Sat, sulfate adenylyl transferase; APS, adenosine phoshosulfate[NiFeSe] hydrogenase; Ohc, octaheme cytochrome complex; ATPS, ATP synthase; MQ, menaquinone; MQH2, menaquinol.
In conclusion, Qrc is the first BRC in the respiratory chain of SRB to have its physiological function clearly elucidated. It is related to bacterial CISM but together with ACIII form a group that does not bind molybdenum and performs electron transfer. This complex is a novel addition to the family of characterized bacterial redox complexes, typically constituted by modular subunits. It provides a striking example of how a totally different physiological function can be achieved with a minimal modification of subunits, a strategy that forms the basis for the diversity and flexibility of bacterial energy metabolism.
Supplementary Material
Acknowledgments
We thank Manuela M. Pereira for helpful discussions and critical reading of the manuscript. We also acknowledge João Carita for cell growth, Manuela Regalla for N-terminal and HPLC analysis, Ana Coelho for mass spectrometry data at the Mass Spectrometry Laboratory, Sofia Silva for formate dehydrogenases, Marta Marques for [NiFeSe] hydrogenase, Isabel Pacheco for TpIc3, Patrícia Refojo for menadiol, and Filipa Sousa for 2,3-dimethyl-1,4-naphthoquinone.
This work was supported by Research Grant PTDC/QUI/68368/2006 funded by Fundação para a Ciência e Tecnologia (FCT, MCTES, Portugal) and FEDER program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1.
- BRC
- bacterial respiratory complex
- ACIII
- alternative complex III
- BN-PAGE
- blue native PAGE
- CISM
- complex iron-sulfur molybdoenzyme
- Dsr
- dissimilatory sulfite reductase complex
- Fdh
- formate dehydrogenase
- FS
- FeS cluster
- Hmc
- high molecular mass cytochrome complex
- HQNO
- 2-heptyl-4-hydroxyquinoline-N-oxide
- Nar
- nitrate reductase
- Qmo
- quinone-interacting membrane oxidoreductase complex
- Qrc
- quinone reductase complex
- SRB
- sulfate-reducing bacteria
- Tmc
- tetraheme cytochrome complex
- TpIc3
- type I cytochrome c3
- Psr
- polysulfide reductase
- HPLC
- high performance liquid chromatography
- Rnf
- putative NAD(H):ferredoxin oxidoreductase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Richardson D. J. (2000) Microbiology 146, 551–571 [DOI] [PubMed] [Google Scholar]
- 2.Falkowski P. G., Fenchel T., Delong E. F. (2008) Science 320, 1034–1039 [DOI] [PubMed] [Google Scholar]
- 3.Jormakka M., Byrne B., Iwata S. (2003) FEBS Lett. 545, 25–30 [DOI] [PubMed] [Google Scholar]
- 4.Simon J., van Spanning R. J., Richardson D. J. (2008) Biochim. Biophys. Acta 1777, 1480–1490 [DOI] [PubMed] [Google Scholar]
- 5.Baymann F., Lebrun E., Brugna M., Schoepp-Cothenet B., Giudici-Orticoni M. T., Nitschke W. (2003) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 358, 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothery R. A., Workun G. J., Weiner J. H. (2008) Biochim. Biophys. Acta 1778, 1897–1929 [DOI] [PubMed] [Google Scholar]
- 7.Pereira M. M., Carita J. N., Teixeira M. (1999) Biochemistry 38, 1268–1275 [DOI] [PubMed] [Google Scholar]
- 8.Yanyushin M. F., del Rosario M. C., Brune D. C., Blankenship R. E. (2005) Biochemistry 44, 10037–10045 [DOI] [PubMed] [Google Scholar]
- 9.Pereira M. M., Refojo P. N., Hreggvidsson G. O., Hjorleifsdottir S., Teixeira M. (2007) FEBS Lett. 581, 4831–4835 [DOI] [PubMed] [Google Scholar]
- 10.Gao X., Xin Y., Blankenship R. E. (2009) FEBS Lett. 583, 3275–3279 [DOI] [PubMed] [Google Scholar]
- 11.Refojo P. N., Teixeira M., Pereira M. M. (2010) Biochim Biophys Acta, in press [DOI] [PubMed] [Google Scholar]
- 12.Muyzer G., Stams A. J. (2008) Nat. Rev. Microbiol. 6, 441–454 [DOI] [PubMed] [Google Scholar]
- 13.Pereira I. A. C. (2008) in Microbial Sulfur Metabolism (Friedrich C., Dahl C. eds) pp. 24–35, Springer-Verlag New York Inc., New York [Google Scholar]
- 14.Li X., Luo Q., Wofford N. Q., Keller K. L., McInerney M. J., Wall J. D., Krumholz L. R. (2009) J. Bacteriol. 191, 2675–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pires R. H., Venceslau S. S., Morais F., Teixeira M., Xavier A. V., Pereira I. A. C. (2006) Biochemistry 45, 249–262 [DOI] [PubMed] [Google Scholar]
- 16.Sebban C., Blanchard L., Bruschi M., Guerlesquin F. (1995) FEMS Microbiol. Lett. 133, 143–149 [DOI] [PubMed] [Google Scholar]
- 17.Pereira P. M., Teixeira M., Xavier A. V., Louro R. O., Pereira I. A. C. (2006) Biochemistry 45, 10359–10367 [DOI] [PubMed] [Google Scholar]
- 18.Jormakka M., Törnroth S., Byrne B., Iwata S. (2002) Science 295, 1863–1868 [DOI] [PubMed] [Google Scholar]
- 19.Bertero M. G., Rothery R. A., Palak M., Hou C., Lim D., Blasco F., Weiner J. H., Strynadka N. C. (2003) Nat. Struct. Biol. 10, 681–687 [DOI] [PubMed] [Google Scholar]
- 20.Jormakka M., Yokoyama K., Yano T., Tamakoshi M., Akimoto S., Shimamura T., Curmi P., Iwata S. (2008) Nat. Struct. Mol. Biol. 15, 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon J., Kern M. (2008) Biochem. Soc. Trans. 36, 1011–1016 [DOI] [PubMed] [Google Scholar]
- 22.Sazanov L. A., Hinchliffe P. (2006) Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 23.Guigliarelli B., Asso M., More C., Augier V., Blasco F., Pommier J., Giordano G., Bertrand P. (1992) Eur. J. Biochem. 207, 61–68 [DOI] [PubMed] [Google Scholar]
- 24.Rothery R. A., Magalon A., Giordano G., Guigliarelli B., Blasco F., Weiner J. H. (1998) J. Biol. Chem. 273, 7462–7469 [DOI] [PubMed] [Google Scholar]
- 25.Manodori A., Cecchini G., Schröder I., Gunsalus R. P., Werth M. T., Johnson M. K. (1992) Biochemistry 31, 2703–2712 [DOI] [PubMed] [Google Scholar]
- 26.Rothery R. A., Weiner J. H. (1996) Biochemistry 35, 3247–3257 [DOI] [PubMed] [Google Scholar]
- 27.Matias P. M., Pereira I. A. C., Soares C. M., Carrondo M. A. (2005) Prog. Biophys. Mol. Biol. 89, 292–32915950057 [Google Scholar]
- 28.Pereira I. A. C., Haveman S. A., Voordouw G. (2007) in Sulphate-Reducing Bacteria: Environmental and Engineered Systems (Barton L. L., Allan Hamilton W. A. eds) pp. 215–240, Cambridge University Press, Cambridge, UK [Google Scholar]
- 29.Cheng V. W., Rothery R. A., Bertero M. G., Strynadka N. C., Weiner J. H. (2005) Biochemistry 44, 8068–8077 [DOI] [PubMed] [Google Scholar]
- 30.Rothery R. A., Seime A. M., Spiers A. M., Maklashina E., Schröder I., Gunsalus R. P., Cecchini G., Weiner J. H. (2005) FEBS J. 272, 313–326 [DOI] [PubMed] [Google Scholar]
- 31.Shen Y., Buick R., Canfield D. E. (2001) Nature 410, 77–81 [DOI] [PubMed] [Google Scholar]
- 32.Rabus R., Hansen T., Widdel F. (2006) in The Prokaryotes (Dworkin M. E. A., eds) 3rd Ed., pp 659–768, Springer-Verlag New York Inc., New York [Google Scholar]
- 33.Pires R. H., Lourenço A. I., Morais F., Teixeira M., Xavier A. V., Saraiva L. M., Pereira I. A. C. (2003) Biochim. Biophys. Acta 1605, 67–82 [DOI] [PubMed] [Google Scholar]
- 34.Oliveira T. F., Vonrhein C., Matias P. M., Venceslau S. S., Pereira I. A., Archer M. (2008) J. Biol. Chem. 283, 34141–34149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heidelberg J. F., Seshadri R., Haveman S. A., Hemme C. L., Paulsen I. T., Kolonay J. F., Eisen J. A., Ward N., Methe B., Brinkac L. M., Daugherty S. C., Deboy R. T., Dodson R. J., Durkin A. S., Madupu R., Nelson W. C., Sullivan S. A., Fouts D., Haft D. H., Selengut J., Peterson J. D., Davidsen T. M., Zafar N., Zhou L., Radune D., Dimitrov G., Hance M., Tran K., Khouri H., Gill J., Utterback T. R., Feldblyum T. V., Wall J. D., Voordouw G., Fraser C. M. (2004) Nat. Biotechnol. 22, 554–559 [DOI] [PubMed] [Google Scholar]
- 36.Rossi M., Pollock W. B., Reij M. W., Keon R. G., Fu R., Voordouw G. (1993) J. Bacteriol. 175, 4699–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira I. A. C., Romão C. V., Xavier A. V., LeGall J., Teixeira M. (1998) J. Biol. Inorg. Chem. 3, 494–498 [Google Scholar]
- 38.Valente F. M., Saraiva L. M., LeGall J., Xavier A. V., Teixeira M., Pereira I. A. C. (2001) ChemBioChem 2, 895–905 [DOI] [PubMed] [Google Scholar]
- 39.Haveman S. A., Brunelle V., Voordouw J. K., Voordouw G., Heidelberg J. F., Rabus R. (2003) J. Bacteriol. 185, 4345–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haveman S. A., Greene E. A., Stilwell C. P., Voordouw J. K., Voordouw G. (2004) J. Bacteriol. 186, 7944–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolyar S., He Q., Joachimiak M. P., He Z., Yang Z. K., Borglin S. E., Joyner D. C., Huang K., Alm E., Hazen T. C., Zhou J., Wall J. D., Arkin A. P., Stahl D. A. (2007) J. Bacteriol. 189, 8944–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira P. M., He Q., Valente F. M., Xavier A. V., Zhou J., Pereira I. A. C., Louro R. O. (2008) Antonie Van Leeuwenhoek 93, 347–362 [DOI] [PubMed] [Google Scholar]
- 43.Zhang W., Gritsenko M. A., Moore R. J., Culley D. E., Nie L., Petritis K., Strittmatter E. F., Camp D. G., 2nd, Smith R. D., Brockman F. J. (2006) Proteomics 6, 4286–4299 [DOI] [PubMed] [Google Scholar]
- 44.McInerney M. J., Rohlin L., Mouttaki H., Kim U., Krupp R. S., Rios-Hernandez L., Sieber J., Struchtemeyer C. G., Bhattacharyya A., Campbell J. W., Gunsalus R. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7600–7605 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.