Abstract
The cohesin complex plays a central role in genome maintenance by regulation of chromosome segregation in mitosis and DNA damage response (DDR) in other phases of the cell cycle. The ATM/ATR phosphorylates SMC1 and SMC3, two core components of the cohesin complex to regulate checkpoint signaling and DNA repair. In this report, we show that the genome-wide binding of SMC1 and SMC3 after ionizing radiation (IR) is enhanced by reinforcing pre-existing cohesin binding sites in human cancer cells. We demonstrate that ATM and SMC3 phosphorylation at Ser1083 regulate this process. We also demonstrate that acetylation of SMC3 at Lys105 and Lys106 is induced by IR and this induction depends on the acetyltransferase ESCO1 as well as the ATM/ATR kinases. Consistently, both ESCO1 and SMC3 acetylation are required for intra-S phase checkpoint and cellular survival after IR. Although both IR-induced acetylation and phosphorylation of SMC3 are under the control of ATM/ATR, the two forms of modification are independent of each other and both are required to promote reinforcement of SMC3 binding to cohesin sites. Thus, SMC3 modifications is a mechanism for genome-wide reinforcement of cohesin binding in response to DNA damage response in human cells and enhanced cohesion is a downstream event of DDR.
Keywords: Checkpoint Control, Chromatin Immunoprecipitation (ChiP), DNA Damage, DNA Repair, Post-translational Modification, SMC3 Acetylation, Sister Chromatid Cohesion
Introduction
Sister chromatid cohesion is a fundamental biological process that the sister chromatids once generated in S-phase are always paired and linked to ensure equal distribution to daughter cells until mitosis. Cohesion plays a central role in genome stability by facilitating spindle bi-orientation, faithful chromosome segregation, homologous recombination, checkpoint activation, and transcriptional regulation (1–3). This process is mediated by a protein complex called cohesin that consists of four core subunits SMC1, SMC3, RAD21 (also known as SCC1 and MCD1), and SA2/SA1 (also known as SCC3, or STAG2 and STAG1) and several accessory factors (4).
To maintain the fidelity of the genome, cells evolve with an elaborate mechanism to deal with DNA damage. DNA damage response (DDR)3 is a signal transduction pathway that coordinates cell cycle arrest, DNA repair, and programmed cell death in the presence of damaged DNA (5, 6). It is regulated by the ATM/ATR, master regulators of the DDR process through phosphorylation of their substrates (6–10). Disruption of DDR results in genomic instability and is the cause for many cancer-prone disorders (11). Cohesin is an important effector in the DNA damage response. SMC1 and SMC3 are phosphorylated in human and mouse cells in an ATM/ATR-dependent manner and these phosphorylation events are required for intra-S checkpoint, cellular survival in response to IR (12–15). Cohesin is recruited to sites of DNA damage for efficient DNA repair (4, 12, 16, 17). However, the molecular mechanism by which cohesin phosphorylation regulates DDR is still unknown.
Acetylation is another post-translational modification mechanism for the regulation of cohesin functions. It has been long recognized that Eco1 (establishment of cohesion 1, also known as Ctf7) is essential for yeast cell viability and sister chromatid cohesion (18, 19). It was also recognized that Eco1 is an acetyltransferase, but how Eco1 regulates cohesion was not understood until recently, when we and others identified Smc3 as the first substrate of Eco1 (20–22). Eco1 and its human ortholog ESCO1 acetylate yeast Smc3 and human SMC3 at two conserved lysine residues (Lys112 and Lys113 in yeast, and Lys105 and Lys106 in human). Mutation of these lysine residues to a non-acetylatable form leads to increased loss of sister chromatid cohesion and genome instability, suggesting that SMC3 acetylation is essential for S-phase sister chromatid cohesion.
Although generation of cohesion is limited to S phase in undamaged cells, double strand DNA break can trigger genome-wide re-establishment of cohesion in the G2/M phase, a process that depends on both the acetyltransferase Eco1 and the DNA damage checkpoint (23, 24). It was suggested that Chk1 phosphorylates Scc1 in response to DDR, which permits Eco1 to acetylate Scc1 instead of Smc3 to re-establish sister chromatid cohesion (25).
The fundamental biological process of sister chromatid cohesin is conserved from yeast to man, but the detailed mechanism and regulation may be different. In yeast, cohesin binds to chromosome DNA in a sequence independent manner, and is often found at intergenic regions where two RNA polymerase II-transcribed genes converge (26, 27), suggesting that cohesin may translocate along the chromosome; in human and mouse cells, it was recently found that cohesin binds to specific genomic loci that in most cases coincide with the CCCTC-binding factor (CTCF) binding sites (28–31). Because CTCF is a transcriptional insulator, binding of cohesin to CTCF sites suggests that cohesin may also contribute to shaping the insulator boundaries that are formed between long range regulatory elements, thus expanding the role of cohesin in mammalian cells beyond pairing of sister chromatids (32, 33).
Here, we report genome-wide reinforcement of cohesin binding at pre-existing sites in response to ionizing radiation in human cells. Like in yeast, this process is regulated by both ATM and ESCO1, but unlike in yeast, SMC3 are targeted for phosphorylation and acetylation. Moreover, phosphorylation and acetylation appear to be in two independent pathways that both regulate reinforcement of cohesin binding. Overall, our data suggest that enhanced cohesin binding in response to DSB is an important downstream event of the DNA damage checkpoint that is conserved between yeast and human, and that targeting different cohesin subunits may reflect functional divergence of the two species.
EXPERIMENTAL PROCEDURES
Antibodies, siRNA, and Generation of Stable Cell Lines
Rabbit polyclonal antibodies against SMC1, SMC1-pS966, SMC3, and SMC3-pS1083 were purchased from Bethyl Laboratories. Mouse monoclonal anti-FLAG M2 antibody was from Sigma. Antibodies against SMC3 AcK105, AcK106, and AcK105/AcK106, and the ESCO1 siRNAs were described previously (22). siRNA transfection was done using Oligofectamine (Invitrogen) according to the manufacturer's instructions.
SMC3-WT, SMC3-AA, and SMC3-S1083A expressing cell lines were generated using “Flp-InTM T-RExTM” system (Invitrogen) in 293T cells as described previously (22). Exogenous SMC3 expression was induced by doxycycline at a concentration of 1 μg/ml.
Stable Isotope Labeling of Cultured Cells (SILAC) Quantification
SILAC quantification was carried out as previously described (15). Cells are grown in a pair of light and heavy SILAC media (Invitrogen) for at least 6 generations. Light and heavy cells are mixed after treatments and prior to cohesin purification. Analyte proteins are resolved on SDS-PAGE and trypsin was digested in gel. A capillary high pressure liquid chromatography (75 μm inner diameter column)-electrospray linear ion trap mass spectrometer (Thermo Fisher) was used for data acquisition. Relative quantification was performed using selective ion monitoring.
Selective Ion Monitoring for Quantitative Mass Spectrometry
The m/z of each pair of expected light and heavy peptide is pre-set in the data acquisition method for MS/MS spectrum acquisition. During data acquisition, the mass spectrometer repetitively acquires MS/MS data with a 3-unit wide window. To quantify each pair of light and heavy peptides, a pair of fragment peaks from the MS/MS spectrum was selected and plotted to extract chromatography. The area of the chromatographic peak of each fragment was calculated using the ICIS peak algorithm in the Qual Browser (version 1.3). The ratio was calculated by dividing the peak area of the fragment peak of the light peptide by that of the heavy counterpart.
Radioresistant DNA Synthesis (RDS) Assay
FRT stable cell lines expressing WT, SMC3-AA, and SMC3-QQ were induced by doxycycline and labeled with 10 nCi/ml of [14C]thymidine (PerkinElmer Life Sciences) for 72 h. Cells were irradiated with 10 Gy of IR and recovered for 1 h and then pulse-labeled with 1 μCi/ml of [3H]thymidine (PerkinElmer Life Sciences) for 30 min. They were then washed with phosphate-buffered saline, fixed with ethanol overnight at −20 °C, and lysed with 0.5 m NaOH. The lysates were counted in a liquid scintillation counter. Radioresistant DNA synthesis was calculated using the ratio of radioactivity of 3H/14C. Overlapping 3H and 14C emissions were corrected with quenched 3H and 14C standard. The RDS checkpoint assay was carried out 72 h after siRNA transfection in siRNA knockdown cells.
Colony Formation Assay for IR Sensitivity
Colony formation was used to determine the IR sensitivity in different SMC3 cell lines and cells depleted of ESCO1. Cells were plated at different densities according to the IR dosage to be used. After the cells were irradiated, they were grown for 1 week to allow colony formation. Cell colonies were then fixed with methanol and stained with Giemsa. The fraction of cell survival was calculated by colony number of treated divided by that of un-treated cells. Three plates of cells were used for each genotype type. All experiments above were repeated at least three times.
Chromatin Immunoprecipitation (ChIP) and Quantitative Real Time PCR
Chromatin immunoprecipitation was performed according to the protocol from Upstate Biotechnology with minor modifications. 107 HeLa cells were used for each reaction. For non-extracted chromatin, cells were treated with 1% formaldehyde for cross-linking and harvested. After sonication, 1% of soluble chromatin fraction was decross-linked by heating at 65 °C overnight and used as an input. The rest of the chromatin fraction was immunoprecipitated with SMC3 antibody and decross-linked by heat. For NETN-extracted chromatin, cells were harvested and lysed with NETN. Chromatin fraction was collected by centrifugation and cross-linked by formaldehyde. DNA was purified with the QiaQuick PCR purification kit (Qiagen) and analyzed by quantitative real time PCR (qPCR).
Quantitative real time PCR was performed on StepOnePlusTM sequence detection system (Applied Biosystems) using SYBR Green master mix (Applied Biosystems). Primers for cohesin sites were designed using Primer Express (Applied Biosystems) and non-cohesin sites were selected from those previously described for qPCR (28).
ChIP-seq Data Analysis
The ChIP-sequencing data were processed using MACS 1.3.5 (34). Non-unique reads and monoclonal mappings were removed. The peaks were called with MACS p values <1 e−8 and fold-change greater than 40. The comparisons between peaks of different samples were performed using custom written python scripts.
RESULTS
Genome-wide Reinforcement of Cohesin Binding at Original Cohesion Sites in Response to IR
Because double strand DNA break can trigger genome-wide re-establishment of cohesion in the G2/M phase in yeast, we asked whether new cohesin binding sites are generated in mammalian cells in response to DSB. To measure genome-wide cohesin binding sites, we carried out ChIP with an antibody against SMC3 from cycling and IR-treated HeLa cells and mapped cohesin binding sites by Solexa sequencing (ChIP-seq) (Fig. 1A). We performed statistic analysis of the sequencing data using the MACman algorithm with parameters of p value <10−8, and >40-fold enrichment as compared with the IgG control to call peaks. We identified more than 7,500 SMC3 common binding sites, and 27 (0.3%) and 32 (0.4%) peaks as unique binding sites to cycling and IR-treated samples, respectively (Fig. 1B). As previously reported, most cohesin binding sites (5,750 peaks, 76.4%) overlap with CTCF binding sites (28–31). Previous work suggests that cohesin may exist in two pools, one is loosely bound to chromatin and the other is tightly bound (35). To investigate whether IR differentially affects the tightly bound pool, we first extracted loosely bound cohesin with NETN and then cross-linked the tightly bound cohesin to chromatin, and analyzed this pool of SMC3 with ChIP-seq. Although the total number of cohesin binding sites detected was reduced, the vast majority of the sites (>2600) were indistinguishable before and after IR, and 0.3 and 0.9% of the peaks were unique to cycling and IR-treated samples, respectively (Fig. 1B). Visual inspection of the unique peaks revealed that they were weak and likely resulted from artifacts of peak calling. From these data we conclude that no new cohesion sites are generated in response to IR.
FIGURE 1.
Genome-wide reinforcement of cohesin binding at original cohesion sites in response to IR. A, cohesin ChIP-Seq enrichment profile for chromosome 2 (top) and an enlarged view of part of chromosome 2 (bottom). ChIP was carried out with anti-SMC3 antibody and anti-pS1083 SMC3 antibody from cycling and IR-treated HeLa cells and mapped cohesin binding sites by ChIP-Seq. B, Venn diagram showing the number of cohesin sites before and after IR treatment. Venn diagram of cohesin sites from non-extracted chromatin (top) and NETN-extracted chromatin (bottom). C, quantitative ChIP assay at two cohesin sites (CBS13 and CBS16) and a non-cohesin site (#32). HeLa cells were harvested 2 h after 10 Gy of IR and analyzed by ChIP-qPCR. Non-irradiated HeLa cells were used as a control and ChIP was carried out with the indicated antibodies. n = 4. Error bars indicate S.D. D, a plot of SMC3 binding in non-irradiated HeLa cells versus HeLa cells exposed to 1.7 Gy IR. The slope and R2 value of linear regression analysis are indicated. Statistically significant differences are marked with the asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
If no new cohesion sites are generated, an alternative mechanism to enforce the cohesion is to strengthen binding at the existing sites. To quantitatively measure cohesin binding, we selected 27 cohesin binding sites from our ChIP-Seq data and 1 cohesin site and 6 non-cohesin sites from a previous study (28). Using specific primers that were validated for sequence specificity, amplification linearity, and efficiency, we quantified the percentage of DNA brought down by the anti-SMC1 or SMC3 ChIP with or without IR treatment. We found that cohesin was specifically localized and highly enriched at all selected cohesion sites (supplemental Fig. S1A and Table S1). Importantly, cohesin binding was increased at all existing cohesion sites (Fig. 1C) and peaked at 2 h after IR (supplemental Fig. S1B), and binding after IR demonstrated excellent linearity to the original binding before IR (Fig. 1D); about a 25% increase of binding was observed with 1.7 Gy of IR at all cohesion sites examined, whereas cohesin binding at non-cohesin sites was not increased at all (Fig. 1D). This suggests that cohesion may be potentiated after IR by enforcing all cohesion sites with a small increase, but not by a large increase at the selected strong binding sites.
Phosphorylated SMC1/3 Binds Cohesion Sites in an ATM-dependent Manner
One critical regulatory event for cohesin in response to IR is the phosphorylation of SMC1 and SMC3 by ATM (12–15). To explore whether phosphorylated cohesin binds to specific chromatin locations, we mapped the localization of pS1083-SMC3 by ChIP-Seq. We treated the HeLa cells with 10 Gy of IR and then performed cross-linking after 2 h, and ChIPed pS1083-SMC3 (Fig. 1A). Although each peak in pS1083-SMC3 ChIP-seq appears to be weaker than that in SMC3, all pS1083-SMC3 peaks overlapped with SMC3 peaks. The weaker signal is likely caused by the lower abundance of phosphorylated SMC3 than total SMC3. Thus, phosphorylated SMC3 at Ser1083 is exclusively detected at cohesion sites after IR treatment.
We validated phospho-SMC1/SMC3 binding at cohesion sites CBS13 and CBS16, and non-cohesion site 32 (Fig. 2A) in IR-treated and untreated cells by ChIP-qPCR. We then chose 16 cohesion sites and 3 non-cohesin sites for further detailed analysis of pS1083-SMC3 binding in response to 10 Gy of IR. Phosphorylated SMC3 was highly enriched at the 16 cohesion sites as compared with the non-cohesion sites (supplemental Fig. S2A and Table S2). As shown in Fig. 2B, pSMC3 at cohesin sites is roughly proportional to the SMC3 amount in non-irradiated cells. Thus, the amount of phosphorylated SMC3 at the particular cohesion site is determined by the amount of SMC3 bound before IR.
FIGURE 2.
Phosphorylated SMC3 at Ser1083 binds cohesion sites and ATM regulates this process. A, quantitative ChIP assay for pS966-SMC1 and pS1083-SMC3 at two cohesin sites (CBS13 and CBS16) and a non-cohesin site (#32) in response to IR. HeLa cells were harvested 2 h after 10 Gy of IR and analyzed by quantitative ChIP assay. n = 4. Error bars indicate S.D. B, a plot of SMC3 binding in non-treated HeLa cells versus phospho-SMC3 binding in HeLa cells exposed to 10 Gy IR. The slope and R2 value of linear regression analysis are indicated. C, kinetics of pS1083-SMC3 at cohesin sites (left) and total pS1083-SMC3 at different time points by Western blotting (WB) (right). HeLa cells were harvested at the indicated time point after 10 Gy of IR and SMC3 phosphorylation at cohesin sites was investigated by quantitative ChIP assay. n = 2. Error bars indicate S.D. D, pS1083-SMC3 level at 4 cohesin sites (CBS14, CBS15, CBS16, and CBS18) and a non-cohesin site (#29) from control HeLa cells (shGFP) and ATM knockdown HeLa cells (shATM). Cells were harvested 2 h after 10 Gy of IR and analyzed by quantitative ChIP assay. n = 4. Error bars indicate S.D. E, SMC3 binding ratio of irradiated to non-irradiated HeLa cells that ATM knocked down by shRNA (shATM). GFP knocked down HeLa cells (shGFP) were used as a control and cells were harvested 2 h after 10 Gy of IR. SMC3 binding at four cohesin sites was investigated by quantitative ChIP assay. n = 4. Error bars indicate S.D. Statistically significant differences are marked with the asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
The kinetics of SMC3 and pSMC3 binding at cohesin sites were measured by ChIP-qPCR (Fig. 2C). The amount of phosphorylated SMC3 at cohesin sites increased during the first 2 h and then decreased thereafter, consistent with the binding kinetics of SMC3 at cohesin sites (supplemental Fig. S1B); however, SMC3 phosphorylation measured by Western blotting peaked at 1 h. It suggests that phosphorylation of SMC3 cannot be fully accountable for increased SMC3 binding at the cohesion sites; other events may also contribute to increased SMC3 binding.
Because SMC3 is phosphorylated by the ATM kinase, the effect of ATM on SMC3 binding at cohesion sites was investigated in HeLa cells in which ATM is stably knocked down by a short hairpin RNA (36). ATM knockdown caused an ∼50% decrease of pS1083-SMC3 binding at cohesin sites compared with that in the control knockdown (Fig. 2D); however, IR-induced SMC3 binding was almost completely abolished (Fig. 2E and supplemental Fig. S2B). Thus, ATM is required for induced SMC3 binding at cohesin sites in response to IR.
SMC3 Acetylation Is Required for DNA Damage Response
Acetylation of SMC3 is another important modification of the cohesin complex that is required for establishment of sister chromatid cohesion in S-phase. To determine whether acetylation of the cohesin subunits is regulated in response to IR, we isolated the cohesin complex from IR-treated HeLa cells and analyzed their modifications on all cohesin subunits (SMC1, SMC3, RAD21, SA1/SA2, and PDS5A/PDS5B) by mass spectrometry. Despite repeated efforts, we did not detect acetylation on any other cohesin subunit including RAD21, whose budding yeast homolog Scc1 was acetylated (25). The only acetylation event we detected was those of SMC3 at Lys105 and Lys106, which occur in the absence of DNA damage, as previously demonstrated (22). We next measured the level of acetylated SMC3 after IR by Western blot and mass spectrometry (Fig. 3A and supplemental Fig. S3A). SMC3 acetylation at Lys105 and Lys106 was increased by 40–60% after cells were treated with IR. Compared with 10–20-fold induction of SMC3 phosphorylation at Ser1083, the induction of acetylation by IR is rather modest, likely because the basal level of acetylation is already high under undamaged conditions.
FIGURE 3.
SMC3 acetylation is required for DNA damage response. A, the relative level of SMC3 double acetylation measured by qMS at the indicated times after 10 Gy of IR treatments. The SMC3 double acetylation level after IR was normalized to that of untreated cells. B, SMC3 acetylation levels before and after IR treatment in cells transfected with siRNA against vimentin or ESCO1 quantified by qMS. Efficiency of ESCO1 knockdown was validated previously (22). C, SMC3 acetylation after IR in HeLa cells transfected with siRNA against ESCO1 and then rescued by the expression of wild-type ESCO1, siRNA-resistant ESCO1 (ESCO1R), or an empty vector. The SMC3 acetylation level was quantified by qMS. D, SMC3 acetylation induction by IR in HeLa cells depleted of ATM or ATR by siRNA treatment. SMC3 acetylation level was quantified by qMS. E, intra-S phase checkpoint measurement with the RDS assay in ESCO1 knockdown cells at the indicated times after 10 Gy of IR. GFP knockdown cells were used as a control. F, colony formation assay of the irradiated ESCO1 knockdown cells. Cells were treated with the indicated dosage of IR and allowed to recover for 1 week. At least three independent experimental triplicates were measured for each data point. G, intra-S phase checkpoint measurement with the RDS assay as described in the legend to Fig. 1A in stable cell lines that express SMC3-WT-2FLAG (WT), Lys105–Lys106 to Ala105–Ala106 (AA), and Gln105–Gln106 (QQ). H, colony formation assay of the stable cell lines described in G. Cells were treated with the indicated IR dosages and the surviving colonies were measured 1 week later. Statistically significant differences are marked with the asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Because SMC3 acetylation during a normal cell cycle depends on ESCO1 (22), we next tested whether IR-induced SMC3 acetylation is also ESCO1 dependent. As reported previously (22), knocking down ESCO1 decreased SMC3 acetylation in the absence of IR treatment (Fig. 3B); more importantly, it almost completely eliminated IR-induced acetylation. Such a defect in SMC3 acetylation caused by ESCO1 knockdown was rescued by overexpression of a siRNA-resistant ESCO1 (Fig. 3C). In addition, expression of ESCO1 potentiated SMC3 acetylation after IR (supplemental Fig. S3B). Thus, IR-induced SMC3 acetylation depends on ESCO1.
Because ESCO1 was identified as a phosphorylated protein in an ATM/ATR substrate screen (7), we investigated whether IR-induced SMC3 acetylation depends on ATM and ATR, master regulators of the DDR process. Although knocking down either ATM or ATR had no effect on SMC3 acetylation under undamaged conditions, both reduced the induction level of SMC3 acetylation upon IR treatment, suggesting that both ATM and ATR play a role in IR-induced SMC3 acetylation (Fig. 3D). Therefore, the increased SMC3 acetylation in response to IR is under control of the DNA damage checkpoint pathway.
Next, we investigated the effects of knocking down ESCO1 on the activation of the intra-S phase checkpoint in response to IR. We used two independent siRNAs to knock down expression of ESCO1 in HeLa cells and measured the rates of RDS and cellular survival in respond to IR (13). Compared with the control (siGFP) knockdown, the rates of RDS were higher in cells experiencing ESCO1 knock down by both ESCO1 siRNAs (Fig. 3E). In addition, these cells were more sensitive to IR than the control cells (Fig. 3F). These results indicate that ESCO1 is indeed required for intra-S phase checkpoint.
Because ESCO1 is important for intra-S phase checkpoint (Fig. 3E), we next tested whether SMC3 acetylation plays a similar role. Using doxycycline-inducible cell lines that stably express FLAG-SMC3-WT (WT), FLAG-SMC3-K105A/K106A (AA, mimicking the unacetylatable form), and FLAG-SMC3-K105Q/K106Q (QQ, mimicking the constitutively acetylated form) (22), we measured activation of the intra-S phase checkpoint in cells expressing these different forms of SMC3 by the RDS assay. Although the host, SMC3-WT, and SMC3-QQ expressing cells exhibited a 25–30% slowdown of DNA synthesis upon IR, cells expressing SMC3-AA showed only about 10% slowdown of their DNA synthesis, indicating a defective intra-S phase checkpoint (Fig. 3G). In contrast, no difference was observed among these different cell lines in a G2/M checkpoint assay (data not shown). We also evaluated whether, like ESCO1, SMC3 acetylation is required for cellular survival in respond to IR. By using a colony formation assay, we found that cells expressing the SMC3-AA mutant were significantly more sensitive to IR than those expressing either SMC3-WT or SMC3-QQ variants (Fig. 3H). Together these results are consistent with the model that acetylation of SMC3 at Lys105 and Lys106 by ESCO1 regulates DDR and is critical for cellular survival.
SMC3 Phosphorylation and Acetylation Are in Two Largely Independent Pathways to Regulate Reinforcement of Cohesion After IR
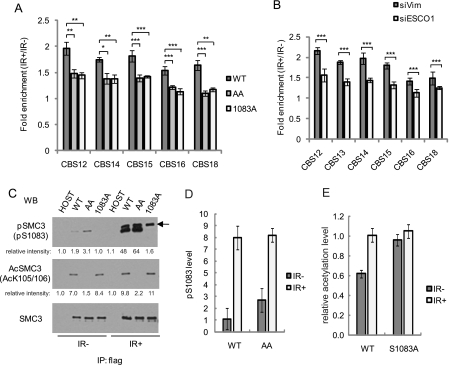
To obtain direct evidence that SMC3 phosphorylation and acetylation are required for increased cohesin binding by IR, we measured binding of the SMC3-AA and SMC3-S1083A mutants. SMC1 antibody was used for the ChIP experiment because the SMC3 antibody that recognizes the extreme C terminus cannot recognize C-terminal FLAG-tagged SMC3. The SMC1 immunoprecipitation contained ∼25% exogenous mutant SMC3 and 75% endogenous WT SMC3, confirming that the mutant protein was incorporated into the cohesin complexes (supplemental Fig. S4A). Importantly, decreased cohesin binding was observed when SMC3-AA or SMC3-S1083A was incorporated into 25% of the cohesin complex (Fig. 4A and supplemental Fig. S4B). This indicates that SMC3 phosphorylation at Ser1083 and acetylation at Lys105 and Lys106 are two essential modifications required for increased binding of cohesin by IR. In addition, we tested whether SMC3 acetyltransferase ESCO1 is required for this process by ChIP-qPCR (Fig. 4B). The increased level of SMC3 is significantly decreased at all investigated cohesin sites by knocking down ESCO1. This supports that SMC3 acetylation is an essential modification for increased binding of cohesin by IR.
FIGURE 4.
SMC3 phosphorylation and acetylation are in two largely independent pathways to regulate reinforcement of cohesion after IR. A, cohesin binding ratio of irradiated to non-irradiated 293T cells expressing SMC3-WT-FLAG (WT), Lys105–Lys106 to Ala105–Ala106 (AA), and Ser1083 to Ala1083 (1083A). Cells were harvested 2 h after 10 Gy of IR and ChIP was carried out with anti-SMC1 antibody. Cohesin binding at five cohesin sites were investigated by quantitative ChIP assay. n = 3. Error bars indicate S.D. B, cohesin binding ratio of irradiated to non-irradiated HeLa cells transfected with siRNA against vimentin (siVim) or ESCO1 (siESCO1). Cells were harvested 2 h after 10 Gy of IR. SMC3 binding at six cohesin sites were investigated by quantitative ChIP assay. n = 4. Error bars indicate S.D. C, SMC3 phosphorylation and acetylation in 293T cells stably expressing SMC3-WT, -AA, and -S1083A. FLAG-SMC3 proteins were immunoprecipitated from cycling cells and IR-treated cells with anti-FLAG antibody and detected by Western blotting. The arrow indicates nonspecific band. D, SMC3 Ser1083 phosphorylation measured by qMS in 293T cells stably expressing SMC3-WT or SMC3-AA. FLAG-SMC3 was immunoprecipitated as described in B. E, relative levels of SMC3 acetylation measured by qMS. SMC3 was immunoprecipitated as in B from the indicated cell lines that were untreated or treated with IR. Statistically significant differences are marked with the asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Because both phosphorylation and acetylation of SMC3 are required for intra-S phase checkpoint activation and both are regulated by the ATM/ATR, we next investigated whether one modification event might be a pre-requisite to the other. To test whether acetylation is required for phosphorylation, we compared Ser1083 phosphorylation levels in cells expressing FLAG-SMC3-WT and those expressing FLAG-SMC3-AA with or without IR treatment (10 Gy). Both fusion proteins were immunoprecipitated with a FLAG-M2 antibody and the level of SMC3 phosphorylation at Ser1083 was measured by Western blot and qMS (Fig. 4, C and D). Such phosphorylation on both molecules was induced by IR and to a level indistinguishable between the two. Similar results were observed with Western blotting using a pS1083-SMC3 specific antibody (Fig. 4C). Thus, SMC3 acetylation is unlikely required for its IR-induced phosphorylation at Ser1083.
We next tested whether phosphorylation of SMC3 at Ser1083 might be required for its IR-induced acetylation. We expressed and purified FLAG-SMC3-WT and FLAG-SMC3-S1083A fusion proteins from cells both before and after IR treatment (10 Gy) and measured their acetylation levels by qMS. Acetylation of both fusion proteins were further induced after IR treatment to similar levels, although the fold of induction in the mutant was less obvious than in the wild-type due to its higher basal level of acetylation (Fig. 4E). These results together suggest that acetylation and phosphorylation of SMC3 occur independently of each other and both are required for activation of the intra-S phase checkpoint in response to DNA damage. In fact, qMS measurements suggest that inactivating either one of the modifications appeared to result in an increased level of the other in the absence of DNA damage (Figs. 4, C–E). It is possible that cells expressing the mutant proteins experience increased endogenous DNA damage, which stimulates DDR. Alternatively, cells might have attempted to compensate for the loss of one modification by increasing the other.
Small Increases in Cohesin Binding at Every Cohesion Site Translate into Larger Increase in Cohesin Binding on Bulk Chromatin
Because the increase of SMC3 binding to every cohesion site after IR appeared to be quite small, we decided to investigate total SMC3 binding to bulk chromatin after IR. Cohesin exists as both a chromatin-bound and soluble complex in cells (35). We have previously shown that acetylated SMC3 molecules preferentially bind to chromosomes in cells grown under unperturbed conditions and these are resistant to extraction with detergents (22). Because IR-induced SMC3 acetylation and phosphorylation are important for SMC3 binding at cohesion sites, we predict that they should be enriched on chromosomes and these modifications make chromatin-bound cohesin more resistant to detergent extraction.
We used a more quantitative assay based on SILAC qMS to test this idea. We treated cells grown in light isotope-labeled medium with 10 Gy of IR and extracted chromatin-bound proteins with a high detergent-containing buffer. After extraction, the non-extracted insoluble fraction contained tightly bound chromatin proteins, whereas the soluble fraction contains weakly bound chromatin proteins and originally soluble proteins. We then mixed both soluble and chromatin-bound fractions with heavy isotope-labeled, untreated whole cell extracts as an internal control and measured SMC3 acetylation levels. We found that acetylated SMC3 was about 12 times more abundant in the chromatin fraction than the soluble fraction (Fig. 5A). This is likely due to tighter binding of acetylated cohesin onto chromatin, preferential acetylation of chromatin-bound SMC3 by ESCO1, or both.
FIGURE 5.
Small increases in cohesin binding at every cohesion site translates into a larger increase in cohesin binding on bulk chromatin. A, relative SMC3 acetylation level measured by qMS in RIPA soluble and insoluble fractions in HeLa cells treated with IR. Acetylation level of the RIPA insoluble fraction is normalized to the acetylation level of RIPA insoluble (= 1). Light isotope-labeled cells were treated with 10 Gy of IR and lysed in RIPA buffer for 40 min on ice to extract loosely bound cohesin. The detergent resistant/insoluble pellet was re-suspended in the RIPA buffer and sonicated to release and solubilize proteins. SMC3 from each fraction was then immunoprecipitated, and mixed with heavy isotope-labeled SMC3 that was isolated from whole cell lysate to normalize the protein amounts. B, stoichiometry of SMC3 Ser1083 phosphorylation measured by qMS as described in Luo et al. (15). The chromatin bound fraction and RIPA soluble fraction of SMC3 was isolated as in A. C, relative RIPA insoluble SMC3 level in ESCO1 knockdown cells. HeLa cells were transfected with siRNA against vimentin (siVim) or ESCO1 (siESCO1) and the relative RIPA-insoluble SMC3 level was measured by qMS.
To obtain more quantitative data for phosphorylated SMC3, we measured the stoichiometry of Ser1083 phosphorylation, the percentage of SMC3 phosphorylated in both soluble and chromatin-bound fractions after IR by a SILAC method as previously reported (15). We found that more than 60% of SMC3 in the chromatin fraction was phosphorylated at Ser1083; in comparison, only ∼25% SMC3 in the soluble fraction was phosphorylated (Fig. 5B). Thus, phosphorylated SMC3 is also enriched on chromatin.
Because DNA damage induces acetylation and phosphorylation of SMC3 and both forms of modified SMC3 are enriched on chromatin, we tested whether IR can increase cohesin binding to chromatin. We treated heavy isotope-labeled culture with IR and mixed the cells with untreated, light isotope-labeled cells. The chromatin fractions were prepared under identical conditions and the relative abundance of heavy- and light-labeled SMC3 was measured by qMS. After normalizing to the amount of H2B, we found that IR induced an increase of SMC3 in the chromatin fraction by about 2.5-fold (Fig. 5C). Furthermore, knocking down ESCO1 expression with a siESCO1 almost completely abolished IR-induced SMC3 binding to chromatin (Fig. 5C). Thus, induction of SMC3 binding to chromatin by IR depends on ESCO1. Taken together, these data show that the extent of SMC3 binding to bulk chromatin after IR can be increased to 2.5-fold of that before IR, and ESCO1 is one of the regulators in human cells.
DISCUSSION
Ionizing Radiation Induces Genome-wide Reinforcement of Cohesin Binding at Pre-existing Sites in Human Cells
In yeast, although cohesion is established exclusively in S-phase in undamaged cells, DSB can induce de novo cohesion establishment in G2/M cells. Remarkably, a single DNA break is able to induce genome-wide cohesion re-establishment. We show here that this is also likely the case in human cells. Using ChIP-seq, we mapped genome-wide cohesin binding sites in cells treated with ionizing radiation and found that more than 99% of the sites overlap with the existing cohesin binding sites. Quantitative analysis by ChIP-qPCR showed an increase of binding by 25–50% at all examined sites. Because 10 Gy of IR induced multiple DSBs that are random and heterogeneous among a large number of cells (∼107), increased SMC3 binding at cohesion sites unlikely correlates with actual damage sites. Cohesin and phosphorylated SMC1/3 are also recruited to the site of damage for efficient repair. In budding yeast, cohesin becomes highly enriched at the DSB and forms an extended “cohesin domain” that spans 50 to 100 kb of DNA around the lesion (16, 37). Phosphorylation of SMC1 and SMC3 is specifically induced by DNA damage. However, our data show that phospho-SMC3 is not limited to DSB sites. SMC3-pS1083 binds to pre-existing cohesin binding sites and its amount displays linear correlation with total SMC3, suggesting that these SMC3 are phosphorylated on the chromatin. In undamaged cells, a large number of cohesin binding sites coincide with transcription insulator CTCF. However, enhanced cohesin binding is not limited to CTCF and transcription function. Thus, our data are consistent with the notion that DSB triggers genome-wide reinforcement cohesin binding at pre-existing sites, which is an evolutionally conserved mechanism for facilitating checkpoint signaling and DNA repair.
ATM and ESCO1 Are Required for Genome-wide Reinforcement of Cohesin Binding
In this work, we show that acetylation of SMC3 by ESCO1 is another post-translational modification that is important to DDR. IR-induced acetylation of SMC3 is regulated by the acetyltransferase ESCO1 and the kinases ATM and ATR. In contrast to phosphorylation, which is induced by >20-fold by IR, acetylation of SMC3 at Lys105 and Lys106 is induced by ∼50%, which can only be accurately measured with the SILAC-based quantitative mass spectrometry. Despite the relatively low levels of induction, ESCO1 and SMC3 acetylation is required for intra-S phase checkpoint and cellular survival in response to IR, as well as for genome-wide reinforcement of cohesin binding in response to IR.
It is not surprising that genome-wide reinforcement of cohesin binding is regulated by ATM. Phosphorylation of SMC1 and SMC3 by ATM/ATR is essential for DDR and recruitment of pSMC1 and pSMC3 to DSB sites has also been shown (12–15, 17). It is important to note that the ATM kinase plays a crucial role as its loss almost completely eliminated IR-induced cohesin binding. It is conceivable that because ATM controls a large number of substrates with diverse functions, it has strong impact on cellular response to DNA damage. Note that ESCO1 was identified as a putative ATM substrate in a proteomics screen (7), thus its function is likely under the regulation of ATM-mediated pathway.
How SMC3 modifications enhance cohesin binding is currently not clear. We speculate that one functional consequence of SMC3 modification is to regulate the ATPase activity of the cohesin complex because the acetylated residues (Lys105 and Lys106) on SMC3 are close to an ATP binding site and the phosphorylated residue (Ser1083) is close to an ATP hydrolysis site. The two events might regulate these two distinct steps of the ATPase cycle. Such a possibility is consistent with the observation that the ATPase function of the SMC1/3 heterodimer is essential for cohesin binding to chromatin and sister chromatid cohesion (38). Another possible mechanism is through modulating its interaction with cohesin regulatory cofactors WAPL and PDS5. SMC3 acetylation in S-phase counteracts the anti-establishment activity of WAPL and PDS5 (39) and is required for replication fork progression (40). An increase in SMC3 acetylation would potentiate its anti-establishment activity, leading to enhanced sister chromatid cohesion. It is currently not known how SMC1 and SMC3 phosphorylation affect cohesion. It will be interesting to test how it affects the binding between cohesin core complex and PDS5-WAPL.
The Biological Process of Cohesion Reinforcement Is Conserved, but the Molecular Details Are Different between Yeast and Human
During preparation of this article, it was reported that budding yeast Eco1p regulates DSB-induced cohesion by acetylating Scc1/Mcd1, a process that depends on Scc1 phosphorylation by the checkpoint kinase Chk1 (41). We did not find any evidence of DNA damage-induced acetylation or phosphorylation of the human homolog RAD21, nor did we detect phosphorylation of Smc3 in yeast; in contrast to Scc1 modification, phosphorylation and acetylation of SMC3 seem to be independent events.
Indeed, regulation of cohesin in yeast and human is different in several aspects. First, whereas yeast cohesin is loaded during S-phase, its human counterpart is loaded in telophase; second, yeast cohesin is enriched within intergenic regions between convergent transcripts, most of human cohesin colocalizes with CTCF, a zinc-finger protein required for transcriptional insulation (26, 28, 31); third, a functional divergence has been observed with WAPL, which seems to play an exclusively negative role in regulating cohesion in human and fission yeast but both positive and negative roles in budding yeast (39, 42–45). Thus, whereas cohesion reinforcement is a conserved biological process used by both yeast and human, the molecular details appear to be different in that distinct cohesin subunit is targeted in response to DNA damage.
Supplementary Material
Acknowledgment
We thank Dr. David J. Chen for providing ATM knockdown HeLa cells.
This work was supported, in whole or in part, by National Institutes of Health Grants CA84199 and CA98500 and the Welch Foundation (to J. Q.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1 and S2.
- DDR
- DNA damage response
- IR
- ionizing radiation
- DSB
- double strand DNA break
- RDS
- radioresistant DNA synthesis
- SILAC
- stable isotope labeling of cultured cells
- ChIP-seq
- chromatin immunoprecipitation sequencing
- qMS
- quantitative mass spectrometry
- MACS
- model-based analysis of ChIP-Seq
- qPCR
- quantitative PCR
- Gy
- gray
- WT
- wild-type
- Eco1
- establishment of cohesion 1
- siRNA
- small interfering RNA
- shGFP
- short hairpin green fluorescent protein.
REFERENCES
- 1.Onn I., Heidinger-Pauli J. M., Guacci V., Unal E., Koshland D. E. (2008) Annu. Rev. Cell Dev. Biol. 24, 105–129 [DOI] [PubMed] [Google Scholar]
- 2.Peters J. M., Tedeschi A., Schmitz J. (2008) Genes Dev. 22, 3089–3114 [DOI] [PubMed] [Google Scholar]
- 3.Losada A., Hirano T. (2005) Genes Dev. 19, 1269–1287 [DOI] [PubMed] [Google Scholar]
- 4.Nasmyth K., Haering C. H. (2009) Annu. Rev. Genet. 43, 525–558 [DOI] [PubMed] [Google Scholar]
- 5.Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 6.Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 7.Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 8.Shiloh Y. (2003) Nat. Rev. Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 9.Cimprich K. A., Cortez D. (2008) Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S. T., Lim D. S., Canman C. E., Kastan M. B. (1999) J. Biol. Chem. 274, 37538–37543 [DOI] [PubMed] [Google Scholar]
- 11.Shiloh Y. (2006) Trends Biochem. Sci. 31, 402–410 [DOI] [PubMed] [Google Scholar]
- 12.Kim S. T., Xu B., Kastan M. B. (2002) Genes Dev. 16, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yazdi P. T., Wang Y., Zhao S., Patel N., Lee E. Y., Qin J. (2002) Genes Dev. 16, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa R., Bakkenist C. J., McKinnon P. J., Kastan M. B. (2004) Genes Dev. 18, 1423–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo H., Li Y., Mu J. J., Zhang J., Tonaka T., Hamamori Y., Jung S. Y., Wang Y., Qin J. (2008) J. Biol. Chem. 283, 19176–19183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ström L., Lindroos H. B., Shirahige K., Sjögren C. (2004) Mol. Cell 16, 1003–1015 [DOI] [PubMed] [Google Scholar]
- 17.Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M. B., Bartek J., Lukas J. (2006) J. Cell Biol. 173, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skibbens R. V., Corson L. B., Koshland D., Hieter P. (1999) Genes Dev. 13, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tóth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. (1999) Genes Dev. 13, 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolef Ben-Shahar T., Heeger S., Lehane C., East P., Flynn H., Skehel M., Uhlmann F. (2008) Science 321, 563–566 [DOI] [PubMed] [Google Scholar]
- 21.Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., Gygi S. P., Koshland D. E. (2008) Science 321, 566–569 [DOI] [PubMed] [Google Scholar]
- 22.Zhang J., Shi X., Li Y., Kim B. J., Jia J., Huang Z., Yang T., Fu X., Jung S. Y., Wang Y., Zhang P., Kim S. T., Pan X., Qin J. (2008) Mol. Cell 31, 143–151 [DOI] [PubMed] [Google Scholar]
- 23.Ström L., Karlsson C., Lindroos H. B., Wedahl S., Katou Y., Shirahige K., Sjögren C. (2007) Science 317, 242–245 [DOI] [PubMed] [Google Scholar]
- 24.Unal E., Heidinger-Pauli J. M., Koshland D. (2007) Science 317, 245–248 [DOI] [PubMed] [Google Scholar]
- 25.Heidinger-Pauli J. M., Unal E., Koshland D. (2009) Mol. Cell 34, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G. P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. (2004) Nature 430, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt C. K., Brookes N., Uhlmann F. (2009) Genome Biol. 10, R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J. M. (2008) Nature 451, 796–801 [DOI] [PubMed] [Google Scholar]
- 29.Rubio E. D., Reiss D. J., Welcsh P. L., Disteche C. M., Filippova G. N., Baliga N. S., Aebersold R., Ranish J. A., Krumm A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stedman W., Kang H., Lin S., Kissil J. L., Bartolomei M. S., Lieberman P. M. (2008) EMBO J. 27, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H. C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., Cobb B. S., Yokomori K., Dillon N., Aragon L., Fisher A. G., Merkenschlager M. (2008) Cell 132, 422–433 [DOI] [PubMed] [Google Scholar]
- 32.Hadjur S., Williams L. M., Ryan N. K., Cobb B. S., Sexton T., Fraser P., Fisher A. G., Merkenschlager M. (2009) Nature 460, 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishiro T., Ishihara K., Hino S., Tsutsumi S., Aburatani H., Shirahige K., Kinoshita Y., Nakao M. (2009) EMBO J. 28, 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerlich D., Koch B., Dupeux F., Peters J. M., Ellenberg J. (2006) Curr. Biol. 16, 1571–1578 [DOI] [PubMed] [Google Scholar]
- 36.Chen B. P., Uematsu N., Kobayashi J., Lerenthal Y., Krempler A., Yajima H., Löbrich M., Shiloh Y., Chen D. J. (2007) J. Biol. Chem. 282, 6582–6587 [DOI] [PubMed] [Google Scholar]
- 37.Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J. E., Koshland D. (2004) Mol. Cell 16, 991–1002 [DOI] [PubMed] [Google Scholar]
- 38.Arumugam P., Gruber S., Tanaka K., Haering C. H., Mechtler K., Nasmyth K. (2003) Curr. Biol. 13, 1941–1953 [DOI] [PubMed] [Google Scholar]
- 39.Rowland B. D., Roig M. B., Nishino T., Kurze A., Uluocak P., Mishra A., Beckouët F., Underwood P., Metson J., Imre R., Mechtler K., Katis V. L., Nasmyth K. (2009) Mol. Cell 33, 763–774 [DOI] [PubMed] [Google Scholar]
- 40.Terret M. E., Sherwood R., Rahman S., Qin J., Jallepalli P. V. (2009) Nature 462, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heidinger-Pauli J. M., Unal E., Guacci V., Koshland D. (2008) Mol. Cell 31, 47–56 [DOI] [PubMed] [Google Scholar]
- 42.Bernard P., Schmidt C. K., Vaur S., Dheur S., Drogat J., Genier S., Ekwall K., Uhlmann F., Javerzat J. P. (2008) EMBO J. 27, 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandhi R., Gillespie P. J., Hirano T. (2006) Curr. Biol. 16, 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kueng S., Hegemann B., Peters B. H., Lipp J. J., Schleiffer A., Mechtler K., Peters J. M. (2006) Cell 127, 955–967 [DOI] [PubMed] [Google Scholar]
- 45.Sutani T., Kawaguchi T., Kanno R., Itoh T., Shirahige K. (2009) Curr. Biol. 19, 492–497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.