Abstract
ABCC6 mutations are responsible for the development of pseudoxanthoma elasticum, a rare recessive disease characterized by calcification of elastic fibers. Although ABCC6 is mainly expressed in the liver the disease has dermatologic, ocular, and cardiovascular symptoms. We investigated the transcriptional regulation of the gene and observed that hepatocyte growth factor (HGF) inhibits its expression in HepG2 cells via the activation of ERK1/2. Similarly, other factors activating the cascade also inhibited ABCC6 expression. We identified the ERK1/2 response element in the proximal promoter by luciferase reporter gene assays. This site overlapped with a region conferring the tissue-specific expression pattern to the gene and with a putative hepatocyte nuclear factor 4α (HNF4α) binding site. We demonstrated that HNF4α regulates the expression of ABCC6, acts through the putative binding site, and determines its cell type-specific expression. We also showed that HNF4α is inhibited by the activation of the ERK1/2 cascade. In conclusion we describe here the first regulatory pathway of ABCC6 expression showing that the ERK1/2-HNF4α axis has an important role in regulation of the gene.
Keywords: ABC Transporter, MAP Kinases (MAPKs), Oxidative Stress, Transcription, Vitamin K, MRP6, Pseudoxanthoma Elasticum
Introduction
Pseudoxanthoma elasticum (PXE)3 is a rare recessive genetic disease characterized by generalized elastic fiber calcification and fragmentation (for a recent review, see Ref. 1). The most characteristic symptom of the disease is the presence of yellowish papules primarily on the neck. In the eye, the Bruch's membrane becomes fragmented and subsequent neovascularization might lead to severe visual loss. The soft tissue calcification typically concerns the arteries, leading to hypertension, intermittent claudication, stroke, and coronary artery disease (1).
It has been demonstrated that mutations in the ABCC6 gene are responsible for PXE (2–4) and more than 200 disease-causing mutations have been reported to date (5). Although the 5′ end of the gene harbors some typical mutations due to gene conversion events between ABCC6 and its two pseudogenes (6, 7), most of the mutations are located in the functionally more important 3′ part. Identified mutation carriers were shown to have blood serum alterations and suffer from subclinical symptoms such as calcification of elastic fibers in the dermis (8–10).
The ABCC6 gene is highly expressed in the liver and at a lower extent in the kidney and intestine (11). This expression pattern and some elegant transplantation experiments carried out between knock-out and control mice have demonstrated that PXE is a metabolic disease (12) and the loss of function of the gene product is responsible for the development of the disease. ABCC6 encodes MRP6, which is a member of the ATP-binding cassette (ABC) protein family (11). The pathological and physiological role of MRP6 is still not understood, although our previous work demonstrated that similarly to other MRP ABC transporters MRP6 is able to transport small organic anions from the cells to the bloodstream and this function is lost in some tested disease-causing mutations (13). However, the physiological substrate, which is most probably a metabolite of hepatic origin, still remains to be identified.
We have previously characterized the proximal promoter of the human ABCC6 gene (14). In this initial study on the regulation of the gene we reported an inverse correlation between the CpG island hypermethylation located in the conserved proximal promoter and the expression of the gene in various cell lines (14). This finding was confirmed in mouse tissues (15). We have also identified by luciferase assay a silencer and a DNA methylation-dependent activator sequence in this conserved region. Several transcription factors have been implicated later in the regulation of the human and mouse ABCC6 genes based on luciferase reporter gene experiments (15–19). More recently, it has been proposed, that the ABCC6 promoter polymorphisms may be functionally linked to PXE (20) and our results suggested that these polymorphisms are modulating the binding capacity of previously identified transcription factors (21). However, to date no cellular signaling pathway has been convincingly linked to the regulation of ABCC6 expression.
In this study we describe the involvement of a physiological kinase cascade in the regulation of the ABCC6 gene expression. We demonstrate that the activation of extracellular-signal related kinase 1/2 (ERK1/2) cascade by growth factors in HepG2 hepatoma cell line reduces the transcription of ABCC6 by preventing the activator role of HNF4α identified in the present study as the major, tissue-specific regulator of ABCC6.
EXPERIMENTAL PROCEDURES
Cell Culture
HepG2 and Hep3B human hepatoma cell lines were obtained from ATCC, and cultured according to the manufacturer's instructions. Before treatment, cells were passaged in medium without serum, and in the case of co-treatment the different inhibitors were added 1 h before addition of the activators. The different molecules have been used at the following concentration: hepatocyte growth factor (HGF, human, recombinant), 40 ng/ml; epidermal growth factor (EGF), 100 ng/ml; insulin, 5 μg/ml; transforming growth factor β (TGFβ), 5 ng/ml; phorbol myristate acetate (PMA), 100 nm; menadione, 5 μm; tert-butyl hydroquinone (tBHQ), 75 nm (Sigma); U0126, 2 μm; bisindolylmaleimide I (BIMI), 500 nm; LY294002, 20 μm (Calbiochem).
Quantitative PCR
ABCC6 and URG7 mRNA expression levels were measured by quantitative PCR. Total RNA was extracted using TRI Reagent (Molecular Research Center, Inc.), and 500 ng of RNA of each condition was reverse transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Synthesized cDNA served as template in a 10-μl reaction, quantitative PCR was performed with LightCycler FastStart DNA Master SYBR Green I (Roche) in an LC480 quantitative PCR machine (Roche). Primer sequences are the followings: ABCC6 forward, GGCCCGGGCATCCAGGTT, and reverse, TTTCATCTACGCGAGCATTGTTCT; URG7 forward, TACCTCCTCTTCATCCACCACCAT, and reverse, CCCTGCCTCCCCCGAACATTG; and ABL forward, GGGCTCATCACCACGCTCCA, and reverse CTGCCGGTTGCACTCCCTCA. Primers were tested for specificity by BiSearch (22). The expression of the target gene was normalized to the ABL gene used as an internal control. Relative concentrations of each template were calculated according to calibration curves as described previously (23).
Western Immunoblotting Analysis
Western blot experiments were carried out as described previously (24). Antibodies against ERK1/2 (K23) (1/1000 dilution) or phospho-ERK (E4) (1/250 dilution) (Santa Cruz) were used to detect ERK1/2. Immunoblots were subjected to quantitative measurements by densitometry.
Promoter Constructs and Transfections
All reporter constructs and luciferase measurements were described previously (21). Standardization has been carried out as described (14). Treatments were carried out 24 h after transfection and for 24 h. Co-transfections of reporter constructs and plasmids carrying cDNA for selected transcription factors were performed analogously to previous work (21). Dominant-negative (K71R) ERK1 expression plasmid was a kind gift from Dr. Melanie Cobb (Dallas, TX), and the constitutively active MEKK1 (caMEKK) expression plasmid (25) was a kind gift from Dr. Dirk Bohmann (Rochester, MA).
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation was performed using the EZ-ChIP kit from Millipore according to the manufacturer's protocol. The following antibodies were used: normal mouse IgG (part of the EZ-ChIP kit) and anti-HNF4 (K9218, Abcam). The transcription factor-bound genomic DNA fragments were quantified by real time PCR as described in Ref. 17. Results were normalized to the total extracted DNA amount in samples taken prior to immunoprecipitation by the ΔCt method.
RESULTS
Growth Factors Inhibit the Expression of ABCC6
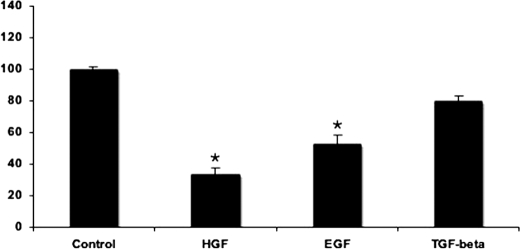
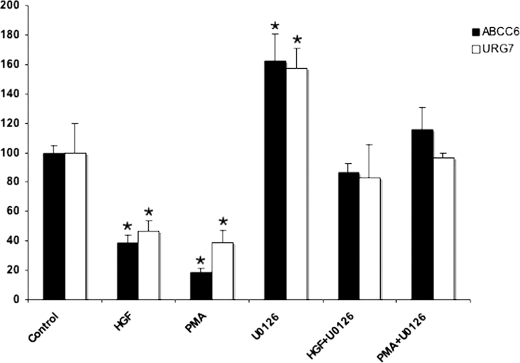
A previous study used the luciferase reporter gene assay to show that transforming growth factor β (TGFβ) is an activator of ABCC6 expression (18). Our aim was to test whether TGFβ and other similar factors such as HGF and EGF, known as major regulators of hepatocytes are also able to induce expression of the ABCC6 gene in its endogenous chromatin environment in HepG2 cells. The effect of growth factors was tested following a 24-h treatment in the absence of serum by quantitative reverse transcription-PCR. In our hands treatment with TGFβ had no effect, whereas HGF and EGF elicited a significant inhibition (60 and 40%, respectively) of ABCC6 expression as Fig. 1 illustrates. Similar but weaker effects were observed on Hep3B cells, another human hepatoma cell line (not shown). In view of these results, we decided to dissect the signaling pathways acting on the expression of ABCC6.
FIGURE 1.
Effect of growth factors on ABCC6 gene expression. HepG2 cells were treated with the indicated compounds or vehicle for controls (dimethyl sulfoxide) in serum-free conditions for 24 h. Relative ABCC6 expression level was determined by quantitative PCR after normalization to the ABL housekeeping gene expression level as described under “Experimental Procedures.” Expression levels are indicated as a percent of untreated control. Four parallels were used for each condition. The experiments were repeated three times ±S.E., *, p < 0.05.
The ERK1/2 Pathway Is Responsible for the Down-regulation of ABCC6
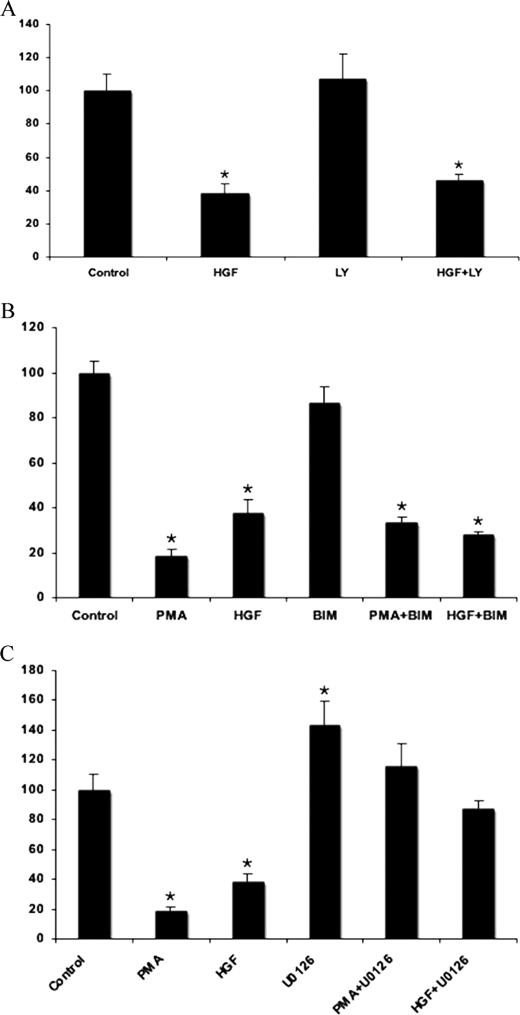
HGF activates PI 3-kinase/Akt, protein kinase C (PKC), and ERK1/2 cascades (26, 27). In the following experiments, we tested the role of the different pathways in the regulation of ABCC6 in HepG2 cells. We found that insulin treatment had only an insignificant effect on ABCC6 expression, suggesting that HGF action is not mediated via the PI 3-kinase/Akt pathway. To confirm this finding, we treated HepG2 cells with LY294002, a specific inhibitor of the PI 3-kinase/Akt pathway, in the presence or absence of HGF. As illustrated in Fig. 2A, no significant effect of the inhibitor could be detected, showing indeed that the PI 3-kinase does not play a major role in the HGF-mediated inhibition of ABCC6.
FIGURE 2.
ERK1/2 cascade is a major regulator of expression of the human ABCC6 gene. HepG2 cells were treated with the indicated compounds or vehicle for controls (dimethyl sulfoxide) in serum-free conditions for 24 h. Relative ABCC6 expression level was determined by quantitative PCR after normalization to the ABL housekeeping gene expression level as described under “Experimental Procedures.” Expression levels are indicated as a percent of untreated control. Four parallels were used for each condition. The experiments were repeated three times ±S.E., *, p < 0.05. A, PI 3-kinase pathway does not modulate ABCC6 expression. Cells were treated with HGF or LY294002 (indicated as LY on the figure), a specific inhibitor of PI 3-kinase, or in combination. When co-treatment was applied, LY294002 was added to the cells 1 h before HGF. B, atypical PKC subfamily member(s) inhibit the expression of ABCC6. Cells were treated with PMA, HGF, or BIM. When co-treatment was applied, BIM was added to the cells 1 h before PMA or HGF. C, inhibition of the ERK1/2 pathway precludes the down-regulation of ABCC6 expression. Cells were treated with PMA, HGF, or U0126, a selective inhibitor of MEK1/2 kinase. When co-treatment was applied, U0126 was added to the cells 1 h before PMA or HGF.
Next we tested the involvement of PKC enzymes in the inhibition of ABCC6 expression by HGF. We treated HepG2 cells with phorbol myristate acetate (PMA), a PKC activator (Fig. 2B). This treatment caused an even increased inhibition of ABCC6 expression relative to that observed with HGF (80%), suggesting the implication of the PKC pathway in the regulation of ABCC6 expression. Similar but weaker effects were observed on Hep3B cells (not shown). Surprisingly, bisindolylmaleimide I (BIMI), an inhibitor of the PKC pathway, was unable to prevent the action of either PMA or HGF (Fig. 2B). To explain this phenomenon, we hypothesized that the PMA-dependent inhibition of ABCC6 expression is mediated via BIMI-resistant PKC subfamily member(s). The BIMI-resistant atypical PKC subfamily members are known to be able to modulate the ERK1/2 pathway (28).
The ERK1/2 cascade is the third kinase cascade to be activated by HGF. Therefore, we tested whether the ERK1/2 pathway is implicated in ABCC6 regulation. We carried out treatment with U0126, a specific inhibitor of ERK1/2 phosphorylation both in the presence and absence of PMA and HGF. When we treated the cells with U0126 alone a 1.6-fold induction of ABCC6 expression was observed (Fig. 2C). When we performed the co-treatments we found that the inhibition of ABCC6 gene expression by PMA or HGF was completely prevented by U0126. Finally, we performed co-treatments with U0126 and EGF (not shown), and observed similar results as for the previous co-treatments further strengthening the major involvement of the ERK1/2 cascade in the inhibition of ABCC6 expression.
ERK1/2 Activation Correlates with the Expression Level of ABCC6
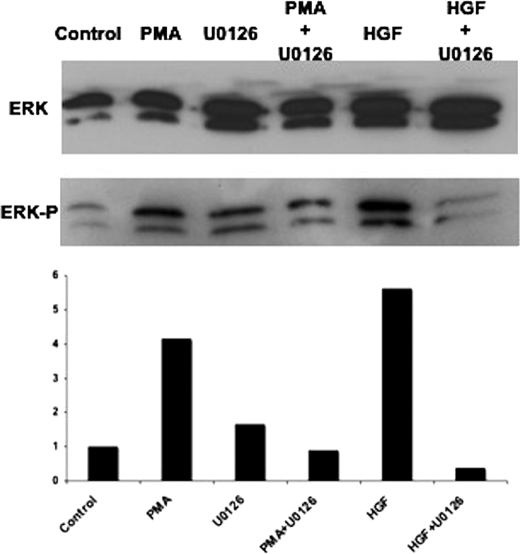
To investigate correlation between the activity of the ERK1/2 pathway and the expression of ABCC6, we performed Western blot experiments with ERK1/2 and phospho-ERK1/2-specific antibodies (Fig. 3A). Although PMA and HGF treatments strongly increased ERK1/2 phosphorylation (4–6 times), co-treatments with U0126 prevented the increase of phosphorylation relative to the control sample as demonstrated by the densitometric analysis of the immunoblot (Fig. 3B). This result emphasizes the involvement of ERK1/2 in the regulation of ABCC6 expression.
FIGURE 3.
The ERK1/2 phosphorylation inversely correlates with the down-regulation of ABCC6 gene expression. HepG2 cells were treated with the indicated compounds or vehicle for controls (dimethyl sulfoxide) in serum-free conditions for 24 h. A, Western blot analysis of ERK1/2 phosphorylation. 50 μg of protein was loaded per lane. The blotted membrane was incubated during 2 h in the primary and subsequently 1 h in the secondary antibody. B, densitometric analysis of the immunoblot shown in panel A (ERK-P/ERK1/2 in arbitrary units).
Oxidative Stress Induced by Vitamin K3 and tert-Butyl-hydroquinone Inhibits ABCC6 Expression via the ERK1/2 Pathway
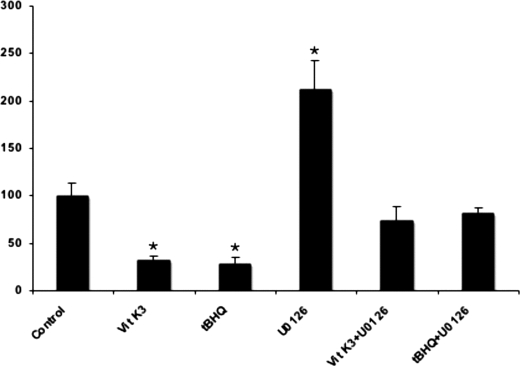
Next we tested whether other activators of the ERK1/2 cascade are also capable of inhibiting the expression of the gene. It has been shown that oxidative stress generated by reactive oxygen species induces ERK1/2 phosphorylation and thereby activation through the direct inhibition of tyrosine phosphatases (29). Thus, we checked two molecules producing reactive oxygen species, menadione (vitamin K3) (30) and tBHQ, if they also inhibit the expression of ABCC6 in HepG2 cells. As shown on Fig. 4, both vitamin K3 and tBHQ inhibited significantly the expression of ABCC6 after a 24-h treatment, whereas co-treatment with U0126 prevented the inhibition. These data strongly suggest that oxidative stress inhibits ABCC6 expression at least partially via the ERK1/2 pathway. This was confirmed by Western blot analysis and a negative correlation between the phospho-ERK and the ABCC6 gene expression was detected similarly to the effect of growth factors (not shown).
FIGURE 4.
Oxidative stress inhibits ABCC6 expression partially via the ERK1/2 cascade. HepG2 cells were treated with the indicated compounds or vehicle for controls (dimethyl sulfoxide) in serum-free conditions for 24 h. The relative ABCC6 expression level was determined by quantitative PCR after normalization to the ABL housekeeping gene expression level as described under “Experimental Procedures.” Expression levels are indicated as a percent of untreated control. Four parallels were used for each condition. The experiments were repeated three times ±S.E., *, p < 0.05.
ERK1/2 Inhibits ABCC6 Expression at the Transcription Initiation Level
After the demonstration of the inhibitory effect of P-ERK1/2 on the expression of ABCC6 we wanted to clarify the mechanism of inhibition. To find out whether P-ERK1/2 acts at the transcriptional or the post-transcriptional level we made use of the URG7 transcript, a splice variant of ABCC6 ending in the 2nd intron of the gene (31). This short, truncated mRNA is transcribed from the same promoter as the ABCC6 transcript but have a different 3′-untranslated region leading to different post-transcriptional regulatory mechanisms (splicing and probably mRNA stability). We hypothesized that if P-ERK1/2 acts at the transcription initiation level, the two transcripts should behave similarly. In contrast, if P-ERK1/2 acts at the post-transcriptional level the inhibition found in the case of ABCC6 would not be observed for URG7.
After the 24-h treatment with PMA, HGF, U0126, and the respective co-treatments, we observed the same effect on the expression level of URG7 as on the ABCC6 transcript (Fig. 5). This result strongly suggests that the ERK1/2 cascade inhibits expression of the ABCC6 gene via regulation of transcription initiation.
FIGURE 5.
ABCC6 expression is regulated via ERK1/2 at the transcriptional level. HepG2 cells were treated with the indicated compounds or vehicle for controls (dimethyl sulfoxide) in serum-free conditions for 24 h. Relative ABCC6 and URG7 expression levels were determined by quantitative PCR after normalization to the ABL housekeeping gene expression level as described under “Experimental Procedures.” Expression levels are indicated as a percent of untreated control. Four parallels were used for each condition. The experiments were repeated three times ±S.E., *, p < 0.05.
Identification of the Regulatory Element in the ABCC6 Promoter Modulated by ERK1/2 Activation
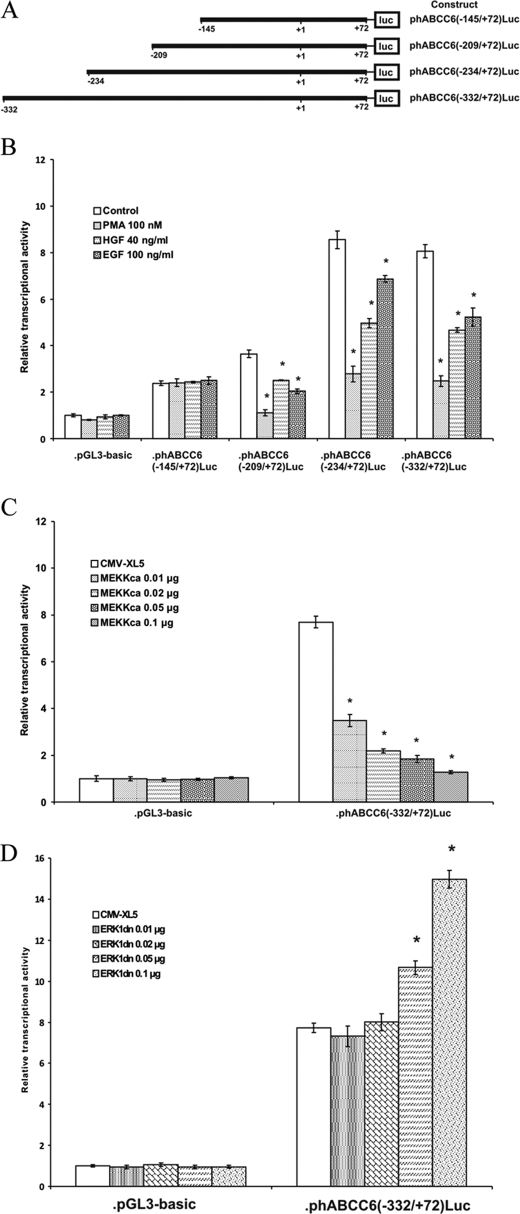
To identify the regulatory elements involved in the ERK1/2 response, we carried out luciferase reporter gene assays. In these experiments we made use of some of our previously tested promoter constructs (Fig. 6A) (17, 21). We observed similar promoter activities in the luciferase assay as in our earlier reports after transient transfection of the constructs into HepG2 cells. Indeed, the −145/+72 construct showed a low basal promoter activity. Although the −209/+72 was slightly more active the −234/+72 construct harbored a strong activator element (17).
FIGURE 6.
Identification of the ERK1/2 response region in the ABCC6 promoter by reporter gene assay. A, reporter plasmids containing serial deletions of the human ABCC6 gene promoter. Numbering starts at the translation start site. B, interaction of the ERK1/2 cascade with the proximal promoter of the ABCC6 gene. Promoter constructs were transiently transfected to HepG2 cells. 24 h after transfection, cells were treated with PMA, EGF, or HGF at the indicated concentrations or vehicle. Cells were treated with the compounds for 24 h and promoter activity was then measured by luciferase assay. Results of the assay were standardized against control reporter activity (pCMV-SEAP) and expressed as fold-induction over the control value (activity of empty vector (pGL3-basic) in control treated cells), mean ± S.E. (n = 5), *, p < 0.05. C, dose-response effect of overexpression of constitutively active MEKK1 (MEKKca) on the transcriptional activity of ABCC6 promoter in HepG2 cells. Results after standardization were expressed as fold-induction over control value (activity of empty vector (pGL3-basic co-transfected with MEKKca or pCMV-XL5 empty expression vector), mean ± S.E. (n = 5), *, p < 0.05. D, dose-response effect of the overexpression of dominant-negative ERK1 (ERK1dn) on the transcriptional activity of the ABCC6 promoter in HepG2 cells. Results after standardization were expressed as fold-induction over control value (activity of empty vector (pGL3-basic) co-transfected with ERK1dn or pCMV-XL5 empty expression vector, mean ± S.E. (n = 5), *, p < 0.05.
When we treated our constructs with PMA, HGF, or EGF we could observe ∼50% inhibition of luciferase activity for all constructs extending in the 5′ region of the promoter beyond position −145 (Fig. 6B). In our following experiments we co-transfected the ABCC6 −332/+72 luciferase promoter construct with a caMEKK construct, which phosphorylates and thereby activates ERK1/2. We demonstrated that caMEKK inhibited in a dose-dependent manner the transcriptional activity of the ABCC6 promoter construct (Fig. 6C). We also tested a dominant-negative ERK1 construct, which inhibits ERK1 activity. Co-transfection of this construct with the ABCC6 −332/+72 construct induced luciferase activity also in a dose-dependent manner (Fig. 6D). These data confirmed that activation of the ERK1/2 cascade inhibits ABCC6 expression by interfering with the transcription initiation of the gene. Furthermore, as the −209/+72 promoter fragment was fully ERK1/2 sensitive in contrast to the −145/+72 construct, we could also locate the interaction site on the proximal promoter between −209 and −145 bp, relative to the translation initiation site.
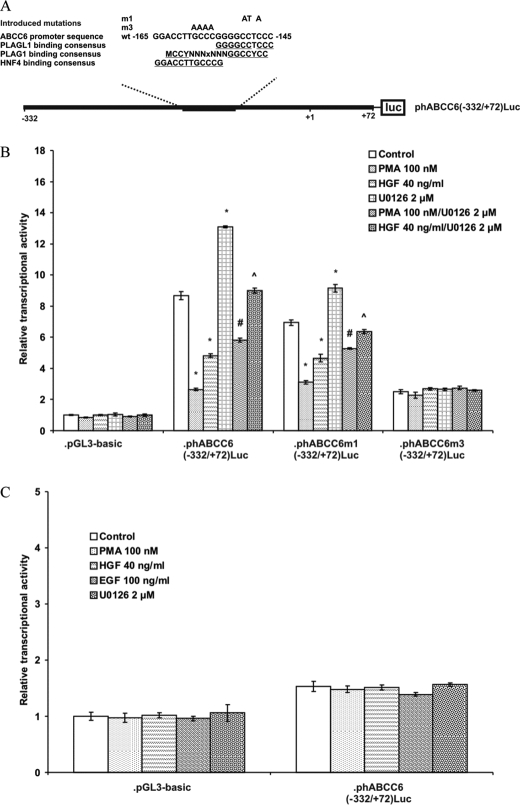
We have shown previously that the −209/−145 region harbors a PLAG transcription factor and another liver-specific transcription factor binding site not characterized in detail (17, 21). Now, we performed a further in silico search to identify liver-specific transcription factor(s), with a response element in the −209/−145 region of the promoter. According to a recent publication hepatocyte nuclear factor 4α (HNF4α) might be a good candidate to be the unidentified liver-specific factor as the study by Bolotin et al. (32) predicts HNF4α binding between −166 and −154 relative to the translation initiation site (Fig. 7A).
FIGURE 7.
Identification of the ERK1/2 response element in the ABCC6 promoter by reporter gene assay. A, part of the proximal promoter fragment of the human ABCC6 and location of PLAG binding sites and introduced mutations. PLAG1 and PLAGL1 consensus binding sequences are overlaid on the wild-type sequence with the conserved nucleotides underlined. A new HNF4α binding site was identified as illustrated on the scheme. m1 and m3 mutation constructs were used to test the role of the region in the ERK1/2 and HNF4α response. B, luciferase assay with wild-type, m1 and m3 constructs. 24 h after transient transfection, HepG2 cells were treated with PMA, HGF, U0126, and the respective co-treatments or vehicle at the indicated concentrations for 24 h. Results of the assay were standardized against control reporter activity (pCMV-SEAP) and expressed as fold-induction over control value (activity of empty vector (pGL3-basic) in control treated cells), mean ± S.E. (n = 4), *, p < 0.05, statistically different from control treated cells; #, p < 0.05, statistically different from cells treated with PMA; ⋀, p < 0.05, statistically different from cells treated with HGF. C, luciferase reporter gene assay in HeLa cells. Wild-type ABCC6 luciferase construct was treated with PMA, HGF, EGF, U0126, or vehicle (dimethyl sulfoxide) at the indicated concentrations for 24 h. Results of the assay were standardized against control reporter activity (pCMV-SEAP) and expressed as fold-induction over control value (activity of empty vector (pGL3-basic) in control treated cells), mean ± S.E. (n = 5).
We used wild-type and mutant promoter constructs to verify whether the ERK1/2 cascade exerts its activity via these factors and/or binding sites. The construct m1(−332/+72) contains a mutated PLAG site, whereas the construct m3(−332/+72) harbors a stretch of mutation between −155 and −158 (21). These mutated nucleotides overlap with the sequence conferring liver-specific expression to ABCC6 and which is a putative HNF4α binding site (see above).
As observed previously, the construct m1(−332/+72) showed an ∼20% reduced basal activity relative to the wild-type (17). However, it remained fully sensitive to treatments with PMA, HGF, EGF, U0126, and the respective co-treatments showed similar effects on the promoter activity as seen for the wild-type construct (Fig. 7B). These results strongly suggested that the ERK1/2-related inhibition of ABCC6 expression is not mediated via the PLAG transcription factor family. In contrast to these data, the m3(−332/+72) construct showed strikingly different behavior. The basal promoter activity of this construct was only 30% of that of the wild-type construct, similar to the −145/+72 wild-type construct (21). The m3(−332/+72) construct was also insensitive to modulation of the ERK cascade (Fig. 7B) suggesting that the liver-specific response element, the putative HNF4α binding site, is mediating the ERK1/2 signal.
To verify this hypothesis we investigated PMA, HGF, EGF, and U0126 sensitivity of the wild-type −332/+72 ABCC6 promoter construct in HeLa cells, where both endogenous ABCC6 (11) and HNF4α expression are absent (33). The promoter activity of the construct was similar to the −145/+72 construct expressed in HepG2 cells, and the construct remained insensitive to the treatments affecting the ERK1/2 pathway (Fig. 7C), suggesting that the regulatory mechanisms deciphered in the present study are indeed cell-type specific.
Identification of the ERK1/2-dependent Factor Responsible for the Regulation of ABCC6 Expression
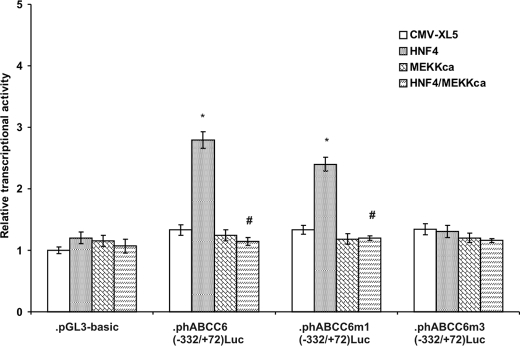
To further prove the regulatory role of HNF4α we chose HeLa as model cells because HepG2 express endogenous HNF4α at a high level. We co-expressed the wild-type −332/+72 luciferase construct and HNF4α in HeLa cells and observed as a result a more than 2-fold induction of the transcriptional activity relative to the promoter construct expressed alone (Fig. 8). The construct abrogating the binding of PLAG factors only slightly diminished the activation by HNF4α. In contrast to the first two constructs, activation of the m3(−332/+72) by HNF4α was completely abolished. These data strongly suggest that the predicted HNF4α binding site is indeed functional and plays a crucial role in regulation in the expression of ABCC6.
FIGURE 8.
HNF4α up-regulates ABCC6 expression in an ERK1/2-dependent manner and confers tissue specificity. Luciferase reporter gene assay in HeLa cells. Wild-type, m1 and m3 constructs were co-transfected with HNF4α, MEKKca, or both or pCMV-XL5 empty expression vector. Results after standardization were expressed as fold-induction over control value (activity of empty vector (pGL3-basic co-transfected with the corresponding expression vectors), mean ± S.E. (n = 5), *, p < 0.05, statistically different from cells transfected with control empty vector (CMV-XL5); #, p < 0.05, statistically different from cells transfected with HNF4α expression vector.
Next we asked whether activation of ABCC6 expression by HNF4α can be prevented by the ERK1/2 pathway. When the luciferase promoter constructs were co-transfected with caMEKK, activation by HNF4α was again prevented, whereas caMEKK alone had no effect on the transcriptional activity of the luciferase constructs (Fig. 8).
In a final set of experiments we performed ChIP assay to prove the ERK1/2 activation-dependent binding of HNF4α to the ABCC6 promoter in its natural chromatin environment (Table 1). HNF4α binding was tested in control, HGF-, and PMA-treated HepG2 cells. In control cells the HNF4α-bound ABCC6 promoter fragments showed a very strong enrichment in the immunoprecipitated fraction in contrast to the control GAPDH promoter. These data demonstrate the specific binding of the transcription factor to ABCC6 in the chromatin environment. The different treatments did not modify HNF4α binding to GAPDH. In contrast, HGF and PMA treatments diminished HNF4α binding to the ABCC6 promoter by 30–50%. Altogether these data show that HNF4α is a major activator of ABCC6 transcription and its action is completely prevented by activation of the ERK signaling cascade.
TABLE 1.
HNF4 binding to the ABCC6 promoter in HepG2 cells demonstrated by chromatin immunoprecipitation
Amount of immunoprecipitated cognate gene promoter sequences was determined by real time PCR and expressed in arbitrary relative values, standardized against amount of input material (mean ± S.E., n = 4).
| Treatment | Gene promoter antibody | Control IgG | Anti-HNF4 |
|---|---|---|---|
| Control | ABCC6 | 2.97 ± 0.12 | 19.97 ± 0.49 |
| GAPDH | 1.26 ± 0.32 | 1.38 ± 0.31 | |
| PMA, 100 nm | ABCC6 | 3.14 ± 0.09 | 10.72 ± 1.02a |
| GAPDH | 1.07 ± 0.34 | 1.39 ± 0.26 | |
| HGF, 40 ng/ml | ABCC6 | 2.96 ± 0.10 | 13.56 ± 0.22a |
| GAPDH | 1.17 ± 0.31 | 1.56 ± 0.18 |
a Statistically significant at p < 0.05.
DISCUSSION
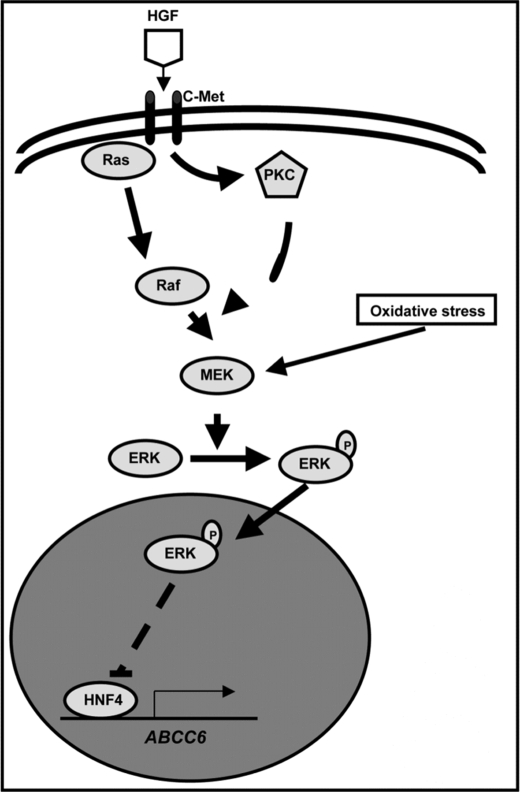
The aim of the present study was to explore the regulation of ABCC6 gene expression by physiologically relevant signaling pathways. Previous reports showed that the transcriptional regulation of ABCC6 depends on various factors, such as NF-κB, RXR, SP1, and PLAG (16–18, 21). However, none of these findings were embedded in a physiological context. Here we demonstrated that various stimuli converging to the activation of the ERK1/2 cascade inhibit the expression of ABCC6 (Fig. 9). We also showed that inhibition of the direct transcriptional activation of the gene by HNF4α is responsible for this effect.
FIGURE 9.
Schematic presentation of ABCC6 expression regulation by the ERK1/2 pathway. The ERK1/2 kinase pathway is represented in circles, as well as the HNF4α transcription factor. Arrows indicate positive regulations.
As a starting point of our study, we observed that both hepatocyte and epidermal growth factors down-regulated the expression of ABCC6 (Fig. 1). Upon binding to its receptor, HGF activates several major highly interconnected cellular signaling cascades, like the PI 3-kinase, and the PKC and ERK cascades (34, 35). Our experiments clearly pointed to the primary role of the ERK1/2 signaling cascade in regulation of ABCC6 gene expression. First, ABCC6 expression was induced by U0126, a specific inhibitor of MEK1/2, the kinase upstream of ERK1/2, whereas U0124, the biologically inactive stereoisomer of U0126, remained ineffective (not shown). Second, HGF- and EGF-induced down-regulation of ABCC6 expression was prevented by co-treatment with U0126 (Fig. 2C). Furthermore, vitamin K3, a source of strong oxidative stress (30) reduced the expression of ABCC6, an effect prevented by U0126 (Fig. 4). Other molecules inducing oxidative stress had similar effects (Fig. 4 and data not shown). These observations were particularly important because they proved that various stimuli converging to activation of the ERK1/2 pathway have the same effect on the expression of ABCC6. These effects were specific to ABCC6 because other members of the ABCC subfamily of ABC transporters, which are evolutionarily closely related to ABCC6, reacted differently to the same stimuli.4 Third, the ABCC6 gene expression level could be correlated to the phosphorylation level of ERK1/2, which indicates the activity of the kinases (Fig. 3). Finally, we have also demonstrated that transfection of a constitutively active MEKK1 inhibited, whereas a dominant-negative ERK1 activated a luciferase construct containing wild-type ABCC6 promoter fragment (Fig. 6, C and D).
Our findings suggested that in the regulation of ABCC6 expression the PKCs are also involved in the signaling cascade downstream of HGF (Fig. 2B). PKCs seem to also act via activation of the ERK1/2 (Fig. 2B). Because the inhibition of ABCC6 by PMA treatment alone was BIMI insensitive, we consider that members of the so-called atypical PKC subfamily are mediating this effect, as resistance to BIMI inhibition is a hallmark of these PKCs (36).
After identification of the ERK1/2 pathway as the major regulator of ABCC6 expression, we determined the responsible molecular mechanisms. As a first step, we used an alternative transcript of ABCC6, called URG7 (31), and showed that the expression level of this splice variant changes in parallel with that of ABCC6 upon the various stimuli (Fig. 5) strongly suggesting that the effect of ERK1/2 is transcriptional.
The luciferase reporter gene assays demonstrated that the ERK1/2 effect is cell type-specific and the response element to the cascade is located in the ABCC6 proximal promoter between −209 and −145 bp from the translation initiation site. This was consistent with our previous findings, revealing that the promoter fragment investigated contains a liver-specific response element (21). This element was identified by mutations in the −165/−150 region. Although PLAG transcription factors bind to an overlapping site with the ERK1/2 response element we disclosed these factors as the final targets of the MAP kinase pathway, because the mutant luciferase construct not binding PLAG remained sensitive to the activation of ERK1/2.
Several lines of evidence strongly suggested after further in silico search that HNF4α might be a good candidate to bind to the tissue-specific ERK1/2 response element. First, an HNF4α binding microarray identified a binding site at −166/−154 of the ABCC6 promoter (32) (Fig. 7A) and second, small interfering RNA against HNF4α significantly decreased the expression level of the ABCC6 gene in HepG2 cells (32). Third, another group by analyzing genome-wide HNF4α binding sites in human hepatocytes identified the ABCC6 promoter by ChIP-on-chip as a functional binding site for the transcription factor (37). Furthermore, HNF4α is known to have strict tissue specificity (33), which mainly overlaps with the expression pattern of ABCC6 (11). Finally, the predicted sequence shows a high degree of evolutionary conservation (14 in a block of 16 nucleotides are identical in four species) (14). All these data pointed to a potential role of HNF4α in the ERK1/2-dependent regulation of ABCC6.
We hypothesized that ABCC6 reporter constructs have low basal expression levels and are not responding to ERK1/2 stimulation in HeLa cells because HNF4α is absent from this cell line. Indeed, co-expression of the wild-type construct and HNF4 in HeLa cells resulted in a substantial induction of luciferase activity, which reached a comparable level to that observed in HepG2 cells. In contrast, the mutated constructs could not be induced by co-expression with HNF4. The stretch of mutation introduced between nucleotides −158 and −155 in our construct destroyed completely the transcriptional activity of the −332/−145 fragment in HepG2 cells by reducing it to the basal activity level of the −145/+72 fragment similar in various cell lines. This suggested that the binding of HNF4 is required for the binding of other activator protein(s) between −234 and −209 and is responsible for tissue-specific expression of the gene.
The induced expression level in HeLa upon co-expression of HNF4α and the wild-type luciferase construct was completely abolished by co-expression of constitutively active MEKK1 (Fig. 8) suggesting that HNF4α is a crucial regulatory factor of ABCC6 expression and is under the negative control of ERK1/2 (Fig. 9). This hypothesis was proven by the ChIP assay performed on untreated or HGF- or PMA-treated HepG2 cells (Table 1).
It has been reported that HNF4α is under the control of various kinases (PKC and PKA) (38, 39) and the role of ERK1/2 in the regulation of HNF4α has also been reported (40, 41). This latter study suggested that the decreased endogenous HNF4α protein level due to various stimuli led to down-regulation of the expression of the target gene. Although we do not know whether the molecular mechanism of inhibition is direct or indirect, the change in the protein level was most probably not the case in our HeLa cell system, as HNF4α was driven from a CMV promoter and not from its own promoter. However, in HepG2 cells, where the endogenous HNF4α was mediating the ERK1/2 effect the changes of its own protein level could also play a role in the regulation of ABCC6 expression.
The major role of HNF4α has also been proposed for regulation of the murine ABCC6, where it acts on the same evolutionarily highly conserved site (19) and the ABCC2 genes (42, 43). Surprisingly, our data do not suggest implication of the ERK1/2 cascade in regulation of ABCC2 expression.4 This is in agreement with the different effect of oxidative stress on expression of the two ABC transporter genes involving ERK1/2 for ABCC6 (this study) and only HNF4α for ABCC2 (43).
Finally, what can we learn from our data about the physiological role and regulation of ABCC6? Under physiological circumstances in resting hepatocytes where the ABCC6 expression reaches its highest level the pathway has a low basal activity, which contributes to attachment of the cells to each other. Most probably in the dividing HepG2 cells, ERK1/2 is much more active leading to a lower ABCC6 expression level. ERK1/2 is activated dynamically by a multitude of signals (e.g. growth factors, G-protein coupled receptors, and cell growth). Similarly, oxidative stress inhibits at least partially the expression of the ABCC6 gene via activation of the pathway, as we have shown here. This suggests that although in the absence of functional ABCC6, leading to PXE, chronic oxidative stress was observed in dermal fibroblast and other cell types (44); the physiological role of MRP6 may not be the protection of the cells from the oxidative stress. Because vitamin K3 strongly inhibited expression of the gene it seems to be unlikely that vitamin K3 itself or one of its derivatives would be the most important physiological substrate of the MRP6 protein, in contrast to a current hypothesis (45).
What might be the consequence of our data with respect to PXE? We presume that some of the mutations do not cause the complete loss of ABCC6/MRP6 protein function. In these cases an increase of gene expression might diminish the severity of the PXE phenotype. Heterozygous carriers were shown to have increased risk of developing coronary artery disease (46, 47). Patients suffering from β-thalassemia frequently develop secondary PXE without mutations in the ABCC6 gene may be due to chronic oxidative stress (48). Both groups of patients may benefit from a therapy based on the induction of ABCC6 expression. According to the observations described in the present study this therapy can be based on available inhibitors of the ERK1/2 pathway. Alternatively, induction of HNF4α, the master regulator of liver metabolism by influencing the metabolic state of the hepatocytes might increase the expression of ABCC6 with fewer side effects.
Acknowledgments
We thank Drs. Faragó, Sladek, Symmons, and Szakács for helpful discussions.
This work was supported in part by National Research Fund of Hungary Grants NI68950 and PD79183, National Development Agency Grant KMOP-1.1.2-07/1-2008-0003, Polanyi Mihaly Grant KFKT-1-2006-0006, a grant called “Lendület” from the Hungarian Academy of Sciences, and a Polish-Hungarian academic cooperation grant.
T. Aranyi, H. de Boussac, G. Köblös, A. Váradi, and T. Aranyi, unpublished data.
- PXE
- pseudoxanthoma elasticum
- ERK1/2
- extracellular signal-regulated kinases 1 and 2
- HNF4α
- hepatocyte nuclear factor 4α
- HGF
- hepatocyte growth factor
- EGF
- epidermal growth factor
- PMA
- phorbol myristate acetate
- tBHQ
- tert-butyl hydroquinone
- BIMI
- bisindolylmaleimide I
- MEK1/2
- MAP kinase or ERK kinase 1/2
- ChIP
- chromatin immunoprecipitation
- TGFβ
- transforming growth factor β
- PI
- phosphatidylinositol
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- caMEKK
- constitutively active MEKK1
- CMV
- cytomegalovirus
- PKA
- protein kinase A.
REFERENCES
- 1.Li Q., Jiang Q., Pfendner E., Váradi A., Uitto J. (2009) Exp. Dermatol. 18, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergen A. A., Plomp A. S., Schuurman E. J., Terry S., Breuning M., Dauwerse H., Swart J., Kool M., van Soest S., Baas F., ten Brink J. B., de Jong P. T. (2000) Nat. Genet. 25, 228–231 [DOI] [PubMed] [Google Scholar]
- 3.Le Saux O., Urban Z., Tschuch C., Csiszar K., Bacchelli B., Quaglino D., Pasquali-Ronchetti I., Pope F. M., Richards A., Terry S., Bercovitch L., de Paepe A., Boyd C. D. (2000) Nat. Genet. 25, 223–227 [DOI] [PubMed] [Google Scholar]
- 4.Ringpfeil F., Lebwohl M. G., Christiano A. M., Uitto J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfendner E. G., Uitto J., Gerard G. F., Terry S. (2008) Exp. Opin. Med. Diagn. 2, 63–79 [DOI] [PubMed] [Google Scholar]
- 6.Cai L., Lumsden A., Guenther U. P., Neldner S. A., Zäch S., Knoblauch H., Ramesar R., Hohl D., Callen D. F., Neldner K. H., Lindpaintner K., Richards R. I., Struk B. (2001) J. Mol. Med. 79, 536–546 [DOI] [PubMed] [Google Scholar]
- 7.Symmons O., Váradi A., Arányi T. (2008) Mol. Biol. Evol. 25, 2601–2613 [DOI] [PubMed] [Google Scholar]
- 8.Hendig D., Schulz V., Arndt M., Szliska C., Kleesiek K., Götting C. (2006) Clin. Chem. 52, 227–234 [DOI] [PubMed] [Google Scholar]
- 9.Hendig D., Zarbock R., Szliska C., Kleesiek K., Götting C. (2008) Clin. Biochem. 41, 407–412 [DOI] [PubMed] [Google Scholar]
- 10.Bacchelli B., Quaglino D., Gheduzzi D., Taparelli F., Boraldi F., Trolli B., Le Saux O., Boyd C. D., Ronchetti I. P. (1999) Mod. Pathol. 12, 1112–1123 [PubMed] [Google Scholar]
- 11.Kool M., van der Linden M., de Haas M., Baas F., Borst P. (1999) Cancer Res. 59, 175–182 [PubMed] [Google Scholar]
- 12.Jiang Q., Endo M., Dibra F., Wang K., Uitto J. (2009) J. Invest. Dermatol. 129, 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iliás A., Urbán Z., Seidl T. L., Le Saux O., Sinkó E., Boyd C. D., Sarkadi B., Váradi A. (2002) J. Biol. Chem. 277, 16860–16867 [DOI] [PubMed] [Google Scholar]
- 14.Arányi T., Ratajewski M., Bardóczy V., Pulaski L., Bors A., Tordai A., Váradi A. (2005) J. Biol. Chem. 280, 18643–18650 [DOI] [PubMed] [Google Scholar]
- 15.Douet V., Heller M. B., Le Saux O. (2007) Biochem. Biophys. Res. Commun. 354, 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratajewski M., Bartosz G., Pulaski L. (2006) Biochem. Biophys. Res. Commun. 350, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 17.Ratajewski M., Van de Ven W. J., Bartosz G., Pulaski L. (2008) Hum. Genet. 124, 451–463 [DOI] [PubMed] [Google Scholar]
- 18.Jiang Q., Matsuzaki Y., Li K., Uitto J. (2006) J. Invest. Dermatol. 126, 325–335 [DOI] [PubMed] [Google Scholar]
- 19.Douet V., VanWart C. M., Heller M. B., Reinhard S., Le Saux O. (2006) Biochim. Biophys. Acta 1759, 426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz V., Hendig D., Henjakovic M., Szliska C., Kleesiek K., Gotting C. (2006) Hum. Mutat. 27, 831. [DOI] [PubMed] [Google Scholar]
- 21.Ratajewski M., de Boussac H., Pulaski L. (2009) Biochem. Biophys. Res. Commun. 383, 73–77 [DOI] [PubMed] [Google Scholar]
- 22.Arányi T., Váradi A., Simon I., Tusnády G. E. (2006) BMC Bioinformatics 7, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bors A., Ribiczey P., Köblös G., Brózik A., Ujfaludi Z., Magócsi M., Váradi A., Tordai A., Kovács T., Arányi T. (2008) Anal. Biochem. 372, 261–263 [DOI] [PubMed] [Google Scholar]
- 24.Buday L., Downward J. (1993) Cell 73, 611–620 [DOI] [PubMed] [Google Scholar]
- 25.Whitmarsh A. J., Shore P., Sharrocks A. D., Davis R. J. (1995) Science 269, 403–407 [DOI] [PubMed] [Google Scholar]
- 26.Sipeki S., Bander E., Buday L., Farkas G., Bácsy E., Ways D. K., Faragó A. (1999) Cell Signal. 11, 885–890 [DOI] [PubMed] [Google Scholar]
- 27.Gujdár A., Sipeki S., Bander E., Buday L., Faragó A. (2004) Cell Signal. 16, 505–513 [DOI] [PubMed] [Google Scholar]
- 28.Schönwasser D. C., Marais R. M., Marshall C. J., Parker P. J. (1998) Mol. Cell. Biol. 18, 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng T. C., Fukada T., Tonks N. K. (2002) Mol. Cell 9, 387–399 [DOI] [PubMed] [Google Scholar]
- 30.Czaja M. J., Liu H., Wang Y. (2003) Hepatology 37, 1405–1413 [DOI] [PubMed] [Google Scholar]
- 31.Lian Z., Liu J., Pan J., Satiroglu Tufan N. L., Zhu M., Arbuthnot P., Kew M., Clayton M. M., Feitelson M. A. (2001) Hepatology 34, 146–157 [DOI] [PubMed] [Google Scholar]
- 32.Bolotin E., Liao H., Ta T. C., Yang C., Hwang-Verslues W., Evans J. R., Jiang T., Sladek F. M. (2010) Hepatology 51, 642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sladek F. M., Zhong W. M., Lai E., Darnell J. E., Jr. (1990) Genes Dev. 4, 2353–2365 [DOI] [PubMed] [Google Scholar]
- 34.You W. K., McDonald D. M. (2008) BMB Rep. 41, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kermorgant S., Parker P. J. (2005) Cell Cycle 4, 352–355 [DOI] [PubMed] [Google Scholar]
- 36.Reyland M. E. (2009) Front. Biosci. 14, 2386–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Volkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., Fraenkel E., Bell G. I., Young R. A. (2004) Science 303, 1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun K., Montana V., Chellappa K., Brelivet Y., Moras D., Maeda Y., Parpura V., Paschal B. M., Sladek F. M. (2007) Mol. Endocrinol. 21, 1297–1311 [DOI] [PubMed] [Google Scholar]
- 39.Viollet B., Kahn A., Raymondjean M. (1997) Mol. Cell Biol. 17, 4208–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy S., Yang W., Taylor D. G., Shen X., Oxender D., Kust G., Leff T. (1999) J. Biol. Chem. 274, 33050–33056 [DOI] [PubMed] [Google Scholar]
- 41.Hatzis P., Kyrmizi I., Talianidis I. (2006) Mol. Cell Biol. 26, 7017–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qadri I., Hu L. J., Iwahashi M., Al-Zuabi S., Quattrochi L. C., Simon F. R. (2009) Toxicol. Appl. Pharmacol. 234, 281–292 [DOI] [PubMed] [Google Scholar]
- 43.Qadri I., Iwahashi M., Kullak-Ublick G. A., Simon F. R. (2006) Mol. Pharmacol. 70, 627–636 [DOI] [PubMed] [Google Scholar]
- 44.Pasquali-Ronchetti I., Garcia-Fernandez M. I., Boraldi F., Quaglino D., Gheduzzi D., De Vincenzi Paolinelli C., Tiozzo R., Bergamini S., Ceccarelli D., Muscatello U. (2006) J. Pathol. 208, 54–61 [DOI] [PubMed] [Google Scholar]
- 45.Borst P., van de Wetering K., Schlingemann R. (2008) Cell Cycle 7, 1575–1579 [DOI] [PubMed] [Google Scholar]
- 46.Köblös G., Andrikovics H., Prohászka Z., Tordai A., Váradi A., Arányi T. (2010) Genet. Test Mol. Biomarkers 14, 75–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trip M. D., Smulders Y. M., Wegman J. J., Hu X., Boer J. M., ten Brink J. B., Zwinderman A. H., Kastelein J. J., Feskens E. J., Bergen A. A. (2002) Circulation 106, 773–775 [DOI] [PubMed] [Google Scholar]
- 48.Hamlin N., Beck K., Bacchelli B., Cianciulli P., Pasquali-Ronchetti I., Le Saux O. (2003) Br. J. Haematol. 122, 852–854 [DOI] [PubMed] [Google Scholar]