Abstract
The tumor suppressor p53 protein is tightly regulated by a ubiquitin-proteasomal degradation mechanism. Several E3 ubiquitin ligases, including MDM2 (mouse double minute 2), have been reported to play an essential role in the regulation of p53 stability. However, it remains unclear how the activity of these E3 ligases is regulated. Here, we show that the HECT-type E3 ligase Smurf1/2 (Smad ubiquitylation regulatory factor 1/2) promotes p53 degradation by enhancing the activity of the E3 ligase MDM2. We provide evidence that the role of Smurf1/2 on the p53 stability is not dependent on the E3 activity of Smurf1/2 but rather is dependent on the activity of MDM2. We find that Smurf1/2 stabilizes MDM2 by enhancing the heterodimerization of MDM2 with MDMX, during which Smurf1/2 interacts with MDM2 and MDMX. We finally provide evidence that Smurf1/2 regulates apoptosis through p53. To our knowledge, this is the first report to demonstrate that Smurf1/2 functions as a factor to stabilize MDM2 protein rather than as a direct E3 ligase in regulation of p53 degradation.
Keywords: E3 Ubiquitin Ligase, p53, Protein Degradation, Ubiquitin, Ubiquitination, MDM2, MDMX, Smurf
Introduction
The p53 protein is a key regulator of cell cycle arrest, DNA repair, and apoptosis and has been characterized as “the guardian of the genome” (1, 2). In quiescent cells, the level of p53 protein is tightly controlled at a low level; however, in response to cellular stress, such as DNA damage and oncogenic insult, p53 is accumulated and activated (2, 3). High levels of p53 protein result in changes of cell growth arrest and apoptosis.
The p53 protein is tightly regulated by ubiquitin-mediated proteasomal degradation pathway, among which ubiquitin ligases (E3s)4 specify the p53 protein as a substrate for ubiquitylation. Several E3s, including MDM2 (4), COP1 (5), Pirh2 (6), ARF-BP1 (7), and CHIP (8), have been reported to mediate p53 ubiquitylation and degradation. Among these E3 ligases, it is well established that MDM2 is a major negative regulator of p53 (9, 10). Homozygous deletion of Mdm2 in mice results in early embryonic lethality, and p53 deficiency rescues the lethal phenotype completely (11, 12). In physiological conditions, MDM2 maintains the p53 protein at an adequate level. However, MDM2 is reported to be frequently amplified or overexpressed in human cancers, many of which lack mutations in the p53 gene (13). The role of overexpressed MDM2 in human cancers is believed to be functionally equivalent to p53 mutation (14).
MDM2 is a RING (really interesting new gene) finger domain E3 ligase and directly interacts with p53 during the ubiquitylation process. The activity of MDM2 is regulated by several mechanisms. First, amounting evidence is shown that co-factors are involved in the regulation of MDM2-mediated p53 degradation. For example, ARF is reported to interfere with the MDM2-p53 interaction, thus to inhibit MDM2-mediated p53 ubiquitylation (15–18). YY1 and PACT are demonstrated to enhance the degradation of p53 by promoting the interaction of MDM2 with p53 (19, 20). Conversely, binding of the ribosomal proteins L5, L11, and L23 to MDM2 inhibits the activity of MDM2 and plays a crucial role in p53 activation upon ribosomal stress (21–23).
Importantly, MDM2 activity is also regulated by a protein degradation mechanism. MDM2 is a short lived protein (9, 10). The stability of MDM2 is tightly regulated by different mechanisms. Although it has been reported that MDM2 protein stability is regulated by other E3 ligases (for example, PCAF) (24), the degradation of MDM2 is mainly induced by a self-ubiquitylation or autodegradation mechanism (25, 26). Furthermore, the self-ubiquitylation or autodegradation is mediated by homodimerization of MDM2 itself.
MDMX (also known as MDM4) has been shown to play a critical role in preventing MDM2 self-ubiquitylation or autodegradation. MDMX is found to interact with MDM2 to form a heterodimer. The heterodimerization of MDM2 with MDMX blocks the homodimerization of MDM2 and inhibits MDM2 self-ubiquitylation (27–29). In such a way, MDMX regulates the activity of MDM2 and furthers the level of p53.
However, it remains unclear how the heterodimerization of MDM2 and MDMX occurs. In this study, we demonstrated that the HECT-type E3 ubiquitin ligases Smurf1 and -2 enhance the heterodimerization of MDM2 and MDMX and block the homodimerization of MDM2. We provide evidence that Smurf1 and -2 play an important role in maintenance of MDM2 stability, by such a way Smurf1 and -2 inhibit p53 activity and block apoptosis.
MATERIALS AND METHODS
Plasmid Constructs
Full-length, truncated, and point mutations of Smurf1, Smurf2, MDM2, MDMX, and Smad5 were constructed by inserting PCR-amplified fragments into the related vectors. Detailed construct information is available upon request. pCMV/p53, pCMV/MDM2, and pcDNA3/poly-HA-tagged ubiquitin were gifts from Dr. Yue Xiong. 6Myc-Smurf1 wild type, 6Myc-Smurf1-C699A, and FLAG-Smurf1 were provided by Dr. Kohei Miyazono. HA-MDMX was provided by Dr. Geoffrey M. Wahl. Constructs of other Nedd4 family members were kindly provided by Dr. Wesley I. Sundquist.
Cell Culture and Transfection
Human embryonic kidney HEK293T cells, human breast cancer MCF7 cells, human colon cancer HCT116 cells (p53+/+ and p53−/−, a kind gift from Dr. Qimin Zhan), and p53−/−Mdm2−/− MEF cells (a kind gift from Dr. Mian Wu) were cultured in Dulbecco's modified Eagle's medium (Hyclone) containing 10% fetal bovine serum (Hyclone). Human lung adenocarcinoma H1299 cells were maintained in RPMI 1640 medium (Hyclone) with 10% fetal bovine serum. Mammalian cells were transfected with Lipofectamine 2000 (Invitrogen) or FuGENE HD (Roche Applied Science) according to the manufacturer's protocols.
Antibodies
Anti-Myc antibody was purchased from Clontech. Anti-FLAG M2 antibody was from Sigma. Anti-MDMX antibody was from Bethyl Laboratories. Anti-Smurf1 and anti-Smurf2 antibodies were from Abcam. The antibodies against p53 (DO-1), MDM2 (SMP14), and glyceraldehyde-3-phosphate dehydrogenase (6C5) were purchased from Santa Cruz Biotechnology. Anti-HA (12CA5) antibody was from Roche Applied Science. Anti-His and anti-GST antibodies were from Tiangen.
Immunoprecipitation and Immunoblotting
Transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. After 48 h, cell lysates were prepared in HEPES lysis buffer (20 mm HEPES (pH 7.2), 50 mm NaCl, 0.5% Triton X-100, 1 mm NaF, and 1 mm dithiothreitol) supplemented with protease inhibitors. Immunoprecipitations were performed using the indicated primary antibody and protein A/G-agarose beads at 4 °C. Lysates and immunoprecipitates were examined using the indicated primary antibodies followed by detection with the related secondary antibody with a Super Signal chemiluminescence kit (Pierce).
GST Pulldown Assay
To detect the direct binding of Smurf1 with MDM2, bacteria-expressed GST or GST-MDM2 proteins were immobilized on glutathione-Sepharose 4B beads (Amersham Biosciences) and then incubated with His-Smurf1 for 8 h at 4 °C under rotation. Beads were washed with GST binding buffer (100 mm NaCl, 50 mm NaF, 2 mm EDTA, 1% Nonidet P-40, and protease inhibitor mixture) and proteins were eluted, followed by immunoblotting.
In Vivo and in Vitro Ubiquitylation Assays
For in vivo ubiquitylation assay, cells were treated with MG132 (20 μm; Sigma) for 8 h before harvesting. The cell lysis were prepared in modified RIPA lysis buffer (10 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5 mm EDTA, 1% (v/v) Nonidet P-40, 1% sodium deoxycholate, 0.025% SDS, protease inhibitors), immunoprecipitated with the indicated antibody, and detected by immunoblotting. For in vitro ubiquitylation assay, E1, UbcH5b, UbcH5c (E2), HA-Ub (all from Boston Biochem), GST-MDM2, and Smurf1 were incubated at 30 °C for 2 h and terminated with sample buffer.
RNA Interference
The siRNAs against Smurf1 (5′-GCAUCGAAGUGUCCAGAGAAG-3′), Smurf2 (5′-CCUUCUGUGUUGAACAUAA-3′), MDM2 (5′-UGGUUGCAUUGUCCAUGGC-3′), MDMX (5′-UCAAUCAGGUACGACCAAA-3′), NEDL1 (5′-GAUGCCAGCUCGUACUUUG-3′), NEDL2 (5′-GAUUGGACUUUAUCAUAUA-3′), and nontargeting control (5′-UUCUCCGAACGUGUCACGU-3′) were synthesized by Shanghai GenePharm. The siRNAs were transfected with a FuGENE HD (Roche Applied Science) reagent, and the interference efficiency was assessed by immunoblotting.
Reporter Assays
The luciferase reporter plasmid pG13-Luc (a gift from Dr. Bert Vogelstein) was transfected into the cells, and the reporter assays were performed as described (30).
Real Time Quantitative PCR Assay
Real time quantitative PCR was performed as described previously (31). Sequences of primers of the p53 targeted genes used in quantitative PCR assays are listed in supplemental Table S1.
Apoptosis Analysis
After transfection of the indicated plasmids or siRNAs in cells for 36 h, cells were collected, washed with phosphate-buffered saline three times, and resuspended in binding buffer and then propidium iodide and annexin-V-PE reagent were added and incubated at room temperature in the dark. Five minutes later, cells were quantified by flow cytometry. The percentage of apoptotic cells was determined by analysis of annexin V-positive cells.
Pulse-Chase Analysis
Pulse-chase metabolic labeling was performed as described (32). Briefly, MCF-7 cells were transfected with the plasmids as indicated. 36 h after transfection, cells were starved with Met-free Dulbecco's modified Eagle's medium (Invitrogen) with 5% dialyzed fetal calf serum (Invitrogen) for 1 h. Cells were then pulse-labeled with 50 μCi/ml [35S]methionine (PerkinElmer Life Sciences) for 30 min and then chased for the indicated times in regular media containing unlabeled methionine. At each time point of the chase, cell lysates were immunoprecipitated with a p53 antibody or an MDM2 antibody, resolved by SDS-PAGE, and visualized by autoradiography.
RESULTS
Smurf1/2 Negatively Regulates p53 Stability and Activity
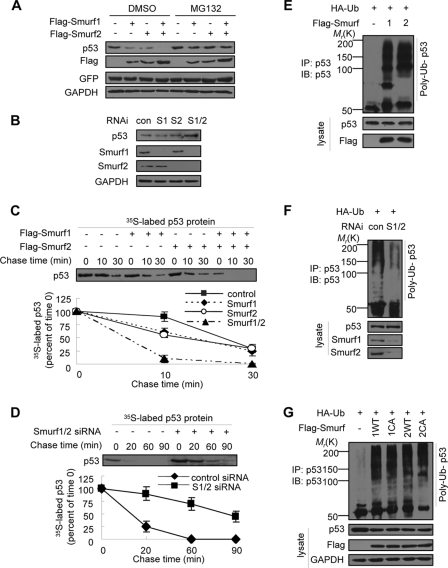
In our attempt to establish a depletion cell line for Smurf1/2, we discovered that the number of colonies was gradually reduced, and finally we failed to culture any clone but we easily obtained stable cell lines for overexpression of Smurf1/2 (data not shown). We speculated that Smurf1/2 might have a role in regulation of apoptosis. Based on the observation that other members of the Smurf family (e.g. WWP1 and NEDL1) regulate p53 activity (33, 34), we attempted to analyze the role of Smurf1/2 on p53 stability. When FLAG-tagged Smurf1 and/or -2 were transfected into MCF7 cells, we observed that the endogenous p53 protein levels were dramatically decreased (Fig. 1A). The effect of Smurf1/2 on p53 steady-state levels was blocked by a treatment with MG132, a proteasome inhibitor, indicating that Smurf1/2 promotes p53 degradation through a ubiquitin-proteasome pathway (Fig. 1A). To address the role of endogenous Smurf1/2, a Smurf1- or Smurf2-specific siRNA was used to deplete the expression of Smurf1/2 in MCF7 cells. A Western blot analysis demonstrated that the levels of endogenous p53 were increased, particularly when both Smurf1 and -2 were depleted by RNA interference (Fig. 1B). Consistently, a pulse-chase assay showed that overexpression of Smurf1/2 shortened (Fig. 1C), whereas depletion of Smurf1/2 prolonged (Fig. 1D), the half-life of p53. Because Smurf1/2 is a HECT (homologous to E6AP C terminus)-type E3 ubiquitin ligase, we questioned whether Smurf1/2 could catalyze the ubiquitylation of p53 in vivo. A Western blot analysis showed that overexpression of Smurf1/2 enhanced the poly-ubiquitylation of endogenous p53 (Fig. 1E), and depletion of Smurf1 and -2 abrogated the p53 poly-ubiquitylation (Fig. 1F).
FIGURE 1.
Smurf1/2 negatively regulates p53 stability. A, effect of exogenous Smurf1/2 on endogenous p53 levels. MCF7 cells were transfected with Smurf1 or Smurf2 constructs as indicated and treated with MG132 (20 μm) for 8 h before harvest. Endogenous p53 level was analyzed by immunoblotting (IB). GFP, green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, effect of Smurf1/2 depletion on endogenous p53 levels. MCF7 cells were transfected with single or double siRNAs against Smurf1 (S1) or Smurf2 (S2). Cell lysates were analyzed. con, control; RNAi, RNA interference. C and D, effect of Smurf1/2 on the half-life of p53 protein. MCF7 cells, transfected with Smurf plasmids or siRNAs, were pulse-labeled with [35S]methionine and then chased for the indicated times in medium containing unlabeled methionine. 35S-Labeled p53 in anti-p53 immunoprecipitates were quantified by phosphorimaging. Data are presented as mean ± S.D. (n = 3). E and F, effect of Smurf1/2 on p53 poly-ubiquitylation. MCF7 cells were transfected with Smurf siRNAs or plasmids and treated with MG132. Ubiquitylated p53 was immunoprecipitated (IP) by anti-p53 and analyzed by immunoblot with p53 antibody. G, Smurf1/2 promotes p53 poly-ubiquitylation independent of their E3 ligase activity. MCF7 cells were transfected with Smurf wild type or ligase-inactive mutant plasmids and treated with MG132. Ubiquitylated p53 was immunoprecipitated by anti-p53 and analyzed by immunoblotting with p53 antibody. Smurf1 CA, C699A; Smurf2 CA, C716A. Green fluorescent protein plasmid was co-transfected into the cells in all of the plasmid transfections. The expression of green fluorescent protein was analyzed to indicate the transfection efficiency, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control.
To further analyze the role of Smurf1/2 on the ubiquitylation of p53, we used a Smurf1 mutant (Smurf1-C669A) and a Smurf2 mutant (Smurf2-C716A) to perform a ubiquitylation assay. Because these two mutants are catalytic inactive for Smurf1/2 activity as the E3 ubiquitin ligase, we expected that the mutants would fail to mediate the ubiquitylation of p53. To our surprise, we observed that these mutants mediated the ubiquitylation of p53 as well as the wild type Smurf1/2 (Fig. 1G). This result suggests that Smurf1/2 promotes p53 ubiquitylation independent of their E3 ligase activity. This unexpected result encouraged us to speculate that Smurf1/2 might function as a co-factor with other E3 ligase to mediate p53 ubiquitylation.
Smurf1/2 Regulates p53 Stability Dependent on MDM2
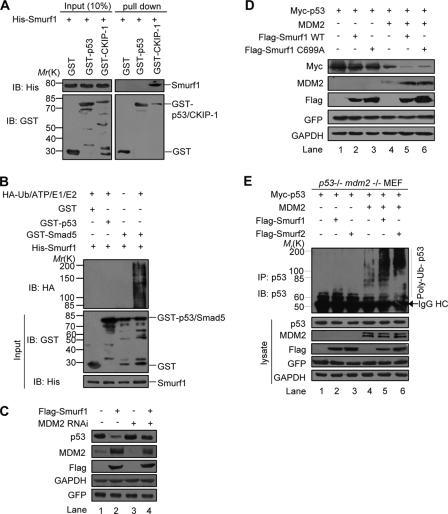
To address whether Smurf1/2 regulates p53 stability through an interaction with the p53 protein directly, we performed an in vitro GST pulldown experiment. The result showed that GST-p53 did not pull down the His-Smurf1 protein, whereas GST-CKIP-1, a known Smurf1-interacting protein (30), strongly pulled down the His-Smurf1 (Fig. 2A), suggesting that no direct interaction between Smurf1 and p53 occurs in vitro.
FIGURE 2.
Regulation of Smurf1/2 on p53 stability is dependent on MDM2. A, interaction between Smurf1 and p53 could not be detectable in GST pulldown assays. Input and pulldown samples were both subjected to immunoblotting (IB) with anti-GST and anti-His antibodies. Input represents 10% of that used for pull down. The interaction between Smurf1 and CKIP-1 was used as the positive control. B, Smurf1 cannot catalyze the ubiquitylation of p53 directly. Purified HA-ubiquitin, E1, E2 (UbcH5c), bacterially expressed and purified Smurf1, p53, and Smad5 were mixed for in vitro p53 ubiquitylation assays and immunoblotted with anti-HA. C, Smurf1 promotes p53 turnover dependently of MDM2. Smurf1, MDM2 siRNA, or control siRNA were expressed in MCF7 cells, and the cell lysates were analyzed by immunoblotting. D, p53−/−Mdm2−/− MEF cells were transfected with Myc-p53, FLAG-Smurf1 (wild type or C699A mutant), MDM2 or together as indicated, and the cell lysates were analyzed by immunoblotting. E, Smurf1/2 promotes MDM2-dependent poly-ubiquitylation of p53. p53−/−Mdm2−/− MEF cells were transfected with p53, Smurf, Ub together with or without MDM2. In vivo ubiquitylation assay was performed. GFP, green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IP, immunoprecipitation; RNAi, RNA interference.
To examine whether Smurf1/2 could mediate ubiquitylation of p53 in vitro, we used purified Smurf1 proteins and performed an in vitro ubiquitylation experiment. The result demonstrated that His-Smurf1 failed to mediate p53 ubiquitylation, although it had a strong activity to mediate the ubiquitylation of Smad5 (Fig. 2B), a known Smurf1 ubiquitylated protein (32). We obtained a similar result for Smurf2 (data not shown). These data clearly indicated that p53 is not a direct substrate of Smurf1/2 ligase, which, however, promotes p53 ubiquitylation and degradation in vivo (Fig. 1).
Given that MDM2 is a major E3 to mediate p53 ubiquitylation and degradation, we then examined whether Smurf1/2 promotes p53 degradation dependent of MDM2. To this end, we transfected an MDM2-specific siRNA and found that this siRNA impaired the ability of overexpressed Smurf1 on the p53 level (Fig. 2C, compare lane 2 with 4). To confirm the dependence of MDM2 on Smurf1/2 in regulation of p53 stability, p53−/−Mdm2−/− MEF were utilized. In these cells, Smurf1 has no effect on p53 protein levels, and only when MDM2 was co-expressed did we observe a decrease of p53 protein levels by both wild type and E3 activity-defective mutant of Smurf1 (Fig. 2D, lanes 5 and 6). Furthermore, we performed an in vivo ubiquitylation experiment by transfection of p53 in p53−/−Mdm2−/− MEF cells, where both endogenous p53 and MDM2 were completely depleted. The results showed that neither Smurf1 nor Smurf2 had any effect on Myc-p53 ubiquitylation in the absence of MDM2 (Fig. 2E, lanes 2 and 3). However, when MDM2 is reintroduced, both Smurf1 and -2 dramatically increased the Myc-p53 ubiquitylation (Fig. 2E, lanes 5 and 6). These data demonstrated that the role of Smurf1/2 on the ubiquitylation of p53 is dependent on MDM2.
Smurf1/2 Interacts with MDM2
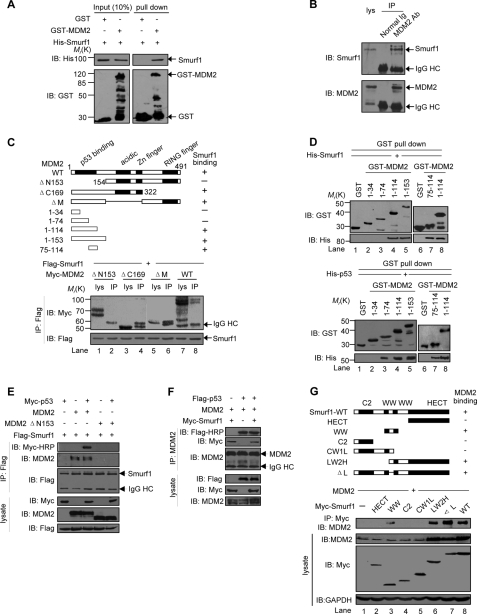
We then examined whether Smurf1/2 interacts with MDM2 directly. A GST pulldown assay result showed a specific interaction of Smurf1 with GST-MDM2 but not with GST alone (Fig. 3A). An immunoprecipitation result showed that the endogenous Smurf1 protein is co-immunoprecipitated down with an antibody against MDM2 in MCF7 cells (Fig. 3B). These results indicate that Smurf1 interacts with MDM2 both in vitro and in vivo.
FIGURE 3.
Smurf interacts with MDM2. A, direct interaction between Smurf1 and MDM2 revealed by GST pulldown assays. Input and pulldown samples were analyzed with anti-GST and anti-His antibodies. Input represents 10% of that used for pull down. IB, immunoblot. B, co-IP of endogenous MDM2 and Smurf1 from MCF7 cells. Whole-cell lysates were immunoprecipitated with MDM2 antibody or control IgG. Both the lysate and the immunoprecipitates were analyzed by immunoblotting with antibodies against Smurf1 or MDM2. HC means heavy chain. C, mapping the Smurf1-binding region on MDM2. The indicated Smurf1 and MDM2 deletion mutants were transfected into HEK293T cells, and co-IP assays were performed. Both the lysate and the immunoprecipitates samples were analyzed. WT, wild type. D, specific Smurf1 (upper panels) and p53 (lower panels) binding region on the N terminus of MDM2 was revealed by a GST pulldown assay. Input and pulldown samples were analyzed with anti-GST and anti-His antibodies. Input represents 10% of that used for pulldown. E, MDM2, p53, and Smurf1 form a ternary complex. p53−/−Mdm2−/− MEF cells were transfected with Myc-p53, MDM2 (wild type (WT) or ΔN153 mutant), and FLAG-Smurf1 as indicated, and co-IP assays were performed with anti-FLAG monoclonal antibody to immunoprecipitate Smurf1 proteins. Both the lysate and the immunoprecipitates samples were analyzed. To avoid the interference of IgG heavy chain on p53 detection, horseradish peroxidase-conjugated Myc antibody was used. F, Smurf1 does not disrupt the MDM2-p53 interaction. p53−/−Mdm2−/− MEF cells were transfected with the indicated plasmids, and co-IP assays were performed. G, mapping the MDM2-binding region on Smurf1. MDM2 and Smurf1 mutants were co-expressed in HEK293T cells. Cell lysates were incubated with an anti-Myc antibody to precipitate Smurf1 mutants. Both the lysate and the immunoprecipitates were analyzed by immunoblot. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To reveal the molecular mechanism for the interaction of Smurf1/2 and MDM2, we generated a series of deletion mutants for MDM2 (Fig. 3C, top panel) and Smurf1 (Fig. 3G, top panel) to map the necessary binding region in vivo. An immunoprecipitation assay result showed that the deletion of 153 residues of MDM2 at the N terminus (ΔN153) abolished the interaction with Smurf1 (Fig. 3C, lane 2). Conversely, deletion of 169 residues, including the RING finger domain at the C terminus (ΔC169), or the central acidic domain, or the zinc finger (ΔM), retained the interaction with Smurf1 (Fig. 3C, lanes 4 and 6).
The results of a detailed mapping experiment demonstrated that the region of amino acids 75–114 but not 1–74 in MDM2 is required for the interaction of Smurf1 with MDM2 (Fig. 3D, upper panel), although p53 was able to interact with both regions of amino acids 75–114 and 1–74 region in MDM2 (Fig. 3D, lower panel). These in vitro binding assays suggested that MDM2 binds to p53 and Smurf1 simultaneously. A co-immunoprecipitation (co-IP) assay result showed that Smurf1 was unable to interact with p53 in the absence of MDM2, although it strongly associated with p53 in the presence of MDM2 (Fig. 3E), suggesting that MDM2 bridges the interaction of Smurf1 and p53. On the other hand, we observed that the affinity for the interaction of Smurf1 and MDM2 was comparable in its absence and presence (Fig. 3E, 2nd versus 3rd lanes), suggesting that p53 had no significant effect on the MDM2-Smurf1 interaction. However, Smurf1 failed to interact with p53 in the presence of the MDM2 ΔN153 mutant, which lost Smurf1 binding ability (Fig. 3E, 4th and 5th lanes). Furthermore, Smurf1 had no significant effect on the interaction of MDM2 and p53 (Fig. 3F). These data indicated that Smurf1 interacts with the N terminus of MDM2 but had no effect on the interaction of MDM2 with p53.
A further immunoprecipitation assay indicated that the second WW domain is required for Smurf1 to interact with MDM2 (Fig. 3G). The interaction pattern between Smurf2 and MDM2 is similar to that of Smurf1 (data not shown). These results indicate that the interaction of Smurf1/2 with MDM2 is via the N terminus of MDM2 and the second WW domain of Smurf1/2.
Smurf1/2 Enhances the Protein Stability of MDM2
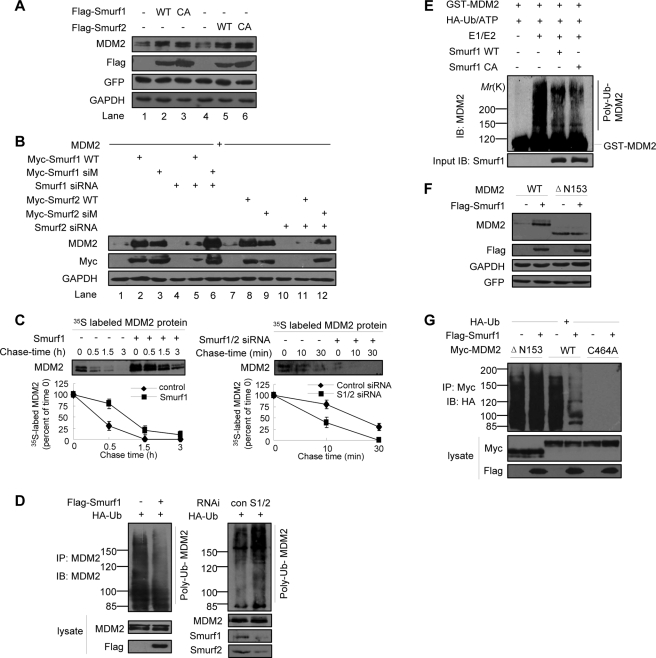
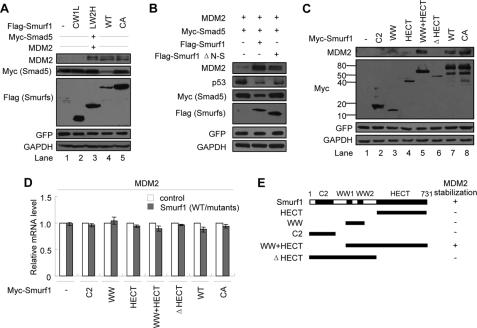
The fact that the second WW domain of Smurf1 mediates the interaction of Smurf1 with MDM2 promoted us to examine whether Smurf1 could regulate the stability of MDM2 because the WW domain has been well defined as a recognition module for ubiquitin-mediated degradation of the targeted protein (35, 36). Interestingly, overexpression of either Smurf1 or Smurf2 results in an enhanced endogenous MDM2 protein level in MCF7 cells (Fig. 4A, compare lanes 2 with 1 and 5 with 4). Remarkably, the E3 ligase activity-defective mutants of Smurf1 and -2 have a similar effect on the MDM2 protein levels (Fig. 4A, lanes 3 and 6). This result implied that Smurf1/2 may stabilize the MDM2 protein. This is a novel function of Smurf1/2 because Smurf1/2 has been characterized as an E3 ligase to mediate protein degradation via its HECT domain (32, 35, 36). To address whether Smurf1/2 plays a role on MDM2 stability in a physiological condition, we co-expressed a Smurf1/2-specific siRNA in MCF7 cells with overexpressed MDM2 proteins. The results indicated that the MDM2 protein level is almost not detectable when the Smurf1/2-specific siRNA is expressed (Fig. 4B, lanes 4 and 10). The stabilizing effect of wild type Smurf1/2 on MDM2 was largely inhibited by the corresponding siRNA (Fig. 4B, compare lane 5 with 2 and lane 11 with 8, respectively). As a control, the siRNA has no effect on the role of an siRNA-resistant mutated (siM) Smurf1/2, and this mutant retains the ability to elevate the MDM2 protein level (Fig. 4B, compare lane 6 with 3 and lane 12 with 9, respectively).
FIGURE 4.
Smurf1/2 inhibits the auto-ubiquitylation of MDM2. A, stabilization of MDM2 by exogenous Smurf1/2. Overexpression of Smurf1, Smurf2, wild-type and the corresponding E3-inactive mutants (i.e. Smurf1-C699A, Smurf2-C716A) up-regulated the levels of endogenous MDM2 in MCF7 cells. B, rescue experiments. The indicated plasmids (including the wild type (WT) Smurf1/2 and the siRNA-resistant Smurf1/2 mutants) and Smurf1/2-specific siRNAs were transfected in MCF7 cells, and cell lysates were analyzed by immunoblot. C, pulse-chase assays. Smurf1 overexpression prolonged the half-life of endogenous MDM2 protein in MCF7 cells (left). Depletion of Smurf1 shortened the half-life of endogenous MDM2 protein in MCF7 cells (right). Data are mean ± S.D. (n = 3). D, Smurf inhibits the poly-ubiquitylation of MDM2 in vivo. MCF7 cells were transfected with HA-Ub, Smurf1 plasmid or Smurf1/2 siRNA, treated with MG132, followed by immunoprecipitation (IP) with MDM2 antibody, and the ubiquitylated endogenous MDM2 was analyzed. IB, immunoblot. E, Smurf1 inhibits the auto-ubiquitylation of MDM2 in vitro independently of its HECT ligase activity. Purified HA-Ub, E1, E2 (UbcH5b), MDM2, and mammalian cell-expressed and immunoprecipitated Smurf1 were mixed for in vitro ubiquitylation assay. Con, control. F, Smurf1 does not regulate the protein level of MDM2 ΔN153 mutant. MCF7 cells were transfected MDM2 (wild type (WT) orΔN153 mutant) and FLAG-Smurf1, and cell lysates were analyzed by immunoblot. G, Smurf1 does not influence the poly-ubiquitylation of MDM2 ΔN153 mutant. MCF7 cells were transfected with HA-Ub, MDM2 (wild type or C464A mutant), FLAG-Smurf1, treated with MG132, followed by immunoprecipitation with anti-Myc antibody, and the ubiquitylated MDM2 was analyzed. GFP, green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RNAi, RNA interference.
To examine whether the effect of Smurf1/2 on the enhanced MDM2 protein levels is through stabilization of the protein, we measured the MDM2 protein half-life by a pulse-chase experiment. The result showed that overexpression of Smurf1 prolonged the MDM2 protein half-life (Fig. 4C, left), although depletion of Smurf1 and -2 shortened the half-life of MDM2 (Fig. 4C, right). We obtained a similar result when Smurf2 is overexpressed (data not shown). Together, all the data indicate that Smurf1/2 plays a role to protect MDM2 from degradation.
Smurf1/2 Inhibits the Auto-ubiquitylation of MDM2
To gain insight into the mechanism how Smurf1/2 regulates MDM2 stability, we examined a possible effect of Smurf1/2 on MDM2 auto-ubiquitylation. An in vivo ubiquitylation assay showed that ectopically expressed Smurf1 attenuated, but depletion of Smurf1 and -2 enhanced, the auto-ubiquitylation of endogenous MDM2 (Fig. 4D). An in vitro ubiquitylation assay also showed that Smurf1 inhibited the auto-ubiquitylation of MDM2 purified from Escherichia coli. Interestingly, Smurf1-C699A, a mutant with no catalytic activity, retained a similar inhibitory effect on MDM2 auto-ubiquitylation (Fig. 4E). These results indicated Smurf1 inhibits MDM2 auto-ubiquitylation.
Furthermore, we observed that the protein level of MDM2 ΔN153, a mutant that failed to bind to Smurf1, was not influenced by Smurf1 (Fig. 4F). An in vivo ubiquitylation assay showed that overexpression of Smurf1 did not decrease the ubiquitylation level of MDM2 ΔN153 (Fig. 4G), suggesting the interaction of MDM2 and Smurf1 is required for Smurf1 to regulate MDM2 auto-ubiquitylation.
E3 Ligase Activity-independent Regulation of Smurf1/2 on MDM2 Stability
The fact that the mutant Smurf1 C699A retained similar effects on the MDM2 protein level (Fig. 4A) and the auto-ubiquitylation (Fig. 4E) implied that the E3 activity of Smurf1/2 is not required for its role on the regulation of MDM2. Further analysis showed that Smurf1 C699A retained the ability to stabilize the MDM2 protein (Fig. 5A, lane 5, top panel), although it significantly decreased the Myc-Smad5 protein level (Fig. 5A, lane 4, 2nd panel). Because the small subdomain of the HECT N-lobe is essential for Smurf2 to interact with E2 (37), we questioned whether the mutant in the N-lobe in Smurf1, which lost the ability to interact with E2, could have any effect on the stability of MDM2. For this purpose, we generated a Smurf1 mutant (Smurf1 ΔN-S) by deletion of the small subdomain of HECT N-lobe. Our data showed that the mutant Smurf1 (ΔN-S) failed to mediate the degradation of Smad5 but still retained the ability to stabilize MDM2 and mediate degradation of p53 (Fig. 5B). These data indicated that the role of Smurf1/2 on the stability of MDM2 and p53 is independent on its E3 ligase activity.
FIGURE 5.
Smurf1/2 stabilizes MDM2 independently of the HECT ligase activity. A, effect of Smurf1 mutants on MDM2 stability. Myc-Smad5, MDM2, and Smurf1 wild type (WT) or mutants were cotransfected into MCF7 cells. Cell lysates were analyzed by immunoblot. The degradation of Smad5 mediated by Smurf1 was analyzed as the control. B, MDM2 stabilized by Smurf1 did not rely on Smurf1 E2 binding activity. Myc-Smad5, MDM2, and Smurf1 wild type or HECT N-lobe small subunit deletion mutant (ΔN-S) were cotransfected into MCF7 cells. Smurf1 CA, C699A; Smurf2 CA, C716A. Cell lysates were analyzed by immunoblot. The degradation of Smad5 mediated by Smurf1 was used as a control. GFP, green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. C, both the WW2 domain and the HECT domain are required for Smurf1 to stabilize MDM2 protein. The indicated deletion mutants of Smurf1 were expressed in MCF7 cells, and the levels of endogenous MDM2 were analyzed by immunoblot. D, Smurf1 overexpression does not influence the mRNA level of MDM2. Total RNA from Smurf1 deletion mutant-transfected MCF7 cells were subjected to a quantitative real time PCR analysis. Data are presented as mean ± S.D. (n = 3). E, schematic representation of the effect of Smurf1 mutants on MDM2 stability.
To reveal the molecular mechanism of the role of Smurf1/2 on the maintenance of MDM2 stability, we examined the MDM2 protein level (Fig. 5C) in the presence of a series of truncated mutants of Smurf1 (Fig. 5E). A Western blot result indicated that the MDM2 protein level was dramatically elevated when the WW+HECT domain protein was co-overexpressed, although expression of each single domain had less effect on the MDM2 protein level (Fig. 5C). However, a quantitative real time PCR analysis indicated that the mRNA level of MDM2 was unchanged when Smurf1 mutants were overexpressed (Fig. 5D). These results indicated that both the WW and the HECT domains are essential for Smurf1 to stabilize MDM2 protein, although its catalytic activity was not required.
Smurf1/2 Enhances the Interaction of MDM2 with MDMX
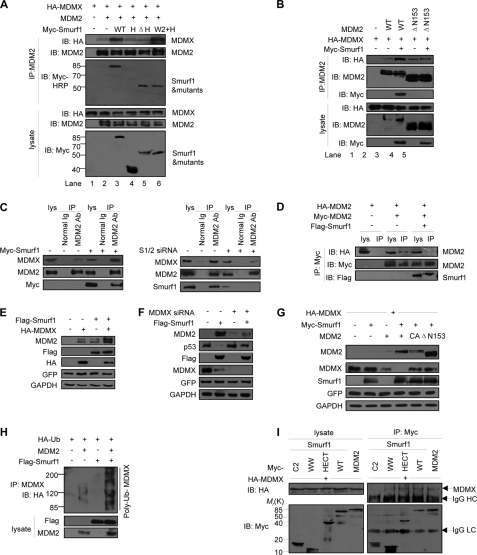
It has been reported that MDM2 stability and activity are maintained by an MDM2/MDMX heterodimer (28, 29). We questioned whether Smurf1/2-stabilized MDM2 protein is through an enhancement of the MDM2-MDMX interaction. Our immunoprecipitation experiment results indicated that the interaction of MDM2-MDMX was significantly enhanced when Myc-Smurf1 was co-expressed (Fig. 6A, lanes 1–3), suggesting that Smurf1 augments the interaction between MDM2 and MDMX. The WW2+HECT protein had a strong ability to maintain the interaction, but either the deletion of the HECT domain or the HECT domain alone abrogated the effects of Smurf1 on the MDM2-MDMX interaction (Fig. 6A, lanes 4–6). Furthermore, a deletion of 153 residues of MDM2 at the N terminus (ΔN153), a region critical for Smurf1-MDM2 interaction (Fig. 3C), abrogated the ability of Smurf1 in the MDM2-MDMX interaction (Fig. 6B). We further observed that overexpression of Smurf1 enhanced the interaction of endogenous MDM2 and MDMX proteins in vivo (Fig. 6C, left, compare lane 6 with 3), although depletion of Smurf1 attenuated MDM2-MDMX interaction (Fig. 6C, right, compare lane 6 with 3). This result suggests that Smurf1 promotes the association of MDM2 with MDMX. Because MDM2 also forms a homodimer, we questioned whether Smurf1/2 has any effect on the formation of the MDM2 homodimer. The result showed that overexpression of Smurf1 inhibited the homodimerization of MDM2 as demonstrated by the interaction of Myc-MDM2 and HA-MDM2 (Fig. 6D), suggesting Smurf1 blocks MDM2 homodimerization.
FIGURE 6.
Smurf1 stabilizes MDM2 by enhancing MDM2-MDMX interaction. A, Smurf1 enhances the interaction between MDM2 and MDMX. MCF7 cells were transfected with MDM2, MDMX, and Smurf1 (or mutants). To avoid the MDM2 self-degradation, MG132 was added. Both the lysates and the immunoprecipitates with MDM2 antibody were analyzed by immunoblot (IB). To avoid the interference of IgG heavy chain, an anti-Myc horseradish peroxidase antibody was used to detect the Smurf1 mutants. B, interaction between Smurf1 and MDM2 was required for Smurf1 to enhance MDM2-MDMX interaction. Myc-Smurf1, HA-MDMX, and MDM2 wild type (WT) or the truncated mutant ΔN153 that lost the ability to interact with Smurf1 were transfected into MCF7 cells singly or in combination as indicated. Thirty six hours later, the cell lysates were prepared, and the immunoprecipitates (IP) with MDM2 antibody were analyzed by immunoblot. C, Smurf1 enhances the endogenous interaction between MDM2 and MDMX. MCF7 cells were transfected with Smurf1 plasmid (left) or Smurf1 siRNA (right) and treated with MG132, and cell lysates were immunoprecipitated with MDM2 antibody. The expression of endogenous MDM2 and MDMX was detected. D, Smurf1 inhibits the auto-interaction between differently tagged MDM2 proteins. Myc-MDM2, HA-MDM2, and Smurf1 were co-expressed as indicated and treated with MG132. Cell lysates were immunoprecipitated with anti-Myc antibody, and both lysate and immunoprecipitates were analyzed. E, Smurf1 synergizes with MDMX to stabilize MDM2. Smurf1 and MDMX were singly or co-expressed in MCF7 cells. The cell lysates were prepared, and endogenous MDM2 level was detected by immunoblot. F, MDMX is required for Smurf1 to stabilize MDM2 and degrade p53. The Smurf1 plasmid and MDMX-specific siRNA were co-expressed in MCF7 cells. The expression levels of MDM2 and p53 in total cell lysates were analyzed. G, Smurf1 promotes MDM2-mediated degradation of MDMX. MCF7 cells were transfected with MDMX, Smurf1, MDM2 (wild type (WT) or mutants) as indicated, and cell lysates were analyzed by immunoblot. H, Smurf1 promotes MDM2-mediated ubiquitylation of MDMX. MCF7 cells were transfected with HA-Ub, MDM2, and Smurf1 as indicated; an in vivo ubiquitylation assay was performed with anti-MDMX antibody, and the immunoprecipitates were analyzed by HA antibody. I, Smurf1 interacts with MDMX through HECT domain. p53−/−mdm2−/− MEF cells were transfected with HA-MDMX, Smurf1 (wild type or deletion mutants), or Myc-MDM2. Cell lysates were immunoprecipitated with anti-Myc antibody, and both lysates and immunoprecipitates were analyzed by immunoblot. The MDMX-MDM2 interaction was used as a positive control. GFP, green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Smurf1 CA, C699A; Smurf2 CA, C716A.
To further confirm whether the role of Smurf1 on the MDM2 stability is through MDMX, we overexpressed Smurf1 under overexpression or depletion of MDMX in the MCF7 cells. Western blot analysis results showed that Myc-Smurf1 had a synergic effect on the MDM2 protein level with overexpression of HA-MDMX (Fig. 6E), although Myc-Smurf1 lost the ability to maintain high levels of MDM2 when MDMX was depleted (Fig. 6F, upper panel). Consistently, the p53 protein level decreased by Smurf1 was recovered in the depletion of MDMX (Fig. 6F, 2nd panel).
To address whether Smurf1 functions on MDMX is a direct or an indirect (via MDM2) event, we performed a degradation assay in p53−/−Mdm2−/− MEF cells. The results showed that Smurf1 alone failed to decrease the protein level of MDMX, although co-expression of Smurf1 and MDM2 strongly decreased the MDMX protein level (Fig. 6G). Intriguingly, Smurf1 lost the ability to decrease the MDMX protein level in the presence of mutants of MDM2, suggesting that Smurf1 mediates the degradation of MDMX via MDM2. Consistently we further observed that Smurf1 enhanced MDM2-mediated ubiquitylation of MDMX, although Smurf1 alone had no effect on the ubiquitylation of MDMX (Fig. 6H).
Furthermore we questioned whether Smurf1 interacts with MDMX. A co-IP assay showed that Smurf1 interacted with MDMX through its HECT domain (Fig. 6I). Taken together, all the data suggest that Smurf1/2 maintains the MDM2 protein level through increasing the MDM2-MDMX interaction.
SRF Motif in Smurf1/2 Is Required for MDM2 Binding
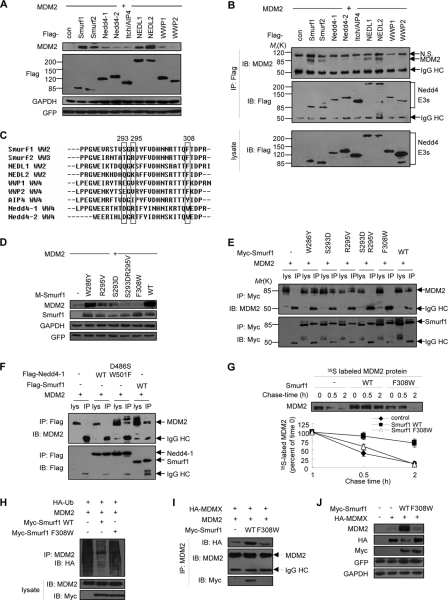
Because all the Nedd4 family E3s share similar structures, we asked whether other non-Smurf members could interact with and stabilize MDM2. Co-expression of Smurf1, Smurf2, NEDL1 (Nedd4-like ubiquitin ligase 1), or NEDL2 significantly stabilized MDM2, whereas five other members had weak or little effect (Fig. 7A). Moreover, we found that Smurf1/2 and NEDL1/2 specifically interacted with MDM2 (Fig. 7B). Alignment of the last WW domain of Nedd4 E3s revealed a motif ((S/T/Q)X(R/K)X12F, where X presents any amino acid) specific to Smurf1/2 and NEDL1/2. We named this motif SRF (serine-arginine-phenylalanine motif) (Fig. 7C) and found it is specific in Smurf1/2 and NEDL1/2. We speculated that this motif is required for the interaction of Smurf1/2 or NEDL1/2 with MDM2.
FIGURE 7.
Identification of an SRF motif in Smurf and NEDL required for MDM2 interaction and stabilization. A, Smurf1/2 and NEDL1/2 increase the protein level of MDM2. All nine members of the human Nedd4 family were individually co-transfected with MDM2 into MCF7 cells. The protein level of MDM2 was determined. B, Smurf1/2 and NEDL1/2 bind to MDM2 protein. An interaction analysis of Nedd4 family E3s with MDM2 was done by co-IP assays in HEK293T cells. Con, control; IB, immunoblot. To avoid the MDM2 self-degradation, MG132 was added. C, sequence alignment of the last WW domain of Smurf1 with those of other members of the Nedd4 family. Numbers indicate the position of the Smurf1 SRF motif. D, mutations of F308W and S293D/R295V abolished the stabilizing effect of Smurf1 on MDM2. MCF7 cells were transfected with Smurf1 (wild type (WT) or mutants) and MDM2, and cell lysates were analyzed by immunoblot. E, interaction analyses of the indicated Smurf1 point mutants with MDM2. HEK293T cells were transfected with MDM2 and Myc-Smurf1 (wild type or mutants) and treated with MG132 for 8 h before harvested. Cell lysates were immunoprecipitated (IP) with an anti-Myc antibody, and both lysates and immunoprecipitates were analyzed by immunoblot. F, mutation of D486S/W501F of Nedd4-1 gains the binding ability to interact with MDM2. HEK293T cells were transfected with MDM2, FLAG-Nedd4-1 (wild type or mutants), or FLAG-Smurf1 and treated with MG132. Cell lysates were immunoprecipitated with an anti-FLAG antibody and analyzed by immunoblot. G, pulse-chase assays. The effect of Smurf1 F308W mutant on the half-life of endogenous MDM2 protein in MCF7 cells is shown. H, effect of Smurf1 F308W mutant on MDM2 ubiquitylation. MCF7 cells were transfected with HA-Ub, MDM2, and Smurf1 (wild type or F308W) and treated with MG132. Cell lysates were immunoprecipitated with an anti-MDM2 antibody and analyzed by immunoblot. I, Smurf1 F308W mutant does not enhance MDM2-MDMX interaction. MCF7 cells were transfected with MDMX, MDM2, and Smurf1 (wild type or F308W), treated with MG132. Cell lysates were immunoprecipitated with an anti-MDM2 antibody and analyzed by immunoblot. J, Smurf1 F308W mutant does not stabilize MDM2 synergically with MDMX. MCF7 cells were transfected with MDMX and Smurf1 (wild type or F308W), and the expression of endogenous MDM2 was analyzed by immunoblot. GFP, green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To address this hypothesis, a series of Smurf1 point mutants on the SRF motif were generated. Our results indicated that the mutants of SR (S293D/R295V) or F (F308W) lost the ability to stabilize the MDM2 protein, but the mutant W286Y, a mutant outside the SRF motif in the WW domain, remained the ability to stabilize the MDM2 protein (Fig. 7D). Consistently, these mutants of the SRF motif lost the ability to interact with MDM2 as demonstrated by a co-IP experiment (Fig. 7E). To further address the role of the SRF motif, we introduced the motif into Nedd4-1, which has no ability to interact with MDM2 (see Fig. 7A), by a mutation of Asp-486 into a serine residue and Trp-501 into a phenylalanine residue (D486S/W501F). A co-IP experiment result indicated that this mutation gained the function of interaction with MDM2 (Fig. 7F). All these data indicated that the SRF motif is critical for the interaction of Smurf1 and MDM2.
Next, we used the mutant (F308W) to examine the role of SRF on the activity of Smurf1 on the regulation of MDM2. Our data indicated that this mutant failed to either stabilize MDM2 (Fig. 7G) or decrease the auto-ubiquitylation of MDM2 (Fig. 7H). Consistently the mutant failed to enhance the interaction of MDM2 with MDMX (Fig. 7I). Finally, this mutant failed to enhance MDMX-mediated stabilization of MDM2 (Fig. 7J). All these data strongly indicate that the SRF motif in the last WW domain is required for Smurf1/2 to interact with MDM2 and then to stabilize the MDM2 protein.
Smurf1/2 Represses p53-mediated Transactivation and Apoptosis
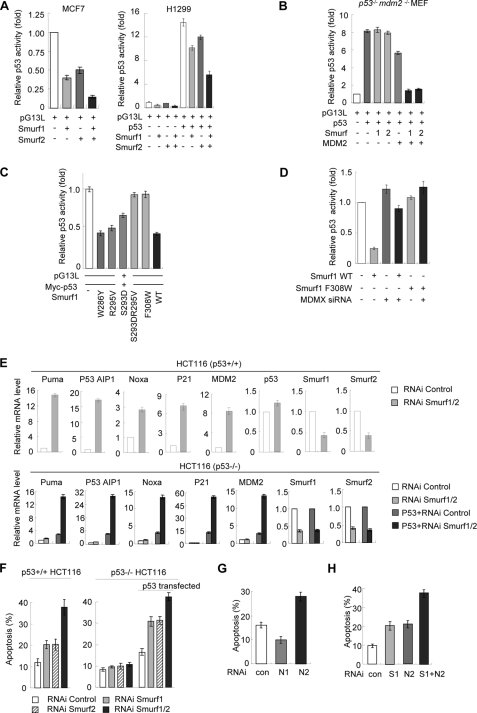
To determine the effect of Smurf1/2 on p53-dependent transactivation, a p53 luciferase reporter plasmid pG13L was transfected into MCF7 cells that are reported to express endogenous p53 (31). Luciferase activity assay results demonstrated that the transcriptional activity of endogenous p53 was suppressed by overexpression of Smurf1/2 (Fig. 8A, left). Moreover, we observed that overexpression of Smurf1/2 also inhibited the transcriptional activity of the exogenously expressed p53 in H1299 cells, a p53-deficient cell line (Fig. 8A, right). To confirm whether Smurf1/2 regulates the p53 transcriptional activity dependently on MDM2, we performed a luciferase reporter experiment in p53−/−Mdm2−/− MEF cells. The results indicated that Smurf1/2 failed to inhibit the transactivation activity of exogenous p53 in these cells (Fig. 8B, 3rd and 4th columns); however, it dramatically inhibited the luciferase activity when MDM2 was re-introduced (Fig. 8B, 6th and 7th columns). Furthermore, the Smurf1 mutants of SRF motif (F308W and S293D/R295V) lost the ability to inhibit p53 activity (Fig. 8C). These results, consistent with the effect of Smurf1/2 on p53 protein stability (Fig. 1), suggest that Smurf1/2 inhibits p53 transcriptional activity dependent of MDM2. Additionally, a luciferase assay result showed that Smurf1 failed to inhibit the p53 transcriptional activity when MDMX is depleted (Fig. 8D), suggesting that MDMX is required for the inhibitory role of Smurf1/2 on the p53 transcriptional activity. All of these results are consistent with the observations in the p53 and MDM2 protein levels.
FIGURE 8.
Depletion of Smurf expression leads to p53 activation and p53-mediated apoptosis. A, inhibitory effects of Smurf1/2 on the transcriptional activity of p53. p53 wild type MCF7 cells (left) were transfected with Smurfs or control vector. The pG13L luciferase activity was assayed as described under “Materials and Methods.” p53-deficient H1299 cells (right) were transfected with exogenous p53 together with Smurf1/2, and the luciferase activity was measured. Representative results of three independent experiments are shown. Data are mean ± S.D. (n = 3). B, Smurf1/2 inhibits p53 activity dependent on MDM2. p53−/−Mdm2−/− MEF cells were transfected with the indicated plasmids, and p53 luciferase activity was measured. C, inhibition of the transcriptional activity of p53 by the indicated Smurf1 mutants. Luciferase reporter assay was performed in H1299 cells. Representative results from three independent experiments are shown. Data are presented as mean ± S.D. (n = 3). D, MDMX is required for Smurf1 to inhibit the activity of p53. Smurf1 plasmid and MDMX siRNA were co-expressed in MCF7 cells and p53 luciferase activity was measured. E, depletion of Smurf1 and -2 significantly up-regulated the expression of p53 target genes. Total RNA from siRNA and/or plasmid-transfected HCT116 cells was subjected to quantitative real time PCR analysis. Data are mean ± S.D. (n = 3). F, Smurf1/2 inhibits cell apoptosis. Smurf1 and Smurf2 were depleted singly or doubly as indicated in the p53+/+ and p53−/− HCT116 cells. Apoptosis was determined by staining with annexin V followed by flow cytometry analysis. In certain cases, exogenous p53 was reintroduced into the HCT116 cells to examine the effect of Smurf1/2 depletion on p53-dependent apoptosis. Data are mean ± S.D. (n = 3). G, apoptosis in NEDL1-(N1) or NEDL2 (N2)-knocked down p53+/+ HCT116 cells was determined by staining with annexin V followed by flow cytometry analysis. Data are presented as mean ± S.D. (n = 3). H, NEDL2 cooperates with Smurf1 in inducing cell apoptosis. p53+/+ HCT116 cells were transfected with Smurf1 siRNA, NEDL2 siRNA, or both. Apoptosis was determined by annexin V assays. Data are presented as mean ± S.D. (n = 3). RNAi, RNA interference.
A quantitative real time PCR analysis demonstrated that depletion of Smurf1 and -2 in HCT116 cells significantly up-regulated the mRNA levels of the p53 targeted genes, including the proapoptotic genes Puma, p53AIP1, and Noxa; the pro-arrest related genes p21; and the negative regulator MDM2 (Fig. 8E, upper panels). Similar results can be observed in the p53−/− cells only when p53 was reintroduced (Fig. 8E, bottom panels).
To uncover the physiological function of Smurf1/2 on the regulation of p53 protein stability, we measured the apoptotic cell numbers when Smurf1/2 was depleted. The data showed that depletion of Smurf1 and/or -2 increased the apoptotic cell number in the wild type HCT116 cells (Fig. 8F, left). However, depletion of Smurf1/2 showed no effect on apoptotic cell number in the p53−/− HCT116 cells but showed a great effect when p53 was re-introduced (Fig. 8F, right). Taken together, all the data suggest that Smurf1/2 regulates apoptosis through destabilizing the p53 protein levels.
To address whether NEDL1/2 function similarly to Smurf1/2, NEDL1/2 were depleted. An apoptosis experiment result indicated that depletion of NEDL2 resulted in enhanced apoptosis (Fig. 8G) and up-regulated expression of p53 target genes (supplemental Fig. S1A). However, depletion of NEDL1 decreased apoptosis (Fig. 8G) and inhibited p53-mediated transcription (supplemental Fig. S1B). The differential effects might be due to the different role of NEDL1 and NEDL2 in the interaction with p53 (34). Our data defined that NEDL2 possesses a similar effect on MDM2 as Smurf1/2 did. Intriguingly, we found that NEDL2 and Smurf1 have an addictive effect on apoptosis (Fig. 8H). Our study revealed the Smurf1/2 plays an important role in regulation of MDM2 activity and thereafter of the apoptosis.
DISCUSSION
p53 plays an important role in the regulation of cell apoptosis and is linked to a variety of human diseases, including cancers. The degradation of the p53 protein is tightly controlled by a ubiquitylation mechanism. MDM2 is regarded as a major E3 ligase to mediate the ubiquitylation and degradation, although several other E3 ligases have been reported (4–8). The activity of MDM2 is critical for the maintenance of the p53 protein level in normal physiological and pathological conditions. Because of the important role of MDM2 to maintain an adequate p53 protein level, cells have to retain a comprehensive network to regulate the activity of MDM2. It has been reported that several factors inhibit or enhance the interaction of MDM2 with p53, and other factors directly regulate MDM2 protein stability (15–24). In this study, we revealed that Smurf1/2, the HECT domain-type E3 ubiquitin ligase, stabilizes the MDM2 protein and enhances its E3 ligase activity to mediate the ubiquitylation and degradation of p53 and thereafter to prevent apoptosis. To our knowledge, this is the first report that Smurf1/2 functions to stabilize the MDM2 protein rather than to mediate the protein degradation. Our study provided a new mechanism for the regulation of MDM2-mediated p53 ubiquitylation and degradation.
Smurf1/2 belongs to the Nedd4 family of E3 ligases, which are characterized by an N-terminal C2 domain, two to four protein-protein interaction WW domains, and a C-terminal catalytic HECT domain (35, 36). The mammalian Nedd4 family consists of nine members, including WWP1 and NEDL1. The functional relationship between this family of E3 ligases and the p53 family of transcriptional factors has been uncovered recently. For example, WWP1 (WW domain-containing protein 1) was reported to mediate mono-ubiquitylation of p53, leading to its nuclear export and a reduction in its transcriptional activity (33). NEDL1 (NEDD4-like E3 ubiquitin-protein ligase 1) was reported to interact directly with p53 and to promote p53-mediated apoptosis (34). In our study, we initially speculated that the role of Smurf1/2 on the p53 protein degradation might be related to its E3 ligase activity. However, to our surprise, we observed that the inactive mutation of Smurf1/2 still retain the activity to promote the ubiquitylation and degradation of p53. Moreover, we found no direct interaction occurred between Smurf1/2 and p53. Based on these basic observations, we attempted to analyze whether Smurf1/2 plays a role on the E3 ligase of p53. Intriguingly, we observed that Smurf1/2 stabilizes the MDM2 protein level. To avoid any possibility of Smurf1/2 functioning as an E3 ligase, we generated a series of Smurf1 and -2 mutants. Our data showed that the E3-inactive mutation of HECT domain retains the function to stabilize the MDM2 protein. Different from WWP1 and NEDL1, we revealed that Smurf1/2 has no direct interaction with p53 but promotes ubiquitylation and degradation of p53 through MDM2.
Although regulation of p53 stability, in particular by the E3 ligase MDM2, has been extensively studied, it remains unclear for the regulation of the activity of the MDM2 protein. In this study, we found that the MDM2 protein is stabilized by Smurf1/2. The detailed molecular mechanism for the Smurf1/2-stabilized MDM2 is attributed to the auto-ubiquitylation of MDM2 by Smurf1/2. Using different methods, we have demonstrated that Smurf1/2 inhibits the auto-ubiquitylation of MDM2. Other proteins such as CARPs (a subfamily of RING E3 ligases), YY1, and PACT/RBBP6 have been reported to modulate the auto-ubiquitylation of MDM2. However, the mechanisms of these factors are quite diverse. For example, CARPs were characterized to interact with MDM2 and to inhibit its auto-ubiquitylation (38), whereas YY1 and PACT/RBBP6 were reported to enhance both MDM2 auto-ubiquitylation and p53 ubiquitylation (19, 20). Although the E3 activity of CARPs is essential to stabilize MDM2, we found that Smurf1 and -2 regulate MDM2 independent of their E3 activity. These studies suggest that MDM2 stability and activity are tightly controlled at multiple levels. The different regulatory factors controlling the stability of p53 provide an important network to ensure the functional homeostasis of cells.
Our study also provided a new insight into the regulation of the MDM2-MDMX complex. We found that Smurf1/2 enhances heterodimerization of the MDM2-MDMX but inhibits homodimerization of MDM2. MDMX is another negative regulator of p53, which represses the transcriptional activity and promotes the MDM2-mediated degradation of p53 (27, 39, 40). Remarkably, Mdmx-deficient mice die during early embryonic development, and p53 null mutation also rescue this lethality phenotype, similar to the case of Mdm2, highlighting the significance of MDMX in the regulation of p53 activity (27, 41). Although MDMX protein contains a C-terminal RING finger domain homologous to that of MDM2, MDMX itself does not possess any intrinsic ubiquitin ligase activity and thus cannot directly catalyze the poly-ubiquitylation of p53 (42). Recent studies demonstrated that MDMX interacts with MDM2 and stabilizes MDM2 (28, 29, 43, 44). The complex of MDM2 with MDMX is more stable than the MDM2-MDM2 complex; therefore, it is believed that the MDM2-MDMX complex plays a prominent role in p53 ubiquitylation in vivo (43–45). In our study, we found that Smurf1/2 enhances the formation of the MDM2-MDMX complex and blocks the MDM2-MDM2 homodimerization. The role of Smurf1/2 on the complex formation may provide a way for cells to adapt to different conditions.
We have observed that a novel SRF motif in the WW2 domain is critical for the interaction of Smurf1/2 with MDM2. The mutants of SRF (S293D/R295V or F308W) impaired the ability of Smurf1/2 to mediate the association of MDM2 and MDMX. Interestingly, although the E3 activity of HECT domain is not required for the Smurf1/2-mediated p53 ubiquitylation, we observed that the HECT domain interacts with MDMX and is required for Smurf1/2 to enhance the association between MDM2 and MDMX. This finding suggests that the HECT domain functions with more than a core of ubiquitin ligase. Based on these results, we propose a model that the WW2 domain of Smurf1/2 binds to the N-terminal part of MDM2, and the HECT domain of Smurf1/2 binds to MDMX, through which Smurf1/2 promotes the heterodimerization of MDM2 with MDMX, thereafter to enhance the poly-ubiquitylation of p53.
Another interesting phenomenon is the complementary function of Smurf1 and -2 on the regulation of p53 degradation. When we deleted both Smurf1 and -2, we observed a strong induction of p53, although the single deletion of Smurf1 or Smurf2 showed less effect (Fig. 1B). This result is reminiscent of the lethal phenotype of the Smurf1/2 double knock-out mice. Both Smurf1 and -2 are reported to play an important role during embryonic development and bone formation (32, 46, 47). Smurf1−/− or Smurf2−/− mouse is born normally, but Smurf1 and -2 double knock-out mouse is embryonic lethal with most dying around E10.5 (46). Our findings indicated that although overexpression of either Smurf1 or Smurf2 was sufficient to stabilize the MDM2 protein, double depletion of both Smurf1 and -2 resulted in a substantial destabilization of MDM2. We reasoned that Smurf1 and -2 may have a complementary effect. When Smurf1 is depleted, the role of Smurf2 retains the function. Indeed, we observed that both Smurf1 and -2 have a similar role in the controlling MDM2 stability and mediating p53 protein degradation. When both Smurf1 and -2 were depleted, we observed very significant effects on p53 degradation.
The finding of the role of Smurf1/2 on the stability of p53 raises many questions. For example, what is the signaling to mediate the interaction of MDM2 with MDMX by Smurf1/2? How is the physiological and pathological role of Smurf1/2 in mediating the stabilization of MDM2 and degradation of p53? Whether the up-regulation of Smurf1 and Smurf2, as has been recently observed in pancreatic and breast cancer cells (48, 49), will result in the inhibition of apoptosis via stabilization of MDM2 and inactivation of p53 is worthy of further investigation.
In conclusion, we have provided strong evidence that Smurf1/2 regulates the p53 protein level by stabilizing the MDM2-MDMX complex. By such a way, Smurf1/2 inhibits apoptosis.
Supplementary Material
Acknowledgments
We thank Drs. B. Vogelstein, Y. Xiong, G. M. Wahl, W. I. Sundquist, M. Wu, and Q. Zhan for providing materials.
The work was supported by National Basic Research Program Grants 2007CB914601, 2006CB910802, 2009CB918402, and 2010CB912202, National Natural Science Foundation Project Grants 30830029 and 30800177, National Key Technologies R & D Program for New Drugs Grants 2009ZX09503-002 and 2009ZX09301-002, and State Key Laboratory of Proteomics Grant K200802.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- E3
- ubiquitin ligase
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- co-IP
- co-immunoprecipitation
- siRNA
- small interfering RNA
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1.Lane D. P. (1992) Nature 358, 15–16 [DOI] [PubMed] [Google Scholar]
- 2.Vousden K. H., Lu X. (2002) Nat. Rev. Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 3.Levine A. J. (1997) Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 4.Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 5.Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G. D., Dowd P., O'Rourke K., Koeppen H., Dixit V. M. (2004) Nature 429, 86–92 [DOI] [PubMed] [Google Scholar]
- 6.Leng R. P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J. M., Lozano G., Hakem R., Benchimol S. (2003) Cell 112, 779–791 [DOI] [PubMed] [Google Scholar]
- 7.Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. (2005) Cell 121, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 8.Esser C., Scheffner M., Höhfeld J. (2005) J. Biol. Chem. 280, 27443–27448 [DOI] [PubMed] [Google Scholar]
- 9.Brooks C. L., Gu W. (2003) Curr. Opin. Cell Biol. 15, 164–171 [DOI] [PubMed] [Google Scholar]
- 10.Michael D., Oren M. (2003) Semin. Cancer Biol. 13, 49–58 [DOI] [PubMed] [Google Scholar]
- 11.Montes de Oca Luna R., Wagner D. S., Lozano G. (1995) Nature 378, 203–206 [DOI] [PubMed] [Google Scholar]
- 12.Jones S. N., Roe A. E., Donehower L. A., Bradley A. (1995) Nature 378, 206–208 [DOI] [PubMed] [Google Scholar]
- 13.Momand J., Jung D., Wilczynski S., Niland J. (1998) Nucleic Acids Res. 26, 3453–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itahana K., Mao H., Jin A., Itahana Y., Clegg H. V., Lindström M. S., Bhat K. P., Godfrey V. L., Evan G. I., Zhang Y. (2007) Cancer Cell 12, 355–366 [DOI] [PubMed] [Google Scholar]
- 15.Kamijo T., Weber J. D., Zambetti G., Zindy F., Roussel M. F., Sherr C. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomerantz J., Schreiber-Agus N., Liégeois N. J., Silverman A., Alland L., Chin L., Potes J., Chen K., Orlow I., Lee H. W., Cordon-Cardo C., DePinho R. A. (1998) Cell 92, 713–723 [DOI] [PubMed] [Google Scholar]
- 17.Stott F. J., Bates S., James M. C., McConnell B. B., Starborg M., Brookes S., Palmero I., Ryan K., Hara E., Vousden K. H., Peters G. (1998) EMBO J. 17, 5001–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Xiong Y., Yarbrough W. G. (1998) Cell 92, 725–734 [DOI] [PubMed] [Google Scholar]
- 19.Sui G., Affar el B., Shi Y., Brignone C., Wall N. R., Yin P., Donohoe M., Luke M. P., Calvo D., Grossman S. R., Shi Y. (2004) Cell 117, 859–872 [DOI] [PubMed] [Google Scholar]
- 20.Li L., Deng B., Xing G., Teng Y., Tian C., Cheng X., Yin X., Yang J., Gao X., Zhu Y., Sun Q., Zhang L., Yang X., He F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7951–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Mol. Cell. Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Mol. Cell. Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linares L. K., Kiernan R., Triboulet R., Chable-Bessia C., Latreille D., Cuvier O., Lacroix M., Le Cam L., Coux O., Benkirane M. (2007) Nat. Cell Biol. 9, 331–338 [DOI] [PubMed] [Google Scholar]
- 25.Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000) J. Biol. Chem. 275, 8945–8951 [DOI] [PubMed] [Google Scholar]
- 26.Chang Y. C., Lee Y. S., Tejima T., Tanaka K., Omura S., Heintz N. H., Mitsui Y., Magae J. (1998) Cell Growth Differ. 9, 79–84 [PubMed] [Google Scholar]
- 27.Finch R. A., Donoviel D. B., Potter D., Shi M., Fan A., Freed D. D., Wang C. Y., Zambrowicz B. P., Ramirez-Solis R., Sands A. T., Zhang N. (2002) Cancer Res. 62, 3221–3225 [PubMed] [Google Scholar]
- 28.Uldrijan S., Pannekoek W. J., Vousden K. H. (2007) EMBO J. 26, 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poyurovsky M. V., Priest C., Kentsis A., Borden K. L., Pan Z. Q., Pavletich N., Prives C. (2007) EMBO J. 26, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu K., Yin X., Weng T., Xi S., Li L., Xing G., Cheng X., Yang X., Zhang L., He F. (2008) Nat. Cell Biol. 10, 994–1002 [DOI] [PubMed] [Google Scholar]
- 31.Tian C., Xing G., Xie P., Lu K., Nie J., Wang J., Li L., Gao M., Zhang L., He F. (2009) Nat. Cell Biol. 11, 580–591 [DOI] [PubMed] [Google Scholar]
- 32.Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]
- 33.Laine A., Ronai Z. (2007) Oncogene 26, 1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., Ozaki T., Kikuchi H., Yamamoto H., Ohira M., Nakagawara A. (2008) Oncogene 27, 3700–3709 [DOI] [PubMed] [Google Scholar]
- 35.Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 36.Ingham R. J., Gish G., Pawson T. (2004) Oncogene 23, 1972–1984 [DOI] [PubMed] [Google Scholar]
- 37.Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005) Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 38.Yang W., Dicker D. T., Chen J., El-Deiry W. S. (2008) Cell Cycle 7, 670–682 [DOI] [PubMed] [Google Scholar]
- 39.Shvarts A., Steegenga W. T., Riteco N., van Laar T., Dekker P., Bazuine M., van Ham R. C., van der Houven van Oordt W., Hateboer G., van der Eb A. J., Jochemsen A. G. (1996) EMBO J. 15, 5349–5357 [PMC free article] [PubMed] [Google Scholar]
- 40.Marine J. C., Jochemsen A. G. (2005) Biochem. Biophys. Res. Commun. 331, 750–760 [DOI] [PubMed] [Google Scholar]
- 41.Parant J., Chavez-Reyes A., Little N. A., Yan W., Reinke V., Jochemsen A. G., Lozano G. (2001) Nat. Genet. 29, 92–95 [DOI] [PubMed] [Google Scholar]
- 42.Linares L. K., Hengstermann A., Ciechanover A., Müller S., Scheffner M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawai H., Lopez-Pajares V., Kim M. M., Wiederschain D., Yuan Z. M. (2007) Cancer Res. 67, 6026–6030 [DOI] [PubMed] [Google Scholar]
- 44.Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 45.Okamoto K., Taya Y., Nakagama H. (2009) FEBS Lett. 583, 2710–2714 [DOI] [PubMed] [Google Scholar]
- 46.Narimatsu M., Bose R., Pye M., Zhang L., Miller B., Ching P., Sakuma R., Luga V., Roncari L., Attisano L., Wrana J. L. (2009) Cell 137, 295–307 [DOI] [PubMed] [Google Scholar]
- 47.Yamashita M., Ying S. X., Zhang G. M., Li C., Cheng S. Y., Deng C. X., Zhang Y. E. (2005) Cell 121, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki A., Shibata T., Shimada Y., Murakami Y., Horii A., Shiratori K., Hirohashi S., Inazawa J., Imoto I. (2008) Cancer Sci. 99, 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin C., Yang Y. A., Anver M. R., Morris N., Wang X., Zhang Y. E. (2009) Cancer Res. 69, 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.