Abstract
CaVβ subunits modulate cell surface expression and voltage-dependent gating of high voltage-activated (HVA) CaV1 and CaV2 α1 subunits. High affinity CaVβ binding onto the so-called α interaction domain of the I-II linker of the CaVα1 subunit is required for CaVβ modulation of HVA channel gating. It has been suggested, however, that CaVβ-mediated plasma membrane targeting could be uncoupled from CaVβ-mediated modulation of channel gating. In addition to CaVβ, CaVα2δ and calmodulin have been proposed to play important roles in HVA channel targeting. Indeed we show that co-expression of CaVα2δ caused a 5-fold stimulation of the whole cell currents measured with CaV1.2 and CaVβ3. To gauge the synergetic role of auxiliary subunits in the steady-state plasma membrane expression of CaV1.2, extracellularly tagged CaV1.2 proteins were quantified using fluorescence-activated cell sorting analysis. Co-expression of CaV1.2 with either CaVα2δ, calmodulin wild type, or apocalmodulin (alone or in combination) failed to promote the detection of fluorescently labeled CaV1.2 subunits. In contrast, co-expression with CaVβ3 stimulated plasma membrane expression of CaV1.2 by a 10-fold factor. Mutations within the α interaction domain of CaV1.2 or within the nucleotide kinase domain of CaVβ3 disrupted the CaVβ3-induced plasma membrane targeting of CaV1.2. Altogether, these data support a model where high affinity binding of CaVβ to the I-II linker of CaVα1 largely accounts for CaVβ-induced plasma membrane targeting of CaV1.2.
Keywords: Calcium Channels, Calmodulin, Cell Sorting, Protein Targeting, Protein-Protein Interactions
Introduction
Voltage-dependent Ca2+ channels (CaV) are membrane proteins that play a key role in promoting Ca2+ influx in response to membrane depolarization in excitable cells. To this date, molecular cloning has identified the primary structures for 10 distinct calcium channel CaVα1 subunits (1–7) that are classified into three main subfamilies according to their high voltage-activated (HVA)2 gating (CaV1 and CaV2) or low voltage-activated gating (CaV3). In addition to the transmembrane pore-forming CaVα1 subunit, CaV1 and CaV2 channels arise from the multimerization of three other proteins (7): a cytoplasmic CaVβ subunit, a mostly extracellular CaVα2δ subunit, and calmodulin constitutively bound to the C terminus of CaVα1 (8–12).
A considerable body of work documents the interaction and modulation of the CaVα1 subunit of CaV1 and CaV2 channels (13–18) by the auxiliary CaVβ. The high affinity CaVα1-CaVβ interaction site on the pore-forming CaVα1 subunit is a conserved 18-residue sequence in the I-II linker called the α interaction domain (AID) (19, 20) that has been structurally resolved by high resolution x-ray crystallography (21–23). Structural work showed that the AID forms a α-helix that binds to the α binding pocket (ABP) in the CaVβ nucleotide kinase (NK) domain. It has been proposed that the MMQKAL cluster of residues within the latter determines the high affinity nanomolar interaction between the two proteins (24–29). Numerous mutational analyses of the AID residues have correlated the CaVβ-induced biophysical modulation with the high affinity binding of CaVβ to the AID peptide in a variety of CaVα1 isoforms for CaV1 and CaV2 channels (25, 29–32).
The association of CaVα1 and CaVβ subunits is also critical for proper channel maturation and cell surface expression of CaV2.2 (17), CaV1.2 (33, 34), and CaV2.3 (35). In CaV2.2, the I-II linker is presumed to play a role in this process (17, 18), and mutations within the AID motif eliminated its cell surface expression and biophysical modulation by CaVβ1b and CaVβ3 (32). In addition, the CaVβ2-induced increase in CaV1.2 whole cell currents was abolished with the AID-defective YWI/AAA mutant (29), suggesting that high affinity binding of CaVβ onto AID is required to traffic CaVα1 to the plasma membrane. Nonetheless, the unique character of the high affinity AID-ABP interface in the membrane targeting of CaVα1 has been questioned (27, 36–40). In particular, it has been suggested that CaVβ-mediated plasma membrane targeting could be uncoupled from CaVβ-mediated modulation of channel gating (26, 41) with important contributions from other intracellular regions (33, 39, 42–44).
In addition to CaVβ, the ancillary subunit CaVα2δ and the ubiquitous calmodulin (CaM) protein have also been proposed to modulate HVA channel maturation and targeting (9). For instance, co-expression of CaVα2δ promoted the trafficking of the CaVα1 subunit of CaV2.2 in COS-7 cells (45), suggesting that CaVα2δ could promote targeting of all HVA CaVα1 subunits. CaM is a soluble, 17-kDa Ca2+-binding protein that serves as a critical Ca2+ sensor for Ca2+-dependent inactivation and facilitation upon Ca2+ binding in many CaV1 and CaV2 channels (46), of which CaV1.2 and CaV2.1 have been best characterized (8, 47). Constitutive apocalmodulin binding was reported on multiple sites in the CaVα1 subunit of CaV1.2 (48) of which the C-terminal pre-IQ and IQ domains are best characterized (49). Mutations (TLF/AAA and I/E) in the pre-IQ and the IQ CaM-binding domains of the C terminus decreased the whole cell current density of CaV1.2, suggesting that Ca2+/CaM could modulate channel trafficking through its interaction with the C terminus (50) as it has been shown for small activated potassium channels (51).
To gauge the synergetic role of intracellular domains and auxiliary subunits in the steady-state plasma membrane expression of CaV1.2, we used a flow cytometry assay with an extracellularly HA-tagged CaV1.2 protein. Co-expression with CaVβ3 produced a robust enhancement in the plasma membrane targeting of the CaVα1 subunit of CaV1.2. The WI residues in the AID helix of the I-II linker of CaV1.2 were critical for CaVβ-stimulated plasma membrane targeting of CaV1.2. No other combination with or without the auxiliary calmodulin and/or the CaVα2bδ subunit produced any significant increase in the plasma membrane targeting of CaV1.2. Hence, CaVβ appears to be the most potent determinant in the plasma membrane targeting of CaV1.2. Altogether, our data support a model where high affinity binding of the ABP of CaVβ to the AID helix of CaVα1 largely accounts for CaVβ-induced plasma membrane targeting of CaV1.2.
EXPERIMENTAL PROCEDURES
Recombinant DNA Techniques
The rabbit CaV1.2 (GenBankTM accession number X15539), the rat CaVβ3 (GenBankTM accession number M88751) (52), the rat brain CaVα2bδ (GenBankTM accession number NM_000722) (53), and the human CaM (GenBankTM accession number M27319) were used. All of the subunits were subcloned in commercial vectors under the control of the cytomegalovirus promoter (see supplemental text for details).
For the CaVβ3 deletion mutants, flanking NotI sites were inserted around the region(s) to be deleted. Following restriction digest of the NotI fragment and religation of the cohesive ends, the resulting NotI site was mutated back to the wild type amino acids. The CaVβ3 fragments (numbered from their deduced amino acid sequence) were subcloned into the NotI sites of the pCMV-Tag5a vector (see supplemental text for details) that is a C-terminal c-Myc tagging vector. A Kozak sequence and an ATG initiation codon were inserted at the 5′-end of the nucleotide sequence.
The calmodulin wild type cDNA was subcloned in the pMT21 vector (54). The dominant negative mutant of CaM (CaM1,2,3,4) that impaired high affinity Ca2+ binding is D20A/D56A/D93A/D129A, which has been described elsewhere (55).
Insertion of the HA Tag in the CaVα1 Subunit
The hemagglutinin (HA) epitope tag (YPYDVPDYA) was inserted in the first extracytoplasmic predicted loop in Domain I at position 574 (nucleotide) for CaV1.2. The biophysical properties of the HA-tagged CaVα1 subunit of CaV1.2 expressed in HEKT cells with the auxiliary CaVβ3 subunit were found not to be significantly different from the wild type CaV1.2 channel expressed under the same conditions (see Fig. 1). In addition, cDNA injection of CaV1.2-HA constructions in concert with CaVα2bδ and CaVβ3 subunits in Xenopus oocytes yielded a biophysical profile not significantly different from that reported previously for the CaV1.2 (56) channels expressed under the same conditions. Hence, the HA-tagged version of the CaVα1 subunit of CaV1.2 will be referred to as CaV1.2 wt throughout the text.
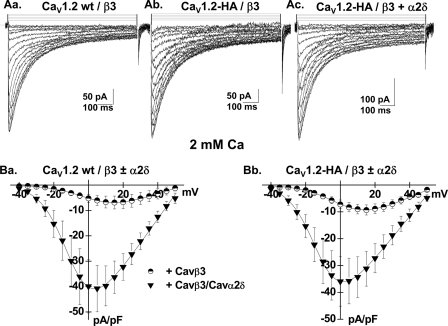
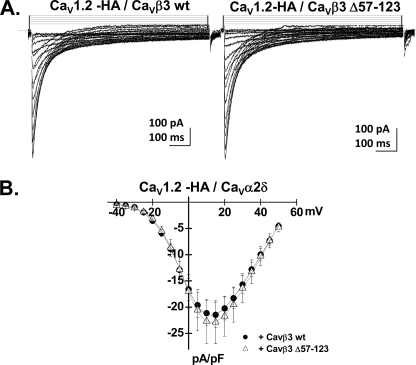
FIGURE 1.
CaVα2bδ stimulated CaV1.2 whole cell currents. A, panel a, whole cell current traces recorded after the transient expression of the CaV1.2 wt channel in the stable CaVβ3 cell line. The charge carrier was 2 mm Ca2+. Panel b, whole cell current traces recorded after the transient expression of the CaV1.2-HA channel in the stable CaVβ3 cell line. Panel c, whole cell current traces recorded after the transient expression of the CaV1.2-HA channel and the CaVα2bδ in the stable CaVβ3 cell line. B, panel a, current-voltage relationships of CaV1.2 wt + CaVβ3 ± CaVα2bδ show a typical voltage-dependent activation with a mean current density of −7 ± 2 pA/pF (n = 6) for the wild type CaV1.2 channel in the stable CaVβ3 stable cell line as compared with a current density of −41 ± 9 pA/pF (n = 7) for the wild type CaV1.2 channel measured in the same cell line after transient transfection with CaVα2bδ subunit. The activation potential of 3 ± 3 mV for the CaV1.2 wt + CaVβ was shifted to −10 ± 2 mV in the presence of CaVα2bδ. Panel b, current-voltage relationships of CaV1.2-HA/CaVβ3 ± CaVα2bδ show a typical voltage-dependent activation with a mean current density of −9 ± 2 pA/pF (n = 5) for the CaV1.2-HA channel in the stable CaVβ3 stable cell line as compared with a current density of −36 ± 8 pA/pF (n = 7) for the CaV1.2-HA channel measured in the same cell line after transient transfection with CaVα2bδ subunit. The activation potential of 2 ± 3 mV for the CaV1.2-HA/CaVβ was shifted to −12 ± 2 mV in the presence of CaVα2bδ. Patch clamp experiments were carried out in the whole cell configuration in the presence of a 2 mm Ca2+ saline solution.
Cell Culture and Transfections
tsA-201 (HEK293T or HEKT), a subclone of the human embryonic kidney cell line HEK-293 that expresses the simian virus 40 T-antigen, and COS1 cells were grown in Dulbecco's high glucose minimum essential medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin at 37 °C under 5% CO2 atmosphere. COS1, HEKT, stable CaVβ3, and CaVα2bδ cells lines were transiently transfected with HA-tagged CaV1.2 cDNA using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. Protein expression of the auxiliary subunits in the stable and transient cell lines were confirmed routinely by Western blotting (see Fig. 2B). Transfection rate of the control pEGFP plasmid was estimated to be 66 ± 2% (n = 4) as assessed by flow cytometry from the fluorescence of the green fluorescent protein. Preliminary tests showed that CaV1.2 protein expression peaked 24–36 h after transfection.
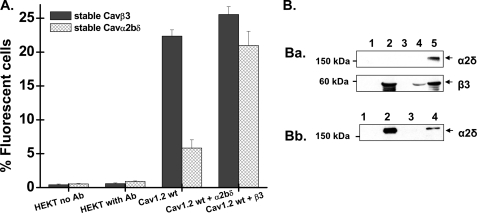
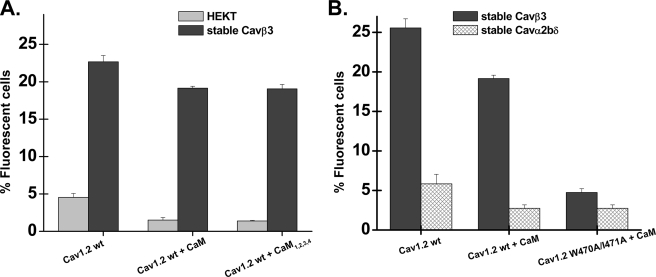
FIGURE 2.
CaVβ stimulated CaV1.2 membrane expression in HEKT cells. A, HA-tagged CaV1.2 wt was co-expressed transiently either in the stable CaVβ3 or the stable CaVα2bδ cell line. Cell surface expression of CaV1.2 wt was determined in intact cells by flow cytometry using the anti-HA FITC conjugate antibody (Ab). The histogram shows the number of fluorescent cells as a function of the experimental conditions. Cell autofluorescence (HEKT no Ab) was <1% throughout, and the addition of the FITC did not significantly increase the level of fluorescence in HEKT cells not transfected with the HA-tagged CaV1.2 (HEKT with Ab). As seen, only co-expression of CaV1.2 with CaVβ3 significantly promoted membrane expression of CaV1.2 (p < 0.001). Co-expression of CaV1.2 with CaVα2bδ did not alter the number of CaV1.2 channels at the membrane (p > 0.1). Co-expression with both auxiliary subunits did not further improve the membrane expression of CaV1.2. The CaV1.2 + CaVβ3 + CaVα2bδ condition (either CaV1.2 + CaVβ3 expressed transiently in the stable CaVα2bδ cell line or CaV1.2 + CaVα2bδ expressed transiently in the CaVβ3 cell line) was not significantly different from the CaV1.2 + CaVβ3 condition (p > 0.1). Similar results were obtained with transient expression systems. The numerical values can be found in Table 1. B, Western blot analyses of HEKT cells transiently or stably transfected with CaVα2bδ or CaVβ3, using CaVα2δ-1 (1:200) and CaVβ3 (1:500) antibodies. Each lane was loaded with 50 μg of protein. Panel a, HEKT cells were transiently or stably transfected with CaVβ3. Lane 1, control nontransfected cells. Lane 2, transient transfection of CaVβ3. Lane 3, control nontransfected cells. Lane 4, stable CaVβ3 cell line. Lane 5, transient co-transfection of CaVβ3 and CaVα2bδ. Panel b, HEKT cells were transiently or stably transfected with CaVα2bδ. Lane 1, control nontransfected cells. Lane 2, transient transfection of CaVα2bδ. Lane 3, control nontransfected cells. Lane 4, stable CaVα2bδ cell line.
Western Blots
Protein expression of all constructs was confirmed by Western blotting in total cell lysates. HA-tagged CaV1.2 constructs were detected with anti-HA. The procedures are detailed in the supplemental text. Briefly, the membranes were incubated with anti-HA (1:500) (Covance Biotechnology, Québec, Canada) and revealed with an anti-mouse horseradish peroxidase secondary antibody (1:10000; Jackson Immunoresearch).
Fluorescence-activated Cell Sorting (FACS) Experiments
Cell surface expression of the CaV1.2 subunits was determined by flow cytometry using a FACScalibur® flow cytometer (Becton Dickinson) at the flow cytometry facility of the Department of Microbiology of the Université de Montréal. The cells expressing the extracellular HA tag were detected using an anti-HA-conjugated FITC fluorophore with a FITC filter (530 nm). The relative intensity of staining provided a metric to quantify cell surface expression of the HA-tagged CaV1.2 proteins (see supplemental Figs. S1 and S2 and supplemental text for details). The HA-tagged CaV1.2 construct was systematically tested as a control with the mutant channels.
Immunofluorescence
For fluorescence microscopy, CaVβ3 stable cells were grown on sterile poly-d-lysine-coated coverslips. The cells were fixed 24 h after transfection in 4% paraformaldehyde, permeabilized with 0.075% saponin for 10 min at room temperature, washed in phosphate-buffered saline, and blocked in IgG-free 2% bovine serum albumin in phosphate-buffered saline for 20 min. The cells were incubated with FITC-conjugated anti-HA antibody (1:100) for 1 h at room temperature prior to the cells being mounted (Prolong antifade kit; Invitrogen) on glass microscope slides. HA-tagged CaV1.2 channels (wild type and mutant) were visualized (×60) using an Olympus microscope IX-81 microscope along with Image-Pro Plus 5.0 software.
Statistical Analysis
Statistical analyses were performed using the built-in one-way analysis of variance fitting routine for two independent populations of Origin 7.0. The data were considered statistically significant at p < 0.01.
RESULTS
CaVα2δ Increases Whole Cell Currents of CaV1.2
Co-expression of CaV1.2 and CaV2.1 with the auxiliary CaVα2δ subunit was shown to stimulate whole cell currents (45) in COS-7 cells, suggesting that CaVα2δ could promote plasma membrane targeting of HVA CaVα1 subunits. In CaV1.2, the gating charge appears to be unaffected by co-expression with CaVα2δ, suggesting that CaVα2δ stimulates channel facilitation by setting CaV1.2 channels in a conformational state very close to the open state without increasing protein density (57). To evaluate the functional role of CaVα2δ, CaV1.2 wt and HA-tagged CaV1.2 α1 subunits were transiently transfected in the CaVβ3 stable HEKT cell line in the absence and in the presence of CaVα2bδ. As shown in Fig. 1A, whole cell currents, recorded in the presence of a physiological solution containing 2 mm Ca2+ (see the supplemental text), were significantly larger when measured in the presence of the CaVa2δ, confirming that CaVα2δ stimulates whole cell currents of CaV1.2 (9). As shown in Fig. 1B, average whole cell current density increased from −7 ± 2 pA/pF (n = 6) for the wild type CaV1.2 channel in the stable CaVβ3 stable cell line as compared with a current density of −41 ± 9 pA/pF (n = 7) for the wild type CaV1.2 channel measured in the same cell line after transient transfection with CaVα2bδ subunit. Similar results were obtained for the HA-tagged CaV1.2 channels (Fig. 1B).
CaVβ Promotes Membrane Targeting of CaV1.2
To determine whether CaVα2bδ stimulates plasma membrane targeting of CaV1.2 channels, protein density of the extracellularly HA-tagged CaV1.2 channel was quantified with an anti-HA-conjugated FITC fluorophore. Fig. 2A shows the histogram of the fluorescent signal measured after transient expression of the CaVα1 and the auxiliary subunit (either CaVβ3 or CaVα2bδ) in nonpermeabilized cells. Protein expression was confirmed by Western blotting (Fig. 2B). As seen, less than 0.5% of the cell population produced autofluorescence, whereas only 1% of the cells were fluorescent after the addition of the FITC antibody to control nontransfected cells (see raw data in supplemental Figs. S1 and S2). Transient co-expression of the HA-tagged CaV1.2 subunit in the stable CaVβ3 cell line increased the number of proteins detected at the membrane from a value of 4.5 ± 0.5% (n = 25) in the nontransfected cell line to 23 ± 1% (n = 29) with CaVβ3 (p < 0.001) (Table 1).
TABLE 1.
Fluorescence-activated cell sorting analysis of CaV1.2 ± auxiliary subunits
FACS results obtained after the transient transfection of CaV1.2-HA wt in either HEKT control cells, stable CaVβ3 cells, or stable CaVα2bδ cells. One day (24 h) after transfection, the cells were incubated with anti-HA FITC conjugate (10 μg/ml) at room temperature for 45 min. FACS separation of FITC-positive cells was performed on a FACScalibur® flow cytometer (Becton Dickinson), and fluorescence was quantified using CellQuest software (Becton Dickinson). The results are reported as percentage values of cells in M2. The data were pooled from experiments carried out over a period of 8 months. The data are shown as the means ± S.E. of the individual experiments, and the number of experiments appears in parentheses. ND, not determined.
| Construct transient expression | Cell lines |
|||
|---|---|---|---|---|
| HEKT | HEKT CaVβ3 stable | HEKT CaVα2bδ stable | HEKT CaVβ3 + CaVα2bδ stable | |
| % | % | % | % | |
| Cells without antibody | 0.3 ± 0.1 (32) | 0.18 ± 0.04 (20) | 0.54 ± 0.07 (3) | 2.8 ± 0.2 (3) |
| Cells with antibody | 1.1 ± 0.3 (19) | 1.0 ± 0.4 (19) | 0.93 ± 0.05 (3) | 4.2 ± 0.4 (3) |
| CaV1.2-HA wt | 4.5 ± 0.5 (25) | 23 ± 1 (29) | 6 ± 1 (3) | 19 ± 2 (3) |
| CaV1.2-HA wt + CaVβ3 | 25 ± 2 (3) | ND | 21 ± 2 (3) | ND |
| CaV1.2-HA wt + CaVβ2a | 14 ± 1 (3) | ND | ND | ND |
| CaV1.2-HA wt + CaVβ4 | 22 ± 3 (3) | ND | ND | ND |
| CaV1.2-HA wt + CaVα2bδ + CaM wt | 2 ± 1 (3) | ND | ND | ND |
| CaV1.2-HA wt + CaVα2bδ | 8 ± 3 (5) | 26 ± 1 (3) | ND | ND |
| CaV1.2-HA wt + CaM wt | 1.5 ± 0.3 (3) | 19.1 ± 0.3 (3) | 2.7 ± 0.4 (3) | ND |
| CaV1.2-HA wt + CaM1,2,3,4 | 1.4 ± 0.1 (3) | 19.1 ± 0.6 (3) | ND | ND |
| CaV1.2-HA wt + CaVβ3 + CaVα2bδ | 23 ± 2 (5) | ND | ND | ND |
The results obtained with CaVβ3 contrast with the effect observed when co-expressing HA-tagged CaV1.2 with CaVα2bδ (p > 0.1). No further increase in the fluorescent signal was observed in the combined presence of the two auxiliary subunits. Similar results were obtained when CaV1.2 was transiently expressed, either in a background of stably transfected CaVβ3 or in a background of stably transfected CaVα2bδ cells (see Table 1 for numerical values). The maximum fluorescence obtained with CaVβ3 confirms that CaVα2bδ has little effect by itself on the CaV1.2 protein density at the plasma membrane. Among CaVβ subunits, transient co-expression of CaV1.2-HA with CaVβ4 caused a similar boost in plasma membrane expression, whereas CaVβ2a was found to be slightly less potent for a CaVβ3 ≈ CaVβ4 > CaVβ2a ranking (Table 1). Altogether, these results validated the fluorescence sorting analysis of HA-tagged CaVα1 proteins to evaluate steady-state protein level in intact cells independently of channel gating.
α Interaction Domain: the Role of the WI Pair
Crystallographic analyses have shown that the AID-CaVβ interaction is anchored through a set of six residues, Asp, Leu, Gly, Tyr, Trp, and Ile, distributed among three α-helical turns of the I-II linker of CaV1.2 (21–23), with the WI pair of residues being most critical for the AID-CaVβ protein interaction (29). To evaluate whether the AID-CaVβ interaction controls the plasma membrane targeting of CaV1.2, the HA-tagged CaV1.2 subunit was transiently co-expressed in HEKT cells or in CaVβ3 stable cells. CaVβ3 stimulated the plasma membrane targeting of N-terminal mutants: L464A, G466A, G466F, Y467G, Y467A, Y467S, and Y467F (Fig. 3A, supplemental Fig. S4, and supplemental Table SII). When compared with the control situation (±CaVβ3), there was a 4–8-fold stimulation in the plasma membrane targeting mutations in the order G466A ≈ G466F > Y467F > Y467G > L464A ≈ Y467A > Y467S. Hence, no single point mutation in the N-terminal region of the AID completely abolished the CaVβ3-induced stimulation in the plasma membrane targeting of CaV1.2. In contrast, the CaVβ3 stimulation effect was completely eradicated in the double mutant G466A/Y467F, even though each individual mutation behaved like the wild type channel, suggesting that each residue contributes to the high affinity interaction with CaVβ3.
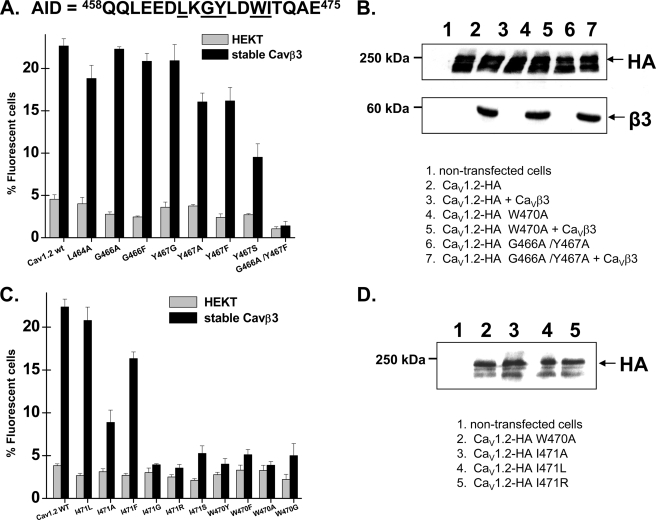
FIGURE 3.
Point mutations within the C-terminal residues on the AID helix disrupted the CaVβ stimulation of CaV1.2 plasma membrane targeting. A, HA-tagged CaV1.2 wt and mutants were expressed transiently either in the HEKT cells or in the stable CaVβ3 cell line. Cell surface expression of the CaV1.2 protein was quantified as described for supplemental Fig. S2. The residues targeted in these experiments are underlined within the primary sequence of the AID region of CaV1.2. The number of fluorescent cells decreased in the order CaV1.2-HA wt ≈ L464A, G466A, G466F, Y467G > Y467A, Y467F, G466A/Y467A > Y467S, G466Y/Y467G ≫ G466A/Y467F. The numerical values can be found in supplemental Table SI. B, Western blot analyses of HEKT cells transiently transfected with CaV1.2 wt or mutants in stable CaVβ3 cells using HA (1:500) and CaVβ3 (1:500) antibodies. Lane 1, control nontransfected cells. Lane 2, CaV1.2-HA. Lane 3, CaV1.2-HA + CaVβ3. Lane 4, CaV1.2-HA W470A. Lane 5, CaV1.2-HA W470A + CaVβ3. Lane 6, CaV1.2-HA G466A/Y467A. Lane 7, CaV1.2-HA G466A/Y467A + CaVβ3. Western blot analyses confirmed that the W470A mutant was expressed in total cell lysates and recognized by the anti-HA (1:500). Each lane was loaded with 50 μg of protein. C, HA-tagged CaV1.2 wt and mutants were expressed transiently either in the HEKT cells or in the stable CaVβ3 cell line. Cell surface expression of the CaV1.2 protein was quantified as described for supplemental Fig. S2. The residues targeted in these experiments are underlined within the primary sequence of the AID region of CaV1.2 shown in A. The number of fluorescent cells decreased in the order CaV1.2-HA wt ≈ I471L > I471F > I471A > I471S > I471R ≫ I471G, W470A, W470F, W470G, W470Y. The numerical values can be found in supplemental Table SI. D, Western blot analyses confirmed that the CaV1.2 W470A, I471A, I471L, and I471R mutants expressed with the expected molecular weight in total cell lysates and were recognized by the anti-HA (1:500). Each lane was loaded with 50 μg of protein. Lane 1, control nontransfected cells. Lane 2, CaV1.2-HA W470A. Lane 3, CaV1.2-HA I471A. Lane 4, CaV1.2-HA I471L. Lane 5, CaV1.2-HA I471R.
Point mutations in the C-terminal WI pair yielded a different picture. I471L was the only mutant that was detected at the membrane to the same extent as the wild type channel in the presence of CaVβ3. However, CaVβ3 stimulated significantly the plasma membrane targeting of I471A and I471F mutants. W470Y, W470F, W470A, W470G, I417G, and I471R were not significantly different in the presence or in the absence of CaVβ3 (Fig. 3C and supplemental Table SII). Western blots carried out in total cell lysates with the anti-HA confirmed that all of the CaV1.2 mutants tested produced proteins with the expected molecular weight (Fig. 3, B and D). Immunofluorescence microscopy confirmed that W470A disrupted the plasma membrane targeting of CaV1.2 in the presence of CaVβ3 (supplemental Fig. S5). Membrane expression of CaV1.2 in cultured hippocampal neurons was also disrupted after mutation of the key tryptophan residue to alanine (58).
Furthermore, double mutations in the same region completely eradicated the CaVβ3 stimulation of CaV1.2 plasma membrane targeting (supplemental Fig. S6 and Table SI). Partial (Δ458–463) or complete (Δ458–475) removal of the AID-binding site within the I-II loop yielded similar results, confirming that no other low affinity CaVβ site within the CaVα1 subunit could promote the plasma membrane targeting of CaV1.2 in the absence of the AID region. Isothermal titration calorimetry assays have substantiated that the affinity of CaVβ2a for the AID region of CaV1.2 decreased after single-point mutations of these residues. There was an >1000-fold increase in the Kd with the W/A and I/A mutants, whereas alanine mutation of the Leu and Gly residues imparted a smaller 5–10-fold decrease in the CaVβ2a affinity (29). Substitution of the tryptophan residue by either tyrosine or phenylalanine only partly compensated for the mutation, confirming the requirement of a residue containing a double aromatic ring at this position. For the neighboring isoleucine position, mutation with the conserved leucine residue was found to preserve the CaVβ3-induced membrane targeting of CaV1.2. For comparison, I387L in CaV2.3 was the only mutant tested in the WI pair that supported CaVβ3 binding and CaVβ3 modulation of gating (31). In contrast, none of the CaV1.2 mutations identified in the short QT syndrome, an inherited form of cardiac arrhythmia (59), was shown to affect the CaVβ3 plasma membrane targeting of CaV1.2 (supplemental Fig. S7 and Table SII). Altogether our results support a strong correlation between CaVβ3 binding affinity to the AID region as determined from fusion proteins and from isothermal titration calorimetry assays (29) and its role in promoting the targeting of CaV1.2 proteins at the plasma membrane. More importantly our results suggest that the molecular determinants that account for CaVβ3 binding to the AID region are also responsible for the CaVβ3-induced stimulation of the plasma membrane targeting of CaV1.2 proteins.
The NK Domain of CaVβ Is Essential for Plasma Membrane Targeting of CaV1.2
The observation that different CaVβs, which all share a conserved core containing the SH3 and NK domains, cause different biophysical effects on CaVα1 subunits suggests that other regions besides the conserved AID-ABP interaction, could influence channel conformational changes (13). The SH3 domain of CaVβ2a was found to bind to the I-II linker of CaV2.1 channels, suggesting that low affinity interactions outside of the AID-ABP interface could contribute to the full functional effects of the CaVβ subunit (40). A few years later, however, the conserved AID-NK domain interaction was found to be necessary for CaVβ-stimulated CaV2.1 channel surface expression (60). To evaluate whether the AID-NK interaction controls the plasma membrane targeting of CaV1.2, the HA-tagged CaV1.2 subunit was transiently co-expressed in HEKT cells in the presence of CaVβ3 full-length or deleted constructs as well as with CaVβ3 fragments.
We found that the NK domain of CaVβ3 (180–364) (Fig. 4A) was required for the plasma membrane targeting of CaV1.2 (Fig. 4B and supplemental Table SIII). Targeted deletion of the SH3 domain between residues 57 and 123 preserved 80% of the CaV1.2 protein detected at the membrane (Fig. 4B). The deleted CaVβ3 Δ57–123 construct preserved the typical CaVβ3 modulation of channel gating and inactivation current kinetics. Peak whole cell current density was not significantly affected (Fig. 5).
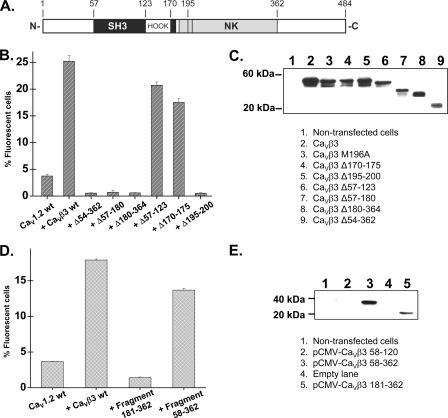
FIGURE 4.
The NK domain of CaVβ3 is critical for the plasma membrane targeting of CaV1.2. A, schematic diagram of the domain organization of the CaVβ3 subunit based on the crystal structure and adapted from (21). B, HA-tagged CaV1.2 wt and CaVβ3 mutants were expressed transiently in the HEKT cells. Cell surface expression of the CaV1.2 protein was quantified as described for supplemental Fig. S2. The residues targeted in these experiments are identified in the primary sequence of CaVβ3. The Δ57–123 deletion removed the SH3 domain; the Δ170–175 removed the PYDVVP sequence; and the Δ195–200 removed the MMQKAL sequence, also termed the ABP domain. The numerical values are provided in supplemental Table SIII. C, Western blot analyses of HEKT cell lysates transiently transfected with CaVβ3 constructs using anti-CaVβ3 (1:500). Each lane was loaded with 10 μg of protein except for CaVβ3 Δ54–362 loaded with 50 μg of protein. Lane 1, control nontransfected cells. Lane 2, CaVβ3 wt. Lane 3, CaVβ3 M196A. Lane 4, CaVβ3 Δ170–175. Lane 5, CaVβ3 Δ195–200. Lane 6, CaVβ3 Δ57–123. Lane 7, CaVβ3 Δ57–180. Lane 8, CaVβ3 Δ180–364. Lane 9, CaVβ3 Δ54–362. Western blot analyses confirmed that the CaVβ3 deleted proteins were detected in total cell lysates with the expected molecular weight. D, HA-tagged CaV1.2 wt and CaVβ3 fragments were expressed transiently in the HEKT cells. Cell surface expression of the CaV1.2 protein was quantified as described for supplemental Fig. S2. The residues targeted in these experiments are identified in the primary sequence of the CaVβ3. The 181–362 fragment is equivalent to the NK domain. The numerical values are provided in supplemental Table SIII. E, Western blot analyses of HEKT cell lysates transiently transfected with the CaVβ3 fragments using anti-c-Myc (1:500). Each lane was loaded with 50 μg of protein. Lane 1, control nontransfected cells. Lane 2, CaVβ3 58–120. Lane 3, CaVβ3 58–362. Lane 4, empty lane. Lane 5, CaVβ3 181–362. The CaVβ3 58–120 fragment formed a 7.4-kDa protein that cannot be seen in this figure. Western blot analyses confirmed that the CaVβ3 fragments were detected in total cell lysates with the expected molecular weight.
FIGURE 5.
CaVβ3 Δ57–123 stimulated CaV1.2 whole cell currents. A, whole cell current traces recorded after the transient expression of the CaV1.2-HA channel with CaVα2bδ and CaVβ3 wt (left panel) or with CaVα2bδ and CaVβ3 Δ57–123 (right panel) in HEKT cells. All of the subunits were transiently expressed. B, current-voltage relationships of CaV1.2-HA + CaVα2bδ + CaVβ3 wt (filled circles) and CaV1.2-HA + CaVα2bδ + CaVβ3 Δ57–123 (open triangles) show a typical voltage-dependent activation with a mean current density of −21 ± 2 pA/pF (n = 5) for CaV1.2-HA + CaVα2bδ + CaVβ3 wt as compared with a current density of −23 ± 4 pA/pF (n = 4) for CaV1.2-HA + CaVα2bδ + CaVβ3 Δ57–123 measured under the same conditions. Patch clamp experiments were carried out in the whole cell configuration in the presence of a 2 mm Ca2+ saline solution.
The CaVα1-CaVβ interaction appears to require the MMKQAL motif in the α3 helix of the NK domain and was identified in the crystal structure (21–23) as critical for the high affinity AID-ABP interaction. Indeed deletion of the 195–200 residue region of CaVβ3 completely abolished plasma membrane targeting of CaV1.2 (Fig. 4B), and the single point mutation M196A in CaVβ3, equivalent to M245 in CaVβ2a (29), significantly decreased plasma membrane targeting with only 13 ± 1% (n = 3) fluorescent cells (supplemental Table SIII). Nonetheless, the NK domain (181–362 fragment) alone was not sufficient for targeting CaV1.2 to the membrane (Fig. 4D). Only the larger fragment (58–362) that includes part of the SH3 domain was found to stimulate significantly the plasma membrane targeting of CaV1.2. The integrity of the constructions was verified by Western blot (Fig. 4, C and E).
Calmodulin in the Plasma Membrane Targeting of CaV1.2
CaM interacts with multiple sites in the CaVα1 subunit of CaV1.2 (48, 61), of which the C-terminal pre-IQ and IQ domains are best characterized. Constitutive CaM binding to the N terminus has also been reported (62). To determine whether low affinity binding of CaM to intracellular regions contributes to trafficking of CaV1.2 channels (50, 63), CaV1.2 was co-expressed with CaM wt or the dominant negative mutant of CaM (CaM1,2,3,4) in HEKT control cells (supplemental Fig. S8) and in CaVβ3 stable cells. Overexpression of CaM wt or its negative dominant mutant in CaVβ3 stable cells did not significantly alter whole cell currents measured in the presence of 2 mm Ca2+ with peak current densities of −8 ± 2 pA/pF (n = 9) (CaM wt) and of −9 ± 3 pA/pF (n = 9) (CaM1,2,3,4), whereas co-expression with the latter significantly decreased calcium-dependent inactivation kinetics (supplemental Fig. S9). Cytometry flux assays also failed to show a change in the plasma membrane expression of CaV1.2 with or without CaVβ3 (Fig. 6 and supplemental Table SIV). These data contrast with previous reports that CaM1,2,3,4 co-expression reduced peak CaV1.2 current amplitude in HEK cells compared with CaM co-expression (8). It suggests that CaVβ3 is the dominant subunit to promote plasma membrane targeting of CaV1.2 and that CaM does not act synergistically with CaVβ3 under these conditions.
FIGURE 6.
Overexpressing CaM wt and CaM1,2,3,4 did not alter the pattern of CaVβ stimulation of CaV1.2 plasma membrane targeting. A, HA-tagged CaV1.2 wt was expressed transiently either in the HEKT cells or in the stable CaVβ3 cell line. Cell surface expression of the CaV1.2 protein was quantified as described for supplemental Fig. S2. The number of fluorescent cells was not significantly influenced by overexpressing the wild type CaM or the dominant negative mutant CaM1,2,3,4. The numerical values can be found in Table 1 and supplemental Table SIV. B, HA-tagged CaV1.2 wt and the double W470A/I471A mutant were expressed transiently either in the stable CaVβ3 or in the stable CaVα2bδ cell line. Cell surface expression of the CaV1.2 protein was quantified as described for supplemental Fig. S2. The number of fluorescent cells was not significantly increased by overexpressing CaM wt in any cell line as compared with the control CaV1.2 cells (p > 0.05). Similar data were obtained with the W470A mutant that abrogated CaVβ3 binding and stimulation of CaV1.2 plasma membrane targeting. The numerical values can be found in Table 1 and supplemental Table SIV.
Overexpression of CaM wt was reported to promote the plasma membrane targeting of CaV1.2 proteins in COS1 cells, provided there was a complete absence of CaVβ (63). To test the hypothesis that CaM could chaperone CaV1.2 to the membrane in the presence of CaVα2bδ in our expression system, FACS experiments were carried out in the stable CaVα2bδ cell line. As seen in Fig. 6B, CaM was unable to increase the number of CaV1.2 proteins at the membrane in the absence of CaVβ3 under these conditions. Overexpression of CaM wt with the double mutant W470A/I471A (supplemental Table SIV) also failed to promote plasma membrane targeting of CaV1.2, thus ruling out a mechanism whereby low affinity binding of CaVβ subunit to the AID region could mask the CaM effect.
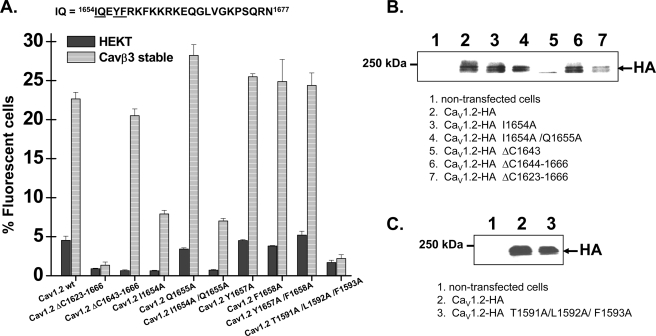
The 1643–1666 fragment in the C terminus forms the high affinity (Kd > 3 nm) IQ-binding domain that co-crystallized with CaM (49). This high affinity binding site overlaps with the C-terminal “targeting domain” identified previously (42, 64). To test the hypothesis that constitutive calmodulin binding to the IQ motif is required for plasma membrane targeting, FACS experiments were carried out after mutations of the aromatic residues responsible for the high affinity (Kd ≈ 3 nm) CaM binding (49). Complete deletion of the 1643–1666 fragment did not alter surface labeling, whereas the W470A mutation in the ΔIQ channel eliminated plasma membrane targeting of CaV1.2 (supplemental Table SIV), suggesting that the IQ domain is not likely to act as a retention signal. Furthermore, point mutations Q1655A, Y1657A, and F1658A, as well as multiple mutations I1654A/F1658A and Y1657A/F1658A, and I1654A/Y1657A/F1658A did not alter plasma membrane targeting of CaV1.2. Plasma membrane targeting was not affected by a triple mutation in the pre-IQ domain (CaV1.2 T1651A/F1652A/L1653A) and was modestly supported in the I1654A and I1654A/Q1655A mutants (Fig. 7 and supplemental Table SIV). It should be remembered that the I/A mutation only moderately affected Ca2+/CaM binding to the C-terminal peptide of CaV1.2 as compared with the I/E mutant (65). Deleting the larger 1623–1666 region, identified as an important targeting domain (42), completely eradicated the plasma membrane expression of CaV1.2 both in the presence and in the absence of CaVβ3 (Fig. 7 and supplemental Table SIV). Furthermore, as shown by others before (50), the CaV1.2 protein could not be detected at the membrane in the presence of the triple T1591A/L1592A/F1593A mutation. The three TLF residues are located in a pre-IQ apocalmodulin-binding site (peptide A) (48, 66), but overexpression with CaM wt or CaM1,2,3,4 did not rescue plasma membrane targeting (supplemental Table SIV). Altogether, these data highlight the role of the C terminus in the plasma membrane targeting of CaV1.2 and suggest that high affinity Ca2+/CaM binding is not critical for the plasma membrane targeting of CaV1.2.
FIGURE 7.
Mutations within the high affinity CaM binding motif did not alter the CaVβ stimulation of CaV1.2 plasma membrane targeting. A, flow cytometry data. HA-tagged CaV1.2 wt and mutants were expressed transiently either in HEKT cells or in the stable CaVβ3 cell line. Cell surface expression of the CaV1.2 protein was quantified as described for supplemental Fig. S2. The number of fluorescent cells decreased significantly for the mutants ΔC1623–1666, I1654A, I1654A/Q1655A, and T1591A/L1592A/F1593A (p < 0.01) as compared with the CaV1.2-HA wt protein under the same conditions. From left to right, the channels were CaV1.2-HA wt, ΔC1623–1666, ΔC1643–1666, I1654A, Q1655A, I1654A/Q1655A, Y1657A, F1658A, Y1657A/F1658A, and T1591A/L1592A/F1593A. The numerical values can be found in supplemental Table SIV. B, Western blot analyses confirmed that the CaV1.2 mutant proteins were detected in total cell lysates by the anti-HA (1:500) with the expected molecular weight. Lane 1, nontransfected cells. Lane 2, CaV1.2-HA. Lane 3, CaV1.2-HA I1654A. Lane 4, CaV1.2-HA I1654A/Q1655A. Lane 5, CaV1.2-HA ΔC1643. Lane 6, CaV1.2-HA ΔC1644–1666. Lane 7, CaV1.2-HA ΔC1623–1666. Each lane was loaded with 50 μg of protein. C, Western blot analyses confirmed that the CaV1.2 mutant proteins were detected in total cell lysates by the anti-HA (1:500) with the expected molecular weight. Lane 1, nontransfected cells. Lane 2, CaV1.2-HA wt. Lane 3, CaV1.2-HA T1591A/L1592A/F1593A. Each lane was loaded with 50 μg of protein.
DISCUSSION
To exhibit functional activity, ion channels must be targeted to the plasma membrane. Co-expression of CaV1.2 with either CaVα2bδ or CaM (alone or in combination) failed to promote significantly the detection of fluorescently labeled CaV1.2-HA channels in intact cells by flow cytometry. Co-expression of CaV1.2 in the presence of CaVβ3 with either CaVα2δ or CaM failed to further increase the number of CaV1.2 proteins detected at the plasma membrane. Furthermore, plasma membrane targeting of AID-disrupted CaV1.2 mutants (thus in the absence of high affinity CaVβ binding) could not be recovered by overexpressing the calmodulin protein alone or in combination with the auxiliary CaVα2bδ subunit, suggesting that CaVβ is the critical auxiliary subunit in the plasma membrane targeting of CaV1.2.
Plasma membrane targeting of CaV1.2 was decreased but not abolished in the double I1654A/Q1655A mutant in the presence of CaVβ3 and was not altered in the absence of CaVβ3, thus ruling out molecular models whereby the IQ motif, containing a polybasic motif, could serve as an endoplasmic reticulum retention signal (67). However, CaVβ-induced membrane expression was unaffected by the complete deletion of the high affinity Ca2+/CaM IQ-binding site (ΔC1643–1666), suggesting that high affinity Ca2+/CaM binding to this domain (1643–1666) is not required. In contrast, deletion of the larger 1623–1666 region in the C terminus and mutation of the TLF site, located in a pre-IQ apocalmodulin-binding site, abolished plasma membrane targeting of CaV1.2 in agreement with previous reports (43, 44, 50). Clearly the C terminus of CaV1.2 harbors key site targeting signals, but our current data do not support a model whereby high affinity Ca2+/CaM binding to the IQ domain (1643–1666) is required. Whether the C terminus is required for proper protein folding or for anchoring apocalmodulin onto low affinity CaM-binding sites on the C terminus (50, 63) as in SK channels (51, 68) will await further structural studies.
CaVα2δ and Surface Targeting of CaV1.2
It is widely acknowledged that CaVα2δ subunits facilitate the voltage dependence of channel gating and potentiate whole cell currents in a number of recombinant HVA CaVα1-CaVβ subunit combinations (69–71). For instance, electrophysiological measurements carried out with CaV2.3 in HEK cells showed a small but significant increase in peak current density in the presence of CaVα2δ that arose, at least in part, from an increase in the number of functional channels (72). Mutating the von Willebrand factor-A domain of CaVα2δ-2 was shown to increase the intracellular localization of CaV1.2 or CaV2.2 throughout the cell (45), suggesting that CaVα2δ could play a role in the plasma membrane targeting of HVA CaVα1 subunits. It can, however, be argued that the 30% decrease of CaV1.2 at the cell surface assessed from cell surface biotinylation assays cannot fully account for the 3-fold decrease in channel peak current density observed with the CaVα2δ-2 mutant (45). Indeed, it has been shown elsewhere that the gating charge of CaV1.2 is unaffected by co-expression with CaVα2δ, supporting a model where CaVα2δ stimulates peak currents by setting CaV1.2 channels in a conformational state very close to the open state without increasing protein density (57). Our current electrophysiological and flow cytometry measurements also support a role for CaVα2δ in the modulation of CaV1.2 channel gating rather than plasma membrane protein density. At this point, however, our data do not exclude a role for CaVα2δ at early steps of protein biosynthesis and/or recycling that could not be detected in our steady-state assay.
CaVβ Stimulated Surface Labeling of CaV1.2
Co-expression with CaVβ subunits produced a 3–5-fold enhancement in the plasma membrane targeting of the CaVα1 subunit of CaV1.2, with CaVβ3 and CaVβ4 being the most potent effects. No other combination with or without the auxiliary CaM and/or the CaVα2bδ subunit produced any significant increase in the plasma membrane targeting of CaV1.2. Hence, CaVβ appears to be the most potent determinant in the plasma membrane targeting of CaV1.2. Our detailed mutational analysis supports a model where high affinity binding of the ABP of CaVβ to the AID helix of CaVα1 largely accounts for CaVβ-induced plasma membrane targeting of CaV1.2.
The mechanism whereby CaVβ antagonizes ER retention of the CaVα1 subunit remains debated (73). In the two-site model, CaVβ stimulation of protein expression and modulation of gating are controlled by distinct sites through a two-to-one stoichiometry. This model opens up the possibility that secondary CaVβ-binding sites could contribute to plasma membrane targeting. Several observations could be suggestive of such a mechanism. Surface expression of CaV1.2-HA channels in Xenopus oocytes was not increased by injection of the CaVβ2a protein, and even decreased gating currents and surface expression of the CaV1.2-ΔAID-expressing oocytes (36, 74). Covalently linking CaVβ2b to the C terminus of CaV1.2 stimulated whole cell currents but failed to modulate channel gating in HEK cells (37, 38). CaVβ2b-induced modulation of trafficking and gating was also uncoupled in N-terminally truncated CaV1.2 (39). Deletion of a low affinity interaction site between the SH3 module of CaVβ and the I-II linker of the CaV2.1 subunit (outside the AID-GK interaction) did not affect CaV2.1 protein trafficking (40). Small fragments of CaVβ2 arising from putative splice variants were also shown to bind to the C terminus of the CaV1.2 subunit where they promoted membrane targeting in the absence of the GK/SH3 module of CaVβ subunits (75, 76). It remains, however, difficult to assess the physiological relevance of these findings, given that they result from in vitro interaction studies between isolated peptides.
In the one-site model, CaVβ interacts sequentially with the CaVα1 subunit through a unique binding site in the I-II linker in a 1:1 stoichiometry to dislodge ER retention signals and modulate gating (25). In the intact channel, high affinity binding of CaVβ onto the AID motif would account for both CaVβ-induced modulation of gating and the CaVβ-plasma membrane trafficking of CaVα1 (32, 77, 78). For CaV1.2 expressed in a mammalian cell system, the Trp470 and Ile471 residues previously shown to account for the high affinity binding of CaVβ (29) onto the CaV1.2 subunit were herein found to account for the CaVβ stimulation of CaV1.2 plasma membrane targeting. As mentioned earlier, disruption of these residues alone or in combination had a dominant effect and abrogated cell surface labeling of CaV1.2. Our data hence support a model whereby high affinity binding of the MMQKAL motif of CaVβ to the AID helix of the CaVα1 subunit is required for chaperoning and modulating HVA CaV channels.
Supplementary Material
Acknowledgments
We thank Serge Sénéchal and Dr. Jacques Thibodeau (Department of Microbiology and Immunology) for help with the fluorescence-activated cell sorting experiments and analysis; Michel Lauzon and Dr. Pierre Bissonnette for help with confocal and fluorescent microscopy; Yolaine Dodier, Alexandra Raybaud, Florian LeCoz, and Guillaume Roussel for preliminary experiments; Julie Verner for cell culture; Michel Brunette for computer maintenance; and Claude Gauthier for artwork.
This work was supported by Grant MOP13390 from the Canadian Institutes of Health Research and a grant from the Canadian Heart and Stroke Foundation (to L. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables SI–SIV, and Figs. S1–S9.
- HVA
- high voltage-activated
- wt
- wild type
- AID
- α interaction domain
- CaM
- calmodulin
- ABP
- α binding pocket
- NK
- nucleotide kinase
- HA
- hemagglutinin
- FACS
- fluorescence-activated cell sorting
- FITC
- fluorescein isothiocyanate.
REFERENCES
- 1.Snutch T. P., Reiner P. B. (1992) Curr. Opin. Neurobiol. 2, 247–253 [DOI] [PubMed] [Google Scholar]
- 2.Zhang J. F., Randall A. D., Ellinor P. T., Horne W. A., Sather W. A., Tanabe T., Schwarz T. L., Tsien R. W. (1993) Neuropharmacology 32, 1075–1088 [DOI] [PubMed] [Google Scholar]
- 3.Birnbaumer L., Campbell K. P., Catterall W. A., Harpold M. M., Hofmann F., Horne W. A., Mori Y., Schwartz A., Snutch T. P., Tanabe T. (1994) Neuron 13, 505–506 [DOI] [PubMed] [Google Scholar]
- 4.Perez-Reyes E., Cribbs L. L., Daud A., Lacerda A. E., Barclay J., Williamson M. P., Fox M., Rees M., Lee J. H. (1998) Nature. 391, 896–900 [DOI] [PubMed] [Google Scholar]
- 5.Cribbs L. L., Lee J. H., Yang J., Satin J., Zhang Y., Daud A., Barclay J., Williamson M. P., Fox M., Rees M., Perez-Reyes E. (1998) Circ. Res. 83, 103–109 [DOI] [PubMed] [Google Scholar]
- 6.Randall A., Benham C. D. (1999) Mol. Cell Neurosci. 14, 255–272 [DOI] [PubMed] [Google Scholar]
- 7.Catterall W. A. (2000) Annu. Rev. Cell Dev. Biol. 16, 521–555 [DOI] [PubMed] [Google Scholar]
- 8.Peterson B. Z., DeMaria C. D., Adelman J. P., Yue D. T. (1999) Neuron 22, 549–558 [DOI] [PubMed] [Google Scholar]
- 9.Dolphin A. C. (2009) Curr. Opin. Neurobiol. 19, 237–244 [DOI] [PubMed] [Google Scholar]
- 10.Dai S., Hall D. D., Hell J. W. (2009) Physiol. Rev. 89, 411–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao T., Puri T. S., Gerhardstein B. L., Chien A. J., Green R. D., Hosey M. M. (1997) J. Biol. Chem. 272, 19401–19407 [DOI] [PubMed] [Google Scholar]
- 12.Carl S. L., Felix K., Caswell A. H., Brandt N. R., Ball W. J., Jr., Vaghy P. L., Meissner G., Ferguson D. G. (1995) J. Cell Biol. 129, 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolphin A. C. (2003) J. Bioenerg. Biomembr. 35, 599–620 [DOI] [PubMed] [Google Scholar]
- 14.Birnbaumer L., Qin N., Olcese R., Tareilus E., Platano D., Costantin J., Stefani E. (1998) J. Bioenerg. Biomembr. 30, 357–375 [DOI] [PubMed] [Google Scholar]
- 15.Van Petegem F., Minor D. L., Jr. (2006) Biochem. Soc. Trans. 34, 887–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidalgo P., Neely A. (2007) Cell Calcium 42, 389–396 [DOI] [PubMed] [Google Scholar]
- 17.Bichet D., Cornet V., Geib S., Carlier E., Volsen S., Hoshi T., Mori Y., De Waard M. (2000) Neuron 25, 177–190 [DOI] [PubMed] [Google Scholar]
- 18.Cornet V., Bichet D., Sandoz G., Marty I., Brocard J., Bourinet E., Mori Y., Villaz M., De Waard M. (2002) Eur. J. Neurosci. 16, 883–895 [DOI] [PubMed] [Google Scholar]
- 19.Pragnell M., De Waard M., Mori Y., Tanabe T., Snutch T. P., Campbell K. P. (1994) Nature 368, 67–70 [DOI] [PubMed] [Google Scholar]
- 20.De Waard M., Scott V. E., Pragnell M., Campbell K. P. (1996) FEBS Lett. 380, 272–276 [DOI] [PubMed] [Google Scholar]
- 21.Chen Y. H., Li M. H., Zhang Y., He L. L., Yamada Y., Fitzmaurice A., Shen Y., Zhang H., Tong L., Yang J. (2004) Nature 429, 675–680 [DOI] [PubMed] [Google Scholar]
- 22.Opatowsky Y., Chen C. C., Campbell K. P., Hirsch J. A. (2004) Neuron 42, 387–399 [DOI] [PubMed] [Google Scholar]
- 23.Van Petegem F., Clark K. A., Chatelain F. C., Minor D. L., Jr. (2004) Nature. 429, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell D. C., Butcher A. J., Berrow N. S., Page K. M., Brust P. F., Nesterova A., Stauderman K. A., Seabrook G. R., Nürnberg B., Dolphin A. C. (2001) J. Neurophysiol. 85, 816–827 [DOI] [PubMed] [Google Scholar]
- 25.Butcher A. J., Leroy J., Richards M. W., Pratt W. S., Dolphin A. C. (2006) J. Physiol. 574, 387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantí C., Davies A., Berrow N. S., Butcher A. J., Page K. M., Dolphin A. C. (2001) Biophys. J. 81, 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geib S., Sandoz G., Cornet V., Mabrouk K., Fund-Saunier O., Bichet D., Villaz M., Hoshi T., Sabatier J. M., De Waard M. (2002) J. Biol. Chem. 277, 10003–10013 [DOI] [PubMed] [Google Scholar]
- 28.Opatowsky Y., Chomsky-Hecht O., Kang M. G., Campbell K. P., Hirsch J. A. (2003) J. Biol. Chem. 278, 52323–52332 [DOI] [PubMed] [Google Scholar]
- 29.Van Petegem F., Duderstadt K. E., Clark K. A., Wang M., Minor D. L., Jr. (2008) Structure 16, 280–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berrou L., Klein H., Bernatchez G., Parent L. (2002) Biophys. J. 83, 1429–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berrou L., Dodier Y., Raybaud A., Tousignant A., Dafi O., Pelletier J. N., Parent L. (2005) J. Biol. Chem. 280, 494–505 [DOI] [PubMed] [Google Scholar]
- 32.Leroy J., Richards M. W., Richards M. S., Butcher A. J., Nieto-Rostro M., Pratt W. S., Davies A., Dolphin A. C. (2005) J. Neurosci. 25, 6984–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao T., Chien A. J., Hosey M. M. (1999) J. Biol. Chem. 274, 2137–2144 [DOI] [PubMed] [Google Scholar]
- 34.Takahashi S. X., Miriyala J., Tay L. H., Yue D. T., Colecraft H. M. (2005) J. Gen. Physiol. 126, 365–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tareilus E., Roux M., Qin N., Olcese R., Zhou J., Stefani E., Birnbaumer L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez-Gutierrez G., Miranda-Laferte E., Neely A., Hidalgo P. (2007) J. Biol. Chem. 282, 2156–2162 [DOI] [PubMed] [Google Scholar]
- 37.Dalton S., Takahashi S. X., Miriyala J., Colecraft H. M. (2005) J. Physiol. 567, 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seu L., Pitt G. S. (2006) J. Gen. Physiol. 128, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanevsky N., Dascal N. (2006) J. Gen. Physiol. 128, 15–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maltez J. M., Nunziato D. A., Kim J., Pitt G. S. (2005) Nat. Struct. Mol. Biol. 12, 372–377 [DOI] [PubMed] [Google Scholar]
- 41.Gerster U., Neuhuber B., Groschner K., Striessnig J., Flucher B. E. (1999) J. Physiol. 517, 353–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao T., Bunemann M., Gerhardstein B. L., Ma H., Hosey M. M. (2000) J. Biol. Chem. 275, 25436–25444 [DOI] [PubMed] [Google Scholar]
- 43.Hulme J. T., Yarov-Yarovoy V., Lin T. W., Scheuer T., Catterall W. A. (2006) J. Physiol. 576, 87–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hulme J. T., Konoki K., Lin T. W., Gritsenko M. A., Camp D. G., 2nd, Bigelow D. J., Catterall W. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5274–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cantí C., Nieto-Rostro M., Foucault I., Heblich F., Wratten J., Richards M. W., Hendrich J., Douglas L., Page K. M., Davies A., Dolphin A. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11230–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang H., DeMaria C. D., Erickson M. G., Mori M. X., Alseikhan B. A., Yue D. T. (2003) Neuron 39, 951–960 [DOI] [PubMed] [Google Scholar]
- 47.Zühlke R. D., Pitt G. S., Deisseroth K., Tsien R. W., Reuter H. (1999) Nature 399, 159–162 [DOI] [PubMed] [Google Scholar]
- 48.Tang W., Halling D. B., Black D. J., Pate P., Zhang J. Z., Pedersen S., Altschuld R. A., Hamilton S. L. (2003) Biophys. J. 85, 1538–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Petegem F., Chatelain F. C., Minor D. L., Jr. (2005) Nat. Struct. Mol. Biol. 12, 1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H. G., George M. S., Kim J., Wang C., Pitt G. S. (2007) J. Neurosci. 27, 9086–9093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee W. S., Ngo-Anh T. J., Bruening-Wright A., Maylie J., Adelman J. P. (2003) J. Biol. Chem. 278, 25940–25946 [DOI] [PubMed] [Google Scholar]
- 52.Castellano A., Wei X., Birnbaumer L., Perez-Reyes E. (1993) J. Biol. Chem. 268, 3450–3455 [PubMed] [Google Scholar]
- 53.Williams M. E., Feldman D. H., McCue A. F., Brenner R., Velicelebi G., Ellis S. B., Harpold M. M. (1992) Neuron 8, 71–84 [DOI] [PubMed] [Google Scholar]
- 54.Klein H., Garneau L., Coady M., Lemay G., Lapointe J. Y., Sauvé R. (1999) J. Membr. Biol. 167, 43–52 [DOI] [PubMed] [Google Scholar]
- 55.Xia X. M., Fakler B., Rivard A., Wayman G., Johnson-Pais T., Keen J. E., Ishii T., Hirschberg B., Bond C. T., Lutsenko S., Maylie J., Adelman J. P. (1998) Nature 395, 503–507 [DOI] [PubMed] [Google Scholar]
- 56.Raybaud A., Dodier Y., Bissonnette P., Simoes M., Bichet D. G., Sauvé R., Parent L. (2006) J. Biol. Chem. 281, 39424–39436 [DOI] [PubMed] [Google Scholar]
- 57.Platano D., Qin N., Noceti F., Birnbaumer L., Stefani E., Olcese R. (2000) Biophys. J. 78, 2959–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Obermair G. J., Schlick B., Di Biase V., Subramanyam P., Gebhart M., Baumgartner S., Flucher B. E. (2010) J. Biol. Chem. 285, 5776–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Antzelevitch C., Pollevick G. D., Cordeiro J. M., Casis O., Sanguinetti M. C., Aizawa Y., Guerchicoff A., Pfeiffer R., Oliva A., Wollnik B., Gelber P., Bonaros E. P., Jr., Burashnikov E., Wu Y., Sargent J. D., Schickel S., Oberheiden R., Bhatia A., Hsu L. F., Haïssaguerre M., Schimpf R., Borggrefe M., Wolpert C. (2007) Circulation 115, 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He L. L., Zhang Y., Chen Y. H., Yamada Y., Yang J. (2007) Biophys. J. 93, 834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fallon J. L., Baker M. R., Xiong L., Loy R. E., Yang G., Dirksen R. T., Hamilton S. L., Quiocho F. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5135–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benmocha A., Almagor L., Oz S., Hirsch J. A., Dascal N. (2009) Channels 3, 337–342 [DOI] [PubMed] [Google Scholar]
- 63.Ravindran A., Lao Q. Z., Harry J. B., Abrahimi P., Kobrinsky E., Soldatov N. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8154–8159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao T., Cuadra A. E., Ma H., Bunemann M., Gerhardstein B. L., Cheng T., Eick R. T., Hosey M. M. (2001) J. Biol. Chem. 276, 21089–21097 [DOI] [PubMed] [Google Scholar]
- 65.Zühlke R. D., Pitt G. S., Tsien R. W., Reuter H. (2000) J. Biol. Chem. 275, 21121–21129 [DOI] [PubMed] [Google Scholar]
- 66.Kim J., Ghosh S., Nunziato D. A., Pitt G. S. (2004) Neuron 41, 745–754 [DOI] [PubMed] [Google Scholar]
- 67.Michelsen K., Yuan H., Schwappach B. (2005) EMBO Rep. 6, 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joiner W. J., Khanna R., Schlichter L. C., Kaczmarek L. K. (2001) J. Biol. Chem. 276, 37980–37985 [DOI] [PubMed] [Google Scholar]
- 69.Parent L., Schneider T., Moore C. P., Talwar D. (1997) J. Membr. Biol. 160, 127–140 [DOI] [PubMed] [Google Scholar]
- 70.Klugbauer N., Lacinová L., Marais E., Hobom M., Hofmann F. (1999) J. Neurosci. 19, 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barclay J., Balaguero N., Mione M., Ackerman S. L., Letts V. A., Brodbeck J., Canti C., Meir A., Page K. M., Kusumi K., Perez-Reyes E., Lander E. S., Frankel W. N., Gardiner R. M., Dolphin A. C., Rees M. (2001) J. Neurosci. 21, 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones L. P., Wei S. K., Yue D. T. (1998) J. Gen. Physiol. 112, 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jarvis S. E., Zamponi G. W. (2007) Curr. Opin. Cell Biol. 19, 474–482 [DOI] [PubMed] [Google Scholar]
- 74.Hidalgo P., Gonzalez-Gutierrez G., Garcia-Olivares J., Neely A. (2006) J. Biol. Chem. 281, 24104–24110 [DOI] [PubMed] [Google Scholar]
- 75.Harry J. B., Kobrinsky E., Abernethy D. R., Soldatov N. M. (2004) J. Biol. Chem. 279, 46367–46372 [DOI] [PubMed] [Google Scholar]
- 76.Lao Q. Z., Kobrinsky E., Harry J. B., Ravindran A., Soldatov N. M. (2008) J. Biol. Chem. 283, 15577–15588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altier C., Dubel S. J., Barrère C., Jarvis S. E., Stotz S. C., Spaetgens R. L., Scott J. D., Cornet V., De Waard M., Zamponi G. W., Nargeot J., Bourinet E. (2002) J. Biol. Chem. 277, 33598–33603 [DOI] [PubMed] [Google Scholar]
- 78.Cohen R. M., Foell J. D., Balijepalli R. C., Shah V., Hell J. W., Kamp T. J. (2005) Am. J. Physiol. Heart Circ. Physiol. 288, H2363–H2374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.