Abstract
The Ccm cytochrome c maturation System I catalyzes covalent attachment of heme to apocytochromes c in many bacterial species and some mitochondria. A covalent, but transient, bond between heme and a conserved histidine in CcmE along with an interaction between CcmH and the apocytochrome have been previously indicated as core aspects of the Ccm system. Here, we show that in the Ccm system from Desulfovibrio desulfuricans, no CcmH is required, and the holo-CcmE covalent bond occurs via a cysteine residue. These observations call for reconsideration of the accepted models of System I-mediated c-type cytochrome biogenesis.
Keywords: Chaperone Chaperonin, Cytochrome c, Cytochromes, Heme, Post-translational Modification, Protein Assembly, Cytochrome c Biogenesis, Cytochrome c Maturation, Heme Chaperone
Introduction
c-Type cytochromes are generally characterized by the covalent attachment of heme to the polypeptide via two thioether bonds formed by reaction of the vinyl groups of heme with the sulfur atoms of two thiols in a CXXCH motif in the protein. This post-translational modification reaction is, surprisingly, catalyzed by several different systems in different cell types and organelles (1–3). Of these, the cytochrome c maturation system (CcmABCDEFGH (4), also called System I) functions in the periplasm of many species of bacteria and in mitochondria of some eukaryotes, including plants (5, 6).
A large number of investigations of the Escherichia coli system have implicated the CcmABC proteins in ATP-dependent handling of heme via covalent attachment to CcmE (e.g. Refs. 7 and 8). CcmD appears to be a single helix protein involved in the interaction of heme with CcmE (9, 10). CcmE has been identified as a heme chaperone, covalently binding heme via an apparently conserved histidine residue (His130 in E. coli) before transfer of the heme to apocytochromes (11, 12); to date, this particular heme-histidine bond has only been identified in CcmE (12, 13). Covalently bound heme on CcmE is transferred to the apocytochrome in a process that is believed to involve CcmF and CcmH (14). E. coli CcmH contains two distinct domains that exist as separate proteins in some organisms. The protein called CcmH in most organisms comprises the N-terminal CXXC-containing globular domain of the E. coli protein. The C-terminal tetratricopeptide repeat-containing domain of E. coli CcmH is analogous to CcmI in other species of bacteria (15). In vitro studies have suggested that peptides carrying the CXXCH motif can interact with the CXXC region of bacterial and plant CcmH (16–18), and two-hybrid analysis indicated that plant CcmH interacts with apocytochrome c (18). CcmG is believed to transfer reductant from the transmembrane DsbD protein to CcmH and/or the apocytochrome (19), and it has been recently proposed, at least for Rhodobacter capsulatus, that CcmG can also interact with the apocytochrome c in a “holdase” role (20). The E. coli Ccm system is inactive in the absence of CcmG (21).
Recent sequence analysis (22) revealed an apparently novel CcmE in a number of archaeal and bacterial species, including two sulfate-reducing bacteria which are rich in, and highly dependent upon c-type cytochromes (e.g. Ref. 23). This CcmE contains a cysteine residue in place of the histidine that, in other CcmE proteins, forms a covalent bond to heme that is currently thought to be an essential intermediate in the cytochrome c maturation pathway (12). In addition, the putative ccm operons from Desulfovibrio desulfuricans and Desulfovibrio vulgaris lack ccmH and ccmG, although a candidate CcmG is encoded elsewhere on the genomes. A ccmI homologue was originally missed in the ccm operon (22) but was subsequently suggested to be encoded at the 3′ end (2). These in silico findings clearly raise important questions about the function of the Ccm proteins. Genetic manipulation of sulfate-reducing bacteria is not straightforward, and c-type cytochromes are important for growth of such organisms. Therefore, we have used E. coli as an heterologous expression system to study c-type cytochrome biogenesis by the D. desulfuricans Ccm system.
Here we report maturation of c-type cytochrome when the ccmEFABCDI genes from D. desulfuricans are expressed in an E. coli strain lacking all endogenous ccm genes. We have termed this variant Ccm machinery “System I*.” This outcome calls for recognition that some current views concerning the function of core Ccm components require revision and illustrates how the acquisition of ever-growing numbers of bacterial genome sequences can generate unexpected challenges to accepted paradigms.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
E. coli strain EC06 (4) contains a chromosomal deletion of the ccm operon and was used to examine holocytochrome formation in the presence of each biogenesis system and its mutants. E. coli strain DH5α (Invitrogen) was used for routine molecular biology. PCR used KOD polymerase from Thermococcus kodakaraensis (Novagen). All oligonucleotides (Sigma-Genosys) used in this study are listed in supplemental Table S1, and all plasmids are listed in supplemental Table S2. All DNA constructs were sequenced before use.
The E. coli ccmABCDEFGH operon (System I) was expressed from pEC86 (24). To create a comparable plasmid lacking any biogenesis system, inverse PCR was performed on pEC86 using AG234 and AG235, and the product self-ligated. This removed the entire ccm operon, and the plasmid created is AD377 (no biogenesis system). To create a suitable plasmid for expression of other biogenesis systems, a XhoI site was introduced immediately after the ATG start codon of ccmA in pEC86 via QuikChange mutagenesis using WC1 and WC2. The resultant construct is pEC86x in which the entire ccm operon can be excised by digestion with XhoI and StuI. The ccm operon (ccmEFABCDI) was amplified from D. desulfuricans genomic DNA using AG81 and AG82 and cloned into the unique XhoI and StuI sites in pEC86x to generate pDD86. The same methodology was utilized to clone the truncated operons ccmEFABCD (using AG81 and AG83 resulting in plasmid AD457) and ccmEFABC (AG81 and AG286 resulting in plasmid AD153). Paracoccus denitrificans cytochrome c550 was expressed from the isopropyl-1-thio-β-d-galactopyranoside (IPTG)4-inducible promoter of pKPD1 (25), a derivative of pKK223-3. Similarly, a CXXCH variant of Trypanosoma brucei cytochrome c was expressed from IPTG-inducible pKK223-TbcytcperiCXXCH as described previously (26). The CXXCK variant of P. denitrificans cytochrome c550 has been described previously (27).
To generate mutants in D. desulfuricans CcmE, the HindIII fragment containing the ccmE open reading frame was excised from pDD86 and cloned into HindIII-digested pTZ19R. Inverse PCR was performed on this construct using AG267 and AG266 (C127H), AG285 (C127M), or AG284 (C127A). The resulting construct was sequenced before the HindIII fragment was reintroduced into pDD86 to create the appropriate mutant plasmids, which are indicated in supplemental Table S2.
To generate a construct for expressing D. desulfuricans membrane-bound CcmE, PCR was performed on D. desulfuricans genomic DNA using oligonucleotides JS3 and JS4. The product was cloned into the NdeI and EcoRI sites of pISC2 to create pDDCcmE. To generate soluble D. desulfuricans CcmE, PCRs were performed using AG298 and, sequentially, AG295, AG296, and AG297. The template for the first PCR was pDD86, and subsequent templates were the purified products of the previous PCR. The final product was cloned into the EcoRI and HindIII sites of pKK223-3 to create pDDCcmEsol. The C127H mutant in this construct was generated via inverse PCR on pDDCcmEsol using AG266 and AG267.
To generate a construct for expressing soluble E. coli CcmE, inverse PCR was performed on pEC415 (12) using FR1 and FR2 to remove the His6 tag. The resultant plasmid is pECCcmEsol. A H130C mutation was created in this construct by QuikChange mutagenesis using oligonucleotides FR3 and FR4. This plasmid is pECcmEsolH130C. To generate the mutation changing the 130HDENY motif of E. coli CcmE to CPSKY, QuikChange mutagenesis was performed on pEC86 using JS1 and JS2 to create pEC86CPSKY.
In each case the plasmid bearing the biogenesis system confers resistance to chloramphenicol, and expression of the biogenesis system is constitutive. The plasmid bearing the cytochrome is IPTG-inducible and confers resistance to carbenicillin. pISC2-derived plasmids are arabinose inducible.
Routine cell growth was conducted using LB (Luria-Bertani) media supplemented with antibiotics where appropriate. Growth on solid media used liquid growth medium supplemented with 1.5% bacteriological agar. For preparation of periplasmic fractions, single colonies containing appropriate plasmids were picked into 500 ml of 2× TY medium (16 g liter−1 peptone, 10 g liter −1 yeast extract, 5 g liter −1 NaCl) supplemented with 1 mm IPTG or 0.1% (w/v) arabinose and appropriate antibiotics in 2 liter flasks. Cultures were grown at 37 °C with shaking at 200 rpm for 20–24 h before harvesting. Carbenicillin was used at 100 μg ml−1, and chloramphenicol was at 34 μg ml−1.
Analysis of Cytochrome Production
Periplasmic extractions were performed as described previously (28). Where appropriate, these fractions were analyzed by SDS-PAGE (Invitrogen pre-cast 10% Bis-Tris gels) followed by heme staining (29) which detects heme covalently bound to protein. Samples were normalized for wet cell weight, and 5–10 μg of protein were loaded per lane. See-Blue Plus 2 (Invitrogen) prestained protein marker was used. UV-visible spectroscopy was performed using a PerkinElmer Life Sciences Lambda 2 UV-visible spectrophotometer; samples were reduced by the addition of a few grains of disodium dithionite (Sigma). The extinction coefficient for reduced P. denitrificans cytochrome c550 at 415 nm is 140 mm−1 cm−1. Pyridine hemochrome spectra were obtained according to the method of Bartsch (30).
Analysis of Covalent Heme Binding by CcmE
Membrane fractions were prepared from cultures containing the appropriate biogenesis system plasmid. Cultures were grown as described above for preparation of periplasmic fractions, and membranes were extracted as described previously (31). 30 μg of total membrane protein was analyzed by SDS-PAGE followed by heme staining (29).
Purification of Soluble D. desulfuricans CcmE
E. coli EC06 cells containing pDDCcmEsol were grown in 2× TY medium containing 1 mm IPTG and carbenicillin for 20 h. Cells were harvested at 5000 × g for 20 min at 4 °C and resuspended in Tris-HCl buffer (50 mm Tris, 150 mm NaCl, pH 7.5) containing a protease inhibitor mixture. Periplasmic extraction was carried out using β-Polymyxin (final concentration of 3 mg ml−1) followed by incubation at 37 °C for 1 h. The spheroplasts were pelleted by centrifugation at 10,000 × g for 40 min at 4 °C. Proteins in the resulting periplasm (supernatant) were purified using a Streptactin (IBA) containing column equilibrated with the above Tris buffer and eluted with the same buffer containing 2.5 mm desthiobiotin. Protein concentration determination was carried out using the bicinchoninic acid (BCA) assay (Pierce).
Western Blot Analysis
Western blotting was carried out following SDS-PAGE by first transferring onto nitrocellulose (Hybond C-Extra, Amersham Biosciences). Blocking was achieved by incubation with 3% bovine serum albumin in Tris-buffered saline (50 mm Tris, 120 mm NaCl, 0.1% Tween 20, pH 7.5). Antibodies were used in the following dilutions: alkaline phosphatase conjugated anti-Strep2 (1:5,000, IBA); anti-CcmE from rabbit (1:1,000, from Prof. Linda Thony-Meyer (12)); alkaline phosphatase conjugated anti-rabbit (1:30,000, Sigma). Development was by a colorimetric reaction using a SigmaFast 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium tablet dissolved in 10 ml of ultrapure water. The reaction was stopped by decanting off the developing solution and washing the membrane with ultrapure water.
Proteomics Analysis
Samples were digested with sequencing-grade trypsin (porcine, Promega), and analysis was done using liquid chromatography-tandem mass spectrometry on a Thermo LTQ Orbitrap mass spectrometer coupled to a Dionex Ultimate 3000 nano-high performance liquid chromatography system or by MALDI-TOF/mass spectrometry on an Ultraflex instrument (Bruker).
RESULTS
Expression of D. desulfuricans ccmEFABCDI Restores c-type Cytochrome Biogenesis in E. coli Δccm
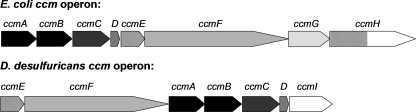
Initial analysis of the ccm operon from D. desulfuricans (22) revealed the presence of homologues of ccmEFABCD. However, we subsequently identified a predicted 22-kDa tetratricopeptide repeat-containing protein at the 3′ end of the operon in which the predicted start codon overlaps the 3′ end of ccmD (Fig. 1) (2). This is a potential homologue of ccmI. Therefore, we cloned the operon, including ccmI, into a modified version of pEC86, a vector routinely used to express the E. coli Ccm proteins, to create pDD86. The predicted ccm operon from D. desulfuricans lacks homologues of ccmH (E. coli N-CcmH) and ccmG; pDD86 allows us to assess if the predicted ccmEFABCDI genes are sufficient to support c-type cytochrome maturation.
FIGURE 1.
Gene organization of the ccm operon in E. coli and D. desulfuricans.
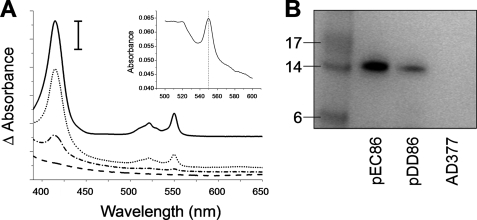
The E. coli strain EC06 lacks the endogenous ccm system and, thus, is unable to make c-type cytochromes (4). Here we have used P. denitrificans cytochrome c550 expressed from pKPD1 as an exogenous c-type cytochrome to assess the ability of the D. desulfuricans Ccm system to restore c-type cytochrome biogenesis in E. coli EC06 cells. To allow comparison of the various systems, the amount of cytochrome produced is expressed as absorbance/g of wet cell pellet where the absorbance is calculated as the absorbance difference between the base line of the spectra at 650 nm and the maximal absorbance of the Soret peak of reduced cytochrome c550 (415 nm). Co-expression of plasmids AD377 (derived from pEC86 and containing no biogenesis genes (32)) and pKPD1 did not result in production of any detectable c-type cytochrome. E. coli EC06 cells containing pEC86 (E. coli ccmABCDEFGH) and pKPD1 produced 1.03 absorbance units/g, equivalent to 3.3 mg of cytochrome c550/g wet cell pellet. Cells containing pDD86 (D. desulfuricans ccmEFABCDI) and pKPD1 produced 0.045 absorbance units/g (0.15 mg cytochrome c550/g wet cell pellet), corresponding to ∼5% of the level produced by pEC86. Fig. 2A shows the visible absorption spectra of the cytochrome c550 produced by cells containing pDD86; it is indistinguishable from material made using the E. coli Ccm system or in P. denitrificans itself (25). Qualitatively similar results to those in this paragraph were obtained when pKPD1 was replaced by a plasmid encoding a CXXCH variant of T. brucei mitochondrial cytochrome c (26), indicating that the D. desulfuricans ccm system is active toward apocytochromes c from different sources. No detectable cytochrome was produced by either of the biogenesis systems when pKPD1 was replaced by pKK223-3, a plasmid lacking the cytochrome gene.
FIGURE 2.
Production of c-type cytochromes by various biogenesis systems. A, shown are visible absorption spectra reflecting the formation of P. denitrificans cytochrome c550 in E. coli catalyzed by E. coli CcmABCDEFGH (solid line), D. desulfuricans CcmEFABCDI (dotted line), D. desulfuricans CcmEFABCD (alternating dashed/dotted line), and as a negative control, no biogenesis system (dashed line). The vertical scale bar represents 0.04 absorbance units. The spectra are vertically offset for clarity. Samples were reduced by the addition of a few grains of disodium dithionite. The periplasmic fraction from cells expressing pEC86 was diluted 20-fold before analysis. The absorbance maxima are at 415, 521.5, and 550 nm. The inset shows the reduced pyridine hemochrome spectrum of cytochrome c550 produced by D. desulfuricans CcmEFABCDI. The vertical line indicates 550 nm. B, detection of c-type cytochromes via SDS-PAGE analysis of periplasmic fractions from cells expressing P. denitrificans cytochrome c550 and the indicated biogenesis system and subsequent heme-staining of the gel. The left-most lane contains See-Blue Plus 2 protein markers of the indicated molecular mass (kDa).
To define the lower limits of detection of P. denitrificans cytochrome c550, we diluted the periplasmic extract from cells containing both pEC86 and pKPD1 (supplemental Fig. S1). The results indicate that UV-visible spectroscopy is slightly more sensitive than heme staining but that both detection methods have very low thresholds. UV-visible spectroscopy can detect P. denitrificans cytochrome c550 via the Soret peak at concentrations down to 3 × 10−8 m, and heme-staining can detect ∼1 × 10−9 mol of cytochrome c550.
To ensure covalent attachment of heme to apocytochrome c550 had occurred, periplasmic fractions from cells expressing pKPD1 together with E. coli ccmABCDEFGH, D. desulfuricans ccmEFABCDI, or AD377 (no biogenesis system) were subjected to SDS-PAGE analysis and subsequent heme staining (Fig. 2B). The presence of a band at ∼14 kDa, corresponding to the mass of holocytochrome c550, indicated covalent attachment of heme. To determine whether the cytochrome c550 produced by the D. desulfuricans system had heme correctly bound, we examined the reduced pyridine hemochrome spectrum. This was characteristic of a c-type cytochrome in which heme is bound via two thioether bonds to the protein, with the maximal absorbance of the α-band at 550 nm (Fig. 2A, inset). A variant of cytochrome c550 that contained the CWSCK motif (rather than the endogenous CKACH motif), found as a heme attachment site in some NrfA nitrite reductases, was not a substrate for the D. desulfuricans Ccm system, paralleling observations with the E. coli system (27).
The Requirement for CcmG, CcmI, and CcmD for Cytochrome c Biogenesis by the D. desulfuricans Ccm System
In addition to lacking a potential homologue of the N-terminal region of E. coli CcmH, the ccm operon in D. desulfuricans also lacks a homologue of ccmG (Fig. 1). It is clear from the evidence presented above that this protein is not required for c-type cytochrome biogenesis by the D. desulfuricans proteins when expressed in E. coli EC06. The endogenous ccmG is absent from EC06.
In E. coli, CcmH appears to be a fusion of two distinct domains, of which only the N-terminal domain is essential for cytochrome c biogenesis (33, 34). The CcmI protein encoded within the D. desulfuricans operon is homologous to the C-terminal domain of E. coli CcmH. Therefore, we created a plasmid containing the D. desulfuricans ccm operon but lacking ccmI (ccmEFABCD). Co-expression of this operon and pKPD1 resulted in production of cytochrome c550 at a level ∼20% (0.0092 absorbance units/g, equivalent to 0.03 mg of cytochrome c550/g of wet cell pellet) that observed in cells containing the ccmEFABCDI operon (Fig. 3). The spectrum did not show any detectable difference from cytochrome c550 produced in the presence of CcmI (Fig. 2A). No cytochrome was observed in the absence of pKPD1. This suggests that, although the presence of D. desulfuricans CcmI significantly enhances cytochrome c production, it is not essential for this process. Therefore, the D. desulfuricans ccmEFABCD genes alone are sufficient for c-type cytochrome biogenesis.
FIGURE 3.
Production of c-type cytochromes by mutant D. desulfuricans Ccm systems. Quantitation of cytochrome c550 production by the indicated biogenesis system (data shown are means of six separate determinations ±S.D. and are expressed as absorbance per gram of wet cell pellet). Details of quantitation are provided under “Results”. For comparison, E. coli EC06 cells containing pEC86 and pKPD1 produced 1.03 absorbance units/g. The “basal” level of absorbance observed in all strains is not due to c-type cytochrome production, as can be deduced from the dashed spectrum in Fig. 2A that was obtained from cell material from which c-type cytochromes were completely absent.
Because of the small size and poor conservation of CcmD proteins, we determined whether the putative ccmD homologue in the D. desulfuricans operon was essential for cytochrome c biogenesis. We created a plasmid containing the D. desulfuricans ccm operon but lacking ccmD and ccmI (ccmEFABC). Co-expression of this operon and pKPD1 or pKK223-3 did not result in detectable production of cytochrome c550 (Fig. 3), suggesting that CcmD is essential for c-type cytochrome biogenesis, as it is in E. coli (9).
Exogenous Reductant Does Not Increase Cytochrome c550 Production
Mutations in E. coli CcmG, CcmH, and DsbD (which supplies reductant to CcmG) can be complemented by provision of exogenous reductant, notably 2-mercaptoethane sulfonic acid, in the growth media (33, 35, 36). However, the addition of 5 mm 2-mercaptoethane sulfonic acid or 1 mm cysteine had no effect on the yield of c-type cytochrome produced by the D. desulfuricans Ccm system when operating in E. coli EC06.
Desulfovibrio CcmE Covalently Binds Heme
E. coli CcmE has been demonstrated to covalently bind heme via His130 (12). However, D. desulfuricans CcmE contains a cysteine at the corresponding position (Cys127; Fig. 4A). To establish whether D. desulfuricans CcmE can covalently bind heme, membrane fractions were prepared from E. coli EC06 cells expressing the D. desulfuricans ccmEFABCDI, ccmEFABCD, and ccmEFABC operons and also those expressing no biogenesis system. The ccmEFABCD operon was included to provide a defect (a lack of CcmI) downstream of CcmE in the cytochrome biogenesis pathway in anticipation of maximizing, by analogy with the work of Reid et al. (37) with E. coli, the proportion of CcmE that may have covalently bound heme. In each case the genes for exogenous c-type cytochromes were absent. Membrane extracts from cells containing pEC86 encoding the entire E. coli ccm operon were included as a positive control (Fig. 4B). Equal protein loading for each extract was confirmed by examination of an equivalent Coomassie Blue-stained gel.
FIGURE 4.
Attachment of heme to CcmE. A, shown are NMR structures of E. coli apoCcmE (PDB entry 1LIZ) (i) and D. vulgaris apoCcmE (PDB entry 2KCT) (ii). Structures were rendered in PyMOL. B, membrane fractions from the indicated strains were prepared and analyzed via SDS-PAGE and subsequent heme-staining for the presence of a covalent complex between E. coli or D. desulfuricans CcmE and heme. The left-most lane contains See-Blue Plus 2 protein markers of the indicated molecular mass (kDa).
As reported previously (12), a heme-staining band was observed at ∼16 kDa from cells expressing pEC86 after separation of the membrane fraction by SDS-PAGE (Fig. 4B, second lane). This is consistent with the presence of the E. coli holo-CcmE covalent complex (molecular weight, ∼18 kDa). This band was not observed in the absence of a biogenesis system (AD377; Fig. 4B, seventh lane). In membrane extracts from E. coli EC06 cells expressing the D. desulfuricans ccmEFABCDI operon, a heme staining band was observed at ∼14 kDa (Fig. 4B, third lane), consistent with the presence of holo-CcmE; D. desulfuricans CcmE has a predicted molecular weight of 15,114 Da, and heme has a mass of 616.5 Da. In both cases the holo-CcmE complexes migrate at a lower molecular weight than predicted; this is not unusual for membrane proteins. We conducted mass spectrometry of trypsin-digested proteins in a gel slice cut from an equivalent Coomassie-stained gel containing the region corresponding to the heme-staining band believed to be D. desulfuricans holo-CcmE. This revealed the presence of several fragments of D. desulfuricans CcmE (17.5% total sequence coverage). No c-type cytochromes were detected in the peptide fragments from the gel slice.
In membranes from cells expressing the D. desulfuricans ccmEFABCD operon (i.e. lacking CcmI) a heme-staining band of the same mass and approximate intensity as that present in the case of ccmEFABCDI was observed (Fig. 4B, fourth lane). This suggests that the absence of CcmI does not affect the accumulation of holo-CcmE. Membranes from cells expressing the ccmEFABC operon had a scarcely detectable amount of holo-CcmE (Fig. 4B, fifth lane), implicating CcmD in transfer of heme to CcmE. Kranz and co-workers recently reported that loss of ccmD from the E. coli system resulted in formation of less holo-CcmE (9).
Cys127 of D. desulfuricans CcmE Is Essential for Heme Binding and Cytochrome c Biogenesis
E. coli CcmE covalently binds heme via His130, and although the H130C variant can covalently bind heme, the heme cannot subsequently be transferred to apocytochrome c (38, 39). The equivalent residue in D. desulfuricans CcmE, Cys127, was mutagenized to histidine, methionine, and alanine. Histidine was chosen as the analogous residue to that in E. coli CcmE. Methionine is a common iron ligand to which heme can be non-covalently bound. In each case E. coli EC06 cells expressing the resultant ccmEmutFABCDI operon and pKPD1 were unable to synthesize cytochrome c550 at a level detectable either by heme staining or spectroscopically (Fig. 3). This demonstrates the importance of cysteine at this position in D. desulfuricans CcmE and that this residue plays an integral role in c-type cytochrome biogenesis. It was also tested whether CcmEC127H could covalently bind heme. SDS-PAGE analysis of membranes prepared from cells expressing the ccmEC127HFABCDI operon did not result in any heme-staining bands (Fig. 4B, sixth lane). Thus, covalent attachment of heme to D. desulfuricans CcmE requires and is most likely directly via Cys127.
The D. desulfuricans Ccm System Attaches Heme Covalently to Cys127 of Soluble D. desulfuricans CcmE
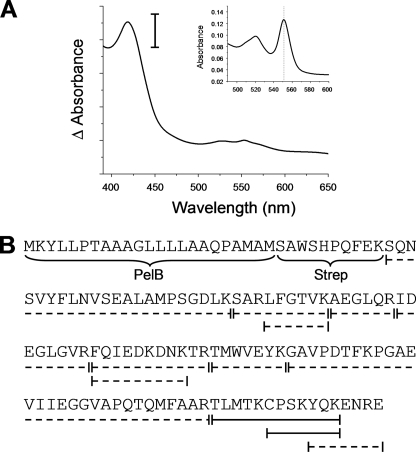
Because of limitations of expression and subsequent analysis of the membrane-anchored D. desulfuricans CcmE, we generated a construct encoding the soluble portion of CcmE (lacking the N-terminal transmembrane helix) preceded by a signal peptide and a Streptavidin II tag, termed CcmEsol. Expression of this construct in the presence of pDD86 (D. desulfuricans ccmEFABCDI) and subsequent UV-visible spectroscopic analysis of the protein purified by affinity chromatography and reduced using dithionite revealed an absorption maximum at 419 nm (Fig. 5A). Pyridine hemochrome analysis showed an α-band at 551.5 nm (Fig. 5A, inset), a value similar to that reported for E. coli holo-CcmE (39). It is notable that single-cysteine c-type cytochromes generally show maxima at 552–553 nm in pyridine hemochrome analysis (e.g. Ref. 26). This difference may reflect the nature of the cysteine-heme bond formed in D. desulfuricans holo-CcmE.
FIGURE 5.
Analysis of D. desulfuricans holo-CcmEsol. A, shown is the visible absorption spectrum reflecting the formation of D. desulfuricans holo-CcmEsol by D. desulfuricans CcmEFABCDI expressed in E. coli. The absorption maximum is at 419 nm. The sample was reduced by the addition of a few grains of disodium dithionite. The vertical scale bar represents 0.2 absorption units. The inset shows the reduced pyridine hemochrome spectrum of D. desulfuricans holo-CcmEsol. The vertical line indicates 551.5 nm. B, shown is a schematic representation of peptide fragments determined by MALDI-TOF mass spectrometry after digestion of purified D. desulfuricans holo-CcmEsol with trypsin. PelB represents the signal peptide, and Strep indicates the position of the Streptavidin II affinity tag. Dotted lines represent peptides with the expected molecular mass detected by mass spectrometry. Solid lines represent peptides with an additional mass of 616 Da, indicating covalent attachment of heme to the peptide.
The purified D. desulfuricans CcmEsol protein was subjected to trypsin digestion followed by mass spectrometry. This revealed a series of peptide fragments encompassing the entire D. desulfuricans CcmE sequence (Fig. 5B). Two peptides corresponding to the C-terminal region of D. desulfuricans CcmE were of particular note. The first of these (CPSKYQK) had a mass 616 Da greater than would be expected for the peptide alone, indicating covalent attachment of otherwise-unmodified heme. A second peptide (YQKENRE) was detected that did not have the additional mass, indicating attachment of the heme to Cys127. Attachment of heme to Pro128, Ser129, or Lys130, the only other possibilities, appears highly unlikely. This is consistent with data above in which no heme binding to membrane-bound D. desulfuricans CcmE was observed upon mutagenesis of Cys127.
D. desulfuricans CcmE and E. coli CcmE Are Not Functionally Interchangeable
Expression of membrane-anchored D. desulfuricans CcmE (pDDCcmE) in E. coli EC65 (ΔccmE (12)) did not restore c-type cytochrome biogenesis of an exogenous c-type cytochrome (data not shown). This indicates that D. desulfuricans CcmE cannot be functionally incorporated into the E. coli Ccm system. Additionally, expression of the E. coli ccm operon in which the 130HDENY CcmE heme-binding motif was replaced with CPSKY (the equivalent residues in D. desulfuricans CcmE; pEC86CPSKY) was unable to restore endogenous c-type cytochrome biogenesis in E. coli EC06 (ΔccmABCDEFGH; data not shown).
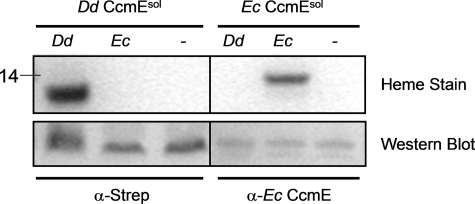
To investigate if the E. coli Ccm system was capable of attaching heme to D. desulfuricans CcmEsol, we coexpressed pEC86 (E. coli ccmABDEFGH) and D. desulfuricans CcmEsol. There was no detectable heme attachment to D. desulfuricans CcmEsol by the E. coli Ccm system (Fig. 6). In the complementary experiment, D. desulfuricans CcmEFABCDI was unable to attach heme detectably to E. coli CcmEsol. The two biogenesis systems were able to attach heme covalently to their own respective soluble CcmE proteins (as indicated by the heme-staining bands in Fig. 6), which could be detected with appropriate antibodies under all conditions.
FIGURE 6.
Specificity of holo-CcmEsol formation. D. desulfuricans CcmEsol or E. coli CcmEsol were co-expressed with the D. desulfuricans ccmEFABCDI operon (Dd), the E. coli ccmABCDEFGH operon (Ec), or no biogenesis system (−). Top panel, formation of holo-CcmEsol was monitored by the detection of heme-staining bands of appropriate molecular masses (weight indicated in kDa). Bottom panel, expression of protein was detected by Western blotting whole cell extracts using an anti-streptavidin antibody for D. desulfuricans CcmEsol and an anti-E. coli CcmE antibody and periplasmic extracts for E. coli CcmEsol.
DISCUSSION
Reported here are several striking results that indicate a need for reappraisal of the functions of at least some of the Ccm proteins. Firstly, the identification of a functional, natural, CcmE variant with cysteine (Cys127) replacing the hitherto invariant and essential histidine residue, which for the E. coli protein (and a plant CcmE (40)), forms a covalent bond to heme. The replacement of Cys127 in D. desulfuricans CcmE by histidine or alanine resulted in loss of any detectable cytochrome c assembly. The failure of CcmEC127A to support activity parallels the result found in the H130A variant of the E. coli protein (12), but the inactivity of C127H shows that the CcmE of D. desulfuricans is a variant adapted to employ cysteine at a crucial site in the protein. It is striking that the E. coli protein is non-functional when its histidine is replaced by cysteine (H130C (38)).
An important question is whether D. desulfuricans CcmE forms a covalent bond to heme. It is clear from our data, in which heme-staining of SDS-PAGE gels (Fig. 4) is supported by spectroscopy and mass spectrometry detecting two heme-containing peptides (Fig. 5), that such a bond is formed. The covalent bond was not detected when Cys127 was mutated to histidine. The conclusion that it is Cys127 which covalently binds heme is confirmed by analysis of peptide fragments (Fig. 5). In the case of E. coli CcmE, His130 has been reported through structural analysis of a tryptic peptide as being joined to the heme via a novel bond between the Nδ1 of the histidine and the β-carbon of a vinyl group of the heme (13). Hence, both the E. coli and D. desulfuricans Ccm systems involve a covalent attachment of heme to CcmE.
The structure of E. coli apoCcmE has been determined by NMR (Fig. 4A, panel i (41)). Briefly, the N-terminal domain forms a β-barrel of six β-sheets, and His130 is surface-exposed near a hydrophobic platform to which heme is proposed to bind. The structure of D. vulgaris CcmE, which is highly homologous to the D. desulfuricans protein, has been determined by NMR (PDB accession number 2KCT; Fig. 4A, panel ii). Unfortunately, the available structure only encompasses residues 44–128, lacking the N-terminal transmembrane helix but also terminating at the proline of the CPSKY motif (where the cysteine is Cys127), thereby excluding the conserved tyrosine that is a heme-iron ligand in E. coli CcmE (42, 43). Nevertheless, the β-barrel structure bears a striking similarity to that of the E. coli protein despite relatively low sequence identity. Cys127 also appears to be surface-exposed, consistent with its role in heme binding. Extensive future work will be required to identify any compensating differences in the Ccm proteins of D. desulfuricans compared with E. coli that permit the use of an essential cysteine rather than histidine in CcmE.
There is no detectable CcmH analogue encoded on the genome of D. desulfuricans. The D. desulfuricans ccm operon confers cytochrome c maturation capability on E. coli lacking its endogenous ccm system, demonstrating that this variant System I can function in the absence of CcmH. This contrasts with the situation in E. coli itself where the N-terminal region of its CcmH protein is essential for c-type cytochrome synthesis (33). Analysis of the D. desulfuricans Ccm proteins did not reveal the presence of additional CXXC motifs or extra domains compared with their E. coli counterparts. Thus, it appears that the reductive role of CcmH is absent and not simply incorporated into another of the D. desulfuricans Ccm components. However, an atypical reductant-transfer motif cannot be excluded. For example, CcmF from D. desulfuricans and D. vulgaris contains several cysteines, although these are not conserved in other System I*-containing organisms. Genomes of some organisms encoding CcmE proteins with cysteine in place of the histidine do contain CcmH homologues (22). Therefore, it would seem that such CcmE proteins can function in the presence of CcmH but that, in contrast to E. coli, CcmH is not essential for c-type cytochrome biogenesis in such a system.
In the present experiments c-type cytochromes have been produced in E. coli without a CcmG protein. This also contrasts with the situation in E. coli where CcmG has been shown to be an essential member of the endogenous Ccm system (21). However, and similarly, it has been shown that expression of c-type cytochrome biogenesis System II in a Δccm E. coli strain leads to cytochrome c production without CcmG (or ResA, the analogous protein from System II) (32, 44). Normally, CcmG transfers reductant from DsbD to the rest of cytochrome c assembly apparatus and/or the apocytochrome. Presumably instances of continued cytochrome c synthesis in the absence of CcmG can be explained on the basis that either reductant from DsbD is not required or that there are other pathways catalyzing reductant transfer from DsbD to the apparatus.
The addition of exogenous reductant has previously been reported to complement for a disruption in DsbD (35) and site-directed mutants of the active cysteines in CcmG (36). However, the addition of either cysteine or 2-mercaptoethane sulfonic acid had no significant effect on cytochrome c550 production in E. coli EC06 strains expressing pDD86 and pKPD1. Thus, the lower yield of c-type cytochrome produced by the D. desulfuricans system in E. coli, relative to that produced by cells containing pEC86, must be attributed to other factors. Among these is a possible lower heterologous expression of D. desulfuricans Ccm proteins compared with the homologous expression of E. coli counterparts. Also, it is possible that other factors in the highly reducing environments in which sulfate-reducing bacteria grow can compensate for the variations in the Ccm system seen in these organisms that depend more extensively on the function of c-type cytochromes than does E. coli.
The results presented here imply that CcmF is the irreducible core of heme attachment via the Ccm system and that CcmG and CcmH may be ancillary, but dispensable, components. We can find only one copy of ccmF on the genomes of Desulfovibrio species despite the fact that these organisms all contain NrfA nitrite reductase which has an active site heme-attachment motif CXXCK (45), known in other organisms to require dedicated variant forms of CcmF for heme attachment (46). A further variant form of CcmF has also been implicated in Shewanella for the attachment of heme to a CX15CH motif (47). Thus, it may be that the CcmF of Desulfovibrio differs from its counterparts in E. coli in that it can participate in heme attachment to both CXXCH and CXXCK. However, we originally noted that D. desulfuricans contains a gene known as resC or ccsA (22). This encodes part of the System II c-type cytochrome assembly apparatus found in various species of bacteria and thylakoids. Homologues of ResC are implicated in substrate recognition of, and heme attachment to, CXXCK and CX15CH in Wolinella succinogenes (48). We have identified a strong correlation between the presence of ResC alongside the Desulfovibrio-type System I* and the presence of a NrfA protein bearing the CXXCK motif. This raises the important possibility that ResC can operate in concert with some parts of System I to attach heme to CXXCK in sulfate-reducing bacteria. Testing this hypothesis is not trivial, although we note that D. desulfuricans CcmEFABCDI cannot mature a CXXCK variant of P. denitrificans cytochrome c550 when expressed in E. coli. This supports involvement of ResC in maturation of CXXCK-containing cytochromes in D. desulfuricans. Clearly this is another unexpected outcome of the present work, which calls for a new perspective on c-type cytochrome assembly pathways.
Crucially, this work raises questions about the role of the histidine-heme bond in CcmE from many organisms. There is strong evidence that a covalent bond between heme and CcmE is a competent intermediate in c-type cytochrome assembly (12), so it is remarkable that the novel heme-histidine chemistry can be substituted by heme-cysteine chemistry. The observation that the histidine-heme bond in CcmE is not essential along with the completely unexpected alterations observed within D. desulfuricans System I* challenge the current paradigm E. coli system as a universal model. Overall, our data suggest that only CcmF and CcmE are at the heart of heme attachment to apocytochromes c and demonstrates a greater degree of variation within System I-mediated cytochrome c biogenesis than previously anticipated.
Supplementary Material
Acknowledgments
We thank Prof. Judy Wall (University of Missouri) for kindly providing D. desulfuricans G20 genomic DNA and Prof. Linda Thöny-Meyer for antibodies, strains, and plasmids. We thank Katalin di Gleria and Benedikt Kessler (Centre for Cellular and Molecular Physiology, Oxford) and Benjamin Thomas (Dunn School of Pathology, Oxford) for expert assistance with proteomics analyses. We acknowledge the Computational Biology Research Group, Medical Sciences Division, Oxford for use of their services in this project.
This work was supported by Biotechnology and Biological Sciences Research Council Grants BB/D523019/1, BB/E004865/1, and BB/D019753/1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Fig. S1.
- IPTG
- isopropyl-1-thio-β-d-galactopyranoside
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Stevens J. M., Daltrop O., Allen J. W., Ferguson S. J. (2004) Acc. Chem. Res. 37, 999–1007 [DOI] [PubMed] [Google Scholar]
- 2.Ferguson S. J., Stevens J. M., Allen J. W., Robertson I. B. (2008) Biochim. Biophys. Acta 1777, 980–984 [DOI] [PubMed] [Google Scholar]
- 3.Bowman S. E., Bren K. L. (2008) Nat. Prod. Rep. 25, 1118–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thöny-Meyer L., Fischer F., Künzler P., Ritz D., Hennecke H. (1995) J. Bacteriol. 177, 4321–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamel P., Corvest V., Giegé P., Bonnard G. (2009) Biochim. Biophys. Acta. 1793, 125–138 [DOI] [PubMed] [Google Scholar]
- 6.Allen J. W., Jackson A. P., Rigden D. J., Willis A. C., Ferguson S. J., Ginger M. L. (2008) FEBS J. 275, 2385–2402 [DOI] [PubMed] [Google Scholar]
- 7.Feissner R. E., Richard-Fogal C. L., Frawley E. R., Kranz R. G. (2006) Mol. Microbiol. 61, 219–231 [DOI] [PubMed] [Google Scholar]
- 8.Christensen O., Harvat E. M., Thöny-Meyer L., Ferguson S. J., Stevens J. M. (2007) FEBS J. 274, 2322–2332 [DOI] [PubMed] [Google Scholar]
- 9.Richard-Fogal C. L., Frawley E. R., Kranz R. G. (2008) J. Bacteriol. 190, 3489–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahuja U., Thöny-Meyer L. (2005) J. Biol. Chem. 280, 236–243 [DOI] [PubMed] [Google Scholar]
- 11.Daltrop O., Stevens J. M., Higham C. W., Ferguson S. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz H., Hennecke H., Thöny-Meyer L. (1998) Science 281, 1197–1200 [DOI] [PubMed] [Google Scholar]
- 13.Lee D., Pervushin K., Bischof D., Braun M., Thöny-Meyer L. (2005) J. Am. Chem. Soc. 127, 3716–3717 [DOI] [PubMed] [Google Scholar]
- 14.Ren Q., Ahuja U., Thöny-Meyer L. (2002) J. Biol. Chem. 277, 7657–7663 [DOI] [PubMed] [Google Scholar]
- 15.Sanders C., Turkarslan S., Lee D. W., Onder O., Kranz R. G., Daldal F. (2008) J. Biol. Chem. 283, 29715–29722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Matteo A., Gianni S., Schininà M. E., Giorgi A., Altieri F., Calosci N., Brunori M., Travaglini-Allocatelli C. (2007) J. Biol. Chem. 282, 27012–27019 [DOI] [PubMed] [Google Scholar]
- 17.Setterdahl A. T., Goldman B. S., Hirasawa M., Jacquot P., Smith A. J., Kranz R. G., Knaff D. B. (2000) Biochemistry 39, 10172–10176 [DOI] [PubMed] [Google Scholar]
- 18.Meyer E. H., Giegé P., Gelhaye E., Rayapuram N., Ahuja U., Thöny-Meyer L., Grienenberger J. M., Bonnard G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabianek R. A., Hennecke H., Thöny-Meyer L. (2000) FEMS Microbiol. Rev. 24, 303–316 [DOI] [PubMed] [Google Scholar]
- 20.Turkarslan S., Sanders C., Ekici S., Daldal F. (2008) Mol. Microbiol. 70, 652–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Throne-Holst M., Thöny-Meyer L., Hederstedt L. (1997) FEBS Lett. 410, 351–355 [DOI] [PubMed] [Google Scholar]
- 22.Allen J. W., Harvat E. M., Stevens J. M., Ferguson S. J. (2006) FEBS Lett. 580, 4827–4834 [DOI] [PubMed] [Google Scholar]
- 23.Pattarkine M. V., Tanner J. J., Bottoms C. A., Lee Y. H., Wall J. D. (2006) J. Mol. Biol. 358, 1314–1327 [DOI] [PubMed] [Google Scholar]
- 24.Arslan E., Schulz H., Zufferey R., Künzler P., Thöny-Meyer L. (1998) Biochem. Biophys. Res. Commun. 251, 744–747 [DOI] [PubMed] [Google Scholar]
- 25.Sambongi Y., Ferguson S. J. (1994) FEBS Lett. 340, 65–70 [DOI] [PubMed] [Google Scholar]
- 26.Allen J. W., Ginger M. L., Ferguson S. J. (2004) Biochem. J. 383, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen J. W., Ferguson S. J. (2003) Biochem. J. 375, 721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen J. W., Tomlinson E. J., Hong L., Ferguson S. J. (2002) J. Biol. Chem. 277, 33559–33563 [DOI] [PubMed] [Google Scholar]
- 29.Goodhew C. F., Brown K. R., Pettigrew G. W. (1986) Biochim. Biophys. Acta 852, 288–294 [Google Scholar]
- 30.Bartsch R. G. (1971) Methods Enzymol. 23, 344–363 [Google Scholar]
- 31.Goddard A. D., Moir J. W., Richardson D. J., Ferguson S. J. (2008) Mol. Microbiol. 70, 667–681 [DOI] [PubMed] [Google Scholar]
- 32.Goddard A. D., Stevens J. M., Rondelet A., Nomerotskaia E., Allen J. W., Ferguson S. J. (2010) FEBS J. 277, 726–737 [DOI] [PubMed] [Google Scholar]
- 33.Fabianek R. A., Hofer T., Thöny-Meyer L. (1999) Arch. Microbiol. 171, 92–100 [DOI] [PubMed] [Google Scholar]
- 34.Robertson I. B., Stevens J. M., Ferguson S. J. (2008) FEBS Lett. 582, 3067–3072 [DOI] [PubMed] [Google Scholar]
- 35.Sambongi Y., Ferguson S. J. (1994) FEBS Lett. 353, 235–238 [DOI] [PubMed] [Google Scholar]
- 36.Fabianek R. A., Hennecke H., Thöny-Meyer L. (1998) J. Bacteriol. 180, 1947–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid E., Eaves D. J., Cole J. A. (1998) FEMS Microbiol. Lett. 166, 369–375 [DOI] [PubMed] [Google Scholar]
- 38.Enggist E., Schneider M. J., Schulz H., Thöny-Meyer L. (2003) J. Bacteriol. 185, 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens J. M., Daltrop O., Higham C. W., Ferguson S. J. (2003) J. Biol. Chem. 278, 20500–20506 [DOI] [PubMed] [Google Scholar]
- 40.Spielewoy N., Schulz H., Grienenberger J. M., Thony-Meyer L., Bonnard G. (2001) J. Biol. Chem. 276, 5491–5497 [DOI] [PubMed] [Google Scholar]
- 41.Enggist E., Thöny-Meyer L., Güntert P., Pervushin K. (2002) Structure 10, 1551–1557 [DOI] [PubMed] [Google Scholar]
- 42.Uchida T., Stevens J. M., Daltrop O., Harvat E. M., Hong L., Ferguson S. J., Kitagawa T. (2004) J. Biol. Chem. 279, 51981–51988 [DOI] [PubMed] [Google Scholar]
- 43.García-Rubio I., Braun M., Gromov I., Thöny-Meyer L., Schweiger A. (2007) Biophys. J. 92, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feissner R. E., Richard-Fogal C. L., Frawley E. R., Loughman J. A., Earley K. W., Kranz R. G. (2006) Mol. Microbiol. 60, 563–577 [DOI] [PubMed] [Google Scholar]
- 45.Cunha C. A., Macieira S., Dias J. M., Almeida G., Goncalves L. L., Costa C., Lampreia J., Huber R., Moura J. J., Moura I., Romão M. J. (2003) J. Biol. Chem. 278, 17455–17465 [DOI] [PubMed] [Google Scholar]
- 46.Eaves D. J., Grove J., Staudenmann W., James P., Poole R. K., White S. A., Griffiths I., Cole J. A. (1998) Mol. Microbiol. 28, 205–216 [DOI] [PubMed] [Google Scholar]
- 47.Hartshorne R. S., Kern M., Meyer B., Clarke T. A., Karas M., Richardson D. J., Simon J. (2007) Mol. Microbiol. 64, 1049–1060 [DOI] [PubMed] [Google Scholar]
- 48.Simon J., Gross R., Einsle O., Kroneck P. M., Kröger A., Klimmek O. (2000) Mol. Microbiol. 35, 686–696 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.