Abstract
L-A is a persistent double-stranded RNA virus commonly found in the yeast Saccharomyces cerevisiae. Isolated L-A virus synthesizes positive strand transcripts in vitro. We found that the 5′ termini of the transcripts are diphosphorylated. The 5′-terminal nucleotide is G, and GDP was the best substrate among those examined to prime the reaction. When GTP was used, the triphosphate of GTP incorporated into the 5′-end was converted to diphosphate. This activity was not dependent on host CTL1 RNA triphosphatase. The 5′-end of the GMP-primed transcript also was converted to diphosphate, the β-phosphate of which was derived from the γ-phosphate of ATP present in the polymerization reaction. These results demonstrate that L-A virus commands elaborate enzymatic systems to ensure its transcript to be 5′-diphosphorylated. Transcripts of M1, a satellite RNA of L-A virus, also had diphosphate at the 5′ termini. Because viral transcripts are released from the virion into the cytoplasm to be translated and encapsidated into a new viral particle, a stage most vulnerable to degradation in the virus replication cycle, our results suggest that the 5′-diphosphate status is important for transcript stability. Consistent with this, L-A transcripts made in vitro are resistant to the affinity-purified Ski1p 5′-exonuclease. We also discuss the implication of these findings on translation of viral RNA. Because the viral transcript has no conventional 5′-cap structure, this work may shed light on the metabolism of non-self-RNA in yeast.
Keywords: RNA Metabolism, RNA Structure, RNA Viruses, Viral Transcription, Yeast, Host Defense, Viral RNA Metabolism
Introduction
The 5′-cap (m7GpppXp) and 3′-poly(A) tail structures are the hallmark of eukaryotic mRNAs and have important biological functions in promoting stability in the nucleus, transport to the cytoplasm, and translation and stability in the cytoplasm. Therefore, many viruses furnish their transcripts with the 5′-cap structure utilizing their own or cellular enzymes or even by snatching the structure from cellular mRNAs (1–3). Transcripts made by T7 RNA polymerase are recognized as foreign and induce innate immune responses in animal cells. Recent studies suggest that the 5′-triphosphate of the transcripts as well as secondary structure, are involved in these responses (4, 5). Therefore, the 5′-end of RNA bears important information concerning on self or non-self for the host cells.
L-A is a persistent double-stranded RNA (dsRNA)2 virus commonly found in the laboratory strains of the yeast Saccharomyces cerevisiae (6). The virus has no extracellular transmission pathway. The viral genome is 4.6-kb-long, and only one strand (the positive strand) encodes proteins; the major 76-kDa coat protein Gag and a minor 170-kDa Gag-Pol (7, 8). The minor protein is made from the two overlapping genes by a −1 ribosomal frameshifting mechanism (9). The Pol domain of the fusion protein has consensus motifs for RNA-dependent RNA polymerases. The RNA genome is packed inside of a 39-nm icosahedral shell consisting of 60 Gag asymmetric dimers (10, 11). One molecule or two of Gag is replaced by Gag-Pol because the fusion protein is essential for RNA encapsidation and replication. L-A virions have transcriptase activity to synthesize full-length positive strands from the dsRNA genome in a conservative way (12). The transcripts are extruded from the virions and serve as templates for translation and also as substrates for encapsidation to form new virions. Once encapsidated, the positive strand is converted to dsRNA by the replicase activity associated with the virion.
M1, a satellite RNA of L-A virus, requires L-A encoded proteins to encapsidate and replicate its RNA genome. M1 dsRNA (1.6–1.8 kb) encodes a secreted protein toxin (killer toxin) that kills M1-free, toxin-sensitive strains (13). Using this killer phenotype as a marker, various host chromosomal genes have been identified that affect replication and maintenance of M1 and L-A. Mutations in SKI genes (SKI1, -2, -3, -4, -6, -7, and -8) cause an elevated copy number of L-A and, in the presence of M1, exhibit a “superkiller” phenotype (14, 15).
Degradation of mRNA in eukaryotes usually begins with shortening of the poly(A) tail at the 3′-end followed by decapping at the 5′-end by the Dcp1p-Dcp2p decapping enzyme. Decapping generates the mRNA body with 5′-monophosphate, which is then degraded by the SKI1/XRN1 5′-exonuclease (16, 17). Alternatively, the deadenylated RNA is digested by a multiprotein complex with 3′-exonuclease activity, called the exosome. Products of SKI2, -3, -4, -6, -7, and -8 are auxiliary or core components of the cytoplasmic exosome (18, 19). These gene products also are known to prevent translation from RNA without the 3′-poly(A) tail (20, 21).
Previously, we observed a strong antiviral activity of SKI1 against L-A virus (22). Deletion of SKI1 elevated the copy number of L-A 5–10-fold and overexpression of the gene cured L-A virus from cells at a high frequency. In the replication cycle of L-A virus, only positive strand transcripts are released from virions into the cytoplasm. We therefore reasoned that the L-A transcript at this stage is vulnerable to the SKI1 5′-exonuclease. The nuclease requires 5′-monophosphorylated RNA as substrate, whereas the 5′-end of L-A transcript has not been well established. It was reported about three decades ago that the 5′ terminus was triphosphorylated (23) or diphosphorylated (24). Here, we establish that L-A has diphosphate at the 5′-end. More significantly, we demonstrate that the phosphate group of GTP or GMP, when the nucleotide was incorporated at the 5′-end of the transcript, was converted to diphosphate. The virus, thus, clearly avoids 5′-triphosphorylated or -monophosphorylated transcripts. Our work may shed light on the metabolism of non-self-RNA in yeast, especially on the role of phosphate groups at the RNA 5′-end.
EXPERIMENTAL PROCEDURES
Strains and Media
Strain 2404 (α kar1-1 his4, L-A-HN, L-BC, M-o) was used to obtain wild type L-A virions. 2927 (a ura3 trp1 his3 ski2-2, L-A-o L-BC-o) was used to prepare an L-A-free cell extract. A ctl1Δ strain was obtained from a haploid deletion series (BY4741 (a ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 20 S RNA-o, 23 S RNA-o, L-A) background) of the EUROFAN collection. The L-A-free killer strain (a trp1 ura3 leu2 his3 pep4::HIS3 nuc1::LEU2, L-A-o, M1, L-BC, pAB2) was obtained from Reed B. Wickner (25). L-A proteins expressed from plasmid pAB2 support replication of M1 in this strain. Cells were grown in rich YPAD medium (1% yeast extract, 2% peptone, 0.04% adenine sulfate, and 2% glucose) or synthetic medium deprived of uracil (H−Ura) or trp (H−Trp) (26). L-A and M1 virions were isolated and purified through CsCl gradient centrifugation as described in Ref. 27.
Transcription Reaction
The standard transcription reaction mixture contained 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 0.1 mm EDTA, 20 mm NaCl, 5 mm KCl, 1 mm dithiothreitol, 2.5 mg/ml bentonite, and 0.5 mm each of ATP, CTP, GTP, and UTP. When a radioactive nucleotide was used, the concentration was reduced to 20 μm. The reaction was started by the addition of purified L-A or M1 virions and incubated at 30 °C. The RNA products were extracted with phenol, phenol-chloroform, and precipitated with ethanol. Then, the RNA was separated in either a 1.3% agarose or a 15% acryalamide gel, depending on the size of the products. Radioactive products were visualized by autoradiography. Nonradioactive RNA was detected by ethidium bromide staining or by Northern hybridization (28) using a 32P-labeled specific probe.
Ski1p-TAP Purification
Tandem affinity purification (TAP)-tagged Ski1p was expressed in strain 2928-4 (a ura3 trp1 his3, L-A-o, L-BC) harboring plasmid pRE1004 by growing the cells in H-ura medium, in which glucose was substituted with 2% galactose. pRE1004 is a derivative of pEMBLyex4 (29) with a 5.3-kb BamHI fragment that contains the entire SKI1 coding sequence directly fused to TAP at the carboxyl terminus. The expression of the protein is under the control of the GAL1-CYC hybrid promoter. Ski1p-TAP was purified in a single-step purification with calmodulin affinity resin (Stratagene) (30, 31). Briefly, a cell lysate was prepared by breaking the cells with a FastPrep instrument (BIO101 Savant) in a buffer containing 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor (GE Healthcare) and clarified by centrifugation at 50,000 rpm for 1 h with a TLA100.2 rotor. The supernatant was dialyzed against IPP 150 calmodulin binding buffer (10 mm β-mercaptoethanol, 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm magnesium acetate, 1 mm imidazole, 2 mm CaCl2, and 0.1% Nonidet P-40) for 1 h and then incubated for 1 h with 200 μl calmodulin affinity resin pre-equilibrated with IPP 150 calmodulin binding buffer. The mixture was transferred to a column, and then the column was washed with 40 ml IPP 150 calmodulin binding buffer. The bound protein was eluted stepwise with IPP 150 calmodulin elution buffer (10 mm β-mercaptoethanol, 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm magnesium acetate, 1 mm imidazole, 2 mm EGTA, and 0.1% Nonidet P-40). The eluted protein was analyzed by SDS-PAGE followed by silver staining. Fraction 3 (see lane 4 in Fig. 6A) contained the highest concentration of Ski1p-TAP (0.09 mg/ml) among elutes, estimated by the Bradford method (32) using bovine serum albumin as standard.
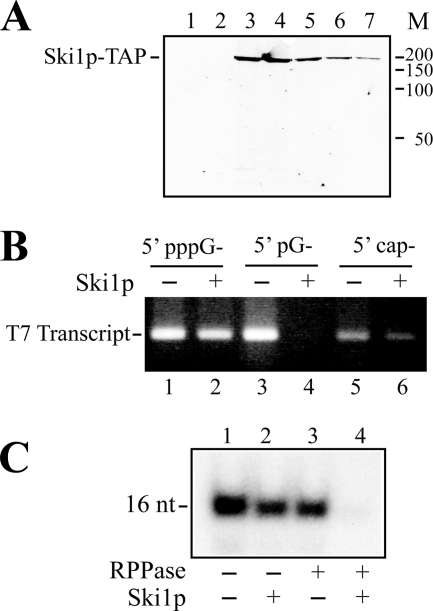
FIGURE 6.
L-A transcript made in vitro is resistant to Ski1p. A, purification of Ski1p. Ski1p-TAP was purified through a calmodulin affinity resin column. The bound protein was eluted stepwise with a buffer containing EGTA. Protein was separated with SDS-PAGE and visualized by silver staining. Lane 1, the last wash of the column. Lanes 2–7, fractions eluted with EGTA. M, molecular standards (kDa). B, specificity of purified Ski1p. Transcripts bearing a 5′-pppG-, 5′-pG-, or 5′-cap structure were made in vitro by T7 RNA polymerase using the same DNA template as described under “Experimental Procedures.” The transcripts were incubated with Ski1p, and the digest was separated in an agarose gel and visualized by ethidium bromide staining. C, the 32P-labeled 16-nt transcript was made in vitro by L-A virions in a CTP-omitted transcription reaction and treated with RNA 5′-polyphosphatase (RPPase) and/or purified Ski1p as indicated below the panel. Then, the transcript was separated in a acrylamide gel and visualized by autoradiography.
T7 Transcripts with Different 5′-Ends
T7 transcripts with different 5′-ends were made by run-off transcription with T7 RNA polymerase as described in Ref. 33. 5′-monophosphorylated and capped RNAs were synthesized by the addition of an excess amount of GMP and m7GpppG cap analogue, respectively, over GTP. We used PvuII-digested pTF60 as template. pTF60 contains only two extra Gs between the T7 promoter and the entire X cDNA sequence (27). X (530 bp) is a mutant of L-A with a large internal deletion (34).
Enzyme Digestion
Terminator 5′-exonuclease or Ski1p-TAP digestion was carried out in a buffer containing 50 mm Tris-HCl, pH 8.0, 2 mm MgCl2, 0.1 m NaCl, and 0.5 units Terminator or 30 ng of Ski1p-TAP. RNA 5′-polyphosphatase consisted of the following: 50 mm HEPES KOH pH 7.5, 0.1 m NaCl, 1 mm EDTA, 0.1% β-mercaptoethanol, 0.01% Triton X-100, and 10 units of the enzyme. S1 nuclease was made up of the following: 30 mm sodium acetate, pH 4.5, 60 mm NaCl, 1 mm ZnCl2, and 20 units of S1. Bacterial alkaline phosphatase (BAP) consisted of the following: 10 mm Tris-HCl, pH 8.0, and 30 units of BAP. Tobacco acid pyrophosphatase consisted of the following: 50 mm sodium acetate, pH 6.0, 1 mm EDTA, 0.1% β-mercaptoethanol, 0.01% Triton X-100, and 5 units of the enzyme. All of the reactions were carried out at 37 °C for 30 min, except that Terminator 5′-exonuclease and Ski1p-TAP were incubated at 30 °C.
Miscellaneous
Radioactive nucleotides were obtained from PerkinElmer Life Sciences. Terminator 5′-exonuclease and RNA 5′-polyphosphatase were from Epicenter. S1 nuclease was from Promega, and BAP was from Invitrogen. Tobacco acid pyrophosphatase, γ-S-GTP, and β-S-GDP were from Sigma-Aldrich. The m7GpppG cap analogue was from Ambion, and malachite Green reagent was from Biomol.
RESULTS
L-A Transcript Has Diphosphate at the 5′-End
Purified L-A virions have RNA polymerase activity and synthesize full-length positive strand transcripts using the dsRNA genome as template. The RNA products were resistant to Terminator 5′-exonuclease (Fig. 1A, lane 2). The enzyme processively hydrolyzes single-stranded RNA with 5′-monophosphate in a 5′- to 3′-direction. Pretreatment with RNA 5′-polyphosphatase, however, converted the RNA to full sensitivity to the 5′-exonuclease (Fig. 1A, lane 4). The polyphosphatase removes the γ- and β-phosphates from 5′-triphosphorylated or -diphosphorylated RNA but has no activity on RNA with a 5′-cap. These results indicate that the L-A transcripts have no 5′-cap structure and suggest that the transcripts are tri- or diphosphorylated at their 5′-ends.
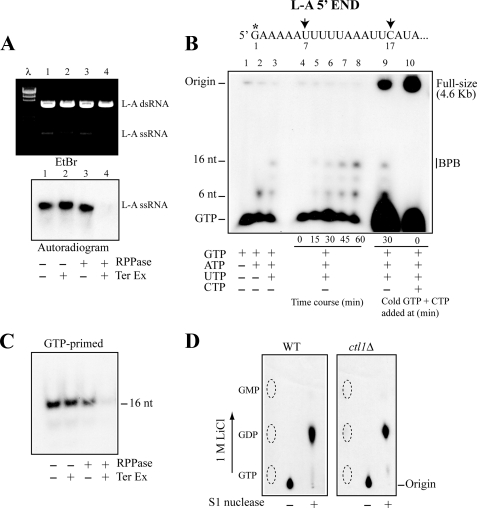
FIGURE 1.
GTP-primed L-A transcript has diphosphate at the 5′-end. A, 32P-labeled, full-sized L-A transcripts made in vitro by L-A virions were treated with Terminator 5′-exonuclease (Ter Ex) and/or RNA 5′-polyphosphatase (RPPase) as indicated below the panel and separated in an agarose gel. L-A single-stranded RNA (ssRNA) was visualized by ethidium bromide staining (upper panel) or by autoradiography (lower panel). λ, λ-HindIII fragments as size markers. B, upper panel shows the 5′-terminal region of the L-A single-stranded RNA transcript. The first U and C appear at positions 7 and 17, respectively (indicated by arrows). The asterisk indicates the position that can be labeled with [α-32P]GTP in an in vitro transcription reaction. The lower panel shows transcription products made in vitro by L-A virions. Transcripts were labeled with [α-32P]GTP, separated in a 15% acrylamide gel, and visualized by autoradiography. Lanes 1–3, L-A virions were incubated for 30 min in a transcription reaction from which ATP, UTP, and CTP (lane 1), or UTP and CTP (lane 2), or CTP (lane 3) was omitted. Lanes 4–8, L-A virions were incubated for the times indicated below the panel in a CTP-omitted transcription reaction. Lane 9, L-A virions were incubated in a CTP-omitted transcription reaction. After a 30-min incubation, excess amounts (1 mm each) of nonlabeled GTP and CTP were added to the mixture, and the reaction was carried out 30 min further. Lane 10, same as in lane 9, except that the nonlabeled GTP and CTP were added at 0 min. The reaction was carried out for 60 min. The mobility of full-sized, 16-nt, and 6-nt transcripts as well as unincorporated GTP is indicated. The 16-nt transcripts comigrated with bromphenol blue (BPB) in this gel system. C, [α-32P]GTP-primed 16-nt transcripts made by L-A virions in a CTP-omitted reaction were treated with RNA 5′-polyphosphatase and/or Terminator 5′-exonuclease as indicated below the panel and separated in a 15% acrylamide gel. An autoradiogram of the gel is shown. D, [α-32P]GTP-primed 16-nt transcripts made by L-A virions isolated from CTL1 (left panel, WT) or ctl1Δ (right panel, ctl1Δ) strain were purified in a 15% acrylamide gel. The transcripts were treated with or without S1 nuclease and analyzed on PEI cellulose with 1 m LiCl. The positions of nonlabeled GTP, GDP, and GMP are indicated by the dotted ovals.
The 5′-region of L-A positive strand starts with a G followed by an AU-rich sequence (Fig. 1B, upper diagram). The first U and C appear at positions 7 and 17, respectively. When L-A virions were incubated only with GTP and ATP, a short transcript (6 nt) was produced (Fig. 1B, lane 2). A further inclusion of UTP in the reaction increased the length to 16 nt (Fig. 1B, lane 3). These short transcripts were apparently released from the RNA polymerase complex because of the lack of incoming nucleotides for elongation. The addition of CTP at 30 min did not convert the 16-nt species into a full-size transcript (Fig. 1B, lane 9), and we did not observe these short transcripts when virions were incubated with four NTPs (Fig. 1B, lane 10). The 16-nt transcript has a single G at the 5′-end; therefore, only the 5′-terminal nucleotide in the molecule can be labeled with [α-32P]GTP. This transcript was purified from an acrylamide gel and treated with 5′-exonuclease. As shown in Fig. 1C, this species was resistant to the 5′-exonuclease but became sensitive to it after treatment with RNA 5′-polyphosphatase, a characteristic similar to the one observed with the full-size transcript. Then the 16-nt transcript labeled with [α-32P]GTP was digested with S1 nuclease and analyzed on PEI cellulose. S1 degrades single-stranded RNA endonucleolytically to yield 5′-phosphoryl terminated products. As shown in Fig. 1D (left panel), the labeled 5′-terminal nucleotide comigrated with GDP, indicating that the L-A transcript has a diphosphate at the 5′-end.
GDP Is Not Essential for Transcription Initiation
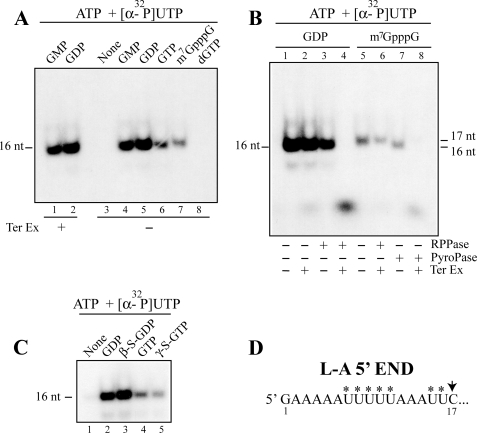
Incubation of L-A virions with ATP and [α-32P]UTP did not produce transcripts (Fig. 2A, lane 3), indicating that the guanine nucleotide is essential for initiation of transcription. dGTP also failed to initiate transcription (Fig. 2A, lane 8). Among the guanine nucleotides examined, GDP was the best substrate for the initiation reaction, consistent with the results that the 5′-end of the transcript is diphosphorylated. GMP also was incorporated well into the 16-nt transcript (Fig. 2A, lane 4). A cap analogue (m7GpppG) could initiate transcription whose product run as a 17-mer because of the extra m7G at the 5′-end (Fig. 2A, lane 7). This 17-mer, unlike the GTP- or GDP-primed transcript, was resistant to the 5′-exonuclease even after treatment with RNA 5′-polyphosphatase (Fig. 2B, lane 6). The 17-mer was converted to 5′-monophosphorylated 16-mer by tobacco acid pyrophosphatase (Fig. 2B, lane 7), and the resulting product was now sensitive to the 5′-exonuclease (Fig. 2B, lane 8). These results clearly demonstrate that a cap analogue (m7GpppG) can be incorporated into the 5′-end of the transcript and that GDP is not essential for transcription initiation. GTP and GDP analogues (γ-S-GTP and β-S-GDP) could initiate transcription, and the amounts of 16-nt transcripts were comparable to those produced by the respective bona fide nucleotides (Fig. 2C). Because γ-S-GTP is non-hydrolyzable, the results again confirm that GDP is not essential for initiation. It also indicates that transcription initiation and conversion of the 5′-triphosphate of the transcript (or GTP) to 5′-diphosphate are two distinct reactions.
FIGURE 2.
Nucleotide specificity of transcription priming. A–C, short transcripts (16 or 17 nt) were made by L-A virions in a CTP-omitted transcription reaction. The transcripts were labeled with [α-32P]UTP and primed without (None) or with 20 μm guanine nucleotides or their analogues as indicated above the panels. The products were separated in a 15% acrylamide gel and visualized by autoradiography. Some of the products were pretreated, before electrophoresis, with RNA 5′-polyphosphatase (RPPase), tobacco acid pyrophosphatase (PyroPase), or Terminator 5′-exonuclease (Ter Ex) as indicated below the panels. It should be noticed that the cap analogue (m7GpppG)-primed transcripts migrate as 17-mer because of an extra G at the 5′-end (A, lane 7). Its size can be reduced to 16-mer by tobacco acid pyrophosphatase treatment (B, lane 7). D, the nucleotide sequence of the L-A positive strand at the 5′-end region is shown. The asterisks indicate the positions in the transcripts, which can be labeled with [α-32P]UTP in vitro.
GMP-primed Transcript Has Diphosphate at the 5′-End
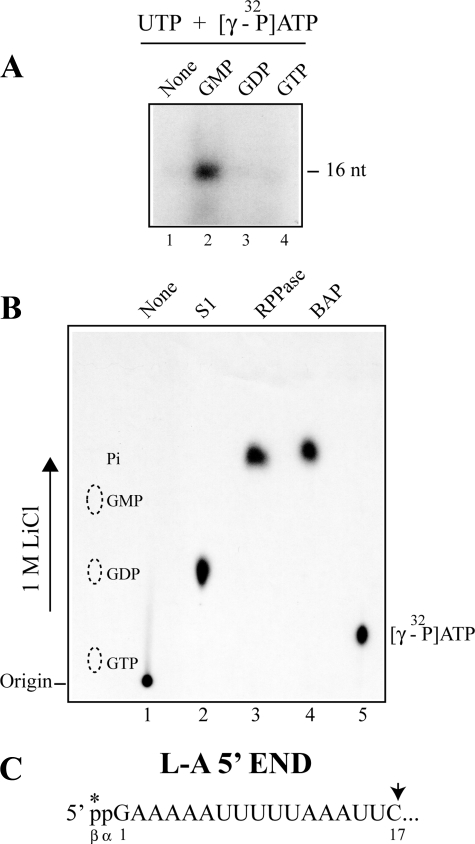
GMP can initiate transcription as efficiently as GDP. The 16-nt product was resistant to Terminator 5′-exonuclease (Fig. 2A, lane 1), indicating that its 5′-end is no longer monophosphorylated. We speculated that the 5′-end of the molecule was modified to diphosphate. To prove this, we carried out the transcription reaction in the presence of [γ-32P]ATP. As shown in Fig. 3A, the 16-nt transcript incorporated radioactivity only when transcription was primed with GMP. The labeled transcript was then isolated from the gel and digested with S1 nuclease. The radioactivity comigrated with GDP in PEI cellulose (Fig. 3B, lane 2), indicating that the GMP-primed transcript has diphosphate at the 5′-end. Furthermore, the transcript released labeled inorganic phosphate when treated with bacterial alkaline phosphatase (Fig. 3B, lane 4) or RNA 5′-polyphosphatase (Fig. 3B, lane 3). Because the latter enzyme cannot act on 5′-monophosphorylated RNA, this result clearly demonstrated that the labeled phosphate derived from [γ-32P]ATP was present in the β-position of the 5′-diphosphate in the transcript. Furthermore, because the GDP-primed transcript was not labeled, this result indicates that the labeled β-phosphate was acquired by a kinase reaction but not by a simple exchange reaction.
FIGURE 3.
GMP-primed transcript has diphosphate at the 5′-end, and its β-phosphate is derived from ATP. A, only the GMP-primed transcript can be labeled with [γ-32P]ATP in in vitro transcription. L-A virions were incubated with UTP and [γ-32P]ATP in the presence of 20 μm GMP (lane 2), GDP (lane 3), or GTP (lane 4) or in the absence of guanine nucleotides (lane 1) as control. The 16-nt transcript was separated in a 15% acrylamide gel and visualized by autoradiography. B, GMP-primed 16-nt transcripts described above were isolated from the gel and treated with S1 nuclease (lane 2, S1), RNA 5′-polyphosphatase (lane 3, RPPase), or bacterial alkaline phosphatase (lane 4, BAP) and analyzed on PEI cellulose with 1 m LiCl. Lane 1, no enzyme-treated 16-nt transcripts. Lane 5, [γ-32P]ATP. An autoradiogram of the thin layer is shown. The positions of nonlabeled GTP, GDP, and GMP are shown by the dotted ovals. C, the 5′-end region of L-A transcript is shown. The asterisk indicates that the β-phosphate at the 5′ terminus is derived from the γ phosphate of ATP when transcription is primed with GMP.
Nucleoside Triphosphatase Activity Associated with L-A Virions
The 5′-triphosphate of GTP was converted to diphosphate when the nucleotide was incorporated into the 5′-end of L-A transcript. It suggests that the L-A virion has RNA or nucleoside triphosphatase activity. It is known that the yeast CTL1 gene product possesses RNA triphosphatase activity (35). It localizes in the cytoplasm, but its physiological role is unknown. A ctl1Δ strain is viable and can harbor L-A virus. L-A virions isolated from this mutant had transcriptase activity and synthesized the 16-nt transcript in the absence of CTP (data not shown). The 16-nt transcript labeled with [α-32P]GTP was gel-purified and treated with S1 nuclease. As shown in Fig. 1D (right panel), the 5′-terminal nucleotide released by S1 digestion was GDP, indicating that the transcript had a diphosphate at its 5′ terminus. Therefore, CTL1 is not responsible for the conversion of triphosphate to diphosphate.
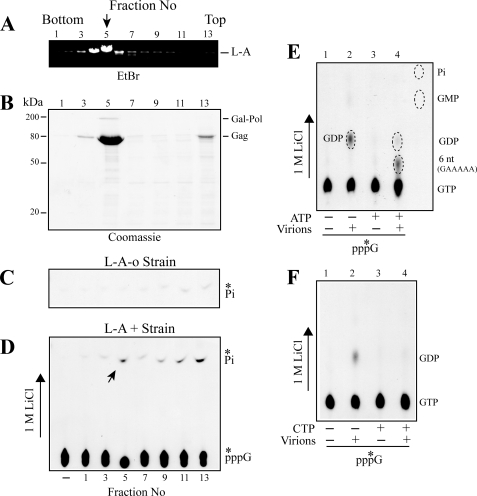
We found a weak nucleoside triphosphatase activity associated with L-A virions in a CsCl gradient (Fig. 4D). A gradient from an L-A-negative strain had no such activity (Fig. 4C). The L-A peak fraction (fraction 5) released a low amount of Pi from ATP or GTP alone, but its activity was stimulated greatly when both nucleotides were present together (supplemental Fig. 1SA). GTP can be replaced by GDP, γ-S-GTP, or β-S-GDP for stimulation, all of which can prime the transcription reaction, whereas a nonpriming nucleotide CTP failed to stimulate Pi release (supplemental Fig. 1S, A and B). A potent inorganic pyrophosphatase activity has been observed to be associated with L-A virions (36). It is likely that the enhanced release of Pi was not caused by nucleoside triphosphatase activity per se but by hydrolysis of inorganic pyrophosphate generated as a byproduct from polymerization of the 6-nt transcript (5′-GAAAAA). Therefore, we carried out the reaction using [α-32P]GTP and directly analyzed the products with PEI cellulose. As shown in Fig. 4E (lane 2), the L-A peak fraction (fraction 5) produced GDP, confirming its triphosphatase activity. In the presence of ATP, the label in GDP decreased, and a new radioactive spot migrating between GDP and GTP appeared (Fig. 4E, lane 4). We confirmed that the latter spot comigrated with the gel-purified 6-nt 5′-GAAAAA transcript. Because this transcript has a single G in the molecule, the results raise the possibility that GDP produced by the nucleoside triphosphatase activity was incorporated into its 5′-end, resulting in the concomitant loss of radioactivity in GDP. When a similar reaction was carried out by substituting ATP with CTP, however, the label in GDP again decreased by the addition of CTP (Fig. 4F, lane 4). Because CTP cannot support polymerization of even short transcripts without ATP and UTP, the results suggest that the decrease of GDP was not caused by its incorporation into RNA transcripts but by the competitive inhibition of the triphosphatase activity by ATP or CTP. It is also possible that unincorporated GDP was reconverted to GTP by virion-associated nucleoside diphosphate kinase (36).
FIGURE 4.
Association of a weak nucleoside triphosphatase activity with L-A virions. A and B, L-A virions were isolated and purified through CsCl gradient centrifugation. Fraction 5 contained the main peak of L-A virions. Fractions 1 and 13 are the bottom and top of the gradient. A, L-A dsRNA was extracted from the gradient, separated in an agarose gel, and visualized with ethidium bromide (EtBr) staining. B, protein in the gradient was separated in a SDS-polyacrylamide gel and visualized by Coomassie brilliant blue staining. Gag and Gag-Pol proteins are indicated. D, CsCl gradient fractions shown in A and B were incubated with [γ-32P]GTP and inorganic phosphate (Pi) released was separated in PEI cellulose with 1 m LiCl. An autoradiogram is shown. The left lane (−) contains no gradient fraction and serves as control. C, same as in D, except that the gradient was made of an extract from an L-A-o strain. Only the upper part of the thin layer is shown. E, [α-32P]GTP was incubated with or without the L-A main peak fraction (fraction 5) (Virions). ATP also was added as indicated below the panel. The products were separated on PEI cellulose and visualized by autoradiography. The positions of GTP, GDP, GMP, inorganic phosphate (Pi), and a 6-nt transcript (GAAAAA) are indicated. F, same as in E, except that ATP was substituted with CTP.
M1 Transcript Has Diphosphate at the 5′-End
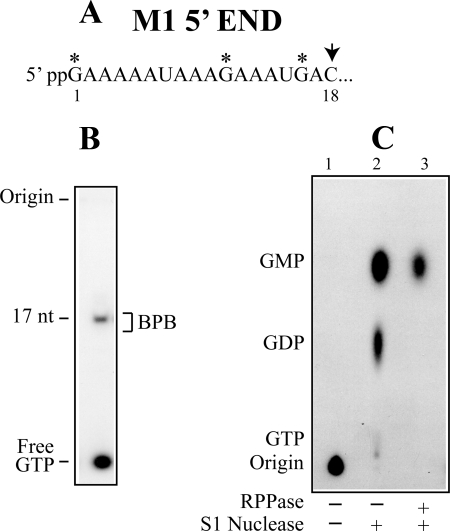
M1, a satellite RNA of L-A virus, utilizes L-A encoded proteins to encapsidate and replicate its RNA genome. M1 can replicate and be maintained in the cell without L-A virus if the viral proteins are provided from a vector. We isolated M1 virions from L-A virus-free cells to avoid the preparation from being contaminated with L-A virus. M1 virions purified through a CsCl gradient synthesized full-length M1 transcripts in the presence of four NTPs and produced a short 17-nt transcript when CTP was omitted (Fig. 5B). The 17-nt transcript labeled with [α-32P]GTP was purified through an acrylamide gel, treated with S1 nuclease, and analyzed by PEI cellulose chromatography. As shown in Fig. 5C, lane 2, the transcript produced two spots corresponding to GDP and GMP, and their ratio was 1:2.1. In addition to the 5′-terminal G, the 17-nt transcript has two additional Gs (Fig. 5A), and these internal Gs are expected to be released as GMP upon S1 digestion. The results, therefore, suggest that GDP was derived from the 5′-end of the transcript and that the transcript thus has diphosphate at the 5′-end. To confirm that GDP was derived from the 5′-end, the 17-nt transcript was treated with RNA 5′-polyphosphatase, ethanol-precipitated, and then digested with S1 nuclease. As shown in Fig. 5C (lane 3), now, the transcript only produced GMP, indicating that GDP was really released from the 5′-end.
FIGURE 5.
M1 transcript also has diphosphate at the 5′-end. A, the 5′-end region of M1 transcript. The first C appears at position 18. M1 virions makes a 17-nt transcript in a CTP-omitted transcription reaction as shown in B. In this gel system (15% acryalmide), the 17-nt transcript comigrates with bromphenol blue (BPB). The asterisks in A indicate three positions in the 17-nt transcript, which can be labeled with [α-32P]GTP. Upon S1 nuclease digestion, the two internal Gs in the transcript will be released as GMP, while the 5′-terminal G would be GDP if the transcript has diphosphate at the 5′-end. C, the [α-32P]GTP-labeled M1 17-nt transcript was isolated from an acrylamide gel and treated with S1 nuclease. The digest was analyzed on PEI cellulose with 1 m LiCl (lane 2). The ratio of radioactive label in GDP and GMP quantified by phosphorimaging was 1:2.1. Lane 1 indicates the non-S1 digested sample. In lane 3, the 17-nt transcript was first treated with RNA 5′-polyphosphatase to eliminate the β-phosphate at the 5′-end. After ethanol precipitation, the transcript was digested with S1 nuclease. RPPase, RNA 5′-polyphosphatase.
5′-Diphosphorylated L-A Transcript Is Resistant to Ski1p 5′-Exonuclease
The L-A transcript has a diphosphate at the 5′-end. We wondered whether the viral transcript is susceptible to Ski1p 5′-exonuclease. A. Stevens purified this enzyme and performed an extensive biochemical analysis on it. It is now well known that the nuclease digests 5′-monophosphorylated RNA as substrate but works poorly on 5′-triphosphorylated or capped RNA (37). However, we did not find information on its reactivity toward 5′-diphosphorylated RNA. We, therefore, purified Ski1p as a TAP fusion protein from yeast cells by one-step purification using calmodulin-affinity resin following the method described in Ref. 31. Fig. 6A shows the elution profile of Ski1p from the affinity resin visualized by silver staining. RNA substrates with different 5′ termini but having the same nucleotide sequence were made in vitro using T7 RNA polymerase and served to test the specificity of the purified enzyme. As shown in Fig. 6B, the protein efficiently digested 5′-monophosphorylated RNA (lane 4) but hardly worked on 5′-triphosphorylated (lane 2) or capped RNA (lane 6), confirming the specificity ascribed to the enzyme. We made the 16-nt L-A transcript in vitro by incubating L-A virions in a CTP-omitted transcription reaction and treated it with purified Ski1p. As shown in Fig. 6C, the transcript was largely resistant to Ski1p (lane 2) but became completely digested with the enzyme after pretreatment with RNA 5′-polyphosphatase (lane 4), the same digestion pattern observed with the commercially available Terminator 5′-exonuclease (Fig. 1C). From these results, we concluded that L-A viral transcript made in vitro is a poor substrate for the Ski1p 5′-exonuclease.
DISCUSSION
In this work, we have demonstrated that L-A transcripts made in vitro by L-A virions are 5′-diphosphorylated. More significantly, irrespective of whether transcription was primed by GTP or GMP, the guanine nucleotide incorporated at the 5′ terminus was diphosphorylated. Thus, the virus deliberately synthesizes RNA transcript with 5′-diphosphate. M1, a satellite RNA of L-A virus, utilizes Gag and Gag-Pol of the helper virus to encapsidate and replicate its RNA genome. Isolated M1 virions likewise synthesized transcripts with 5′-diphosphate. We also have demonstrated that the L-A transcript made in vitro is a poor substrate for the Ski1p 5′-exonuclease. When the 5′-end of the transcript was converted from diphosphate to monophosphate, however, Ski1p efficiently digested it. Previously, we observed that the deletion of SKI1 resulted in a 5–10-fold increase in the copy number of L-A virus and that overexpression of Ski1p cured L-A virus from the cell at a high frequency (22). Because Ski1p is the major exonuclease involved in mRNA degradation in the budding yeast (19), our results suggest that the virus makes its transcript 5′-diphosphorylated apparently to protect it from a direct attack of the potent Ski1p. Our results also suggest that the host cell possesses an RNA diphosphatase activity in the cytoplasm to convert the viral transcript from diphosphate to monophosphate at the 5′-end. Consistent with this view is that in ski1Δ strains L-A transcripts accumulate in large amounts and can be detected easily by Northern hybridization. Furthermore, these transcripts are 5′-monophosphorylated, as judged from their susceptibility to Terminator 5′-exonuclease or purified Ski1p.3 In wild type strains, these transcripts are quickly digested by Ski1p and do not accumulate in large quantities. These observations suggest that in a hypothetical RNA diphosphatase-negative strain, viral transcripts will be stabilized because of the inability of Ski1p to directly attack them. Thus, these mutants are expected to exhibit a superkiller phenotype when harboring the satellite M1 dsRNA.
Another possible role for the 5′-diphosphate status of the viral transcript is on translation and is more speculative. Here, we demonstrated that the cap analogue m7GpppG can be used to prime transcription in vitro by L-A virions. Barbone et al. (38) reported that viral transcripts made in vitro in the presence of the cap analogue are translated in a yeast cell-free translation system 8–15-fold more efficiently than the ones made in its absence, although they failed to directly prove its incorporation into viral transcripts. These data together suggest that viral transcripts with 5′-diphosphate are poor templates for translation but that the incorporation of the cap structure at the 5′-end converted them to more efficient templates. In the mRNA-capping reaction, RNA guanylyltransferase transfers GMP to the diphosphate end of pre-mRNA through a covalent enzyme-GMP intermediate. It has been known that L-A Gag protein binds to 5′-capped mRNAs and forms a covalent bond with m7GMP through His-154 while releasing the mRNA body (25, 39). This raises the possibility that the m7GMP attached to His-154 can be transferred to the diphosphate end of an emerging viral transcript, thus forming a 5′-cap structure on the RNA. A 5′-capped species of viral RNA has not been detected yet (40) presumably because the efficiency of transfer is low. This cap-snatching hypothesis originally was described in Ref. 25. In the conventional capping reaction, guanylyltransferase forms a covalent bond with GMP through Lys but not His. However, it recently has been demonstrated that in the capping reaction of vesicular stomatitis virus, L protein forms an intermediate by covalently binding the 5′-monophosphorylated pre-mRNA through His and transfers the bound pre-mRNA to GDP (41). It appears that members of the alphavirus-like superfamily also utilize His to form a phosphamide bond with m7GMP and transfer the m7GMP moiety to the 5′-diphosphate end of a viral transcript during the capping reaction (42–44). L-A Gag with mutations at His-154 failed to form a covalent bond with m7GMP but did not affect replication and maintenance of M1 (25). Therefore, those reactions, including transcription ((+) strand RNA synthesis), encapsidation of the (+) strand into the virion, and (−) strand synthesis inside the virion, are not affected by the mutations. Furthermore, M1 virions with a mutant Gag (Arg-154) synthesize M1 transcripts with 5′-diphosphate as wild type virions do.3 Interestingly, however, the expression of M1-encoded killer toxin is severely compromised in vivo by the mutations at His-154 (25), suggesting a specific role of His-154 on translation. Therefore, these data fit well within the framework of this capping hypothesis.
We observed a weak nucleoside triphosphatase activity associated with L-A virions. However, it is not clear whether this activity is involved in the synthesis of the 5′-diphosphorylated transcript primed with GTP. RNA polymerase may preferentially incorporate GDP generated by nucleoside triphosphatase over GTP to prime transcription. The polymerase itself may be able to convert GTP to GDP during transcription initiation. Finally, the conversion may occur post-transcriptionally on the nascent transcript during its release from the virion through a pore in the capsid. Our data indicate that the host CTL1 RNA triphosphatase is not involved in this process. The conversion from triphosphate to diphosphate, however, is not essential for the initiation of transcription because a cap analogue (m7GpppG) or γ-S-GTP could prime transcription efficiently. GDP was a better substrate to prime transcription than GTP. This raises the possibility that the conversion of GTP to GDP is slow, and this slow process might be responsible for the low efficiency of the GTP-primed reaction. However, because γ-S-GTP could prime transcription as efficiently as GTP, this possibility is unlikely. We demonstrated that GMP-primed transcript also had diphosphate at the 5′-end, the β-phosphate of which was derived from the γ-phosphate of ATP. This kinase reaction could occur before, during or after transcription initiation, as discussed above for the conversion from triphosphate to diphosphate. It is noteworthy to mention that, when M1 transcription was primed by GMP in the presence of ATP and UTP, we observed not the 10-nt transcript but a longer 17-nt species that had incorporated two internal Gs before termination due to the lack of CTP.3 This suggests that the virion sequentially converted GMP to GDP and then to GTP at the expense of ATP and used this newly made GTP to synthesize the 17-nt species. It is thus conceivable that the GDP produced in this process was incorporated at the 5′-end of the viral RNA during transcription initiation. Georgopoulos et al. (36) has observed that L-A virions are associated with numerous nucleotide-metabolizing enzyme activities, including nucleoside triphosphate phosphotransferase, nucleoside diphosphate and monophosphate kinases, in addition to inorganic pyrophosphatase activity, and speculated that these enzymes may allow the virus to utilize nucleotide pools distinct from those used in host cell transcription. However, the results shown in this work raise the possibility that some of the enzymatic activities associated with the virion are directly involved in the metabolism of phosphate group at the 5′ termini of viral transcripts. L-A virus encodes only two proteins, Gag and Gag-Pol. Gag has decapping activity of mRNA and the Pol domain of the fusion protein has RNA-dependent RNA polymerase motifs and also is involved in packaging of viral RNA into virions (45). It is an open question whether the viral proteins further possess these phosphate-metabolizing enzymatic activities. Alternatively, the virus may recruit host enzymes and specifically incorporate them into the virion to process the 5′-ends during transcription.
Supplementary Material
Acknowledgment
We thank Dr. Reed B. Wickner for providing the L-A-free killer strain.
This work was supported by Grant BFU2007-60057 from the Spanish Ministry of Education and Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1S.
T. Fujimura and R. Esteban, unpublished results.
- dsRNA
- double-stranded RNA
- nt
- nucleotides
- BAP
- bacterial alkaline phosphatase
- TAP
- tandem affinity purification
- PEI
- polyethyleneimine
- nt
- nucleotide
- Pol
- polymerase.
REFERENCES
- 1.Furuichi Y., Shatkin A. J. (2000) Adv. Virus Res. 55, 135–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuman S. (2001) Prog. Nucleic Acid Res. Mol. Biol. 66, 1–40 [DOI] [PubMed] [Google Scholar]
- 3.Koonin E. V., Moss B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3283–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehwinkel J., Tan C. P., Goubau D., Schulz O., Pichlmair A., Bier K., Robb N., Vreede F., Barclay W., Fodor E., Reis e Sousa C. (2010) Cell 140, 397–408 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt A., Schwerd T., Hamm W., Hellmuth J. C., Cui S., Wenzel M., Hoffmann F. S., Michallet M. C., Besch R., Hopfner K. P., Endres S., Rothenfusser S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickner R. B. (2007) in Fields Virology (Knipe D. M., Howley P. M. eds) 5th Ed., pp. 737–768, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 7.Fujimura T., Wickner R. B. (1988) Cell 55, 663–671 [DOI] [PubMed] [Google Scholar]
- 8.Icho T., Wickner R. B. (1989) J. Biol. Chem. 264, 6716–6723 [PubMed] [Google Scholar]
- 9.Dinman J. D., Icho T., Wickner R. B. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng R. H., Caston J. R., Wang G. J., Gu F., Smith T. J., Baker T. S., Bozarth R. F., Trus B. L., Cheng N., Wickner R. B., Steven A. C. (1994) J. Mol. Biol. 244, 255–258 [DOI] [PubMed] [Google Scholar]
- 11.Castón J. R., Trus B. L., Booy F. P., Wickner R. B., Wall J. S., Steven A. C. (1997) J. Cell Biol. 138, 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimura T., Esteban R., Wickner R. B. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 4433–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tipper D. J., Bostian K. A. (1984) Microbiol. Rev. 48, 125–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toh-E A., Guerry P., Wickner R. B. (1978) J. Bacteriol. 136, 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridley S. P., Sommer S. S., Wickner R. B. (1984) Mol. Cell. Biol. 4, 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker R., Song H. (2004) Nat. Struct. Mol. Biol. 11, 121–127 [DOI] [PubMed] [Google Scholar]
- 17.Wilusz C. J., Wormington M., Peltz S. W. (2001) Nat. Rev. Mol. Cell Biol. 2, 237–246 [DOI] [PubMed] [Google Scholar]
- 18.Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. (1997) Cell 91, 457–466 [DOI] [PubMed] [Google Scholar]
- 19.Anderson J. S., Parker R. P. (1998) EMBO J. 17, 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Widner W. R., Wickner R. B. (1993) Mol. Cell. Biol. 13, 4331–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Searfoss A. M., Wickner R. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9133–9137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteban R., Vega L., Fujimura T. (2008) J. Biol. Chem. 283, 25812–25820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruenn J., Keitz B. (1976) Nucleic Acids Res. 3, 2427–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nemeroff M. E., Bruenn J. A. (1987) J. Biol. Chem. 262, 6785–6787 [PubMed] [Google Scholar]
- 25.Blanc A., Ribas J. C., Wickner R. B., Sonenberg N. (1994) Mol. Cell. Biol. 14, 2664–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickner R. B. (1980) Cell 21, 217–226 [DOI] [PubMed] [Google Scholar]
- 27.Fujimura T., Wickner R. B. (1992) J. Biol. Chem. 267, 2708–2713 [PubMed] [Google Scholar]
- 28.Fujimura T., Esteban R., Esteban L. M., Wickner R. B. (1990) Cell 62, 819–828 [DOI] [PubMed] [Google Scholar]
- 29.Dente L., Sollazzo M., Baldari C., Cesareni G., Cortese R. (1985) in DNA Cloning (Glover D. M. ed) Vol. 1, pp. 101–107, IRL Press, London [Google Scholar]
- 30.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 31.Cheng C. P., Serviene E., Nagy P. D. (2006) J. Virol. 80, 2631–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 33.Pellegrini O., Mathy N., Condon C., Bénard L. (2008) Methods Enzymol. 448, 167–183 [DOI] [PubMed] [Google Scholar]
- 34.Esteban R., Fujimura T., Wickner R. B. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4411–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez C. R., Takagi T., Cho E. J., Buratowski S. (1999) Nucleic Acids Res. 27, 2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgopoulos D. E., Leibowitz M. J. (1987) Yeast. 3, 117–129 [DOI] [PubMed] [Google Scholar]
- 37.Stevens A. (2001) Methods Enzymol. 342, 251–259 [DOI] [PubMed] [Google Scholar]
- 38.Barbone F. P., Williams T. L., Leibowitz M. J. (1992) Virology. 187, 333–337 [DOI] [PubMed] [Google Scholar]
- 39.Blanc A., Goyer C., Sonenberg N. (1992) Mol. Cell. Biol. 12, 3390–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masison D. C., Blanc A., Ribas J. C., Carroll K., Sonenberg N., Wickner R. B. (1995) Mol. Cell. Biol. 15, 2763–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino T., Yadav S. P., Banerjee A. K. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3463–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahola T., Laakkonen P., Vihinen H., Kääriäinen L. (1997) J. Virol. 71, 392–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahola T., Ahlquist P. (1999) J. Virol. 73, 10061–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y. L., Han Y. T., Chang Y. T., Hsu Y. H., Meng M. (2004) J. Virol. 78, 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujimura T., Ribas J. C., Makhov A. M., Wickner R. B. (1992) Nature 359, 746–749 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.