Abstract
The eukaryotic DNA replication protein Mcm10 associates with chromatin in early S-phase and is required for assembly and function of the replication fork protein machinery. Xenopus laevis (X) Mcm10 binds DNA via a highly conserved internal domain (ID) and a C-terminal domain (CTD) that is unique to higher eukaryotes. Although the structural basis of the interactions of the ID with DNA and polymerase α is known, little information is available for the CTD. We have identified the minimal DNA binding region of the XMcm10-CTD and determined its three-dimensional structure by solution NMR. The CTD contains a globular domain composed of two zinc binding motifs. NMR chemical shift perturbation and mutational analysis show that ssDNA binds only to the N-terminal (CCCH-type) zinc motif, whose structure is unique to Mcm10. The second (CCCC-type) zinc motif is not involved in DNA binding. However, it is structurally similar to the CCCC zinc ribbon in the N-terminal oligomerization domain of eukaryotic and archaeal MCM helicases. NMR analysis of a construct spanning both the ID and CTD reveals that the two DNA binding domains are structurally independent in solution, supporting a modular architecture for vertebrate Mcm10. Our results provide insight in the action of Mcm10 in the replisome and support a model in which it serves as a central scaffold through coupling of interactions with partner proteins and the DNA.
Keywords: DNA-binding Protein, DNA Replication, NMR, Protein DNA-interaction, Protein Motifs, Protein Structure
Introduction
DNA synthesis at the eukaryotic replication fork requires coordination of enzymatic activities through a network of interactions within the dynamic multiprotein replisome. During replication initiation, the individual components of the replisome are assembled at each origin of replication in a sequential and regulated fashion to ensure that the genome is copied only once and at the proper time during each cell cycle. During G1, each origin is licensed for replication through origin recognition complex (ORC)3-dependent loading of a pre-replicative complex (pre-RC), which consists of the ORC, Cdc6, Cdt1, and the Mcm2–7 helicase (reviewed in Ref. 1). As cells transition into S-phase, Mcm10 is recruited to the origin (2, 3) and the pre-RC is activated through a series of phosphorylation events by cyclin- and Dbf4-dependent kinases (CDK and DDK) (4, 5). Chromatin association of Mcm10 is required for loading Cdc45 and GINS (2, 6), which function with Mcm2–7 as a Cdc45-GINS-Mcm2–7 (CMG) helicase complex (7–9). The mechanism of initial denaturation of duplex DNA at the origin is unknown, but is signaled by the presence of single-stranded (ss) DNA-binding protein, replication protein A (RPA) (7, 10–12). DNA synthesis is initiated by DNA polymerase α-primase (pol α), which associates with chromatin and the CMG complex via several factors, including Mcm10, Cdc45, RPA, And-1/Ctf4, and RecQL4 (3, 6, 13–17). Association of replicative DNA polymerases δ and ϵ, along with RPC loading of PCNA completes the replisome and initiates the elongation process (reviewed in Ref. 18).
Mcm10 is essential for the formation of an active replication fork (19) and participates in numerous interactions with components of the replisome (8). Mcm10 interacts with single-stranded and duplex DNA, consistent with a possible role as a protein-DNA mediator during origin melting (20–22). In early S-phase Mcm10 interacts with subunits of Mcm2–7, Cdc45, and is necessary for the assembly of the CMG helicase complex (8, 17, 19, 23–26). In addition, Mcm10 interacts with DNA pol α, preventing its degradation in vivo and possibly serving to recruit pol α to the replisome (3, 21, 27–29). Mcm10 interactions with DNA, Mcm2–7, and pol α suggest it may function as a scaffold to physically link helicase and polymerase machinery within the replisome during the stages of replication initiation and primer elongation (3, 30).
Despite the identification of Mcm proteins from the same genetic screen for mutants defective in minichromosome maintenance (31), Mcm10 is evolutionarily distinct from Mcm2–7, and no sequence or structural homology has been identified between them (32). We previously established that the 95-kDa XMcm10 protein contains at least three structured domains: an N-terminal coiled-coil domain believed to facilitate protein oligomerization, and zinc-binding internal (ID) and C-terminal (CTD) domains that both bind DNA and pol α (21). Interestingly, an electron micrograph structure of human Mcm10 revealed a hexameric ring structure (33), although no other reports of Mcm10 hexamerization exist in the literature. To date, the only high resolution structural information available for Mcm10 exists for the highly conserved ID (34, 35).
The vertebrate orthologs of Mcm10 contain a second DNA and pol α binding domain at the extreme C terminus (21). The sequence of the vertebrate CTD, not identifiable in yeast or land plants, contains putative winged-helix and zinc binding domains that are predicted to facilitate DNA binding. To better understand the role of the vertebrate CTD and its interactions with DNA and other replication proteins, we mapped the DNA binding site to the zinc-coordinating region of XMcm10-CTD and determined the NMR solution structure of a globular domain containing this activity. Structural and mutational data support a separation of function for the two CTD zinc motifs comprising this domain. NMR studies of a tandem ID plus CTD construct revealed the modular architectural organization. These results support a model in which Mcm10 functions as a scaffold through essential protein and DNA interactions within the replisome.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The cDNA for all CTD-containing fragments were PCR-amplified from a previously described plasmid (2) and ligated into a modified pET-32a (Novagen) expression vector to generate N-terminal thioredoxin (Trx)-His6 fusion proteins. XMcm10230–860 was purified as previously described (34). XMcm10755–842, XMcm10690–842, XMcm10596–794, XMcm10596–757, and XMcm10596–860 proteins were overexpressed in Escherichia coli BL21(DE3) cells in LB medium supplemented with 100 μg/ml ampicillin, 7.5 μm ZnSO4, and 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 3 h at 37 °C. Isotopically enriched Mcm10 samples for NMR were overexpressed in M9 minimal medium supplemented with 15NH4Cl and/or [13C6]glucose (Cambridge Isotope Laboratories) as the sole sources of nitrogen and/or carbon. Cells were resuspended in 50 mm Tris-HCl (pH 7.5), 500 mm NaCl, and 10% glycerol (Buffer L) and lysed with an EmulsiFlex-C3 homogenizer (Avestin). Cell lysates were centrifuged at 35,000 × g for 20 min. Trx-His6-Mcm10 proteins were purified from the supernatant by nickel-nitrilotriacetic (Ni-NTA, Qiagen) affinity chromatography using a Buffer L wash and Buffer L/250 mm imidazole elution. Fractions were visualized by SDS-PAGE and Coomassie Blue staining. Those containing Mcm10 were pooled and incubated overnight at 4 °C with PreScission Protease (GE Healthcare) at a 1:50 protease:Mcm10 mass ratio to remove the Trx-His6 affinity tag. After cleavage, this mixture was diluted 10-fold in 50 mm Tris-HCl (pH 7.5), and 10% glycerol (Buffer A) and passed through a 5-ml Q Sepharose HP anion exchange column directly onto a 5-ml SP Sepharose HP cation column (GE Healthcare). Proteins were eluted using a linear gradient from Buffer A/0.1 m NaCl to Buffer A/1 m NaCl. Fractions containing Mcm10 proteins were pooled, concentrated using an Amicon spin concentrator (Millipore) and purified over a 320-ml Superdex 200 gel filtration preparative grade column (GE Healthcare) that had been equilibrated in 20 mm Tris-HCl (pH 7.5), 100 mm NaCl, 5 mm β-mercaptoethanol, and 5% glycerol.
NMR Spectroscopy
All NMR data were collected at 25 °C using Bruker DRX600 and DRX800 spectrometers equipped with cryoprobes. Data were processed with Topspin 2.0b and further analyzed using Sparky (36). All XMcm10 samples were buffer exchanged using an Amicon Spin Concentrator (Millipore) into 25 mm NaH2PO4 (pH 6.5) in 90% H2O/10% D2O and 100 mm NaCl (XMcm10690–842 and XMcm10755–842) or 175 mm NaCl (XMcm10230–860). XMcm10690–842 samples were concentrated to 305 μm for {1H}-15N heteronuclear NOE experiments and to 190 μm for 15N-1H HSQC titrations. XMcm10755–842 was concentrated to 920 μm for structure determination experiments and to 300 μm for HSQC titrations. XMcm10230–860 samples for 15N-1H TROSY-HSQC were at a concentration of 80 μm.
Backbone resonance assignments for XMcm10690–842 and XMcm10755–842 were obtained using a combination of two-dimensional 15N-1H HSQC and 3D HNCACB, CBCA(CO)NH and HNCO spectra. Side chain aliphatic resonances were assigned using H(CCCO)NH, (H)CC(CO)NH, and HBHANH three-dimensional experiments. Aromatic side chain resonances were assigned using (HB)CB(CGCD)HD and two-dimensional homonuclear COSY, TOCSY and NOESY (Tm = 120 ms) experiments. NOE-based distance restraints were assigned from a homonuclear two-dimensional NOESY experiment, as well as three-dimensional 13C-NOESY-HSQC and 15N-NOESY-HSQC (Tm = 120 ms) experiments using 13C,15N-enriched samples. Steady-state {1H}-15N heteronuclear NOE data were collected with and without 3 s of 1H saturation and 7 s of recycle delay. NOEs for 144 resolved resonances were determined using the ratio of signal intensities with and without 1H saturation. Additional details on the acquisition parameters are provided in supplemental Table S1.
Structure Calculation
Starting structures were determined using CYANA (37). Seven iterative cycles of calculations were carried out starting with a set of manually assigned NOEs. 100 structures were created per cycle with backbone torsion angle restraints obtained from 1H, 13Cα, 13Cβ, and 15N chemical shifts using TALOS with a minimum range of ±35°. The 50 structures with lowest values of the CYANA target function were further refined using restrained molecular dynamic simulations in AMBER (38) following published protocols (39). The twenty conformers with the lowest restraint violation energy were selected as the final representative ensemble. PROCHECK-NMR and MolProbity were used to assess the quality of the final ensemble (40, 41). The final ensemble and distance restraints have been deposited in the PDB under accession code 2KWQ.
DNA Binding
Mcm10 DNA binding affinities were measured by following the increase in fluorescence polarization as the protein was added to DNA oligonucleotides labeled at the 3′-end with 6-carboxyfluorescein (FAM, Integrated DNA Technologies). Binding data for CTD deletion mutants shown in Fig. 3C were measured using d(ATGGTAGGCAACCAT)-FAM. Oligonucleotides used to determine the length dependence of DNA binding are shown in supplemental Fig. S5. Mcm10 proteins were added over a concentration range of 0–20 μm to a solution of 50 nm fluorescein-DNA, and polarized fluorescence intensities were measured at excitation and emission wavelengths of 495 and 538 nm, respectively. The experiments were performed at 25 °C in 25 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 5% glycerol. Each measurement was made in triplicate and the apparent dissociation constants (Kd) were calculated by fitting the binding curve using a two-state binding model using Kaleidagraph 3.6 (Synergy Software).
FIGURE 3.
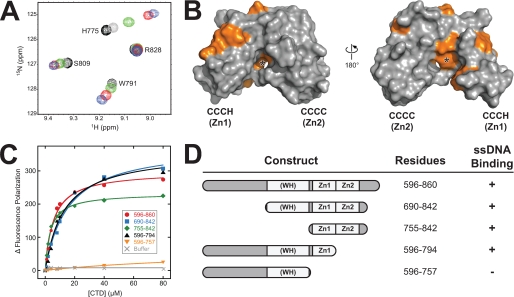
ssDNA binding to the CCCH zinc motif. A, overlays of a representative region of the 15N-1H HSQC spectra of 15N-XMcm10755–842 in the absence (black) and presence of 0.5 (gray), 1 (green), 2 (red), and 4 (blue) fold molar excess of ssDNA. B, residues perturbed by ssDNA binding from the HSQC titration are colored orange against the molecular surface of XMcm10755–842. The asterisk denotes the position of Phe776. C, ssDNA binding to CTD deletion constructs was monitored by the change in fluorescence polarization as protein was added to fluorescein (FAM)-labeled d(ATGGTAGGCAACCAT). Addition of buffer only to FAM-DNA is shown as gray Xs. Data shown are from one representative experiment and were reproduced in triplicate. D, schematic representation of the data shown in panel C.
Chemical shift perturbation experiments with CTD constructs were performed using d(ATGGTAGGCAACCAT) at Mcm10:DNA molar ratios of 1:0, 1:0.5, 1:1, 1:2, and 1:4. Chemical shift perturbations were quantified using the equation Δδave = (((Δδ1H)2 + (Δδ15N/5)2)/2)1/2. Values of Δδave greater than one standard deviation from the mean were considered significant.
RESULTS
Structural Characterization of the C-terminal Domain of Mcm10
We previously identified residues 596–860 of Xenopus laevis Mcm10 as a structured C-terminal domain (CTD) containing two Zn2+ ions and encompassing both DNA and pol α binding activities (21). Hence structural studies were initiated using intact XMcm10596–860. However, this and a library of constructs spanning this region proved refractory to crystallization presumably as a result of flexible and/or disordered polypeptide segments within this domain. To test this hypothesis, limited proteolysis experiments were performed on XMcm10596–860. A stable ∼20-kDa fragment was observed (supplemental Fig. S1), which was identified to span residues 690–842 by MALDI-TOF mass spectrometry and Edman degradation. Sequence analysis of this subdomain revealed it contains a putative winged-helix (residues 692–755) and two clusters of conserved cysteine and histidine residues (residues 768–821) previously predicted to coordinate Zn2+ ions (Fig. 1) (21, 33). No structural motifs and little secondary structure were predicted within residues 596–620. Importantly, purified XMcm10690–842 bound ssDNA with roughly the same affinity as the larger XMcm10596–860 construct, as measured by a fluorescence polarization assay (see below). Thus, XMcm10690–842 appeared to be a stable subdomain of the CTD that retains DNA binding activity.
FIGURE 1.
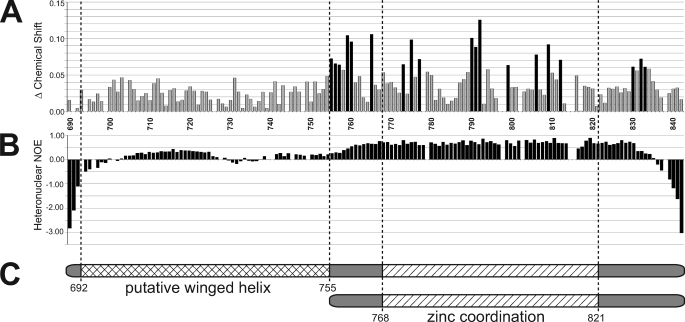
Mcm10-CTD contains a DNA binding subdomain. A, chemical shift differences between 15N-XMcm10690–842 alone and in the presence of a 4-fold molar excess of ssDNA are plotted for each residue. Residue numbers are displayed along the x axis. Black bars represent chemical shift differences greater than 1 standard deviation from the mean. B, {1H}-15N heteronuclear NOE values, which correlate with the degrees of freedom for individual backbone residues. Positive values indicate more structural restraint, while negative values indicate more mobility. C, schematic of the two CTD constructs used in structural studies, XMcm10690–842 (top) and XMcm10755–842 (bottom) and the locations of putative motifs.
The structural features of XMcm10690–842 were investigated by heteronuclear NMR spectroscopy (Fig. 1), and to this end, over 90% of the backbone amide resonances were assigned. Using these backbone assignments, we first probed the DNA binding region using an NMR chemical shift perturbation experiment. DNA binding was monitored by perturbation of the 15N-1H HSQC spectrum as unlabeled ssDNA was titrated into 15N-enriched XMcm10690–842. Signals from 19 residues showed a significant perturbation in the HSQC spectra in response to the binding of ssDNA, indicative of a specific binding event (Fig. 1A). All of the perturbed residues localized to the putative Zn2+ binding region within residues 755–833. No signals corresponding to the putative winged helix were affected from addition of even a 4-fold molar excess of ssDNA (Fig. 1A). Thus, DNA binding in the CTD is localized exclusively to the zinc binding region.
We next assayed the relative flexibility of the backbone of XMcm10690–842 using {1H}-15N heteronuclear NOE experiments (Fig. 1B). Strikingly, residues 758–834 encompassing the zinc binding region generated an average NOE value of 0.66, indicative of a well-folded, globular structured domain. In contrast, the putative winged-helix region (residues 690–757) had an average NOE of 0.14, indicative of much higher backbone flexibility. Taken together, the high NOE values and chemical shift perturbation data demonstrate that the zinc binding region is a well-folded, DNA binding motif (Fig. 1C). The high sequence conservation and eight invariant cysteine and histidine residues in the C-terminal 100 residues of Mcm10 from higher eukaryotes suggest XMcm10690–842 contains a functionally important domain suitable for structure determination.
Solution NMR Structure of XMcm10755–842
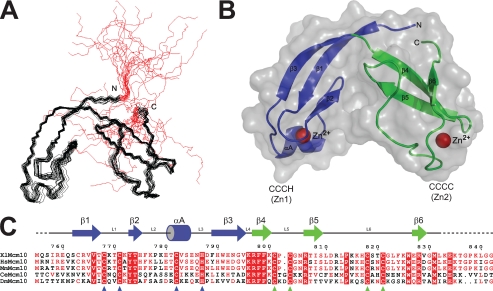
The solution structure of XMcm10755–842, a construct encompassing the zinc binding region, was determined by multidimensional heteronuclear NMR (42). Nearly complete resonance assignments were obtained for this construct using standard double and triple resonance experiments, along with an (HB)CB(CGCD)HD spectrum that was acquired to assign sidechain resonances of the aromatic residues. NMR structures were generated using a combination of CYANA distance geometry calculations (37) and restrained molecular dynamics refinements in AMBER (38). Because of the large number of long-range restraints identified, a high precision structure was obtained with low total violation energies, no distance violations greater than 0.2 Å, no torsion angle violations greater than 5°, and low molecular energies (Table 1). The 20 conformers with the lowest restraint violation energy were selected for the final representative ensemble and are shown in Fig. 2A.
TABLE 1.
Structural statistics for XMcm10755–842
| Restraints for calculation | |
| Total NOE restraints | 2436 |
| Intraresidue | 487 |
| Sequential | 632 |
| Medium range | 348 |
| Long range | 969 |
| Dihedral angle restraint | 28 |
| Constraint violations, mean ± S.D. | |
| Distance violations | |
| 0.1 Å < d < 0.2 Å | 1.79 ± 1.03 |
| d > 0.2 Å | 0 |
| Average maximum distance violations (Å) | 0.14 ± 0.02 |
| Torsion angle violations > 5.0° | 0 |
| Average maximum torsion angle violations (°) | 0 |
| AMBER energies, mean ± S.D. (kcal mol−1 | |
| Restraint | 2.25 ± 0.40 |
| van der Waals | −645 ± 11 |
| Total molecular | −2804 ± 11 |
| Precision, RMSD from mean (Å), ordered regiona | |
| Backbone | 0.63 ± 0.21 |
| All heavy atoms | 1.04 ± 0.15 |
| Ramachandran statisticsb (%) | |
| Most favored | 86.0 |
| Additionally allowed | 13.1 |
| Generously allowed | 0.4 |
| Disallowed | 0.4 |
a Residues 761–832.
b PROCHECK nomenclature.
FIGURE 2.
The structure of the dual zinc cluster in XMcm10-CTD. A, backbone superposition of the twenty lowest energy NMR structures of XMcm10755–842. Regions in red correspond to unrestrained terminal regions, which have been omitted from subsequent figures. B, XMcm10755–842 depicted as a ribbon and superimposed onto a transparent gray molecular surface. Individual CCCH and CCCC zinc motifs are colored blue and green, respectively, and Zn2+ ions are depicted as red spheres. C, sequence alignment of the C-terminal region of Mcm10 proteins from X. laevis (Xl), Homo sapiens (Hs), Mus Musculus (Mm), Caenorhabditis elegans (Ce), and Drosophila melanogaster (Dm). The secondary structure is shown schematically above the sequence, and Zn2+-coordinating residues are highlighted with triangles below.
Because zinc had been previously shown to bind to this domain, we examined the NOE-based structures to determine if the zinc coordinating residues could be identified. In fact, the eight invariant cysteine/histidine residues predicted to coordinate zinc ions were positioned into two distinct clusters in the structure (supplemental Fig. S2). Moreover, the side chains within each cluster were posed in a tetrahedral geometry consistent with zinc binding. In addition, we observed that proton and carbon chemical shifts for many of these conserved residues deviated from the expected range represented in the Biological Magnetic Resonance Bank. Based on these observations and on our previous identification of two zinc atoms binding in the CTD (21), we incorporated zinc ions and imposed distance restraints in both clusters in the final stages of structure refinement. As anticipated, these additional restraints significantly increased the precision of the structure in and around the Zn2+ sites.
The three-dimensional structure of XMcm10755–842 is comprised of two independent zinc motifs tethered closely together in the shape of a V, with the Zn2+ ions bound at the ends of each arm (Fig. 2B). Superposition of the individual zinc motifs within the ensemble of NMR structures revealed that there is a high degree of similarity and that each is slightly better defined than the entire globular domain (supplemental Fig. S3). The N-terminal CCCH zinc motif (Zn1) spans residues 755–795 and consists of a three-stranded antiparallel β-sheet (β1-β3) capped with a short perpendicular α-helix (αA). The Zn2+ ion is coordinated between the helix and sheet by Cys-768 and Cys-771 on the L1 loop, Cys-782 on the αA helix and His-787 on the L3 loop. The C-terminal CCCC zinc motif (Zn2) adopts a twisted antiparallel β-sheet (β4-β6) with the zinc coordinated by Cys-801 and Cys-803 on the short loop between strands β4 and β5, and by Cys-818 and Cys-821 on the extended loop between β5 and β6. Overall, XMcm10755–842 adopts a relatively globular fold as a result of the short linker and side chain interactions between the two zinc motifs.
ssDNA Binding Is Localized to the CCCH Zinc Motif
Close inspection of the chemical shift perturbation data shown in Fig. 1A suggested that DNA binding to the CTD was dominated by the Zn1 arm. Taking advantage of the complete sequence specific NMR assignments of XMcm10755–842 amides, we repeated the NMR titrations with this shorter construct to map the ssDNA binding site onto our structure (Fig. 3). DNA binding was determined by monitoring perturbations in the 15N-1H HSQC spectrum as unlabeled ssDNA was titrated into 15N-enriched XMcm10755–842 (Fig. 3A). Eleven signals shifted significantly in response to DNA, while the others remained unaffected (supplemental Fig. S4). Mapping the positions of the perturbed residues onto the structure of XMcm10755–842 revealed that ssDNA binding is indeed localized almost exclusively to the Zn1 motif (Fig. 3B). The perturbed residues trace a continuous ∼35 Å path around the Zn1 arm, raising the possibility that ssDNA partially encircles this motif. In support of this, the optimal length of ssDNA needed to fully engage the CTD was between 10 and 15 nucleotides, as measured by the in vitro fluorescence polarization assay (supplemental Fig. S5). Interestingly, Phe-776 at the Zn1-Zn2 interface showed a dramatic resonance shift (Fig. 3B and supplemental Fig. S4), suggesting DNA contacts extend to this region or that an allosteric hinge-like movement of the Zn1 arm accompanies binding of ssDNA.
Given the lack of chemical shift perturbation outside of the Zn1 motif, we examined the contribution of Zn2 and winged-helix motifs to DNA binding by mutational analysis. A series of CTD deletion constructs were tested for their ability to bind 15mer ssDNA by fluorescence polarization (Fig. 3, C and D). Addition of the full-length CTD (XMcm10596–860) to fluorescein-labeled ssDNA resulted in a robust change in fluorescence polarization and an apparent Kd of 5.2 ± 0.1 μm. Deletion of Zn2 from the CTD (XMcm10596–794) had only a modest 2-fold effect on ssDNA binding (Kd = 10.8 ± 1.1 μm). Likewise, XMcm10690–842 and XMcm10755–842, constructs lacking the region N-terminal to Zn1, bound ssDNA with similar affinity (Kd = 14.4 ± 2.5 and 3.4 ± 0.2 μm, respectively). In contrast, removal of both Zn1 and Zn2 motifs from the CTD (XMcm10596–757) completely abrogated DNA binding (Fig. 3C). We therefore conclude that Zn1 is necessary for ssDNA binding by XMcm10-CTD and that Zn2 and the putative winged-helix motif do not significantly contribute to the binding affinity.
Spatial Separation of Mcm10 DNA Binding Motifs
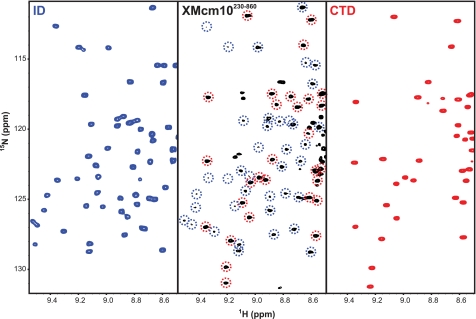
The large separation in DNA binding regions from the ID and CTD raises the question of how the two domains work together to bind DNA with relatively high affinity (21, 34). We previously suggested that the proteolytic sensitivity and lack of secondary structure in the region between the ID and CTD was the result of inherent flexibility that may provide Mcm10 with the ability to adapt to different structural states during replisome assembly and progression (21). To gain insight into the extent of the interaction between the two domains, we took advantage of the sequence specific NMR assignments for both the ID and CTD. NMR is a powerful technique for studying protein structural dynamics, and has been applied recently to the highly modular 116-kDa RPA heterotrimer (43). The high protein concentrations required for NMR experiments prevented structural analysis of full-length XMcm10. However, we were able to obtain a high-quality 15N-1H TROSY-HSQC spectrum for XMcm10230–860, which encompasses both the ID and CTD as well as the intervening linker region (Fig. 4 and supplemental Fig. S6). The central (∼8 ppm) region of the XMcm10230–860 spectrum corresponding primarily to residues in random coil and α-helical conformation contains numerous overlapping signals. However, signals outside of this region are well resolved and can be readily compared with signals nearly identical in the spectra of the individual ID and CTD constructs (35). We found 94 resonances in nearly identical positions to those in the isolated ID and CTD domains. Because the NMR chemical shift is exclusively sensitive to structural perturbations, these data provide convincing evidence that the structures of the ID and CTD are structurally independent of one another in the XMcm10230–860 construct. The conclusion is supported by the absence in chemical shift perturbations of individually 15N-enriched ID and CTD domains when added together in trans (data not shown).
FIGURE 4.
Structural analysis of XMcm10230–860. 15N-1H HSQC spectra of 15N-XMcm10230–427 (ID, left panel) and 15N-XMcm10755–842 (CTD, right panel), and the 15N-1H TROSY-HSQC spectrum of 15N-XMcm10230–860 (center panel). Signals in the XMcm10230–860 spectrum that correspond to residues in the ID or CTD have been circled in blue and red, respectively.
NMR line widths and signal intensities imply a modular organization of XMcm10230–860. Indeed, three regimes were observed. CTD signals were more intense than those of the larger ID (supplemental Fig. S6), consistent with the smaller size of the globular portion of CTD relative to the ID. The third set of signals corresponded to unassigned resonances with 1H chemical shifts of ∼8.2 ppm, which can be attributed to the linker between ID and CTD. The location of these signals in the random coil region of the spectrum combined with their extraordinarily high intensity implies the linker residues are dynamically disordered. Taken together with the low sequence conservation and high proteolytic sensitivity of the linker (residues 430–595) (21), the NMR studies of XMcm10230–860 strongly support the existence of a flexible linker between the two DNA binding domains of XMcm10.
DISCUSSION
A Novel DNA Binding Motif in the Mcm10 C Terminus
In this study, we determined the structure of the zinc cluster within Mcm10-CTD and identified the CCCH zinc motif (Zn1) as the predominant DNA binding region. A search for structural homologs to Zn1 using the Dali server (44) returned no results, suggesting Mcm10 is structurally distinct from other replication proteins. Interestingly, no C-terminal zinc motifs are found in yeast Mcm10 sequences, implying that Mcm10 in lower eukaryotes has a different functional architecture and mode of action. Differences between yeast and vertebrate Mcm10 are also evident from the mapping of DNA binding regions. Schizosaccharomyces pombe Mcm10 binds ssDNA with nanomolar affinity through a domain corresponding to the ID and has no affinity in the extreme C-terminal 180 residues (20). In contrast, XMcm10 utilizes two relatively low-affinity DNA binding domains to attain nanomolar affinity for the full-length protein (34). The hypothesis that the Mcm10 proteins do not function similarly is consistent with the differences in composition between yeast and vertebrate replisomes and fundamental differences between other replication proteins such as DNA primases.
Conservation of Zinc Motif Sequence and Structure but Not Function
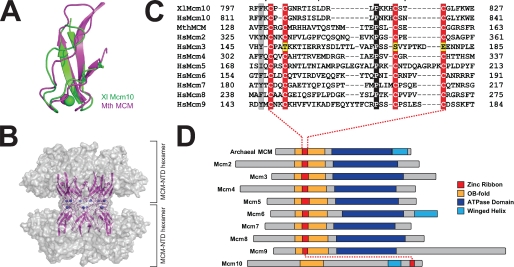
Unlike the novel Zn1 motif, a Dali search performed on the Zn2 motif identified a clear hit from the crystal structure of Methanobacterium thermoautotrophicum MCM helicase N-terminal domain (MthMCM-NTD, PDB: 1LTL) (45). The zinc motifs in the two structures are very similar, with an RMSD of 2.7 Å for all backbone atoms between MthMCM residues 125–166 and XMcm10 residues 795–830 (Fig. 5A). In MthMCM, this zinc motif mediates a head-to-head interaction between two hexameric rings (Fig. 5B) and is important for double hexamer formation and proper helicase function (45–48). As shown in Fig. 5C, the sequence of this motif is conserved in the six eukaryotic Mcm2–7 subunits (45, 47, 49, 50) and in the recently identified Mcm8 and Mcm9 proteins (51–53). Thus, the structure of the Mcm10 CCCC zinc ribbon presented here reveals a homology between the evolutionarily distinct Mcm10 and Mcm2–9 protein families (54).
FIGURE 5.
Conservation of the CCCC zinc ribbon among MCM proteins. A, structural alignment of the CCCC zinc ribbons from X. laevis (Xl) Mcm10755–842 (green) and Methanobacterium thermoautotrophicum (Mth) MCM (magenta, PDB: 1LTL). B, crystal structure of the MthMCM N-terminal domain double hexamer is shown as a transparent gray molecular surface with the head-to-head CCCC motifs at the hexamer-hexamer interface rendered as magenta ribbons and blue Zn2+ spheres. C, sequence alignment of the CCCC zinc motifs from Xenopus and human Mcm10, MthMCM, and human Mcm2–9 proteins. Conserved residues are shaded gray, invariant residues black, and zinc-coordinating residues red and yellow. D, schematic alignment of archaeal MCM and eukaryotic Mcm2–10 proteins colored according to domain (AAA+, blue) or supersecondary motif (CCCC zinc ribbon, red; OB-fold, gold; winged helix, cyan).
Unlike the CCCC zinc motifs, Mcm10 and MthMCM contain OB-folds that are in the same location in the primary structures (Fig. 5D). In the MCM helicase, the CCCC zinc ribbon is inserted into the L12 loop of the OB-fold to form a continuous domain important for helicase activity, likely by stabilizing the overall fold of the MCM-NTD (supplemental Fig. S7) (45, 48). In contrast, the L12 loop in Mcm10 does not contain a zinc motif and is directly involved in ssDNA binding (34, 35). In fact, the OB-fold in the ID and the zinc motif in the CTD are separated in sequence by over 400 residues and are completely independent (Figs. 4 and 5D). Thus, although Mcm10 and Mcm2–7 share common motifs, including putative winged-helix regions (Fig. 5D), it is unlikely that the two proteins share a similar architecture.
Implication for Mcm10 Function
Like MthMCM, the yeast Mcm2–7 replicative helicase was recently shown to load onto DNA as a double hexamer connected through its NTD (55). Given the conservation of the CCCC zinc motif (Fig. 5C), the double hexamer in the eukaryotic helicase likely occurs in a manner to MthMCM. Indeed, mutations within this zinc motif disrupt double hexamer formation in MthMCM (46) and result in lethality or temperature sensitivity in yeast (56, 57). Interestingly, replication progression complexes contain only one copy of Mcm4, which suggests that a single Mcm2–7 ring is sufficient to unwind DNA during elongation. Because the NTD zinc motif mediates double hexamer formation, the zinc motif surface would be available to bind other proteins. It is enticing to speculate that Mcm10 engages Mcm2–7 through interactions between zinc motifs. Indeed, Mcm10 has been shown to interact directly with various subunits of the Mcm helicase (8, 24, 25) and has been implicated in physically linking the helicase and pol α (3, 30). The observation that Mcm10 remains associated with the polymerases upon uncoupling the helicase-polymerase complex (10) suggests a higher affinity interaction between Mcm10 and pol α. Although poorly folded in the context of the isolated CTD, the putative winged-helix domain may provide an additional protein interaction module for Mcm10. Together, the modular architecture and myriad of protein and DNA interaction sites support a model in which Mcm10 functions as a scaffold, serving to co-localize critical elements of the replisome during the initiation and elongation phases of replication.
Supplementary Material
Acknowledgments
We thank Sivaraja Vaithiyalingam for assistance with NMR and Borden Lacy for use of the fluorometer.
This work was supported, in whole or in part, by National Institutes of Health Grants GM080570 (to B. F. E.) and GM065484 (to W. J. C.) and the Vanderbilt Discovery Grant Program.
The atomic coordinates and structure factors (code 2KWQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S7.
- ORC
- origin recognition complex
- ID
- internal domain
- CTD
- C-terminal domain
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation.
REFERENCES
- 1.Bell S. P., Dutta A. (2002) Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- 2.Wohlschlegel J. A., Dhar S. K., Prokhorova T. A., Dutta A., Walter J. C. (2002) Mol. Cell 9, 233–240 [DOI] [PubMed] [Google Scholar]
- 3.Ricke R. M., Bielinsky A. K. (2004) Mol. Cell 16, 173–185 [DOI] [PubMed] [Google Scholar]
- 4.Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007) Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- 5.Zegerman P., Diffley J. F. (2007) Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]
- 6.Xu X., Rochette P. J., Feyissa E. A., Su T. V., Liu Y. (2009) EMBO J. 28, 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyer S. E., Lewis P. W., Botchan M. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 9.Ilves I., Petojevic T., Pesavento J. J., Botchan M. R. (2010) Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C. (2006) Mol. Cell 21, 581–587 [DOI] [PubMed] [Google Scholar]
- 11.Tanaka T., Nasmyth K. (1998) EMBO J. 17, 5182–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou L., Stillman B. (2000) Mol. Cell. Biol. 20, 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter J., Newport J. (2000) Mol Cell 5, 617–627 [DOI] [PubMed] [Google Scholar]
- 14.Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel J. A., Nutt L. K., Kornbluth S., Dutta A. (2007) Genes Dev. 21, 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka H., Katou Y., Yagura M., Saitoh K., Itoh T., Araki H., Bando M., Shirahige K. (2009) Genes Cells 14, 807–820 [DOI] [PubMed] [Google Scholar]
- 16.Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., Labib K. (2009) EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Im J. S., Ki S. H., Farina A., Jung D. S., Hurwitz J., Lee J. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgers P. M. (2009) J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merchant A. M., Kawasaki Y., Chen Y., Lei M., Tye B. K. (1997) Mol. Cell. Biol. 17, 3261–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fien K., Cho Y. S., Lee J. K., Raychaudhuri S., Tappin I., Hurwitz J. (2004) J. Biol. Chem. 279, 16144–16153 [DOI] [PubMed] [Google Scholar]
- 21.Robertson P. D., Warren E. M., Zhang H., Friedman D. B., Lary J. W., Cole J. L., Tutter A. V., Walter J. C., Fanning E., Eichman B. F. (2008) J. Biol. Chem. 283, 3338–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisenberg S., Korza G., Carson J., Liachko I., Tye B. K. (2009) J. Biol. Chem. 284, 25412–25420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homesley L., Lei M., Kawasaki Y., Sawyer S., Christensen T., Tye B. K. (2000) Genes Dev. 14, 913–926 [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi M., Yatagai F., Hanaoka F. (2001) J. Biol. Chem. 276, 48526–48531 [DOI] [PubMed] [Google Scholar]
- 25.Lee J. K., Seo Y. S., Hurwitz J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2334–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen T. W., Tye B. K. (2003) Mol. Biol. Cell 14, 2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X., Gregan J., Lindner K., Young H., Kearsey S. E. (2005) BMC Mol Biol 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricke R. M., Bielinsky A. K. (2006) J. Biol. Chem. 281, 18414–18425 [DOI] [PubMed] [Google Scholar]
- 29.Chattopadhyay S., Bielinsky A. K. (2007) Mol. Biol. Cell 18, 4085–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee C., Liachko I., Bouten R., Kelman Z., Tye B. K. (2010) Mol. Cell. Biol. 30, 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maine G. T., Sinha P., Tye B. K. (1984) Genetics 106, 365–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Richards T. A., Aves S. J. (2009) BMC Evol. Biol. 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okorokov A. L., Waugh A., Hodgkinson J., Murthy A., Hong H. K., Leo E., Sherman M. B., Stoeber K., Orlova E. V., Williams G. H. (2007) EMBO reports 8, 925–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren E. M., Huang H., Fanning E., Chazin W. J., Eichman B. F. (2009) J. Biol. Chem. 284, 24662–24672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren E. M., Vaithiyalingam S., Haworth J., Greer B., Bielinsky A. K., Chazin W. J., Eichman B. F. (2008) Structure 16, 1892–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goddard T. D., Kneller D. G. (2006) SPARKY 3, University of California, San Francisco [Google Scholar]
- 37.Güntert P. (2004) Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 38.Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu H., Chazin W. J. (2003) J. Mol. Biol. 330, 473–484 [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 41.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavanagh J., Fairbrother W. J., Palmer A. G. I., Rance M., Skelton N. J. (2007) Protein NMR Spectroscopy: Principles and Practice, Academic Press Inc., New York [Google Scholar]
- 43.Brosey C. A., Chagot M. E., Ehrhardt M., Pretto D. I., Weiner B. E., Chazin W. J. (2009) J. Am Chem. Soc. 131, 6346–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 45.Fletcher R. J., Bishop B. E., Leon R. P., Sclafani R. A., Ogata C. M., Chen X. S. (2003) Nat. Struct. Biol. 10, 160–167 [DOI] [PubMed] [Google Scholar]
- 46.Fletcher R. J., Shen J., Gómez-Llorente Y., Martín C. S., Carazo J. M., Chen X. S. (2005) J. Biol. Chem. 280, 42405–42410 [DOI] [PubMed] [Google Scholar]
- 47.Poplawski A., Grabowski B., Long S. E., Kelman Z. (2001) J. Biol. Chem. 276, 49371–49377 [DOI] [PubMed] [Google Scholar]
- 48.Kasiviswanathan R., Shin J. H., Melamud E., Kelman Z. (2004) J. Biol. Chem. 279, 28358–28366 [DOI] [PubMed] [Google Scholar]
- 49.Kearsey S. E., Labib K. (1998) Biochim. Biophys. Acta 1398, 113–136 [DOI] [PubMed] [Google Scholar]
- 50.Tye B. K. (1999) Annu. Rev. Biochem. 68, 649–686 [DOI] [PubMed] [Google Scholar]
- 51.Gozuacik D., Chami M., Lagorce D., Faivre J., Murakami Y., Poch O., Biermann E., Knippers R., Bréchot C., Paterlini-Bréchot P. (2003) Nucleic Acids Res. 31, 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lutzmann M., Maiorano D., Méchali M. (2005) Gene 362, 51–56 [DOI] [PubMed] [Google Scholar]
- 53.Yoshida K. (2005) Biochem. Biophys. Res. Commun. 331, 669–674 [DOI] [PubMed] [Google Scholar]
- 54.Liu W., Pucci B., Rossi M., Pisani F. M., Ladenstein R. (2008) Nucleic Acids Res. 36, 3235–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Remus D., Beuron F., Tolun G., Griffith J. D., Morris E. P., Diffley J. F. (2009) Cell 139, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dalton S., Hopwood B. (1997) Mol. Cell. Biol. 17, 5867–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan H., Gibson S., Tye B. K. (1991) Genes Dev. 5, 944–957 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.