Abstract
Glycerolipid synthesis in plants involves two major metabolic pathways compartmentalized in the chloroplasts and cytosol, respectively. Although these two parallel pathways are regulated with considerable flexibility, the factors mediating this process remain unclear. To investigate the influence of glycerol 3-phosphate (Gly-3-P) on the interactions of the glycerolipid pathways, we generated transgenic Arabidopsis lines with a feedback-resistant Gly-3-P dehydrogenase gene (gpsAFR) from Escherichia coli. gpsAFR was detected in the cytosol, but augmented Gly-3-P levels were observed in the cytosol as well as in chloroplasts. Glycerolipid composition and fatty acid positional distribution analyses revealed an altered fatty acid flux that affected not only the molar ratios of glycerolipid species but also their fatty acid composition. To decipher this complex pathway, a transgenic line was subjected to lipidomic analysis and a global gene-expression survey. The results revealed that changes in Gly-3-P metabolism caused altered expression of a broad array of genes. When viewed from the perspective of glycerolipid metabolism, coherent networks emerged, revealing that many enzymatic components of the glycerolipid pathways operate in a modular manner under the influence of Gly-3-P. Transcript levels of the enzymes involved in the prokaryotic pathway were mostly induced, whereas genes of the eukaryotic pathway enzymes were largely suppressed. Hence, the gene-expression changes were consistent with the detected biochemical phenotype. Our results suggest that Gly-3-P modulates the balance of the two glycerolipid pathways in Arabidopsis by influencing both metabolic flux and gene transcription.
Keywords: Fatty Acid Metabolism, Gene Expression, Gene Regulation, Glycerolipid, Metabolic Regulation, Microarray, Plant, Glycerol 3-Phosphate, Glycerol-3-phosphate Dehydrogenase, Glycerolipid Pathways
Introduction
In plant cells, fatty acids are generated primarily in plastids, but glycerolipid biosynthesis takes place both in chloroplasts and in the cytosol (1–3). The glycerolipid pathway in chloroplasts is believed to be crucial for generating phosphatidylglycerol (PG),2 but in species such as Arabidopsis, it also contributes partially to the production of monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglcyerol (DGDG) in chloroplasts. The glycerolipid pathway in the cytosolic compartment, known as the eukaryotic pathway, sustains the synthesis of all membrane phospholipids in the cytosol, and it also generates some of the diacylglycerol (DAG) moieties required for the synthesis of MGDG and DGDG (3). The chloroplast pathway, which incorporates fatty acid groups from acyl-ACP to generate glycerolipid molecules resembling cyanobacteria glycerolipids with C16 fatty acids were esterified almost exclusively at the sn-2 position (4–7), is termed the prokaryotic pathway. Because the sn-2 C16 fatty acid in MGDG originating from the prokaryotic pathway is subsequently desaturated to form cis-7,10,13-hexadecatrienoic acid (C16:3) (8, 9), the presence of 16:3 is indicative of a contribution from the prokaryotic pathway. Species that possess a significant portion of 16:3 MGDG are generally called 16:3 plants.
In Arabidopsis, a typical 16:3 plant, close to half of the DAG moieties originating from the eukaryotic pathway are channeled to the chloroplast for membrane glycerolipid synthesis (10). However, the flux of DAG is flexible, being frequently readjusted under different growth conditions (11–13) and in the presence of genetic lesions (14–19). Understanding this metabolic adjustment is important because impairments in balancing the two glycerolipid pathways have major consequences for plant performance (20–22).
Gly-3-P is an obligate precursor at the initial step of glycerolipid synthesis. Experiments using isolated chloroplast and leaf explants have all revealed Gly-3-P as a factor influencing the balance of the two glycerolipid pathways (1, 3, 7, 23, 24). A deficiency in Gly-3-P in chloroplasts reduces the carbon flux through the prokaryotic pathway (25). In previous reports, the role of Gly-3-P was explained chiefly by its capacity to affect fatty acylation processes in chloroplasts. In this study, we generated transgenic Arabidopsis plants with increased Gly-3-P levels by overexpressing an Escherichia coli gene encoding a Gly-3-P dehydrogenase (gpsAFR) insensitive to feedback inhibition (26). The transgenic lines had Gly-3-P levels severalfold higher than wild-type controls under standard growth conditions, and they exhibited a glycerolipid phenotype consistent with an adjusted metabolic balance between the two glycerolipid pathways. We conducted lipidomic as well as global gene-expression analyses with the transgenic plants. Our data provide a comprehensive view of the metabolic and enzymatic components involved in balancing the two glycerolipid pathways in response to Gly-3-P level changes. Our results also reveal many transcriptional changes in the enzymatic components of the glycerolipid pathways associated with Gly-3-P metabolic perturbation.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Plants were grown in growth chambers with a 16-h light and 8-h dark regime with a photosynthetic photon flux density of 75 μmol of photons m−2 s−1. The day/night temperature was controlled at 22/17 °C. All biochemical analyses were performed with leaves harvested at the rosette stage from 3-week-old plants.
Construction of a Plant Transformation Vector for gpsAFR
Bacterial strain BB26-36R2 (gpsAFR, plsB) was a generous gift from Professor John E. Cronan, Jr. (University of Illinois, Urbana-Champion). The gpsAFR allele was PCR amplified and fully sequenced. Primers GAGAGCTCTTAGTGGCTGCTGCGCTC (GPSA31) and GAAGAAGGATCCAACAATGAACCAACGTAA (GPSA5) were designed according to the sequence of gpsAFR (accession P37606) with SacI and BamHI restriction sites, respectively, at each end. The primers were used to perform PCR amplification of the gpsAFR sequence. The PCR products were purified with a QIAquick PCR purification Kit (Qiagen) and digested with SacI/BamHI. The SacI/BamHI-digested gpsAFR DNA fragment was subsequently inserted into the Agrobacterium binary vector pBI121 (Clontech) to replace the GUS gene. The gpsAFR gene-expression cassette thus contains the gpsAFR gene under control of the constitutive 35S-promoter and flanked at the 3′ end with a NOS terminator. The junction region between the 35S-promoter and the gpsAFR coding sequence was confirmed through sequencing. Transformation of Arabidopsis was performed through vacuum infiltration, and T1 transgenic plants were selected through germinating seeds in ½ MS medium containing 50 mg/liter of kanamycin. Biochemical analysis was performed with tissues harvested from T2 plants.
Chloroplast Preparation
Chloroplasts were isolated from 3-week-old Arabidopsis rosette leaves according to the method of Rensink et al. (27) using a Percoll gradient consisting of 2 ml of 70% (v/v) Percoll (Sigma), 4 ml of 50% (v/v) Percoll, and 4 ml of 40% (v/v) Percoll/grinding buffer in a 15-ml polycarbonate tube. The gradient was centrifuged for 15 min at 5000 × g in an HB-4 rotor with the brake off. The lower band at the 50 to 70% interface was isolated for Gly-3-P content and Western blot analysis. All operations were carried out at 4 °C. Protein content was measured according to the method of Bradford (28).
Metabolite Extraction and Analysis
Gly-3-P in leaf tissues was extracted according to the method of Shen et al. (29). Gly-3-P in chloroplasts was extracted by the protocol of Sauer and Heise (30). Ice-cold chloroplast suspensions were immediately centrifuged through a layer of silicon oil into 20 μl of 1 m HClO4 after isolation. The concentration of Gly-3-P in the neutralized leaf and chloroplast extracts was measured enzymatically via Gly-3-P dehydrogenase based on the method of Shen et al. (29).
Fatty Acid and Lipid Analysis
For determining the fatty acid composition of leaf tissues, Arabidopsis leaves (200 mg) in 2 ml of 10% KOH in methanol were heated at 80 °C for 2 h. After the mixture was cooled to room temperature and 1 ml of 50% HCl was added, the mixtures were extracted twice with 2 ml of hexane and dried under N2. Each sample was then supplemented with 2 ml of 3 n methanolic HCl and heated at 80 °C for 2 h. After the addition of 2 ml of 0.9% NaCl solution and 2 ml of hexane, fatty acid methyl esters were extracted into the organic phase and determined by gas chromatography. Fatty acid double bond indices were calculated as described by Falcone et al. (31).
Lipid extraction and purification by two-dimensional thin layer chromatography (TLC) on Silica Gel 60 (EMD Chemical, Germany) was performed according to the method of Miquel and Browse (18). Leaf tissues were frozen rapidly by immersion in liquid N2 and ground under liquid N2 before being extracted. The plate was first developed in chloroform, methanol, 50% aqueous ammonia (65:35:2, by volume). After being allowed sufficient time to dry, the plate was then developed, at right angles to the first development, in chloroform, methanol, acetic acid, water (85:15:10:3, by volume). Lipids were visualized with iodine vapor. To determine the fatty acid composition and the relative amounts of individual lipids, the silica gel from each lipid spot was transferred to a screw-capped tube, and fatty acid methyl esters were prepared as described above. The methyl esters were quantified by gas chromatography using 17:0 fatty acid as an internal standard.
The fatty acid composition at the sn-2 position of individual lipids was determined by lipase digestion according to Miquel et al. (25). After TLC, lipids were extracted from the silica gel by the method of Bligh and Dyer (32). The protocol for digestion with Rhizopus sp. lipase, including purification of the lyso-derivatives and fatty acids, was described by Siebertz and Heinz (33), except that 50 mm H3BO4 was added to the buffer used for lipase digestion to minimize intramolecular acyl transfer on the lyso-lipids produced (25). Fatty methyl esters were formed from untreated lipids, lyso-lipids, and fatty acids as described above, and the fatty acid composition of each compound was determined by gas chromatography.
Lipid extraction for lipidomic analysis was performed according to the method of Welti et al. (34). For each sample, the rosettes of two or three plants were cut at the sampling time and transferred to 3 ml of isopropyl alcohol with 0.01% butylated hydroxytoluene at 75 °C. After 15 min, 1.5 ml of chloroform and 0.6 ml of water were added. The tubes were shaken for 1 h, followed by removal of the extract. The plants were re-extracted with chloroform/methanol (2:1) with 0.01% butylated hydroxytoluene five times with 30 min of agitation each time until all of the remaining plant tissue appeared white. The remaining plant tissues were heated overnight at 105 °C and weighed. The weights of these dried, extracted tissues were the “dry weights” of the plants. The dry weights ranged from 9 to 47 mg. The combined extracts were washed once with 1 ml of 1 m KCl and once with 2 ml of water. The solvent was evaporated under nitrogen, and the lipid extract was dissolved in 1 ml of chloroform. Electronspray tandem mass spectrometry (ESI-MS/MS) analysis was performed at the Kansas Lipidomic Research Center.
Immunoblot Analysis of gpsAFR Protein from Leaves and Chloroplasts
Polyclonal antibodies were raised in rabbits to two polypeptides, GLEAETGRLLQDVAREALGDQIPLAVISGP and LASTDQTFADDLQQLLHCGKSFRVYSNPDF, synthesized based on the deduced amino acid sequence of E. coli gpsAFR. Immunoblot analysis of gpsAFR protein from total leaf extracts and purified chloroplasts was performed according to Millenaar et al. (35). Proteins (10 μg) were separated by SDS-PAGE and subsequently electrotransferred to nitrocellulose filters using blot transfer buffer (25 mm Tris, 192 mm glycine, 20% (v/v) methanol). Antiserum for gpsAFR was used as the primary antibody with a dilution factor of 1:1000. Anti-rabbit IgG Fab fragments conjugated to peroxidase (Roche Applied Science) were used as the secondary antibody (1:25,000). The blot was detected for 1.5 min using the protein gel blotting detection reagent ECL+ Plus (Amersham Biosciences) and developed with BioMax MR film (Kodak).
RNA Extraction and Microarray Analysis
Total RNA of leaf tissues was extracted using an RNeasy Mini Kit (Qiagen) under the manufacturer's recommendations and subjected to RNase-free DNase I treatment (Invitrogen) at 25 °C for 15 min to remove contaminating DNA. The yield and RNA purity were determined spectrophotometrically with a spectrophotometer DU@740 (Beckman), and the quality of the RNA was verified by electrophoresis on a denaturing formaldehyde-agarose gel. Purified total RNA was precipitated and resuspended in diethyl pyrocarbonate-treated water to a final concentration of 500 ng/μl.
Synthesis of cDNA, cRNA labeling, hybridization, and scanning were performed at the Botany Affymetrix Genechip Facility, University of Toronto (Toronto, ON, Canada). Three independent biological replicates were performed for both the wild-type and the transgenic line. A total of six. CEL files containing the raw probe intensity values were imported into the R-package Affy (36). Background adjustment and quantile normalization were performed using the robust multianalysis package implemented in Bioconductor (37). The quantile normalization method was used because it has better precision than MicroArray Suite 5.0 (Affymetrix) and dChip for low expression values (38). Using the R-package LIMMA (by reading the data and creating an expression set of the data after log2 transformation), a linear model was fitted and contrast of “transgenic versus wild-type” was extracted. For each gene, the mean log2 fold-change and corresponding p value were calculated from an empirical Bayes approach to compute moderated t-statistics (39). False discovery rate correction (40) was applied to account for testing of multiple genes. Genes were considered differentially regulated if the p value was less than 0.05 and the fold-change in the expression level was greater than 1.5. Functional annotations of genes were obtained from the Bio-Array Resource for Arabidopsis Functional Genomics data base (BAR) using the probe set numbers provided by the GeneChip manufacturer (Affymetrix Co.).
Real Time Quantitative RT-PCR Analyses
Total RNA was extracted from the leaves of wild-type and transgenic plants with the Plant RNeasy Mini kit (Qiagen). The amount of RNA was determined by spectrophotometry at 260 nm, and its integrity was assessed by gel electrophoresis.
For real time quantitative RT-PCR (qRT-PCR), 1 μg of total RNA was used for cDNA synthesis with a QuantiTect Reverse Transcription kit (Qiagen). Specific primers (Tm, 57 °C-63 °C) were designed to generate PCR products between 75 and 130 bp. The specificity of all primers was checked with BLASTn searches. Actin 2 (At3g18780) was used as an endogenous control for standardization. The primers used for qRT-PCR are listed in supplemental Table S5.
Real time qRT-PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) and amplification was monitored with ABI StepOne Real Time PCR Systems (Applied Biosystem). A standard thermal profile was used for all PCRs: 50 °C for 2 min; 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 57 °C for 30 s, 72 °C for 30 s. Data acquisition and analysis were performed using StepOne software 2.0 (Applied Biosystems). Genes picked for verification with real time PCR were chosen based on their possible significance in glycerolipid pathways. Results from three replicates are shown.
Statistical Analysis
Statistical significance was determined by the Student's unpaired t test (two-tailed).
RESULTS
Transgenic Plants with a Feedback-insensitive E. coli Gly-3-P Dehydrogenase Gene (gpsAFR) Had Elevated Gly-3-P Content in Both the Cytosol and Chloroplasts
Bell and Cronan (26) discovered a mutant E. coli strain, BB26-36R2 (gpsAFR, plsB), in which the Gly-3-P auxotrophic phenotype, due to a mutation in Gly-3-P acyltransferase (plsB), is suppressed by a second mutation in the structural gene (gpsA, accession P37606) encoding the biosynthetic Gly-3-P dehydrogenase. The mutated version of the Gly-3-P dehydrogenase, gpsAFR, is about 20-fold less sensitive to feedback inhibition, and as a result, E. coli strains harboring this allele had Gly-3-P contents 12 times higher than that of the control. We amplified the coding region of the gpsAFR allele and found a point mutation in the cordon for amino acid 255 (GAS → GAO), which resulted in a conversion of Asp255 in the wild-type enzyme (PID, g91790037) to Glu255 in gpsAFR. The structural significance of this amino acid substitution is unclear, but the residue is located very close to the active site pocket based on the structure of human Gly-3-P dehydrogenase (41). This gpsAFR allele was inserted into a plant transformation vector under the control of the constitutive CaMV 35S-promoter and introduced into Arabidopsis thaliana (ecotype Columbia).
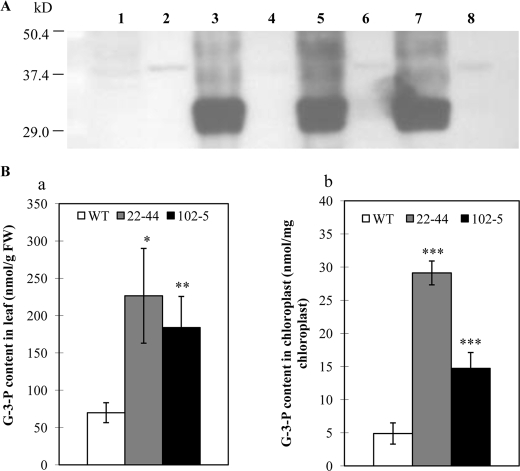
The gpsAFR transgenic plants were indistinguishable from wild-type plants in growth and development under standard growth conditions. We assessed gpsAFR protein levels in several transgenic lines by immunoblot analysis. Total leaf protein extracts and protein prepared from purified chloroplasts were subjected to SDS-PAGE as described under “Experimental Procedures.” High levels of gpsAFR protein were detected in the total leaf extract, but not purified chloroplast extract (Fig. 1A). Thus, as expected, the gpsAFR protein appeared to be localized in the cytosol.
FIGURE 1.
Immunoblot analysis of gpsAFR protein and measurement of Gly-3-P levels in the gpsAFR transgenic lines and wild-type (WT). A, immunoblot analysis of the gpsAFR protein. Immunoblots of total leaf and chloroplast proteins from both gpsAFR transgenic and wild-type lines probed with polyclonal antibodies raised against two polypeptides from gpsAFR. Lane 1, wild-type total leaf protein extract; lane 2, wild-type chloroplast protein extract; lane 3, total leaf protein extract from gpsAFR transgenic line 84-10; lane 4, chloroplast protein extract from gpsAFR transgenic line 84-10; lane 5, total leaf protein extract from gpsAFR transgenic line 102-5; lane 6, chloroplast protein extract from gpsAFR transgenic line 102-5; lane 7, total leaf protein extract from gpsAFR transgenic line 22–44; and lane 8, chloroplast protein extract from gpsAFR transgenic line 22–44. Approximately 10 μg of protein was loaded in each lane. B, Gly-3-P levels in the gpsAFR transgenic lines and wild-type. Gly-3-P levels were assayed as described under “Experimental Procedures.” Panel a, leaf Gly-3-P levels in the gpsAFR transgenic lines and wild-type plants; panel b, chloroplast Gly-3-P levels in gpsAFR transgenic lines and wild-type plants. Error bars represent mean ± S.D. (n = 3). Statistical significance compared with the wild-type was determined by the two-tailed Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Gly-3-P content was assessed using rosette leaves of plants growing in soil. Leaf tissues of wild-type Arabidopsis plants had Gly-3-P levels of 60 nmol/g of fresh weight, whereas the transgenic lines had Gly-3-P level of 200 nmol/g of fresh weight (Fig. 1B, panel a). We also isolated chloroplasts and determined Gly-3-P levels in this particular compartment. Wild-type Arabidopsis chloroplasts under our growth conditions had a Gly-3-P level of 5 nmol/mg of chlorophyll, a level within the range reported for spinach (11 nmol/mg of chlorophyll) (1). However, chloroplasts of the transgenic lines had a Gly-3-P level severalfold higher than that of the wild-type control (Fig. 1B, panel b). The chloroplast Gly-3-P content might have been underestimated due to possible leakage during organelle fractionation. Nevertheless, by comparing the wild-type plants and the transgenic lines that underwent the same procedure, it became apparent that the Gly-3-P content in the chloroplasts of the transgenic lines was much higher. Thus, consistent with the presence of a chloroplast membrane carrier that is capable of accepting 3-carbon compounds with a phosphate molecule attached to it (42), our findings suggest that Gly-3-P in the cytosol was likely transported across the chloroplast membrane.
The gpsAFR Transgenic Plants Exhibit Altered Fatty Acid Profiles in Leaves but Have No Changes in Double Bond Indices
Surveys of total lipid fatty acid profiles in leaves from a large number of transgenic lines consistently showed increased 16:0 and 16:3 content (data not shown). Table 1 presents data from some of the selected lines. The molar ratios of C18:1, C18:2, and C18:3 fatty acids were all substantially lower. Consequently, the general alteration of the fatty acid profile in leaf tissues can be best described as an elevated C16:C18 fatty acid ratio. Thus, in keeping with chloroplast feeding experiments (1), the elevated Gly-3-P levels appeared to reduce palmitoyl-ACP elongation in planta as well. However, these fatty acid compositional changes do not seem to alter the general membrane unsaturation level based on assessment of double bond indices (31). We also analyzed the fatty acid composition and content of the seed oil. Despite of repeated analyses with multiple lines, we failed to detect any significant changes in either seed trait when compared with those of the wild-type plants raised together under standard growth conditions. Further analysis presented below was focused on rosette leaf tissues of two transgenic lines.
TABLE 1.
Fatty acid composition of total leaf lipids of gpsAFR transgenic lines and wild-type (WT) Arabidopsis
Values are expressed as mean ± S.D. (n = 3).
| Fatty acid composition |
C16:C18 | Double bond indices | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 16:0 | 16:1a | 16:2 | 16:3 | 18:0 | 18:1 | 18:2 | 18:3 | |||
| mol % | ||||||||||
| WT | 15.0 ± 0.3 | 3.9 ± 0.2 | 0.9 ± 0.1 | 12.6 ± 0.5 | 1.2 ± 0.1 | 3.1 ± 0.2 | 14.7 ± 0.3 | 48.6 ± 0.6 | 0.48 | 2.16 |
| 15–6 | 18.2 ± 0.8b | 3.8 ± 0.2 | 0.7 ± 0.6 | 22.6 ± 2.2b | 2.2 ± 0.3b | 0.9 ± 0.8b | 9.0 ± 0.1c | 42.4 ± 0.7d | 0.83 | 2.18 |
| 22–44 | 19.8 ± 1.6b | 3.2 ± 0.3b | 0.9 ± 0.1 | 18.0 ± 1.0c | 1.6 ± 1.0 | 2.2 ± 1.3 | 10.8 ± 0.8b | 43.5 ± 0.9c | 0.72 | 2.15 |
| 54–8 | 18.0 ± 0.9b | 2.9 ± 0.2c | 0.8 ± 0.1 | 17.8 ± 0.8c | 1.7 ± 0.3 | 1.2 ± 0.1d | 11.3 ± 0.7c | 46.3 ± 1.4 | 0.65 | 2.31 |
| 84–10 | 16.2 ± 0.3c | 3.6 ± 0.1c | 1.4 ± 0.1c | 23.0 ± 0.5d | 1.6 ± 0.1c | 1.5 ± 0.1d | 9.2 ± 0.3d | 43.5 ± 0.4d | 0.79 | 2.22 |
| 102–5 | 17.2 ± 0.4c | 3.1 ± 0.1c | 0.6 ± 0.5 | 21.1 ± 1.3c | 1.7 ± 0.1c | 0.7 ± 0.6b | 10.1 ± 0.6c | 45.6 ± 1.4b | 0.72 | 2.25 |
a Sum of 16:1 cis and trans fatty acid.
b Statistical significance compared with WT was determined by the two-tailed Student's t test, p < 0.05.
c Statistical significance compared with WT was determined by the two-tailed Student's t test, p < 0.001.
d Statistical significance compared with WT was determined by the two-tailed Student's t test, p < 0.01.
The gpsAFR Transgenic Plants Have Altered Glycerolipid Composition in Leaves
We separated major glycerolipid species using two-dimensional TLC and analyzed their profiles in leaf tissues. The results presented in Table 2 include several changes. First, substantial increases in 16:0 across all lipid species, but particularly in DGDG and phosphatidylcholine (PC), were noticeable. Second, the proportion of MGDG was substantially elevated, whereas the levels of DGDG and phospholipid molecules (PC and PE) were reduced. Third, the compositions of the polyunsaturated fatty acids in galactolipid molecules were modified, with both MGDG and DGDG displaying less C18:3; in the case of MGDG, the reduction of C18:3 was almost entirely counterbalanced by an increase in C16:3 and thus yielded no changes in the level of trienoics; in DGDG, increases in both C16:3 and C16:0 were detected. Fourth, both PC and PE in the transgenic lines contained less 18:2, and they consequently had an elevated 18:3/18:2 ratio.
TABLE 2.
Glycerolipid profile and fatty acid composition of leaf lipids from wild-type (WT) and gpsAFR transgenic lines
Lipids were extracted from leaf material of 3-week-old plants. The polar lipids were separated by thin layer chromatography, and fatty acid compositions were determined by gas chromatography of methyl esters obtained by derivatization of individual lipids using 3 n methanolic HCl. Lipids were quantified by adding 10 μ g of 17:0 as an internal standard prior to derivatization. Values are expressed as mean ± S.D. (n = 3 to 5).
| Glycerolipid | Total polar lipids | Fatty acid composition |
C16:C18 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 16:0 | 16:1a | 16:2 | 16:3 | 18:0 | 18:1 | 18:2 | 18:3 | |||
| % | mol % | |||||||||
| MGDG | ||||||||||
| WT | 39.1 ± 3.6 | 0.9 ± 0.3 | 1.3 ± 0.1 | 1.7 ± 0.1 | 38.8 ± 2.0 | 0.2 ± 0.1 | 1.0 ± 0.1 | 2.9 ± 0.5 | 53.3 ± 3.2 | 0.74 ± 0.07 |
| 22–44 | 45.6 ± 3.1b | 2.2 ± 0.4b | 1.0 ± 0.3 | 1.7 ± 0.2 | 43.0 ± 3.4b | 0.3 ± 0.1 | 0.6 ± 0.3 | 1.8 ± 0.4 | 49.4 ± 1.9 | 0.92 ± 0.09b |
| 102–5 | 49.3 ± 2.1c | 2.2 ± 0.5b | 1.3 ± 0.2 | 1.7 ± 0.6 | 41.6 ± 2.1b | 0.5 ± 0.2 | 0.7 ± 0.6 | 3.5 ± 2.1 | 48.6 ± 3.1 | 0.88 ± 0.03b |
| DGDG | ||||||||||
| WT | 17.1 ± 1.3 | 14.6 ± 0.2 | 0.3 ± 0.2 | 0.7 ± 0.1 | 3.0 ± 0.3 | 1.2 ± 0.5 | 0.9 ± 0.3 | 4.9 ± 0.7 | 74.4 ± 1.1 | 0.23 ± 0.01 |
| 22–44 | 13.3 ± 0.9b | 25.0 ± 1.5d | 0.4 ± 0.1 | 1.0 ± 0.1 | 5.0 ± 0.8b | 2.6 ± 0.1b | 0.9 ± 0.1 | 4.8 ± 0.5 | 60.2 ± 0.9d | 0.46 ± 0.01d |
| 102–5 | 14.9 ± 1.7b | 24.0 ± 3.0c | 0.5 ± 0.1 | 0.9 ± 0.2 | 4.4 ± 0.5b | 2.5 ± 1.2 | 1.2 ± 0.4 | 5.4 ± 0.9 | 61.1 ± 4.1c | 0.43 ± 0.06c |
| Sulfoquinovosyl diacylglycerol | ||||||||||
| WT | 2.7 ± 0.7 | 46.7 ± 1.2 | 2.5 ± 0.6 | 2.0 ± 0.4 | 7.2 ± 0.9 | 41.6 ± 0.7 | 0.88 ± 0.04 | |||
| 22–44 | 2.5 ± 0.2 | 62.3 ± 0.5d | 4.2 ± 0.3b | 0.7 ± 0.7b | 4.2 ± 0.3c | 28.6 ± 1.3d | 1.66 ± 0.04d | |||
| 102–5 | 2.5 ± 0.4 | 54.6 ± 4.1b | 6.2 ± 3.4 | 1.7 ± 0.8 | 5.2 ± 1.3 | 32.4 ± 5.3b | 1.20 ± 0.21b | |||
| PG | ||||||||||
| WT | 11.7 ± 1.4 | 30.2 ± 5.9 | 26.8 ± 2.3 | 1.1 ± 0.3 | 4.9 ± 0.4 | 9.9 ± 1.4 | 27.1 ± 3.3 | 1.33 ± 0.20 | ||
| 22–44 | 12.8 ± 1.8 | 42.0 ± 6.5 | 26.5 ± 4.9 | 2.8 ± 0.3c | 3.2 ± 0.6b | 5.4 ± 1.2c | 20.1 ± 2.1b | 2.18 ± 0.15c | ||
| 102–5 | 13.8 ± 0.8 | 35.4 ± 3.8 | 28.4 ± 1.3 | 2.6 ± 0.6b | 4.0 ± 1.2 | 6.8 ± 2.0 | 22.8 ± 2.9 | 1.78 ± 0.23b | ||
| PC | ||||||||||
| WT | 16.6 ± 0.4 | 25.9 ± 2.4 | 2.3 ± 0.6 | 4.6 ± 0.7 | 30.2 ± 4.1 | 36.9 ± 1.0 | 0.35 ± 0.04 | |||
| 22–44 | 13.4 ± 1.4c | 35.4 ± 4.8b | 5.9 ± 1.5b | 2.1 ± 0.1c | 21.2 ± 3.1b | 35.4 ± 3.5 | 0.55 ± 0.11b | |||
| 102–5 | 9.5 ± 0.9d | 33.3 ± 4.5b | 5.5 ± 2.4b | 2.6 ± 0.3b | 23.1 ± 4.1c | 35.6 ± 3.5 | 0.5 ± 0.10b | |||
| PE | ||||||||||
| WT | 10.4 ± 1.9 | 32.9 ± 2.9 | 2.1 ± 0.5 | 3.3 ± 1.4 | 34.6 ± 3.6 | 27.2 ± 1.5 | 0.49 ± 0.06 | |||
| 22–44 | 9.6 ± 1.5 | 38.4 ± 4.1 | 3.5 ± 0.7b | 1.1 ± 0.6b | 26.1 ± 3.3b | 30.9 ± 1.8b | 0.62 ± 0.10 | |||
| 102–5 | 8.4 ± 0.9b | 36.7 ± 5.1 | 5.2 ± 1.1 | 1.4 ± 0.6 | 27.6 ± 4.8b | 29.0 ± 4.6 | 0.58 ± 0.12 | |||
| PI | ||||||||||
| WT | 3.4 ± 0.7 | 50.1 ± 3.5 | 2.8 ± 1.3 | 1.6 ± 0.2 | 20.8 ± 3.6 | 24.7 ± 3.1 | 1.00 ± 0.15 | |||
| 22–44 | 3.5 ± 0.5 | 54.0 ± 0.9 | 3.7 ± 1.1 | 0.9 ± 0.1b | 17.2 ± 2.1 | 24.2 ± 0.1 | 1.18 ± 0.04 | |||
| 102–5 | 3.2 ± 1.2 | 49.8 ± 5.2 | 5.4 ± 1.5b | 1.9 ± 0.9 | 18.5 ± 6.3 | 24.5 ± 4.9 | 0.99 ± 0.21 | |||
a Represents sum of 16:1 cis and trans in MGDG and DGDG, in PG represent 16:1 trans.
b Statistical significance compared with WT was determined by the two-tailed Student's t test, p < 0.05.
c Statistical significance compared with WT was determined by the two-tailed Student's t test, p < 0.01.
d Statistical significance compared with WT was determined by the two-tailed Student's t test, p < 0.001.
Glycerolipid Composition Changes Resulted from a Modified Flux between the Two Glycerolipid Pathways
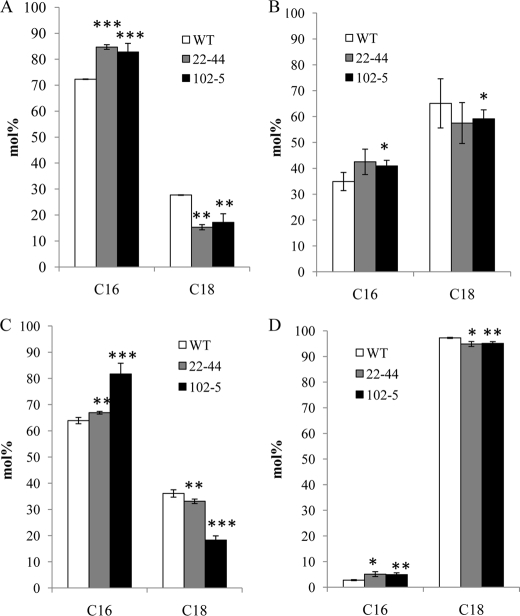
The increased C16:3 and augmented ratio of total 16- and 18-carbon fatty acids in MGDG and DGDG suggested that the prokaryotic glycerolipid pathway was enhanced in the transgenic lines. To verify this, we performed a compositional analysis of fatty acids at the sn-2 position of MGDG and DGDG. As seen in Fig. 2, the molar percent of sn-2 C-16 fatty acid was significantly higher in MGDG and DGDG of the transgenic lines. Our analysis with PG also showed a similar trend. These results thus confirmed that in the transgenic lines, the enhanced Gly-3-P production was associated with a metabolic adjustment augmenting the relative contribution of the prokaryotic glycerolipid pathway. Based on the firmly established concept that diacylglycerol moieties with an sn-2 C16 fatty acid originate from the prokaryotic pathway (10), we were able to deduce the relative contribution of the two glycerolipid pathways in the transgenic lines. As shown in Table 3, the relative contribution from the prokaryotic pathway was clearly enhanced. Interestingly, our analysis also showed that there was an increased level of C16 at sn-2 of PC, suggesting that DAG formed by the prokaryotic pathway may also be channeled back for PC synthesis (17).
FIGURE 2.
Fatty acid composition at the sn-2 position of leaf glycerolipids (lyso-derivatives). Individual lipids were purified by two-dimensional TLC on Silica Gel 60, eluted from the gel, and incubated with Rhizopus lipase as described under “Experimental Procedures.” The resulting lyso lipids (representing the sn-2 position of the parent lipid) were purified by TLC and derivatized using 3 n methanolic HCl, and the resulting fatty acid methyl esters were analyzed by gas chromatography. C16 represents the sum of 16:l, 16:2, and 16:3; C18 represents the sum of 18:0, 18:1, 18:2, and 18:3. A, MGDG; B, DGDG; C, PG; D, PC. Error bars represent mean ± S.D. (n = 3). Statistical significance compared with the wild-type (WT) was determined by the two-tailed Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
TABLE 3.
Comparison of fatty acid flux sharing in wild type (WT) and gpsAFR Arabidopsis leaves
| Mass of fatty acida |
sn-2 positionb |
Flux to prokaryotic pathwayc | Flux to eukaryotic pathwayc | ||
|---|---|---|---|---|---|
| C16 | C18 | ||||
| mol/1000 mol | % | mol/1000 mol of fatty acid | |||
| MGDG | |||||
| WT | 391 | 72.3 | 27.7 | 283 | 108 |
| 22–44 | 456 | 84.7 | 15.3 | 386 | 70 |
| 102–5 | 493 | 82.8 | 17.2 | 408 | 85 |
| DGDG | |||||
| WT | 171 | 34.9 | 65.1 | 60 | 111 |
| 22–44 | 133 | 42.5 | 57.2 | 56 | 77 |
| 102–5 | 149 | 40.9 | 59.1 | 61 | 88 |
| PG | |||||
| WT | 117 | 63.9 | 36.1 | 75 | 42 |
| 22–44 | 128 | 66.9 | 33.1 | 86 | 42 |
| 102–5 | 138 | 81.7 | 18.3 | 112 | 26 |
| PC | |||||
| WT | 166 | 2.8 | 97.2 | 5 | 161 |
| 22–44 | 134 | 5.1 | 94.9 | 7 | 127 |
| 102–5 | 95 | 4.9 | 95.1 | 5 | 90 |
a An original input of 1000 mol of fatty acids synthesized in the chloroplast as acyl-ACP species.
b The tissue complement of each lipid was divided between the prokaryotic and eukaryotic pathways on the basis of content of C16 fatty acid at the sn-2 position.
Lipidomic Analysis Recapitulates Features of an Enhanced Prokaryotic Glycerolipid Pathway
Comparative analysis of individual molecular species within subsets of glycerolipids may shed new light on the alterations of lipid metabolism in the gpsAFR lines. To this end, a complete lipid compositional profile of rosette leaf tissues of 3-week-old seedlings was generated from a lipidomic analysis performed at the Kansas Lipidomic Research Center (43). The analysis, which is based on ESI-MS/MS, yields information on phospholipid and glycolipid molecules to the level of the head group, carbon chain length, and degree of unsaturation of the acyl groups. Data presented in supplemental Fig. S1 were uncorrected for specific response factors of individual molecular species, and the analysis was focused on a comparison of relative mol % of the individual glycerolipid species. In MGDG, there was a significant increase in C34:6, which presumably had a composition of C18:3/16:3 and came from the prokaryotic pathway. Some increase of C34:6 DGDG was also observed, but in DGDG the major change was a decrease in C36:6 (C18:3/18:3) originating from the eukaryotic pathway. Together with the aforementioned sn-2 fatty acyl composition analysis, these results further confirmed the reduced channeling of DAG moieties from the eukaryotic pathway to chloroplasts. In PG, the most significant change was increases in C32:1, C32:0, and C34:0.
Altered Gly-3-P Metabolism Influences Gene-expression Networks
To investigate a potential regulatory role of Gly-3-P metabolism in the context of global gene-expression networks, we surveyed the transcriptome of 3-week-old rosette leaves using the Affymetrix ATH1 genechip. To identify genes that are differentially regulated in gpsAFR transgenic plants, the data were preprocessed using robust multiarray analysis for background adjustment and normalization (37). A total of 389 differentially expressed genes were identified (1.5-fold change cut-off; false discovery rate, p value < 0.05), including 78 up-regulated and 311 down-regulated genes (supplemental Table S1). Interestingly, the microarray data suggested that introduction of the gpsAFR gene had no apparent effect on the transcript levels of plant genes directly involved in Gly-3-P metabolism. These include the glycerol kinase, GLI1 (At1g80460), and several known Gly-3-P dehydrogenase isoforms in the cytosolic and plastidic compartments (At2g40690, At2g41540, At3g07690, and At5g40610).
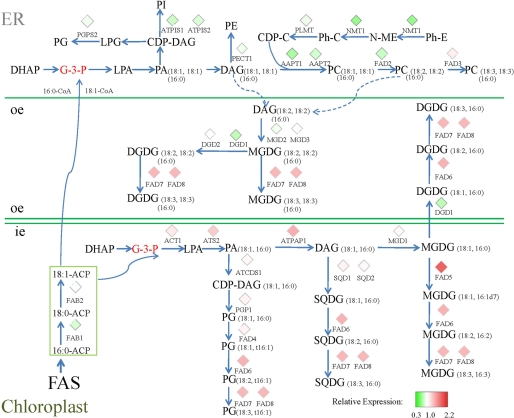
We specifically inspected lipid metabolism genes and the microarray data for all of these genes are provided in supplemental Table S2 (44). Genes with expression changes reaching 1.5-fold represented only a small portion. This was expected because perturbation of transcripts encoding basal components of the biochemical pathways should be small. However, there were a number of genes displaying an expression ratio that significantly deviated from 1 when compared with the wild-type control. A total of 29 glycerolipid biosynthesis-related genes were selected according to their significance in the glycerolipid pathways (supplemental Table S3) and then mapped onto the biochemical pathways compiled from the AraCyc data base and Refs. 2 and 45. This revealed a pattern illustrating a coordinated adjustment of gene expression between enzymes of the two glycerolipid pathways (Fig. 3). Many genes involved in the prokaryotic pathway displayed some degree of enhanced expression, among which the most induced was acyl carrier protein 4 (ACP4), which, in addition to carrying nascent acyl chains during the synthesis of 16- and 18-carbon acyl groups, also serves as the acyl donor for glycerolipid biosynthesis within chloroplasts. Although the plants were raised under identical conditions, the expression of the plastidic desaturase genes, including fatty acid desaturase 5 (FAD5), fatty acid desaturase 6 (FAD6), fatty acid desaturase 7 (FAD7), and fatty acid desaturase 8 (FAD8), were all elevated somewhat in the gpsAFR line. The transcript levels of the eukaryotic pathway enzyme genes, on the other hand, were mostly reduced, most notably the lysophosphatidic acid transferase LPAAT2 (At3g57650) and the ER-bound desaturase FAD2 (At3g12120). Moreover, enzymes involved in PC synthesis, including CDP-DAG synthase (At4g22340), the amino alcohol phosphotransferase, AAPT1 (At1g13560) and AAPT2 (At3g25585), phosphoethanolamine N-methyltransferase, NMT1 (At3g18000), and choline kinase (At4g09760), were all suppressed in the gpsAFR line.
FIGURE 3.
Schematic view of glycerolipid biosynthesis pathways integrated with microarray data. To visualize the relative transcript levels between gpsAFR transgenic plants and the wild-type control, gene-expression data were mapped to the pathway diagrams compiled from the AraCyc data base and Refs. 2 and 45. Different lipid species are derived either from the ER pathway (sn-1/sn-2, C18:C18 or C16:C18) or from the chloroplast pathway (sn-1/sn-2, C18:C16). Dashed lines represent possible channeling between the ER and chloroplasts. Red diamonds represent induced genes, whereas green diamonds denote repressed genes based on the transcript level ratio. Ph-E, phosphorylethanolamine; Ph-C, phosphorylcholine; CDP-C, CDP-choline; oe, outer envelope; ie, inner envelope; FAS, fatty acid biosynthesis.
Exceptions to the contrasting changes in the two glycerolipid pathways were also found, but they too were consistent with the lipid phenotype. Transcripts of digalactosyl diacylglycerol deficient (DGD1), which mediates the synthesis of DGDG from MGDG in chloroplasts, was not elevated, but rather was substantially reduced. This may in part contribute to the lower total amount of DGDG in the gpsAFR line. We also detected some reduction in the transcript level of β-ketoacyl-ACP synthase II (FAB1_KAS II). Because KAS II is involved in fatty acid elongation from 16:0-ACP to 18:0-ACP, its reduction may be relevant to the increased C16:C18 ratio of the gpsAFR lines. In the ER compartment, fatty acid desaturase 3 (FAD3), which generates 18:3 from 18:2 in the PC and PE, exhibited a modest increase in transcript level in contrast to the reduced transcript level of FAD2. This may lead to an enhanced conversion of 18:2 to 18:3, thereby explaining the higher 18:3/18:2 ratio detected in PC and PE.
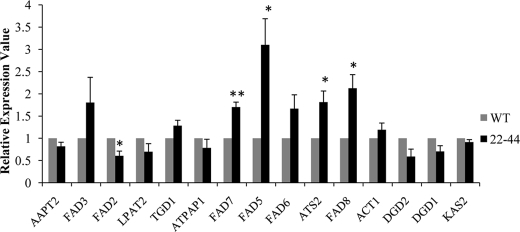
Because the expression changes of most genes in the glycerolipid pathways were less than 1.5, we conducted quantitative RT-PCR assays on 15 genes to verify the microarray results (supplemental Table S4). The fold-change values from both techniques were highly correlated (Pearson correlation value = 0.83) (Fig. 4). These results confirmed the overall reliability of the microarray data.
FIGURE 4.
Real time quantitative RT-PCR validation of the microarray data. The relative expression levels of representative genes from the microarray data as revealed by real time gene-expression analysis. Real time qRT-PCR results using cDNA from WT (wild-type, gray bar) and 22–44 (gpsAFR transgenic line, black bar) as templates. For comparison, the results from wild-type control plants were normalized to 1 using StepOne software 2.0 (Applied Biosystems). The values represent the average of three independent replicates. Statistical significance compared with the WT was determined by the two-tailed Student's t test (*, p < 0.05; **, p < 0.01).
DISCUSSION
Gly-3-P is an intermediary metabolite linking several pathways in the metabolic network (29, 46–49). In addition to glycerolipid synthesis, Gly-3-P metabolism concerns glycolysis, gluconeogenesis (49), homeostasis of the cytosolic NADH/NAD+ ratio, and mitochondrial respiration (29). Routes of Gly-3-P generation exist both in the cytosolic and plastidic compartments through glycerol-3-phosphate dehydrogenases (46, 48). Direct glycerol phosphorylation by glycerol kinase (47) also produces Gly-3-P. The cellular Gly-3-P pool may thus be influenced by a multitude of metabolic factors. In comparison to glycerol/Gly-3-P feeding experiments and mutant studies, transgenic enhancement of Gly-3-P level offers the advantage of not disrupting the associated regulatory circuit. The feedback-resistant gpsAFR gene appears to be particularly potent at increasing Gly-3-P levels, because it was recently reported that overexpression of Gly1 (At2g40690), a Gly-3-P dehydrogenase from Arabidopsis, yielded only a wild-type basal level of Gly-3-P (50). The apparent lack of impact of gpsAFR on the transcript level of the Arabidopsis genes directly leading to Gly-3-P production is intriguing. However, our data does not rule out the possibility that these metabolic steps are affected at the enzyme activity level, as exemplified by Gly-3-P feedback inhibition of the gpsAFR protein in E. coli.
Metabolic networks are often studied as systems downstream of the control of the transcriptome. How an intermediary metabolite influences gene expression in the metabolic network has rarely been studied. Although it is not possible to ascertain whether Gly-3-P serves as a direct or indirect signal in affecting the regulatory architecture of the glycerolipid pathways, our study uncovered clear evidence that an increase in the Gly-3-P level was associated with many transcriptional changes in the glycerolipid pathway enzymes. More importantly, the changes at the transcript level were found to be entirely consistent with the biochemical phenotype, i.e. expression of the prokaryotic pathway enzymes was induced, whereas enzymes of the eukaryotic pathway were suppressed. Thus, although biochemical machinery defines the details as to how the two glycerolipid pathways function as units with distinct underlying biological activities, the Gly-3-P level seems to coordinate their components at the transcript level.
Previously, the Gly-3-P levels in chloroplasts were found to be a limiting factor at the second acylation step of the prokaryotic pathway (30). It was proposed that an elevated stroma Gly-3-P level stimulates the consumption of C16 onto the sn-2 position of the 1-oleoyl glycerol-3-P, thereby reducing the chain elongation of C16 to C18 fatty acids (30). Hence, increases in the C16 level of glycerolipids in the gpsAFR lines are in general agreement with a proposition that in planta the overall C16:C18 ratio is determined by competition between alternative pathways of C16:0 metabolism (15). But the transcript level changes detected in the gpsAFR transgenic lines provide new insight into how this biochemical phenotype was brought about. That the prokaryotic pathway enzymes, particularly the plastidic lysophosphatidic acid acyltransferase (ATS2, At4g30580), were induced indicates that the prokaryotic pathway is enhanced through regulation at the transcript level as well. Interestingly, we also found that ACP4 was one of the most induced genes in the gpsAFR line. It is noteworthy that the km of the spinach chloroplast Gly-3-P acyltransferase for ACP-II, an ortholog of the Arabidopsis ACP4, was shown to be 5 times less than that of ACP-I; whereas the thioesterase has a much lower km for ACP-I (51, 52). Based on these reports, a higher ACP4 expression is expected to boost the fatty acylation process in the chloroplast. We thus suggest that the preferential expression of ACP4 is yet another contributing factor that effected favorable partitioning of fatty acids to the prokaryotic pathway under the modulation of Gly-3-P. Moreover, we observed that the change in the chain length ratio could also be partly explained by a modest reduction at the transcript level of KAS II (At1g74960), which was detected at 0.85 of the wild-type control. This reduction in KAS II on the one hand shifted the partitioning of lipid synthesis to the prokaryotic pathway as demonstrated in the fab1 mutant (19), whereas on the other hand it also reduced the elongation of 16:0-ACP to 18:0-ACP.
In summary, our study reveals connections among Gly-3-P levels, metabolic flux, and the expression of enzymes involved in glycerolipid metabolism in Arabidopsis. Given the current state of genomic and metabolic tools and resources, future studies will be able to determine whether it is Gly-3-P that acts as the de facto signal regulating gene transcription or the changes in the dynamics of the lipid pathway instigated by Gly-3-P metabolism that transduce the signals for gene expression.
Supplementary Material
Acknowledgments
We thank Dr. John E. Cronan, Jr. for providing the bacterial strain BB26-36R2 (gpsAFR, plsB). We are grateful to Mary Roth and Dr. Ruth Welti of the Kansas Lipidomics Research Center for the lipidomic analysis, Michael Giblin and Darwin Reed for technical assistance in GC analysis, Dr. Adrian Cutler, Mark Smith, and Patrick Covello for critical reading of the manuscript, and Dr. Wilf Keller and Dr. Faouzi Bekkaoui for project management.
This work was supported in part by the Saskatchewan Canola Development commission, the National Research Council Canada-Genomic and Health Initiatives, Genome Prairie, and Genome Canada. This research is National Research Council Canada Publication 50160.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Fig. S1.
- PG
- phosphatidylglycerol
- Gly-3-P
- glycerol 3-phosphate
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- MGDG
- monogalactosyldiacylglycerol
- DGDG
- digalactosyldiacylglycerol
- DAG
- diacylglycerol
- qRT
- quantitative reverse transcription
- ER
- endoplasmic reticulum
- ACP
- acyl carrier protein
- ESI-MS
- electronspray tandem mass spectrometry.
REFERENCES
- 1.Gardiner S. E., Roughan P. G., Slack C. R. (1982) Plant Physiol. 70, 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohlrogge J., Browse J. (1995) Plant Cell 7, 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roughan P. G., Slack C. R. (1982) Annu. Rev. Plant. Physiol. 33, 97–132 [Google Scholar]
- 4.Frentzen M., Heinz E., McKeon T. A., Stumpf P. K. (1983) Eur. J. Biochem. 129, 629–636 [DOI] [PubMed] [Google Scholar]
- 5.Heinz E. (1977) in Lipids and Lipid Polymers in Higher Plants (Telvini M., Lichtentaler H. K. eds) pp. 102–120, Springer-Verlag, Berlin [Google Scholar]
- 6.Heinz E., Roughan P. G. (1983) Plant Physiol. 72, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roughan P. G., Holland R., Slack C. R. (1979) Biochem. J. 184, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardiner S. E., Roughan P. G. (1983) Biochem. J. 210, 949–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt H., Heinz E. (1993) Biochem. J. 289, 777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browse J., Warwick N., Somerville C. R., Slack C. R. (1986) Biochem. J. 235, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson G., Williams J. P. (1989) Plant Physiol. 91, 924–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockman J. A., Norman H. A., Hilderbrand D. F. (1990) Phytochemistry 29, 1447–1453 [Google Scholar]
- 13.Härtel H., Dormann P., Benning C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browse J., McCourt P., Somerville C. (1986) Plant Physiol. 81, 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunst L., Browse J., Somerville C. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browse J., Kunst L., Anderson S., Hugly S., Somerville C. (1989) Plant Physiol. 90, 522–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst L., Browse J., Somerville C. (1989) Plant Physiol. 90, 943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miquel M., Browse J. (1992) J. Biol. Chem. 267, 1502–1509 [PubMed] [Google Scholar]
- 19.Wu J., James D. W., Jr., Dooner H. K., Browse J. (1994) Plant Physiol. 106, 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kachroo P., Shanklin J., Shah J., Whittle E. J., Klessig D. F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. (2003) Plant Cell 15, 2952–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kachroo A., Venugopal S. C., Lapchyk L., Falcone D., Hildebrand D., Kachroo P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKee J. W., Hawke J. C. (1979) Arch. Biochem. Biophys. 197, 322–332 [DOI] [PubMed] [Google Scholar]
- 24.Roughan P. G., Holland R., Slack C. R. (1980) Biochem. J. 188, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miquel M., Cassagne C., Browse J. (1998) Plant Physiol. 117, 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell R. M., Cronan J. E., Jr. (1975) J. Biol. Chem. 250, 7153–7158 [PubMed] [Google Scholar]
- 27.Rensink W. A., Pilon M., Weisbeek P. (1998) Plant Physiol. 118, 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 29.Shen W., Wei Y., Dauk M., Tan Y., Taylor D. C., Selvaraj G., Zou J. (2006) Plant Cell 18, 422–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauer A., Heise K. P. (1983) Z. Naturforsch. 38c, 399–404 [Google Scholar]
- 31.Falcone D. L., Ogas J. P., Somerville C. R. (2004) BMC Plant Biol. 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 33.Siebertz H. P., Heinz E. (1977) Z. Naturforsch. 32c, 193–205 [Google Scholar]
- 34.Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H. E., Rajashekar C. B., Williams T. D., Wang X. (2002) J. Biol. Chem. 277, 31994–32002 [DOI] [PubMed] [Google Scholar]
- 35.Millenaar F. F., Roelofs R., Gonzàlez-Meler M. A., Siedow J. N., Wagner A. M., Lambers H. (2000) Plant J. 23, 623–632 [DOI] [PubMed] [Google Scholar]
- 36.Gautier L., Cope L., Bolstad B. M., Irizarry R. A. (2004) Bioinformatics 20, 307–315 [DOI] [PubMed] [Google Scholar]
- 37.Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 39.Smyth G. K. (2004) Statistical Applications in Genetics and Molecular Biology, Vol. 3, Iss. 1, Article 3 [Google Scholar]
- 40.Benjamini Y., Hochberg Y. (1995) J. R. Stat. Soc. 57B, 289–300 [Google Scholar]
- 41.Ou X., Ji C., Han X., Zhao X., Li X., Mao Y., Wong L. L., Bartlam M., Rao Z. (2006) J. Mol. Biol. 357, 858–869 [DOI] [PubMed] [Google Scholar]
- 42.Fliege R., Flügge U. I., Werdan K., Heldt H. W. (1978) Biochim. Biophys. Acta 502, 232–247 [DOI] [PubMed] [Google Scholar]
- 43.Devaiah S. P., Roth M. R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006) Phytochemistry 67, 1907–1924 [DOI] [PubMed] [Google Scholar]
- 44.Beisson F., Koo A. J., Ruuska S., Schwender J., Pollard M., Thelen J. J., Paddock T., Salas J. J., Savage L., Milcamps A., Mhaske V. B., Cho Y., Ohlrogge J. B. (2003) Plant Physiol. 132, 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly A. A., Froehlich J. E., Dörmann P. (2003) Plant Cell 15, 2694–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frentzen M. (1993) Lipid Metabolism in Plants, pp. 195–231, CRC Press, Boca Raton, FL [Google Scholar]
- 47.Ghosh S., Sastry P. S. (1988) Arch. Biochem. Biophys. 262, 508–516 [DOI] [PubMed] [Google Scholar]
- 48.Wei Y., Shen W., Dauk M., Wang F., Selvaraj G., Zou J. (2004) J. Biol. Chem. 279, 429–435 [DOI] [PubMed] [Google Scholar]
- 49.Quettier A. L., Shaw E., Eastmond P. J. (2008) Plant Physiol. 148, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chanda B., Venugopal S. C., Kulshrestha S., Navarre D. A., Downie B., Vaillancourt L., Kachroo A., Kachroo P. (2008) Plant Physiol. 147, 2017–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guerra D. J., Ohlrogge J. B., Frentzen M. (1986) Plant Physiol. 82, 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonaventure G., Ohlrogge J. B. (2002) Plant Physiol. 128, 223–235 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.