Abstract
Plant vacuolar Na+/H+ antiporters play important roles in maintaining cellular ion homeostasis and mediating the transport of Na+ out of the cytosol and into the vacuole. Vacuolar antiporters have been shown to play significant roles in salt tolerance; however the relatively low Vmax of the Na+/H+ exchange of the Na+/H+ antiporters identified could limit its application in the molecular breeding of salt tolerant crops. In this study, we applied DNA shuffling methodology to generate and recombine the mutations of Arabidopsis thaliana vacuolar Na+/H+ antiporter gene AtNHX1. Screening using a large scale yeast complementation system identified AtNHXS1, a novel Na+/H+ antiporter. Expression of AtNHXS1 in yeast showed that the antiporter localized to the vacuolar membrane and that its expression improved the tolerance of yeast to NaCl, KCl, LiCl, and hygromycin B. Measurements of the ion transport activity across the intact yeast vacuole demonstrated that the AtNHXS1 protein showed higher Na+/H+ exchange activity and a slightly improved K+/H+ exchange activity.
Keywords: Evolution/Protein, Exchange/Sodium/Proton, DNA, Membrane Proteins, Yeast, DNA Shuffling, Na+/H+ Antiporter, Salt Tolerance
Introduction
To cope with salt stress and the toxic effect of sodium, plants have developed several adaptive mechanisms including osmotic adjustment and ion homeostasis (1). Because plants lack Na+-ATPases, Na+ homeostasis depends mainly on the expression and activities of Na+/H+ antiporters. Plants use two mechanisms to remove the excess of cytosolic Na+; (i) plasma membrane Na+/H+ antiporters (SOS1-type) that remove Na+ out of the plant cells (2), (ii) the transport of Na+ into the vacuolar compartments, mediated by vacuolar Na+/H+ antiporters (NHX1-type) (3). The plasma membrane and the vacuolar Na+/H+ antiporters are driven by the proton-motive force generated by the plasma membrane H+-adenosine triphosphatase and the vacuolar H+-adenosine triphosphatase and H+-inorganic pyrophosphatase, respectively (3). The sequestration of Na+ into the vacuoles is a significant and cost-effective strategy for reducing cytosol Na+ content that also contributes to the cellular osmotic adjustment for water uptake, cell turgor, and expansion (4). Vacuolar Na+/H+ antiporters have been also shown to play significant roles in endosomal pH regulation (5), Na+ homeostasis, vesicular trafficking, and protein targeting (6–8).
The first plant vacuole Na+/H+ antiporter gene AtNHX1 was cloned and characterized from Arabidopsis thaliana (9). Showing significant sequence homology with the Na+/H+ antiporters ScNHX1 of Saccharomyces cerevisiae, AtNHX1 is functionally analogous to ScNHX1 and localized to the plant vacuole membrane fractions (9, 10). Moreover, Na+/H+ exchange activity could be measured in plant vacuoles (11, 12), vacuolar membrane fraction of yeast expressing the AtNHX1 protein and reconstituted liposomes (13, 14). The overexpression of AtNHX1 conferred salt tolerance in the transgenic plants and the salt tolerance was correlated with increased vacuolar Na+/H+ exchange activity and vacuolar sodium accumulation. These studies suggested that plant vacuole Na+/H+ antiporter genes play a significant role in plant salt tolerance (11, 12, 15).
Some advances have been achieved toward understanding the structure-function relationships of the Na+/H+ antiporter in yeast (16–20). In contrast, studies on the plant vacuole Na+/H+ antiporters are scanty. Studies indicated that the C terminus of plant Na+/H+ antiporter play an important role in the regulation of the antiporter activity. The AtNHX1 C terminus was found to regulate the activity of the Na+/H+ antiporter via its interaction with AtCaM15 in a pH- and Ca2+-dependent manner (21, 22), and several amino acid residues were shown to be critical for the antiporter cation specificity (23). Although the overexpression of plant vacuolar Na+/H+ antiporters conferred enhanced salt tolerance to the transgenic plants, the relatively low Vmax of the cloned Na+/H+ antiporters are a limitation for the application of the Na+/H+ antiporter genes for crop molecular breeding.
DNA shuffling is a powerful process for directed evolution, which generates diversity by recombination, combining useful mutations from individual genes (24). DNA shuffling followed by screening or selection has proved to be a useful approach for the evolution of single gene products with enhanced activity, altered substrate specificity, or improving protein folding and of entire operons with improved function (25). In this study, we applied DNA shuffling methodology combined with the heterologous expression of the shuffled gene products in yeast to screen novel Na+/H+ antiporter genes conferring improved activity and enhanced tolerance of yeast to high NaCl concentrations. One shuffled Na+/H+ antiporter gene obtained showed a 1-fold improved NaCl resistance. The yeast strain harboring the shuffled Na+/H+ antiporter gene accumulated more sodium than that expressing AtNHX1. Furthermore, The shuffled Na+/H+ antiporter displayed higher Vmax of the Na+/H+ exchange across the yeast vacuole membrane, thus indicating that the novel Na+/H+ antiporter gene possesses enhanced sodium transport activity.
EXPERIMENTAL PROCEDURES
Yeast Strains and Medium
S. cerevisiae mutant Δena1–2 (Δena1–2::HIS3) and Δena1–4Δnhx1 (Δena1–4::HIS3, Δnhx1:: TRP1) were derived from W303–1B(MATα'ura3–1′leu2–3′trp1–1′his3–11′ade2–1,can1–100), NHX1 deletion mutant TY001 (Δnhx1::TRP1) were derived from the vacuolar protease-deficient strain MM476 (MATα′pep4–3′leu2′trp1′ura3–52′prb1–1122). Yeast cells were grown in YPD (1% yeast extract/2% peptone/2% glucose). Salt tolerance tests were performed in APG medium, which is synthesis medium containing 10 mm arginine, 8 mm phosphoric acid, 2 mm MgSO4, 1 mm KCl, 0.2 mm CaCl2, 2% (w/v) glucose, trace vitamins, and minerals (26). Where indicated, NaCl, LiCl, KCl, or hygromycin B was added, or the pH was adjusted to 5.5 with acetic acid.
DNA Shuffling of AtNHX1
The cDNA of AtNHX1 was amplified from the pGM-T-AtNHX1 construct with the two primers: Atr1 and Atf1 (Table 1), and the product was purified using Wizard PCR Preps DNA purification (Promega).
TABLE 1.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| At f1 | TCCCCCGGGATGTTGGATTCTCTAGTG |
| At r1 | GCGTCGACTCAAGCCTTACTAAGATCAGG |
| At f2 | TCCCCCGGGATGTTGGATTCTCTAG |
| At r2 | CCGGAATTCAGCCTTACTAAGATCAGG |
| GFP3 f | CCGGAATTCATGAGTAAAGGCGAAGAAC |
| GFP3 r | CGGGGTACCTTATTTGTATAGTTCATCCATG |
| AtS1 r2 | CCGGAATTCGTCATCAGATAACATGCTC |
| At L29P f | GTTGAATCTCTTTGTTGCACCtCTTTGTGC |
| At L29P r | aGGTGCAACAAAGAGATTCAACGCAACC |
| At S158P f | CCATATTTGCTGCAACAGATcCAGTATGTAC |
| At S158P r | TGgATCTGTTGCAGCAAATATGGCACC |
| At CDL296 r | GCGTCGACTCAGTCATCAGATAACATGCTC |
a New restriction sites are underlined, mutated nucleotides are in lowercase letters.
The shuffling procedure was performed as described (24, 27). Total 2-μg products were digested by DNase I for 2 min, and the fragments between 100 and 200 bp were confirmed on a 2% agarose gel before purification by Wizard SV Gel and PCR Clean-up system (Promega).
The purified fragments (1 μg) were reassembled by PCR without primer. The PCR program was as follow: 60 s, 94 °C followed by 40 cycles of 30 s 94 °C, 30 s 50 °C, 30 s 72 °C, followed by 7 min at 72 °C.
1 μl of this reaction was used as template in a 30-cycle PCR reaction to amplify the reassembled products. PCR program was conducted as follow: 60 s, 94 °C followed by 30 cycles of 30 s 94 °C, 60 s 58 °C, 30 s 72 °C, followed by 7 min at 72 °C. This program produces a single band with the correct size.
The Construction of Shuffled Gene Library and Screening for High Salt Tolerance Yeast Clones
The reassembled fragment of the correct size were purified by Wizard SV Gel and PCR clean-up system, and digested by SalI and SmaI before purification. The purified products were ligated into the yeast express vector pYPGE15, which was digested by the same enzymes. The constructs were transformed into the double mutant yeast strain Δena1–4::HIS3 Δnhx1::TRP1 using lithium acetate method as described before (28). The strains were screened on the APG selective medium supplemented with 100 mm NaCl at pH 5.5. The growth was at 30 °C for 4 days. For the second selection, the yeast clones obtained from the first screening were grown to A600 = 0.5, and 5 μl of the cells were loaded onto APG-selective medium supplemented with 125 mm NaCl at pH 5.5.
Yeast Transformation, Drop Tests, and Intracellular Ion Content Determination
To compare salt tolerance of shuffled Na+/H+ antiporter gene with that of the wild-type gene, the open-reading frame of AtNHX1 was cloned into the yeast express vector pYPGE15 using SalI and SmaI. The construct was introduced into the yeast double mutant strain W303–1B Δena1–4::HIS3 Δnhx1::TRP1. Whereas the yeast wild-type strain W303–1B, yeast mutant strains W303–1B Δena1–2::HIS3 and W303–1BΔena1–4::HIS3 Δnhx1::TRP1 were all transformed with empty pYPGE15.
For drop tests, the cells of different strains were harvested by centrifugation and resuspended into APG medium, adjusted to A600 of 0.5. Serial 5-time dilutions were made, and 5 μl of the cells were loaded onto APG medium with different salt supplements.
The total Na+ and K+ content of the different yeast cells grown in APG medium supplemented with 70 mm NaCl was determined as described before (29, 30). The yeast cells were collected by centrifugation and then washed twice with ice-cold 10 mm MgCl2, 10 mm CaCl2, 1 mm HEPES. Intracellular ions were extracted by HCl and analyzed with an atomic absorption spectrometer.
AtNHX1-GFP Localization and Confocal Microscopy
C terminus GFP3-tagged AtNHX1 and AtNHXS1 were constructed separately. GFP3 with stop codon was subcloned into the pYPGE15 vector, digested by EcoRI and KpnI enzymes. The new construct was named pYPGE15-GFP3. The AtNHX1 and AtNHXS1 open-reading frames without the stop codon were separately inserted between EcoRI and SmaI site of pYPGE15-GFP3. These constructs were introduced into the yeast mutant strain TY001, respectively.
Drop tests were performed as described above, and cells were placed onto YPD medium with 10 μg/ml hygromycin B. Exponentially growing cells were labeled with 16 μm FM4-64 for 30 min and mounted on the coverslips. Confocal images were acquired with a Leica TCSP5 confocal laser microscope system and a 63× oil immersion objective.
Isolation of Intact Yeast Vacuoles
The empty pYPGE15, pYPGE15-AtNHX1, and pYPGE15-AtNHXS1 were introduced into the vacuolar protease-deficient strain TY001. Intact vacuoles were isolated as described before (21). The intact vacuoles were collected at the top of the gradient and resuspended in 10 mm MES-Tris2 (pH 7.5). The highly purified vacuoles were almost free of contamination by unbroken spheroplasts and lipid granules. Protein concentrations were determined by the Protein Assay (Tiangen) according to the manufacturer's protocol.
Transport Assay
The fluorescence quenching of acridine orange was used to monitor the establishment and dissipation of vacuolar inside acidic pH gradients as described before (21). The time-dependent fluorescence changes were monitored on a Hitachi F4500 fluorescence spectrophotometer with excitation and emission wavelengths of 495 and 540 nm, respectively, and a slit width of 5 nm with a 1% transmittance filter. Initial rates were measured as the slope of the relaxation of the quench over a period of 25 s. Rates were reported as percent quench per min per mg of protein. Curves were fitted to the mean rate values at each concentration.
Mutagenesis and Deletion of AtNHX1, Yeast Transformation, and Growth Test
To explore the effects of the amino acids mutations and the deleted C terminus on the functions of Na+/H+ antiporters, the L29P, S158P mutants and the C-terminal-deleted mutant were constructed. The pGM-T-AtNHX1 was used as the template for site-directed mutagenesis. All amino acid substitutions were generated by PCR methods using the appropriate base changes in the synthetic primers. The C-terminal deletion was obtained by PCR amplification with the primer Atf1 and AtCDL296r (Table 1). Mutagenesis and deletion mutants were cloned into the expression vector pYPGE15 digested by SalI and SmaI enzymes and were confirmed by sequencing. These constructs were introduced into the yeast double mutant strain W303–1B Δena1–4::HIS3 Δnhx1::TRP1, drop tests were performed as described above, onto APG plates with different NaCl or KCl or LiCl supplements at pH 5.5 or onto YPD medium with 10 μg/ml hygromycin B. The growth of these strains in the liquid APG medium with salt or the liquid YPD medium with hygromycin B was also investigated for quantitation. Vectors expressing AtNHX1 bearing the mutations L29P or S158P or CDL296 were introduced into the vacuolar protease-deficient strain TY001. Vacuoles were isolated as described above and cation-dependent proton transport was performed by monitoring the fluorescence quenching of acridine orange as described.
RESULTS
Screening and Validation of Shuffled AtNHX1
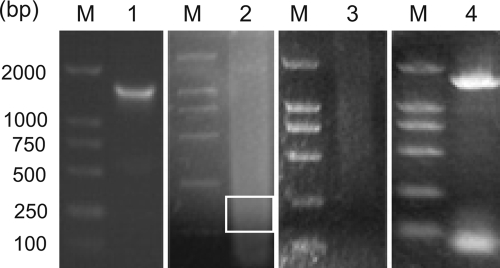
The wild-type AtNHX1 gene was amplified by PCR when the random mutants were produced and was fragmented by DNaseI. After the combination, the column-purified 100–200-bp fragments were assembled through PCR without primer to a single gene product of the correct size. The results of the gene fragmentation, reassembly, and amplification are shown as Fig. 1. After being ligated into the yeast expression vector pYPGE15 and then introduced into the yeast double mutant strain W303–1BΔena1–4Δnhx1, which lacks Na+-ATPase genes ENA1–4 (31) and the endosomal Na+/H+ antiporter gene NHX1 (32), the shuffled gene library was expressed and screened on APG medium supplied with 100 mm NaCl at pH 5.5. The transformation rate was about 103-104/μg plasmid DNA, and about 105 shuffled gene mutants were constructed by a one-time operation. For screening, 25 plates containing 400–600 clones per plate were screened simultaneously, allowing for an efficient large scale method for the identification of yeast carrying improved forms of the vacuolar antiporter With the NaCl selective pressure, more than two hundred clones grew normally on the plates with NaCl selective pressure, and the clone showing best grow (clone 25) during the second selection was selected for further characterization (Fig. 2A). The shuffled gene expressed in this yeast clone was named AtNHXS1.
FIGURE 1.
DNA agarose gel (2.5%) showing process of high fidelity DNA shuffling of wild-type AtNHX1 gene. Lane M, DL2000, from top to the bottom: 2000, 1000, 750, 500, 250, and 100 bp. Lane 1, 1.6-kb fragment of AtNHX1 acquired by PCR; lane 2, DNA fragments after 2 min of DNaseI digestion in the presence of Mn2+, white box indicates the 100–200-bp fragments that are recycled; lane 3, fragment reassembly with Pfu polymerase; and lane 4, PCR amplification of the reassembly with Pfu polymerase.
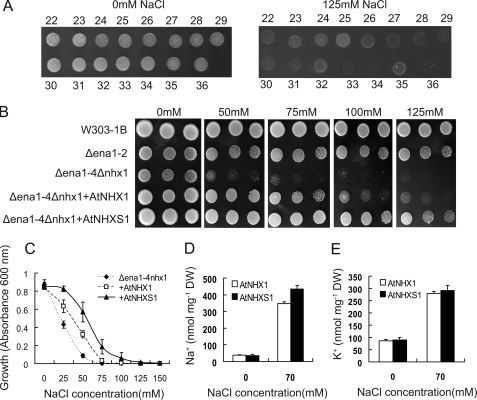
FIGURE 2.
NaCl tolerance of AtNHX1 and shuffled AtNHXS1 gene in expressed in the Δean1–4 Δnhx1 yeast double mutant. A, second selection of shuffled AtNHX1 gene. Several yeast clones (numbered 22–36) obtained from the first screening were grown on an APG plate containing 125 mm NaCl for 4 days. B, serial dilutions (at 105, 2 × 104 and 4 × 103 cells) of the yeast strains. Cells were grown on selective plates as described under “Experimental Procedures.” C, growth of the yeast strains in liquid medium. Absorbance measured at 600 nm after culturing for 96 h at 30 °C Results are the mean ± S.D. (n = 3). D and E, Na+ and K+ content of cells grown in the presence of NaCl. Intracellular Na+ and K+ contents were determined in cells grown in APG medium with 70 mm NaCl until the mid-exponential growth phase. Results are the mean ± S.D. (n = 3).
Increased Salt Tolerance Conferred by Shuffled AtNHX1
To compare the salt tolerance of yeast strains harboring and expressing the shuffled and wild-type genes, the pYPGE15-AtNHX1 was introduced into the yeast mutants. The different yeast strains were grown on the APG medium supplemented with different NaCl concentrations.
As shown in Fig. 2B, the yeast mutant strain Δena1–4Δnhx1 lacking the endogenous Na+/H+ antiporter was more sensitive to NaCl than that of the wild-type W303–1B and single mutant strain Δena1–2. Expression of AtNHX1 in the yeast mutant partially recovered the mutant phenotype, indicating that AtNHX1 functions as a Na+/H+ antiporter. The shuffled AtNHX1 conferred the yeast mutant a tolerance to salt to up to 125 mm NaCl, whereas the yeast cells expressing the wild-type gene was only able to tolerate the presence of 75 mm NaCl in the growth medium. The liquid salt tolerance test showed that the salt tolerance of yeast expressing AtNHXS1 was improved about 1-fold as compared with the yeast carrying AtNHX1 (Fig. 2C).
The total ion contents of the yeast cells were determined. In the presence of 70 mm NaCl in the growth medium, the yeast strain harboring AtNHXS1 was able to accumulate more Na+ than the wild type (Fig. 2D). The K+ contents of the yeast expressing AtNHXS1 was slightly higher than that of yeast harboring AtNHX1 (Fig. 2E). These results would suggest that the yeast strain expressing AtNHXS1 have a higher ability to sequester Na+ into the vacuole, and a concomitant improved NaCl tolerance.
Vacuole Localization of Mutant AtNHX1
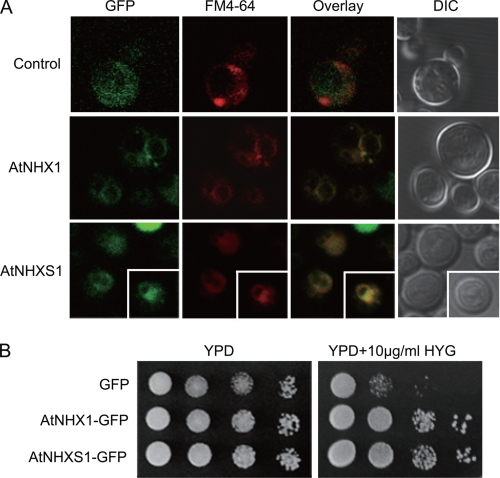
Previous studies have been shown that AtNHX1 localized in the vacuolar membrane fraction of yeast cells (9, 10). To verify the localization of shuffled AtNHX1, GFP was fused downstream with the Na+/H+ antiporter, which did not change the protein function. The expression of AtNHX1-GFP fused version in S. cerevisiae TY001 resulted in the improved tolerance to hygromycin B (Fig. 3B). Similar to that of AtNHX1-GFP3, the fluorescence of the GFP3-tagged AtNHXS1 was predominant in membranes of the vacuole and endosomes, whereas GFP3 distributed around the yeast cells, indicating its cytosolic localization (Fig. 3A).
FIGURE 3.
Confocal microscopy of GFP-tagged AtNHX1 and AtNHXS1 in yeast. A, TY001 cells were transformed with GFP, AtNHX1-GFP, and AtNHXS1-GFP constructs, respectively seen under differential interference contrast (DIC) and confocal microscopy. Vacuoles and endosomes were stained with FM4-64. B, growth of the same strains on the YPD supplemented with or without hygromycin B.
Transport Assays
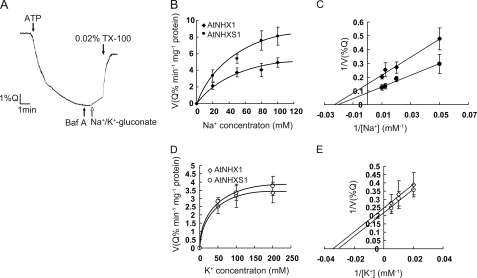
To test the effect of the C terminus and mutations of some AtNHXS1 amino acids on the regulation of the antiporter activity, proton-dependent sodium movements were monitored by monitoring the fluorescence quenching of acridine orange. As shown in Fig. 4A, vacuolar vesicle acidification was initiated by the addition of ATP, and after a steady-state acidic-inside pH gradient was attained, the activity of the H+-ATPase was partially inhibited by the addition of Bafilomycin A. The addition of Na+-gluconate or K+-gluconate resulted in a cation-dependent H+ movement and the alkalinization of the vacuole lumen (recovery of the fluorescence). The addition of 0.02% Triton X-100 resulted in the collapse of the ΔpH across the tonoplast.
FIGURE 4.
Ion transport activities of AtNHX1 and AtNHXS1. Na+- or K+-dependent proton movements were monitored by following the fluorescence quenching of acridine orange as described under “Experimental Procedures.” A, vacuole Na+-dependent H+ transport. ATP, Bafilymycin A (Baf A), Na+-gluconate or K+-gluconate, and Triton X-100 were added at the indicated times. B, AtNHX1- or AtNHXS1-mediated Na+/H+ exchange kinetics. C, double reciprocal plot of the Na+-dependent H+ fluxes. AtNHX1 (♦), AtNHXS1 (●). D, AtNHX1- or AtNHXS1-mediated K+/H+ exchange kinetics. E, double reciprocal plot of the K+-dependent H+ fluxes. AtNHX1 (◇), AtNHXS1 (○). Values are the mean ± S.D. (n = 3).
AtNHX1 and AtNHXS1 displayed both Na+- and K+-coupled H+ transport activity. In all cases, these two transporters showed a Michaelis-Menten-type saturation kinetics. The shuffled AtNHX1 displayed a significantly higher apparent Vmax of the Na+/H+ with about 1-fold increase (from 6.65 to 11.4 Q % min−1 mg−1 protein) (Fig. 4, B and C), while the Vmax of the K+/H+ was slightly higher (from 4.05 to 4.56 Q % min−1 mg−1 protein) compared with that of the wild-type AtNHX1 (Fig. 4, D and E). AtNHXS1 displayed a higher Na+/K+ selectivity (Vmax(Na+)/Vmax(K+) = 2.5) than AtNHX1 (Vmax(Na+)/Vmax(K+) = 1.6) and a minor decrease in affinity for Na+ (48.5 mm and 43 mm in AtNHXS1 and AtNHX1, respectively) and K+ (32.5 mm and 29.2 mm).
Sequence Analysis of Shuffled AtNHX1
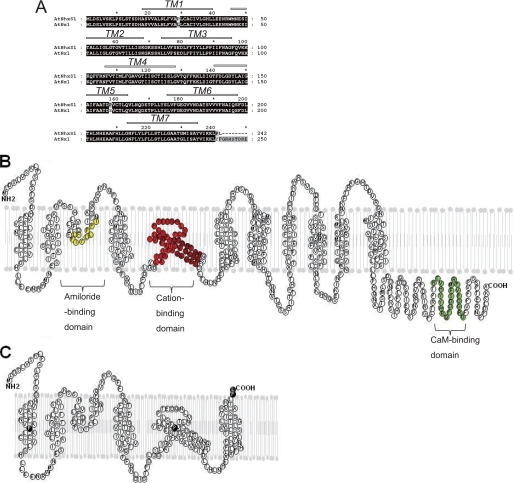
A comparison of AtNHXS1 with AtNHX1 showed that the AtNHXS1 gene has seven nucleic acids mutations and forty-two nucleic acids deletions, which resulted in the presence of a premature termination code (Table 2). In the amino acid sequence of AtNHXS1, there are four substitutions, leucine for proline at the first putative transmembrane segment (L29P), proline for serine at the fifth transmembrane domain (S158P), tyrosine for praline (Y241P), and phenylalanine for leucine (F242L) at the end of the shuffled protein, and there is one truncation of 296 amino acids in the C terminus (Fig. 5).
TABLE 2.
DNA substitutions and deletion in shuffled AtNHX1 gene
| Base | Base substitution |
|---|---|
| 86 | T → C |
| 472 | T → C |
| 691 | C → A |
| 850 | T → C |
| 1099 | G → A |
| 1498 | T → C |
| 1555 | T → C |
| 720–753 | Deletion |
FIGURE 5.
Sequence analysis of AtNHXS1. A, comparison of deduced amino acid sequences of shuffled AtNHX1 and wild-type AtNHX1. The partial topology of the AtNHX1 protein, as proposed by protease protection assays (21) is shown above the sequence with open bars (predicted transmembrane segments). B, topological model of AtNHX1 based on Yamaguchi et al. (21). C, topological model of AtNHXS1; black residues indicate single amino acid substitutions.
Salt Tolerance Conferred by Mutagenesis in AtNHX1 and Truncated AtNHX1
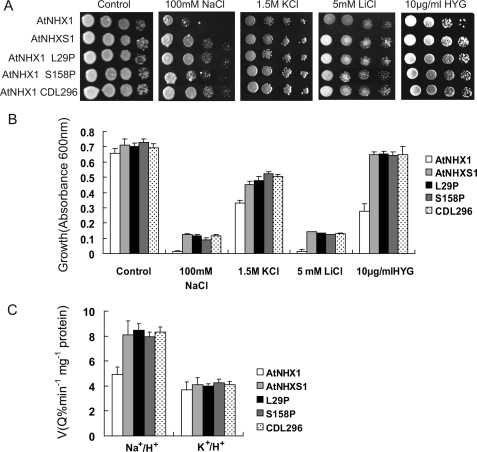
To elucidate which amino acid residues and/or the partial deletion of the partial C terminus in AtNHXS1 is indispensable to attain the increased activity of the shuffled Na+/H+ antiporter, the different mutagenesis, and deletion mutants of AtNHX1 were constructed and expressed in the yeast double mutants. As shown in Fig. 6, yeast cells expressing AtNHX1 protein with the mutation L29P, or the mutation S158P, or with a 296 amino acids C-terminal deletion, conveyed NaCl tolerance as effectively as that resulting from the expression of the AtNHXS1 protein. Also, the expression of the NHX1 mutations L29P, S158P, and the C-terminal truncation displayed similar enhancement of tolerance to KCl, LiCl, and hygromycin B, a compound that its toxicity has been correlated with yeast membrane depolarization (33).
FIGURE 6.
Growth of yeast containing either wild-type or mutant AtNHX1. A, Δena1–4Δnhx1 cells expressing AtNHX1, AtNHXS1, L29P, S158P, and 296 amino acid-truncated C-terminal versions were serially diluted 10-fold from A600 = 1.0 and spotted onto the APG medium supplemented with NaCl, KCl, LiCl, at the indicated concentrations or YPD medium supplemented with hygromycin B (HYG). Plates were incubated for 3 days at 30 °C. B, same strains (20 μl of a saturated seed culture) were grown in 3 ml of APG medium (pH 5.5) supplemented with or without NaCl, KCl, LiCl, or YPD medium supplemented with hygromycin B for 72 h with shaking. Growth was measured as absorbance at 600 nm. Values are the mean ± S.D. (n = 3). C, ion transport activities of AtNHX1, AtNHXS1, and AtNHX1 bearing L29P or S158P or CDL296 mutations. Na+- or K+-dependent proton movements were monitored by following the fluorescence quenching of acridine orange as described before. Na+/H+ and K+/H+ exchange was measured by addition of 100 mm Na+-gluconate or 200 mm K+-gluconate, respectively. Values are the mean ± S.D. (n = 3).
The cation-dependent H+ transport was also measured in vacuoles isolated from yeast expressing AtNHX1 molecules bearing L29P or S158P or CDL296 mutations. Notably, the initial rates of Na+/H+ and K+/H+ exchange of each antiporter was similar to that of AtNHXS1 (Fig. 6C).
DISCUSSION
DNA shuffling is a powerful tool for molecular directed evolution which has been adopted widely for the improvement of the stability and activity of enzymes, changing the affinity of the substrate, enzyme turnover, etc (25, 34). Because plant Na+/H+ antiporters play a very important role in ion homeostasis and salt tolerance (35), the molecular evolution of Na+/H+ antiporters not only could potentially increased the application of these genes in salt tolerance-crop breeding, but can be also helpful to understand the mechanism(s) of transport and regulation of the Na+/H+ antiporters.
In this study, AtNHX1 was the substrate for DNA shuffling. Because the gene is about 1.6 kb in length, 100–200-bp fragments were recycled and were successfully recombined through primerless PCR. An appropriate digested fragment length is crucial for a successful shuffling. In addition, a suitable selective model and expression system are key factors for successful mutant screening. We applied DNA shuffling methodology combined with heterologous expression in yeast to screen novel Na+/H+ antiporter shuffled genes with improved activity and enhanced NaCl tolerance. The selection of transformed yeast in medium containing high NaCl allowed for the simultaneous large-scale screening of a large number of shuffled genes (≈104). More than 200 clones were obtained after the first screening cycle, and one clone (25) was chosen to further analysis after the second selection test. Yeast complementation tests showed that this shuffled clone displayed enhanced tolerance to NaCl, KCl, LiCl, and hygromycin B (Figs. 2 and 6).
When expressed in yeast, C-terminal green fluorescent protein-tagged AtNHXS1 remained localized to the vacuole (Fig. 3). The yeast expressing the shuffled Na+/H+ antiporter accumulated more sodium than that containing the wild type form of AtNHX1 (Fig. 2D), and AtNHXS1 had higher sodium transport activity across the intact yeast vacuole as compared with that of AtNHX1 (Fig. 4). These results suggested that the amino acid mutations and C-terminal partial deletion in AtNHXS1 did not alter the localization of the antiporter and increased the antiporter Na+/H+ exchange activity, thus enabling a higher NaCl tolerance. On the other hand, AtNHXS1 displayed a lesser increment in the Vmax of K+/H+ exchange, and there was some increase in yeast KCl tolerance.
Some progress has been made in understanding the mechanism of ion transport and regulation of Na+/H+ antiporter in Escherichia coli, yeast, and mammals (36). The C terminus of the mammal NHE1 contains an autoinhibitory region, and the interaction of this region with CaM-regulatory proteins restored optimal Na+/H+ antiporter activity (37). A similar phenomenon was found in yeast (16). In A. thaliana, the deletion of vacuolar AtNHX1 C terminus doubled the Na+/K+ selectivity ratio in yeast, also suggesting a regulatory role of the C terminus on the antiporter activity (21). Further studies indicated that the cation selectivity of AtNHX1 can be modified through the interaction of its C terminus with AtCaM15, a calmodulin (CaM)-like protein in a pH- and Ca2+- dependent manner (22). Increased pH values weakened the binding of CaM to the C terminus and favored Na+ transport without affecting significantly K+ transport into the vacuole.
In this study, AtNHXS1 contains four amino acid substitutions; L29P in the first transmembrane domain, S158P in the fifth transmembrane domain, Y241P and F242L at the end of the shuffled protein and the deletion of 296 amino acids of its C terminus. Taking into consideration the topology proposed for AtNHX1 (21), AtNHXS1 would comprise only seven transmembrane domains (Fig. 5), lacking the 8–12 transmembrane domains and the hydrophilic tail of the C terminus containing the CaM-binding domain. L29P resides in the first transmembrane domain of AtNHX1. Leu29 and Leu30 in AtNHX1 are similar to Leu110 and Leu111 in the second transmembrane domain of NHE1, a region postulated to participate in the formation of path for cation transport (36). S158P is located in a region containing amino acids 147–193, a highly conserved region among Na+/H+ antiporters of mammals (NHE1), yeast (Nhx1), and plants (AtNHX1). This region has been postulated to contain the putative cation-binding domain as mapped by homology to other antiporters (23). Y241P and F242L would not play any role in cation or proton transport. The shuffled Na+/H+ antiporter, AtNHXS1, displayed an improved Na+/H+ exchange activity, and its expression conferred a significantly higher NaCl tolerance to yeast. Our data support the notion that the C-terminal deletion and the mutations in the putative cation transport path and the mutations in the cation-binding domain of the Na+/H+ antiporter increased Na+/H+ exchange activity and Na+ selectivity, while decreasing Na+ and K+ affinity.
A few critically conserved charged amino acids may be responsible for the cation binding and coordination by Na+/H+ exchanger proteins. For example, the polar amino acids are usually pH sensors and mutants of these amino acids usually results in the change of the Na+/H+ activities under different pH conditions (38–40). Asp138 of the Na+/H+ antiporter from Synechocystis species PCC6803 was found to be necessary for the cation transport activity. The related mutants lead to the lost of transport function (41). In this study, the L29P and S158P mutations, localized in the predicted transmembrane domains, were found to increase the salt tolerance of AtNHX1, significantly. As proline is a helix breaker that can disrupt the α-helical structure (42), it is possible that the L29P or S158P substitutions resulted in the alteration of Na+/H+ antiporter structure, favoring Na+/H+ exchange. Notably, when each of the mutations L29P, S158P, and the C-terminal deletion CDL296 were expressed, the cation-H+ exchange activity did not differ from that displayed by AtNHXS1. These results would suggest that similar to the mammalian antiporter NHE1, there is an interaction between the TM1 and the cation-binding domain in AtNHX1. Moreover, the effects seen in the mutant bearing the C-terminal deletion confirmed the proposed regulatory role of the C terminus on antiporter cation selectivity and transport activity (22).
In conclusion, we used DNA shuffling methodology combined with a large scale yeast complementation system to evolve the plant Na+/H+ antiporter gene AtNHX1 and obtained one novel Na+/H+ antiporter gene AtNHXS1, containing four amino acid substitutions and a C-terminal deletion. These mutations and deletion did not alter the vacuolar localization of the Na+/H+ antiporter but increased the Na+/K+ selectivity. Furthermore, AtNHXS1 displayed a higher Na+/H+ transport activity, which enabled yeast to accumulate more Na+ and K+ in the vacuole and increased its salt tolerance.
This work was supported by the National Natural Science Foundation of China (30671336).
- MES
- 4-morpholineethanesulfonic acid
- NHX1
- Na+/H+ antiporter
- GFP
- green fluorescent protein.
REFERENCES
- 1.Bohnert H. J., Nelson D. E., Jensen R. G. (1995) Plant Cell 7, 1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi H., Lee B. H., Wu S. J., Zhu J. K. (2003) Nat. Biotechnol. 21, 81–85 [DOI] [PubMed] [Google Scholar]
- 3.Apse M. P., Blumwald E. (2007) FEBS Lett. 581, 2247–2254 [DOI] [PubMed] [Google Scholar]
- 4.Chinnusamy V., Jagendorf A., Zhu J. K. (2005) Crop Sci. 45, 437–448 [Google Scholar]
- 5.Yamaguchi T., Fukada-Tanaka S., Inagaki Y., Saito N., Yonekura-Sakakibara K., Tanaka Y., Kusumi T., Iida S. (2001) Plant Cell Physiol. 42, 451–461 [DOI] [PubMed] [Google Scholar]
- 6.Apse M. P., Sottosanto J. B., Blumwald E. (2003) Plant J. 36, 229–239 [DOI] [PubMed] [Google Scholar]
- 7.Bowers K., Levi B. P., Patel F. I., Stevens T. H. (2000) Mol. Biol. Cell 11, 4277–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sottosanto J. B., Gelli A., Blumwald E. (2004) Plant J. 40, 752–771 [DOI] [PubMed] [Google Scholar]
- 9.Gaxiola R. A., Rao R., Sherman A., Grisafi P., Alper S. L., Fink G. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintero F. J., Blatt M. R., Pardo J. M. (2000) FEBS Lett. 471, 224–228 [DOI] [PubMed] [Google Scholar]
- 11.Apse M. P., Aharon G. S., Snedden W. A., Blumwald E. (1999) Science 285, 1256–1258 [DOI] [PubMed] [Google Scholar]
- 12.Zhang H. X., Blumwald E. (2001) Nat. Biotechnol. 19, 765–768 [DOI] [PubMed] [Google Scholar]
- 13.Venema K., Quintero F. J., Pardo J. M., Donaire J. P. (2002) J. Biol. Chem. 277, 2413–2418 [DOI] [PubMed] [Google Scholar]
- 14.Darley C. P., van Wuytswinkel O. C., van der Woude K., Mager W. H., de Boer A. H. (2000) Biochem. J. 351, 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H. X., Hodson J. N., Williams J. P., Blumwald E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinclová O., Ramos J., Potier S., Sychrová H. (2001) Mol. Microbiol. 40, 656–668 [DOI] [PubMed] [Google Scholar]
- 17.Kamauchi S., Mitsui K., Ujike S., Haga M., Nakamura N., Inoue H., Sakajo S., Ueda M., Tanaka A., Kanazawa H. (2002) J. Biochem. 131, 821–831 [DOI] [PubMed] [Google Scholar]
- 18.Dibrov P., Young P. G., Fliegel L. (1998) Biochemisry 37, 8282–8288 [DOI] [PubMed] [Google Scholar]
- 19.Simon E., Barceló A., Ariño J. (2003) FEBS Lett. 545, 239–245 [DOI] [PubMed] [Google Scholar]
- 20.Mitsui K., Kamauchi S., Nakamura N., Inoue H., Kanazawa H. (2004) J. Biochem. 135, 139–148 [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi T., Apse M. P., Shi H., Blumwald E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12510–12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi T., Aharon G. S., Sottosanto J. B., Blumwald E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernández A., Jiang X., Cubero B., Nieto P. M., Bressan R. A., Hasegawa P. M., Pardo J. M. (2009) J. Biol. Chem. 284, 14276–14285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stemmer W. P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10747–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crameri A., Raillard S. A., Bermudez E., Stemmer W. P. C. (1998) Nature 391, 288–291 [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez-Navarro A., Ramos J. (1984) J. Bacteriol. 159, 940–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H., Arnold F. H. (1997) Nucleic Acids Res. 25, 1307–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gietz D., St. Jean A., Woods R. A., Schiestl R. H. (1992) Nucleic Acids Res. 20, 1425–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venema K., Belver A., Marin-Manzano M. C., Rodríguez-Rosales M. P., Donaire J. P. (2003) J. Biol. Chem. 278, 22453–22459 [DOI] [PubMed] [Google Scholar]
- 30.Montiel V., Ramos J. (2007) FEMS Yeast Res. 7, 102–109 [DOI] [PubMed] [Google Scholar]
- 31.Blumwald E., Aharon G. S., Apse M. P. (2000) Biochim. Biophys. Acta Biomembr. 1465, 140–151 [DOI] [PubMed] [Google Scholar]
- 32.Nass R., Cunningham K. W., Rao R. (1997) J. Biol. Chem. 272, 26145–26152 [DOI] [PubMed] [Google Scholar]
- 33.Madrid R., Gómez M. J., Ramos J., Rodríguez-Navarro A. (1998) J. Biol. Chem. 273, 14838–14844 [DOI] [PubMed] [Google Scholar]
- 34.Stemmer W. P. C. (1994) Nature 370, 389–391 [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi T., Blumwald E. (2005) Trends Plant Sci. 10, 615–620 [DOI] [PubMed] [Google Scholar]
- 36.Landau M., Herz K., Padan E., Ben-Tal N. (2007) J. Biol. Chem. 282, 37854–37863 [DOI] [PubMed] [Google Scholar]
- 37.Kemp G., Young H., Fliegel L. (2008) Channels 2, 329–336 [DOI] [PubMed] [Google Scholar]
- 38.Wiebe C. A., DiBattista E. R., Fliegel L. (2001) Biochem. J. 357, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiebe C. A., Rieder C., Young P. G., Dibrov P., Fliegel L. (2003) Mol. Cell. Biochem. 254, 117–124 [DOI] [PubMed] [Google Scholar]
- 40.Kinclova-Zimmermannova O., Zavrel M., Sychrova H. (2006) Mol. Membr. Biol. 23, 349–361 [DOI] [PubMed] [Google Scholar]
- 41.Hamada A., Hibino T., Nakamura T., Takabe T. (2001) Plant Physiol. 125, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barlow D. J., Thornton J. M. (1988) J. Mol. Biol. 201, 601–619 [DOI] [PubMed] [Google Scholar]