Abstract
Growth factor stimulation induces c-Jun-dependent survival of primary endothelial cells. However, the mechanism of c-Jun anti-apoptotic activity has not been identified. We here demonstrate that in response to growth factor treatment, primary human endothelial cells as well as mouse fibroblasts respond with an increased expression of c-Jun that forms a complex with ATF2. This complex activates the expression of the anti-apoptotic protein Bcl-XL. By site-directed mutagenesis experiments, we identified two AP-1-binding sites located within the proximal promoter of the Bcl-X gene. Site-directed mutagenesis demonstrated that these AP-1 sites are required for the transcriptional activation of the promoter. Chromatin immunoprecipitation experiments show that in response to growth factor treatment, the heterodimer c-Jun·ATF2 binds to these functional AP-1 sites. Silencing of either c-Jun or ATF2 demonstrated that both nuclear factors are required for the activation of the proximal Bcl-X promoter. Taken together, our experiments provide evidence that growth factor-independent signaling pathways converge in the formation of an active c-Jun·AFT2 dimer, which induces the expression of the anti-apoptotic factor Bcl-XL that mediates a pro-survival response.
Keywords: AP-1 Transcription Factor, Coregulator Transcription, Endothelium, Jun Transcription Factor, Transcription Factors, Transcription Regulation
Introduction
Activator protein-1 (AP-1)3 refers to a family of dimeric transcription factors composed of a mixture of homo- and heterodimers between the Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, FosB, Fra1, and Fra2) proteins. Jun and Fos proteins can also dimerize other basic leucine zipper proteins such as ATF, CCAAT enhancer-binding protein, Maf, and NF-E2, all of which were characterized by a basic DNA-binding domain followed by an amphipatic dimerization domain defined basic leucine zipper (1). They all bind to a common consensus sequence-defined AP-1-binding site. Jun-Jun and Jun-Fos dimers bind preferentially to a 12-O-tetradecanoylphorbol-13-acetate-responsive element, whose consensus is TGACTCA, whereas Jun·ATF dimers prefer to bind to the c-AMP-responsive element whose consensus is TGAGCTCA (2). Because the AP-1 factor is not a protein but a mixture of different factors, each of them regulated independently, the AP-1 factors regulate different target genes executing different biological functions such as cell proliferation, differentiation, response to stress, or cell death (3, 4).
c-Jun sequence shows a high degree of identity among species (1). Its activation domain is at the N-terminal part, whereas its C-terminal domain contains the basic leucine zipper DNA-binding domain. In serum-starved cells, the c-Jun protein level decreases, and its expression is strongly stimulated by growth factor treatment. In response to serum or growth factors, c-Jun mRNA is induced as an early gene, and the protein is phosphorylated at serine residues 63 and 73 and at threonine 91 and 93 of its N-terminal domain by the Jun N-terminal kinase (JNK). This phosphorylation event functions in a cell context-specific and cell type-specific manner to integrate proliferation, differentiation, survival, and migration signals (5) affecting c-Jun stability (6) and its transcriptional activity (7). Cellular and animal models demonstrated that c-Jun is involved in the control of cell proliferation (1). c-Jun deregulation leads to transformation in different cell types, and Ras-dependent transformation is impaired in Jun−/− fibroblasts or by the dominant negative form of c-Jun (1). Elevated levels of c-Jun may lead to apoptosis in neuronal cells, whereas embryos lacking c-Jun show a higher rate of apoptotic cells in liver (4).
We previously demonstrated that in primary human endothelial cells, growth factor stimulation induced c-Jun expression. Reduction of c-Jun expression in these cells either by inhibition of JNK activity or by c-Jun silencing impaired the growth factor-induced pro-survival signaling, demonstrating that in primary endothelial cells c-Jun is a key molecule in growth factor-mediated cell survival (8).
In the present study we investigated the mechanism by which c-Jun exerts its pro-survival activity in response to growth factor signaling. We provide compelling evidence that c-Jun activates the expression of the anti-apoptotic Bcl-XL protein. By chromatin immunoprecipitation experiments, we demonstrated the presence of two functional AP-1-binding sites in both the murine and human Bcl-X genes, which are recognized by the heterodimer c-Jun·ATF2, and silencing experiments demonstrated that c-Jun and ATF2 are required for both the expression of Bcl-XL as well as protection from apoptosis, thus demonstrating that Bcl-XL is a critical factor regulated by c-Jun·ATF2 required for cell survival.
EXPERIMENTAL PROCEDURES
Plasmid DNA Constructs
The full-length cDNA of human Bcl-XL was amplified with the oligonucleotides 5′-GCCACCATGTCTCAGAGCAACCGGGAG-3′ (forward) and 5′-ATTTCCGACTGAAGAGTGAG-3′ (reverse) from a human muscle cDNA library. The resulting fragment was cloned in TOPO® PCR cloning vector (Invitrogen) and then subcloned in the expression retroviral vector MIGR1 (kindly provided by Dr. Guido Franzoso). The full-length cDNA of mouse c-Jun was subcloned in the MIGR1 retroviral vector.
Silencing experiments were performed using retroviral vectors pLKO.1 from the TRC lentiviral shRNA library (Open Biosystems, Huntsville, AL) expressing specific shRNAs for human c-Jun (oligonucleotide TRCN0000010366), ATF2 (oligonucleotide TCRN0000013713, referred to as C13; and oligonucleotide TCRN0000013714, referred to as C14), and Bcl-XL (oligonucleotide TRCN0000004682, referred to as 82; and oligonucleotide TRCN0000004684, referred to as 84). Bcl-XL silencing vectors express shRNAs carrying nucleotide sequences conserved in mouse and human Bcl-X genes. Silencing of mouse c-Jun was performed by annealing and cloning the oligonucleotides 5′-TCGAGAACGCAGCAGTTGCAAACGTTTAACTTGAGAAAACGTTTGCAACTGCTGCTTTTTCTGCA-3′ (sense) and 5′-GAAAAAGCAGCAGTTGCAAACGTTTTCTCAAGTTAAACGTTTGCAACTGCTGC-3′ (antisense) into the ClaI-SalI sites of the cassette for the expression of shRNA under the U6 promoter in a lentiviral vector as previously described (9).
Human Bcl-X proximal promoter was amplified from genomic DNA with the oligonucleotides L586 (5′-ACCAACTAAATCCATACCA-3′) and L753 (5′-TTTTATAATAGGGATGGGC-3′) cloned in the in TOPO® PCR cloning vector and then subcloned in the luciferase reporter plasmid pGL3 (Promega, Madison, WI) to obtain the pGL3-hP1wt construct. The pGL3 plasmids carrying the Bcl-X promoter with mutated AP-1 sites were obtained by using a QuikChange mutagenesis site-directed mutagenesis kit (Promega, Madison, WI) changing the AP-1 (1) sequence from TGAGTTAC to TCTAGAAC (pGL3-hP1m1) and AP-1 (2) sequence from TGACTTAA to AAGCTTAA (pGL3-hP1m2). Double mutant (pGL3-hP1m1/2) was obtained introducing both of the described sequences substitutions. All of constructs were confirmed by sequencing.
Cell Cultures Transfection and Transduction
For each experiment at least three independent extractions of human umbilical vein endothelial cells (HUVEC) from umbilical cords were used. HUVEC were grown on gelatin in M199 medium (Invitrogen) supplemented with 20% fetal bovine serum, 50 units/ml penicillin-streptomycin, 10 units/ml heparin, and 100 μg/ml brain extract filtered through an 0.22-μm sterile filter (SCGPU05RE, Stericup-GP filter unit; Millipore). Virus preparation and HUVEC infections were previously described (8).
GPG293 adenovirus 5-transformed human embryonic kidney 293GPG packaging cells were used for the generation of retroviral vector particles (10). The 293GPG cells were grown at 37 °C and 5% CO2 in high glucose Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum and 1 μg/ml tetracycline (Sigma).
Mouse fibroblasts were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 50 units/ml penicillin-streptomycin. c-Jun+/+ and Jun−/− fibroblasts were kindly provided by Dr. Bruce Spiegelman (11). HUVEC were transfected by electroporation using 30 μg of luciferase plasmid DNA, 5 μg of SV40 Renilla plasmid DNA (Promega), and 4 × 106 cells for each point. Mouse fibroblasts were transfected using polyfectamine reagent (Qiagen). Twenty-four hours after transfection, the cells were analyzed for luciferase and Renilla activity using the Dual-Glo® luciferase assay (Promega).
Protein Extracts, Immunoblotting, and Immunoprecipitation
Following stimulation, the cells were lysed in radioimmune precipitation assay plus buffer (50 mm Tris-HCl, pH 7.2, 100 mm NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 50 mm NaF, 2 mm sodium orthovanadate, 1 mm dithiothreitol, anti-protease, and anti-phosphatase cocktails (Sigma), and cell debris was eliminated by centrifugation at 21,000 × g at 4 °C for 10 min. The following antibodies were used for immunoblotting: anti-Bcl-XL (antibody 2762; Cell Signaling), anti-c-Jun (antibody sc-45X), anti-SP1 (antibody sc-59), anti-ATF2 (antibody sc-187) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-ATF2 Thr(P)69/71 (antibody 05-891; Upstate), and anti-β-actin (antibody A5441; Sigma). For immunoprecipitations, the protein extracts prepared as described above were incubated for 1 h at 4 °C with anti-ATF2 (sc-187) coupled to Dynabeads® protein A (Dynal Biotech, Oslo, Norway).
Apoptosis Assay
5000 cells/cm2 were grown in complete medium for 16 h on a 1% gelatin-coated 8-well chamber slide. The cells were starved for 24 h in 0.25% bovine serum albumin with and without growth factor and fixed in 2% paraformaldehyde in phosphate-buffered saline. Apoptosis was assessed by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining (Promega), following the manufacturer's instructions. The percentage of apoptotic cells was calculated from the number of TUNEL-positive cells divided by the total number of counted 4′,6-diamidino-2-phenylindole-stained cells.
RNA Extraction and Quantification
Total mRNA was extracted with PureLinkTM RNA kit (Invitrogen), and quantitative real time RT-PCR was performed with a SuperScript® III Platinum® one-step quantitative RT-PCR kit (Invitrogen) and a Rotor-Gene 6000 real time rotary analyzer (Corbett Life Science). For the specific RT-qPCR of human and mouse Bcl-XL, we used the oligonucleotides L461 (5′-GAGTTTGAACTGCGGTACC-3′) and L462 (5′-CCAGCCGCCGTTCTCCTGG-3′).
Chromatin Immunoprecipitation (ChIP) Assay
Each ChIP experiment was performed in at least two independent biological samples and performed as previously described (12). Immunoprecipitation was performed with the following antibodies: anti-ATF2 (antibody sc-N96), c-Fos (antibody sc-52X), anti-c-Jun (antibody sc-45X), anti-ATF-3 (antibody sc-188X), anti-Fra1 (antibody sc-605X), anti-Fra2 (antibody sc-171X), and anti-FosB (antibody sc-48X) (Santa Cruz Biotechnology).
DNA was analyzed by quantitative real time PCR by using the SYBR GreenER kit (Invitrogen). All of the experimental values were normalized to those obtained with a nonimmune serum and divided by input, using the procedure previously described (13). The data shown represent triplicate real time quantitative PCR measurements of the immunoprecipitated DNA. The data are expressed as ‰ and express 1/1000 of the DNA inputs. For the amplification of immunoprecipitated DNA, we used the following oligonucleotides: M179 (5′-AAGCTTCGCAATTCCTCTGT-3′) and M180 (5′-GCCTTTCTCCAAAAGTCACC-3′) for the mouse Bcl-X promoter region; M554 (5′-CAATTCCTGTGTCGCCTT-3′) and M555 (5′-GAAAAGGCTGGTGGGAGATTCAG-3′) for the human Bcl-X promoter region; M550 (5′-TACTTCGTCTGTCTCCCTCACT-3′) and M551 (5′-AGCACCCGTTCCTTCCCTTAT-3′) for the human Snai1 promoter region; N268 (5′-TTGGGCCAACTTCCCAAGCA-3′) and N270 (5′-AGAGAAGGCCTTTCCACAGGT-3′) for the mouse Snai1 promoter region; and M537 (5′-CTAGCAAAATAGGCTGTCCC-3′) and M273 (5′-ATCACAGGTCGGGAGAGG-3′) for transfected plasmid DNA.
RESULTS
c-Jun Protects Endothelial Cells from Apoptosis Regulating Bcl-XL Expression
We previously demonstrated that c-Jun, induced by VEGFR-3/JNK signaling, is a pro-survival factor in primary human endothelial cells. However, the molecular targets of c-Jun involved in this signaling pathway are yet unknown. First we analyzed whether in endothelial cells c-Jun is also induced by VEGF, which has been shown to protect endothelial cells from apoptosis via VEGFR-2 (14). Western blot analysis demonstrated that VEGF treatment induces the up-regulation of c-Jun and its sustained expression in HUVEC (Fig. 1A). To identify downstream effectors of c-Jun in its pro-survival activity, we analyzed the expression of anti- and pro-apoptotic proteins. Our analysis revealed the up-regulation of Bcl-XL, whereas we did not observe variation of other proteins involved in the apoptotic pathway (Fig. 1A and data not shown). Accordingly, real time RT-qPCR analysis using specific primers for the transcripts coding for the Bcl-XL isoform revealed its induction at the mRNA level with a peak at 2 h and its down-regulation at later time points (Fig. 1B). The sustained expression of Bcl-XL protein at later time points might reflect protein accumulation and/or post-translational modifications.
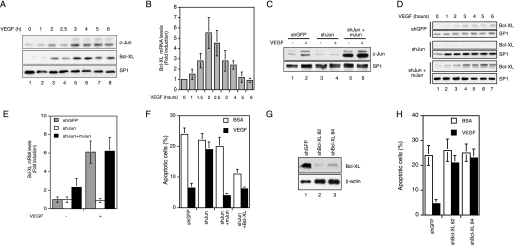
FIGURE 1.
c-Jun is required for VEGF-dependent survival in human endothelial cells. A, c-Jun and Bcl-XL expression was detected by Western blot analysis in HUVEC serum-starved and treated with 200 ng/ml VEGF for the indicated times. SP1 expression was used to confirm equal loading. B, analysis of Bcl-XL mRNA levels by quantitative real time RT-PCR. RNA was extracted from primary endothelial cells, serum-starved, and treated with VEGF. The gene expression was normalized to the amount of glyceraldehyde-3-phosphate dehydrogenase on the same sample. C, HUVEC were infected with lentiviral vectors expressing unrelated (shGFP) or c-Jun shRNA (shJun). c-Jun silenced HUVEC were infected with a viral vector expressing mouse c-Jun (shJun+mJun). c-Jun expression was analyzed by Western blotting in serum-starved (−) or VEGF-treated cells (+). SP1 expression was used to verify equal loading. D, infected primary endothelial cells as in C were treated with VEGF. Bcl-XL expression was analyzed by Western blotting, and SP1 expression was used to confirm equal loading. E, analysis of Bcl-XL mRNA levels by quantitative real time RT-PCR. RNA was extracted from HUVEC infected and treated as in C. The gene expression was normalized to the amount of glyceraldehyde-3-phosphate dehydrogenase on the same sample. F, quantification by TUNEL staining of apoptotic serum-starved (bovine serum albumin (BSA)) or VEGF-treated (VEGF) HUVEC. The cells were infected with a lentiviral vector expressing unrelated (shGFP) or c-Jun shRNA (shJun). c-Jun silenced HUVEC were infected with a viral vector expressing mouse c-Jun (shJun+mJun) or human Bcl-XL (shJun+Bcl-XL). G, HUVEC were infected with lentiviral vectors expressing either the control shRNA (shGFP) or two independent shRNA for Bcl-XL (shBcl-XL 82 and shBcl-XL 84). Bcl-XL expression was analyzed by Western blotting. H, quantification by TUNEL staining of apoptotic serum-starved (bovine serum albumin (BSA)) or VEGF-treated (VEGF) HUVEC of cells infected as in G. The results were expressed as the means ± S.E. of four independent experiments each in triplicate.
To verify whether c-Jun expression is required for Bcl-XL induction, we silenced the expression of c-Jun by infecting endothelial cells with a lentivirus expressing c-Jun shRNA, which determined over 90% reduction of c-Jun expression (Fig. 1C). Western blot analysis revealed a strong reduction of Bcl-XL in the absence of c-Jun (Fig. 1D). As a control for specificity c-Jun silenced HUVEC were transduced with a viral construct expressing the mouse c-Jun, which is not affected by the shRNA used in these experiments. Mouse c-Jun was constitutively expressed (Fig. 1C, lanes 5 and 6) and able to restore Bcl-XL expression (Fig. 1D). The regulation of Bcl-XL by c-Jun was confirmed at the RNA level. In fact, RT-qPCR analysis showed that in c-Jun silenced HUVEC, VEGF treatment did not lead to the up-regulation of Bcl-XL expression, whereas the reintroduction of mouse c-Jun in these cells restored Bcl-XL expression and significantly increased the Bcl-XL mRNA levels also in unstimulated conditions (Fig. 1E).
To analyze whether in endothelial cells Bcl-XL induced by c-Jun was inducing a pro-survival effect, we performed a TUNEL assay both in serum-deprived conditions and after growth factor treatment. We observed that c-Jun silencing strongly reduced the survival effect mediated by VEGF, whereas the expression of either mouse c-Jun or Bcl-XL in c-Jun silenced HUVEC restored VEGF-dependent cell survival (Fig. 1F). Because the Bcl-XL overexpression might protect from apoptosis independently from VEGF/Jun pathway, we tested the effective function of Bcl-XL survival activity in response to VEGF treatment by using two independent lentiviral clones expressing Bcl-XL silencing shRNAs, which induced a strong reduction of Bcl-XL expression (Fig. 1G). TUNEL assay showed that Bcl-XL silencing abrogates the survival response in HUVEC (Fig. 1H). Taken together, these results suggest that in endothelial cells, Bcl-XL expression is under the control of c-Jun and mediates growth factor-dependent cell survival.
c-Jun Regulates Bcl-XL Expression in Mouse Fibroblasts
To test whether this growth factor survival response through c-Jun was a general phenomenon, we analyzed Bcl-XL regulation and survival response in wild type and c-Jun knock-out mouse fibroblasts. Western blot assay showed that in Jun+/+ cells derived from wild type sibling Bcl-XL expression increased following EGF treatment, whereas in Jun−/− cells Bcl-XL was barely detectable. The reintroduction of c-Jun in Jun−/− cells under the control of a constitutive promoter restores Bcl-XL induction both at the protein and mRNA level after EGF treatment (Fig. 2, A–C). Importantly, TUNEL assay demonstrated that in mouse fibroblasts, the absence of c-Jun strongly reduced the survival effect mediated by the growth factor, whereas the reintroduction of c-Jun in Jun−/− increased cell survival (Fig. 2D).
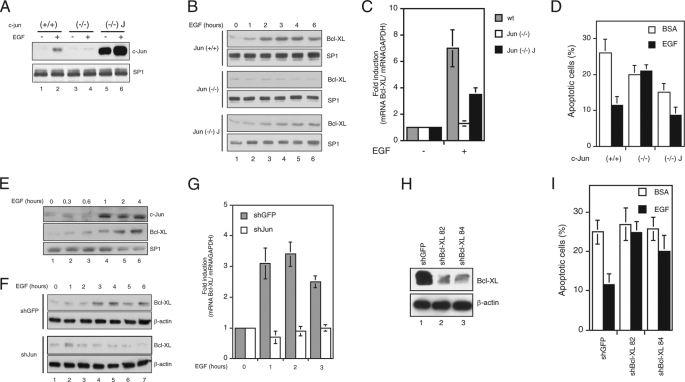
FIGURE 2.
c-Jun expression is involved in EGF-dependent survival of mouse fibroblasts. A, Jun−/−-deficient fibroblasts, obtained from c-Jun knock-out mice, Jun+/+ fibroblasts, derived from their wild type siblings, and Jun−/− J fibroblasts, which express constitutively mouse c-Jun, were serum-starved (−) or induced with 10 ng/ml EGF for 3 h. c-Jun expression was analyzed by Western blotting, and SP1 expression was used to confirm equal loading. B, Bcl-XL expression was detected by Western blot analysis in Jun+/+, Jun−/−, and Jun−/− J fibroblasts serum-starved and treated with 10 ng/ml EGF at the times indicated. SP1 expression was used to confirm equal loading. C, analysis of Bcl-XL mRNA levels by quantitative real time RT-PCR. RNA was extracted from Jun+/+, Jun−/−, and Jun−/− J fibroblasts serum-starved (−) or treated with 10 ng/ml EGF for 3 h. The gene expression was normalized to the amount of glyceraldehyde-3-phosphate dehydrogenase on the same sample. wt, wild type. D, quantification by TUNEL staining of apoptotic serum-starved (bovine serum albumin (BSA)) or EGF-treated (EGF) Jun+/+, Jun−/−, and Jun−/− J cells. The results were expressed as the means ± S.E. of at least three independent experiments each in triplicate. E, c-Jun and Bcl-XL were detected by Western blot analysis in mouse embryo fibroblasts serum-starved and treated with 10 ng/ml EGF at the times indicated. SP1 expression was used to confirm equal loading. F, primary mouse fibroblasts were infected with a lentiviral vector expressing unrelated (shGFP) or c-Jun shRNA (shJun), serum-starved, and treated with 10 ng/ml EGF at the times indicated. Bcl-XL expression was analyzed by Western blotting, and β-actin expression was used to confirm equal loading. G, analysis of Bcl-XL mRNA levels by RT-qPCR. RNA was extracted from mouse embryo fibroblasts infected as in B, serum-starved, and treated with 10 ng/ml EGF at the times indicated. The gene expression was normalized to the amount of glyceraldehyde-3-phosphate dehydrogenase on the same sample. H, wild type fibroblasts were infected with lentiviral vectors expressing either the control shRNA (shGFP) or two independent shRNA for Bcl-XL (shBcl-XL 82 and shBcl-XL 84). Bcl-XL expression was analyzed by Western blotting. I, quantification by TUNEL staining of apoptotic serum-starved (bovine serum albumin (BSA)) or EGF-treated (EGF) cells infected as in H. The results were expressed as the means ± S.E. of three independent experiments each in triplicate.
Similar results were obtained by silencing c-Jun expression in primary mouse embryo fibroblasts from CD1 mice. In fact serum-starved mouse embryo fibroblasts showed increasing expression of both c-Jun and Bcl-XL expression after EGF treatment (Fig. 2E), whereas mouse embryo fibroblasts infected with a lentivirus expressing c-Jun shRNA showed a reduced Bcl-XL expression at both protein and mRNA levels, compared with cells infected with a lentivirus expressing an unrelated shRNA (Fig. 2, F and G), thus excluding the possibility that the low Bcl-XL expression observed in Jun−/− cells was due to clonal variation.
To confirm the pro-survival role of Bcl-XL observed in HUVEC, we silenced Bcl-XL in fibroblasts and measured their apoptotic response following EGF treatment. We observed that Bcl-XL silencing abolished the pro-survival response to EGF in mouse fibroblasts (Fig. 2, H and I). Taken together, these results suggest that the activation of a Jun-dependent pro-survival response by inducing Bcl-XL expression is a general cell response to growth factor treatment.
c-Jun Regulates Bcl-X Transcription
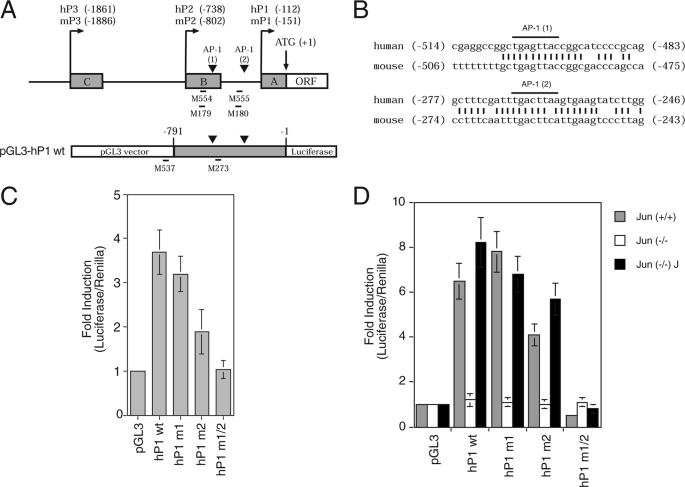
Next we analyzed whether c-Jun could directly regulate the Bcl-X promoter to express Bcl-XL. The Bcl-X promoter region is highly conserved between mouse and human and contains different promoters (Fig. 3A) (15–18). Inspection of the Bcl-X promoter sequences with the TRANSFAC data base (19) revealed the presence of two putative AP-1-binding sites within the ubiquitous proximal promoter. Alignment of the murine and human Bcl-X genes showed that these sites were conserved both at the nucleotide sequences and in relative position with respect to the translation start site between mouse and human genes (Fig. 3, A and B). To analyze whether these sites play a regulatory role in Bcl-X transcription, we fused the human P1 promoter region with a luciferase reporter gene (hP1wt) and mutagenized the AP-1 consensus sites. Transient transfections of the wild type and mutant constructs in HUVEC treated with VEGF revealed that the mutation of the more upstream AP-1 consensus site alone (hP1m1) did not significantly affect the promoter response, whereas the mutation of the downstream AP-1 consensus site (hP1m2) had a stronger effect. Mutation of both AP-1 sites (hP1m1/2) completely abolished promoter activity (Fig. 3C), demonstrating that both of these AP-1 sites contribute to the regulation of Bcl-XL expression. To verify whether c-Jun activates Bcl-X promoter via these AP-1 sites, the same reporter constructs were transfected in Jun−/− or wild type fibroblasts. We observed that the wild type promoter induced the expression of the reporter gene in wild type fibroblasts but not in Jun−/− cells. The expression of the luciferase was rescued in knock-out fibroblasts expressing ectopic c-Jun obtained by retroviral infection (Fig. 3D). Moreover, the mutations in both AP-1 consensus sites abolished the promoter activity both in wild type as well as in knock-out cells irrespective of the expression of ectopic c-Jun. Taken together, these experiments suggest that c-Jun activates the transcription of the Bcl-X gene via the conserved AP-1 sites on its proximal promoter.
FIGURE 3.
Two AP-1-binding sites contained in proximal Bcl-X promoter are required to respond to growth factor induction. A, schematic representation of the human and mouse Bcl-X promoter region covering the proximal promoters (P1, P2, and P3), and the alternative noncoding exons (boxes A, B, and C). The numbers shown refer to the distance upstream of the translation initiation codon (ATG). Untranslated exons are indicated in gray. The putative AP-1 sites are indicated by arrowheads. ORF, open reading frame. The genomic region containing the human P1 promoter (hP1), from nucleotides −791 to −1, was cloned upstream of the luciferase reporter gene in pGL3 plasmid; the construct obtained is indicated in the lower part of the panel. The locations and names of the oligonucleotides used in the ChIP assay are indicated. B, the AP-1 sites are conserved between mouse and human Bcl-X genes as both nucleotide sequences and positions into the genes. C, reporter constructs carrying the P1 promoter of the human Bcl-X gene as either wild type or mutated in the upstream AP-1, indicated as (1), downstream AP-1, indicated as (2), or at both sites (hP1wt, hP1 m1, hP1 m2, and hP1 m1/2) were cotransfected in HUVEC together with a constitutively expressing Renilla. Promoter activity values were normalized using Renilla activity, and the fold induction was calculated with respect to basal luciferase vector pGL3. D, the same constructs as in C were transfected in fibroblasts derived from c-Jun knock-out mouse (Jun (−/−)) and wild type control mouse (Jun (+/+)). As control, rescue of c-Jun was obtained by infection of knock-out fibroblasts with retrovirus for the overexpression of c-Jun (Jun (−/−) J). The mean values for three independent experiments ± S.D. of the mean are shown.
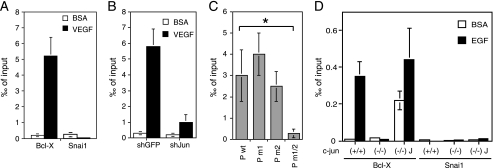
To verify the direct binding of c-Jun to the Bcl-X AP-1 sites, we performed ChIP followed by real time quantitative PCR. Quantitative ChIP analysis revealed the recruitment of c-Jun to the Bcl-X proximal promoter containing the AP-1 sites after cell treatment with VEGF, whereas we could not observe c-Jun binding to the Snai1 promoter (Fig. 4A). As a further control we tested c-Jun binding to Bcl-X promoter in endothelial cells silenced for c-Jun (Fig. 4B). Because on the endogenous promoter we could not distinguish whether c-Jun was bound to the upstream or downstream AP-1 site, HUVEC were transfected with the Bcl-X luciferase reporter constructs carrying the wild type regulatory region or the mutated AP-1 sites, and we analyzed the recruitment of c-Jun to the plasmid DNA by quantitative ChIP assays. As shown in Fig. 4C, the binding of c-Jun was not significantly reduced on the constructs carrying the single mutations, whereas it was abolished when both the AP-1 elements were mutated, thus suggesting that both sites are recognized in vivo by c-Jun. The binding of c-Jun on the AP-1 elements was also observed in mouse wild type fibroblasts following EGF treatment but not in c-Jun knock-out cells nor on the control Snai1 promoter (Fig. 4D), demonstrating that the Bcl-X promoter is induced by c-Jun binding to the conserved AP-1 sites also in mouse fibroblasts.
FIGURE 4.
c-Jun binds the AP-1 sites of Bcl-X proximal promoter. A, chromatin was isolated from HUVEC starved for 16 h and subsequently treated with VEGF, and ChIP assays were performed with specific anti-c-Jun antibody, control IgG, and primers M554 and M555 for the amplification of the genomic region surrounding AP-1(1) site (primer position is reported in Fig. 3A) and primers M550 and M551 for human Snai1. B, the panel documents ChIP assays performed as in A with HUVEC infected with lentivirus for the silencing of c-Jun and GFP as control. c-Jun binding on Bcl-X proximal promoter significantly diminished in c-Jun silenced cells, demonstrating the specificity of the assay. C, HUVEC were transfected by electroporation with luciferase reporter constructs carrying the wild type or mutated Bcl-X promoter region and subsequently used in ChIP assays with specific anti-c-Jun antibody, control IgG, and primers M537 and M273 for the amplification of the cross-linked plasmid DNA. The results are expressed as the means ± S.D. of three independent experiments (*, p < 0.01). D, ChIP assays performed in fibroblasts showed the c-Jun binding on the Bcl-X promoter region upon EGF induction in wild type or knock-out fibroblasts. No binding of c-Jun was observed with the Snai1 promoter.
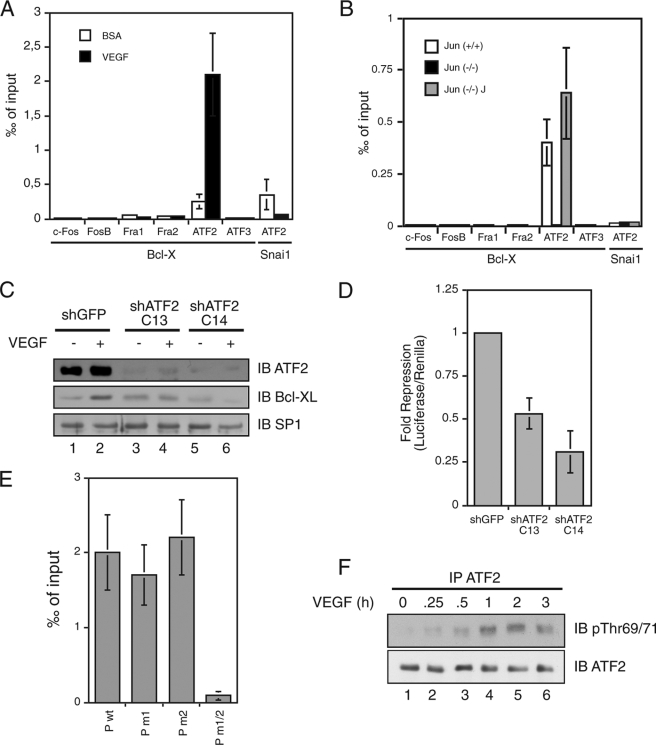
ATF2 Binds the AP-1-binding Sites and Is Necessary for Activation of Bcl-X Transcription
The above results demonstrate that c-Jun directly activates the transcription driven by the Bcl-X proximal promoter. Because c-Jun can either homo- or heterodimerize, we tested whether c-Jun binds the promoter AP-1 regulatory elements alone or with a partner. By ChIP we observed a significant binding of the factor ATF2 to the DNA fragment containing the AP-1 sites following VEGF treatment, whereas none of the Fos family members or ATF3 associated to Bcl-X promoter (Fig. 5A). The recruitment of ATF2 to the AP-1 sites was dependent on c-Jun binding because we could observe ATF2 binding in EGF-treated Jun+/+ but not in Jun−/− mutant fibroblasts (Fig. 5B). Moreover, ATF2 silencing further demonstrated that ATF2 is required for the growth factor-dependent Bcl-XL expression and the proximal promoter activity (Fig. 5, C and D). ChIP analysis on the wild type and mutant promoters in either one or both the AP-1 sites demonstrated that ATF2 binds, like c-Jun, to both sites, and its binding is abolished when both sites are mutated (Fig. 5E).
FIGURE 5.
ATF2 binds the Bcl-X proximal promoter and is necessary for the full activation of transcription. A, chromatin was isolated from HUVEC starved for 16 h and subsequently VEGF-treated for 2 h. ChIP assays were performed with HUVEC chromatin and specific antibodies recognizing the indicated c-Jun heterodimerization partners, control IgG, and primers M554 and M555. ATF2 bound the Bcl-XL promoter region, and its binding was increased after VEGF stimulation. B, the same experiment was performed with chromatin obtained from EGF treated fibroblasts wild type (Jun (+/+)) and knock-out for c-Jun (Jun (−/−)). The ATF2 binding on the Bcl-X proximal promoter was c-Jun-dependent. C, HUVEC were transfected with two independent ATF2 shRNA vectors (shATF2 C13 and shATF2 C14) and for GFP as control (shGFP). 30 h later, the cells were starved for 16 h and subsequently induced with VEGF. Whole cell extracts were immunoblotted with specific antibodies for ATF2, Bcl-XL, and SP1 as loading control. D, HUVEC were cotransfected with luciferase reporter construct Bcl-X promoter region and shRNA expressing vectors targeting ATF2 and GFP as control. Knockdown of ATF2 repressed Bcl-X proximal promoter activity. E, HUVEC were transfected with luciferase reporter constructs carrying the wild type or mutated Bcl-X proximal promoter and subsequently used in ChIP assays with specific anti-c-ATF2 antibody, control IgG, and primers M537 and M273. F, HUVEC were induced at the indicated time points with VEGF, and whole cell extract was immunoprecipitated (IP) with specific anti-ATF2 antibody and immunoblotted (IB) with anti-ATF2 and anti-Thr(P)69/71 ATF2 antibodies.
Finally, because ATF2 is constitutively expressed in endothelial cells, we tested whether growth factor treatment induces ATF2 phosphorylation. It has been previously shown that ATF2 is activated by ERK- and p38-dependent phosphorylation of threonine 69 and 71, which enhances ATF2 intrinsic acetylase activity (20, 21); we therefore analyzed whether ATF2 was phosphorylated in HUVEC upon VEGF treatment. As shown in Fig. 5F, a time course experiment demonstrated that in endothelial cells ATF2 is indeed activated in response to VEGF treatment by the phosphorylation at threonine 69 and 71, and this phosphorylation corresponds to Bcl-XL mRNA up-regulation. Taken together, these data suggest that in response to growth factor stimulation, cells induce both the up-regulation of c-Jun and the activation of ATF2, which bind together to Bcl-X proximal promoter and converge in the activation of its transcription and promote cell survival.
DISCUSSION
Growth factor stimulation induces cell survival. We previously demonstrated that in primary endothelial cells growth factor signaling induces c-Jun that is required for the cell survival response (8). Here we studied the mechanism by which c-Jun protects from apoptosis. We found that c-Jun together with ATF2 binds to the Bcl-X proximal promoter inducing the expression of the anti-apoptotic protein Bcl-XL leading to cell survival.
The Bcl-X gene is a Bcl-2 family member spliced in different isoforms. The major product of Bcl-X is the ubiquitous large isoform Bcl-XL that protects from apoptosis, whereas the short isoform Bcl-XS, because of alternative splicing, functions as a pro-apoptotic protein interacting and inhibiting the function of Bcl-XL and Bcl-2 (22–25).
Both human and mouse Bcl-X regulatory regions are highly conserved. The Bcl-X promoter region is complex because it contains different promoters that generate different 5′ noncoding exons and different proportions of Bcl-XL and Bcl-XS mRNAs (15–18). It has been previously demonstrated that various nuclear factors including NFκB, STAT, Ets, Ikaros, and steroid hormones activate Bcl-X transcription acting on its different promoters in various cell types (15, 17, 18, 26–28). Little is known about the transcriptional regulation of Bcl-X in response to growth factors.
We here demonstrated that growth factor signaling induces a c-Jun-dependent up-regulation of the pro-survival Bcl-XL isoform. Our experiments provide much evidence to support this model. We observed that in primary human endothelial cells and in mouse embryo fibroblasts growth factor treatment induces Bcl-XL expression and that this expression is dependent on c-Jun in both cell types. The up-regulation of Bcl-XL is due to increased transcription of the Bcl-X gene. In fact, we found that the more proximal ubiquitous Bcl-X promoter responds both to VEGF in endothelial cells and to EGF in fibroblasts, and this response is impaired in c-Jun silenced endothelial cells or in Jun−/− fibroblasts. This promoter contains two noncanonical AP-1 sites that are conserved in human and mouse. Deletion of these AP-1 sites eliminates the promoter response to growth factors, demonstrating that these sites are required for the promoter activity. These AP-1 sites belong to the subgroup of the consensus of eight bases, which are preferred by the c-Jun·ATF dimers (2). In agreement with this prediction, the experiments of in vivo binding by chromatin immunoprecipitations demonstrated that this promoter is recognized by both c-Jun and ATF2, whereas other putative c-Jun partners do not bind to the promoter. In support of this, ATF2 silencing experiments demonstrated that this factor is also required for the promoter activity. Site-directed mutagenesis demonstrated that both c-Jun and ATF2 recognize both sites. Importantly, in c-Jun silenced cells ATF2 does not bind, suggesting that each AP-1 site is recognized by the c-Jun·ATF2 heterodimer. Furthermore, we observed that in endothelial cells following VEGF signaling, ATF2 is phosphorylated at threonines 69 and 71. These phosphorylation events on ATF2 have been previously demonstrated to be important for ATF2 transcriptional activity and to require the activity of ERK and p38, whereas JNK is not involved (20). We previously demonstrated that the survival signaling activates both the expression of c-Jun and its activation/stabilization via ERK and JNK signaling (8). Thus following growth factor treatment the c-Jun·ATF2 complex is activated by the convergence of independent pathways, revealing a further level of control that is mediated by the cross-talk between cellular signaling.
Numerous studies demonstrated that c-Jun acts as a pro-survival as well as a pro-apoptotic factor (4). In neuronal cells the Jun·ATF2 dimer, induced by potassium deprivation, has been shown to induce a pro-apoptotic response by regulating the death protein 5/harakiri (29, 30). We have here demonstrated that following growth factor stimulation c-Jun·ATF2 dimer, by activating the proximal Bcl-X promoter, induces a pro-survival response by specifically up-regulating the expression of the pro-survival Bcl-XL. In fact in our experiments we could not detect the expression of the short pro-apoptotic isoform Bcl-XS, although the antibody used could reveal it. This result is in agreement with the fact that the proximal Bcl-X promoter has been described to drive the transcription almost exclusively of the mRNA coding for Bcl-XL isoform (15). In conclusion, c-Jun·ATF2 complexes are key regulators that by regulating different effector genes can exert different cellular response from pro-survival to pro-apoptotic, depending on the different cell contexts and the type of stimulus.
Acknowledgment
We thank Beatrice Grandi for technical support.
This work was supported by grants from Associazione Italiana Ricerca sul Cancro, Istituto Tosscano Tumori, and Ministero Italiano dell'Istruzione, dell'Università e della Ricerca.
- AP-1
- activator protein-1
- HUVEC
- human umbilical vein endothelial cell(s)
- VEGF
- vascular endothelial growth factor
- EGF
- epidermal growth factor
- JNK
- Jun terminal kinase
- shRNA
- small hairpin RNA
- TUNEL
- terminal deoxynucleotidyl transferase dUTP nick end labeling
- RT
- reverse transcription
- qPCR
- quantitative PCR
- ChIP
- chromatin immunoprecipitation
- STAT
- signal transducers and activators of transcription
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Mechta-Grigoriou F., Gerald D., Yaniv M. (2001) Oncogene 20, 2378–2389 [DOI] [PubMed] [Google Scholar]
- 2.Karin M. (1995) J. Biol. Chem. 270, 16483–16486 [DOI] [PubMed] [Google Scholar]
- 3.Angel P., Karin M. (1991) Biochim. Biophys. Acta 1072, 129–157 [DOI] [PubMed] [Google Scholar]
- 4.Ameyar M., Wisniewska M., Weitzman J. B. (2003) Biochimie 85, 747–752 [DOI] [PubMed] [Google Scholar]
- 5.Wagner E. F., Nebreda A. R. (2009) Nat. Rev. Cancer 9, 537–549 [DOI] [PubMed] [Google Scholar]
- 6.Sabapathy K., Hochedlinger K., Nam S. Y., Bauer A., Karin M., Wagner E. F. (2004) Mol. Cell 15, 713–725 [DOI] [PubMed] [Google Scholar]
- 7.Weiss C., Schneider S., Wagner E. F., Zhang X., Seto E., Bohmann D. (2003) EMBO J. 22, 3686–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salameh A., Galvagni F., Bardelli M., Bussolino F., Oliviero S. (2005) Blood 106, 3423–3431 [DOI] [PubMed] [Google Scholar]
- 9.Zippo A., De Robertis A., Bardelli M., Galvagni F., Oliviero S. (2004) Blood 103, 4536–4544 [DOI] [PubMed] [Google Scholar]
- 10.Ory D. S., Neugeboren B. A., Mulligan R. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson R. S., van Lingen B., Papaioannou V. E., Spiegelman B. M. (1993) Genes Dev. 7, 1309–1317 [DOI] [PubMed] [Google Scholar]
- 12.Zippo A., De Robertis A., Serafini R., Oliviero S. (2007) Nat. Cell Biol. 9, 932–944 [DOI] [PubMed] [Google Scholar]
- 13.Kouskouti A., Talianidis I. (2005) EMBO J. 24, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrara N. (2004) Endocr. Rev. 25, 581–611 [DOI] [PubMed] [Google Scholar]
- 15.Pecci A., Viegas L. R., Baranao J. L., Beato M. (2001) J. Biol. Chem. 276, 21062–21069 [DOI] [PubMed] [Google Scholar]
- 16.Grillot D. A., González-García M., Ekhterae D., Duan L., Inohara N., Ohta S., Seldin M. F., Nuñez G. (1997) J. Immunol. 158, 4750–4757 [PubMed] [Google Scholar]
- 17.Viegas L. R., Vicent G. P., Barañao J. L., Beato M., Pecci A. (2004) J. Biol. Chem. 279, 9831–9839 [DOI] [PubMed] [Google Scholar]
- 18.Habens F., Lapham A. S., Dallman C. L., Pickering B. M., Michels J., Marcusson E. G., Johnson P. W., Packham G. (2007) Oncogene 26, 1910–1919 [DOI] [PubMed] [Google Scholar]
- 19.Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E. (2006) Nucleic Acids Res. 34, D108–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouwens D. M., de Ruiter N. D., van der Zon G. C., Carter A. P., Schouten J., van der Burgt C., Kooistra K., Bos J. L., Maassen J. A., van Dam H. (2002) EMBO J. 21, 3782–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki H., Schiltz L., Chiu R., Itakura K., Taira K., Nakatani Y., Yokoyama K. K. (2000) Nature 405, 195–200 [DOI] [PubMed] [Google Scholar]
- 22.Boise L. H., González-García M., Postema C. E., Ding L., Lindsten T., Turka L. A., Mao X., Nuñez G., Thompson C. B. (1993) Cell 74, 597–608 [DOI] [PubMed] [Google Scholar]
- 23.Fang W., Rivard J. J., Mueller D. L., Behrens T. W. (1994) J. Immunol. 153, 4388–4398 [PubMed] [Google Scholar]
- 24.Shiraiwa N., Inohara N., Okada S., Yuzaki M., Shoji S., Ohta S. (1996) J. Biol. Chem. 271, 13258–13265 [DOI] [PubMed] [Google Scholar]
- 25.Yang X. F., Weber G. F., Cantor H. (1997) Immunity 7, 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khoshnan A., Tindell C., Laux I., Bae D., Bennett B., Nel A. E. (2000) J. Immunol. 165, 1743–1754 [DOI] [PubMed] [Google Scholar]
- 27.Ezzat S., Zhu X., Loeper S., Fischer S., Asa S. L. (2006) Mol. Endocrinol. 20, 2976–2986 [DOI] [PubMed] [Google Scholar]
- 28.Grad J. M., Zeng X. R., Boise L. H. (2000) Curr. Opin Oncol. 12, 543–549 [DOI] [PubMed] [Google Scholar]
- 29.Yuan Z., Gong S., Luo J., Zheng Z., Song B., Ma S., Guo J., Hu C., Thiel G., Vinson C., Hu C. D., Wang Y., Li M. (2009) Mol. Cell. Biol. 29, 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma C., Ying C., Yuan Z., Song B., Li D., Liu Y., Lai B., Li W., Chen R., Ching Y. P., Li M. (2007) J. Biol. Chem. 282, 30901–30909 [DOI] [PubMed] [Google Scholar]