Abstract
We report the identification of McpS as the specific chemoreceptor for 6 tricarboxylic acid (TCA) cycle intermediates and butyrate in Pseudomonas putida. The analysis of the bacterial mutant deficient in mcpS and complementation assays demonstrate that McpS is the only chemoreceptor of TCA cycle intermediates in the strain under study. TCA cycle intermediates are abundantly present in root exudates, and taxis toward these compounds is proposed to facilitate the access to carbon sources. McpS has an unusually large ligand-binding domain (LBD) that is un-annotated in InterPro and is predicted to contain 6 helices. The ligand profile of McpS was determined by isothermal titration calorimetry of purified recombinant LBD (McpS-LBD). McpS recognizes TCA cycle intermediates but does not bind very close structural homologues and derivatives like maleate, aspartate, or tricarballylate. This implies that functional similarity of ligands, such as being part of the same pathway, and not structural similarity is the primary element, which has driven the evolution of receptor specificity. The magnitude of chemotactic responses toward these 7 chemoattractants, as determined by qualitative and quantitative chemotaxis assays, differed largely. Ligands that cause a strong chemotactic response (malate, succinate, and fumarate) were found by differential scanning calorimetry to increase significantly the midpoint of protein unfolding (Tm) and unfolding enthalpy (ΔH) of McpS-LBD. Equilibrium sedimentation studies show that malate, the chemoattractant that causes the strongest chemotactic response, stabilizes the dimeric state of McpS-LBD. In this respect clear parallels exist to the Tar receptor and other eukaryotic receptors, which are discussed.
Keywords: Bacteria, Bacterial Signal Transduction, Calorimetry, Chemotaxis, Protein Self-assembly, Receptor Structure-function, Receptors, Signal Transduction, Ultracentrifugation
Introduction
Chemotaxis allows motile bacteria to migrate toward or away from different environmental signals. The major components of the bacterial chemotaxis apparatus include methyl-accepting chemotaxis receptor proteins (MCPs),3 the sensor kinase CheA, and the response regulator CheY. Attractant binding to the chemoreceptor modulates CheA autophosphorylation activity and the subsequent transphosphorylation of CheY, which interacts directly with the flagellar motor (1). The specificity of a chemotactic response is determined by the MCP, which is typically composed of a periplasmic ligand-binding domain (LBD) and a cytosolic signaling domain. The molecular mechanism of chemotaxis has been studied primarily in Escherichia coli, which was shown to possess 4 MCPs (2).
The chemotactic behavior of soil and aquatic microorganisms is poorly understood. Genome analyses revealed that soil bacteria generally have a large number of MCPs (3). This is exemplified by the complete genomes of Pseudomonas and Clostridium strains, which typically have more than 20 MCPs. It is therefore likely that these bacteria show chemotactic behavior to an increased number of compounds, which might reflect a major physiological importance. However, most of these MCPs are of unknown function. To compensate for nutritional shortages, many soil microorganisms are able to use TCA cycle intermediates present in plant root exudates or cell debris for growth (4, 5). This, combined with the fact that TCA cycle intermediates are abundantly present in plant tissue and root exudates (4, 6) might explain why many bacteria show a chemotactic response toward TCA cycle intermediates (7–13). However, the molecular basis of this taxis remains poorly understood and so far only two receptors have been identified, which are Tcp of Salmonella typhimurium (9) mediating attraction to citrate and the malate-specific PA2652 of Pseudomonas aeruginosa (7).
The inspection of the SMART data base (14) reveals that the large majority of MCP LBDs remains un-annotated. In the case of MCPs with annotated LBDs, these domains belong to the TarH (15), PAS, GAF (16), CACHE (17), and CHASE (18) families. However, there is a clear need for research to study MCPs with un-annotated LBD.
Ligand binding at the LBD causes an alteration of CheA activity bound at the other extreme of the chemoreceptor. MCP function is thus based on the transmission of the ligand-mediated molecular stimulus over very large distances (19). Some insight into the molecular consequences of ligand binding on the chemoreceptor has been obtained. These studies have primarily used the Tar and Tsr receptors, which have TarH-type LBDs that form a 4-helix bundle structure (20). Murphy et al. (21) have studied the influence of serine binding to Tsr. The authors have demonstrated that serine binding causes a backbone shift by around 1 Å in the distance between α1 and α4 of the TarH LBD. This supports a molecular mechanism by which ligand binding causes a piston shift of the final α-helix of the LBD (22).
Milliagan and Koshland (23) have studied the effect of aspartate binding to the recombinant LBD of Tar. In the absence of aspartate a dynamic equilibrium between LBD monomers and dimers is described. Interestingly, in the presence of saturating concentrations of aspartate the KD of the monomer-dimer equilibrium was at least 2 orders of magnitude lower, indicating a significant stabilization of the LBD dimer by aspartate (23). Central to understanding of the signal transduction processes is the question whether the affinity of a signal molecule for its receptor determines the final response. There are several examples of one- and two-component regulator systems that show that ligand affinity for its receptor does not determine the final regulatory output (24, 25). Ligand binding was proposed to trigger differentially another molecular event that in turn determines the final regulatory output (24).
In this study we have identified a chemoreceptor, termed McpS that mediates chemotaxis specifically toward 6 TCA cycle intermediates and butyrate. The LBD of McpS is unusually large and predicted to contain 6 helices. Using titration calorimetry (26) of purified protein, the precise thermodynamic profile for the binding of all ligands was determined. All ligands were found to compete in vivo and in vitro for the same receptor binding site. The chemotactic response triggered by these compounds varied largely. Compounds that triggered a strong response were also most efficient in increasing the thermal stability of the recombinant LBD. Data are presented that show that malate stabilizes the LBD dimer of McpS. This was found to be a feature observed in other prokaryotic and eukaryotic receptors.
EXPERIMENTAL PROCEDURES
Agarose-plug assays were carried out as previously described (10). Bacteria were grown in M9 minimal medium supplemented with 15 mm succinate. Plugs containing chemotaxis buffer (KH2PO4/K2HPO4, 0.05% (v/v) glycerol, 10 mm EDTA, pH 7.0) or 5 mm toluene were used as negative and positive controls, respectively.
Qualitative capillary assays were carried out as described previously (10). Cultures of Pseudomonas putida KT2440RmcpS::Tn5 (pRK415) and KT2440RmcpS::Tn5 (pRK415-mcpS) were grown in MSB medium containing 5 mm succinate and 20 μg/ml of tetracycline. Cells were harvested when the A600 was between 0.15 and 0.35, washed once in chemotaxis buffer, and resuspended to an A600 of 0.1. Capillaries contained 2% low-melting temperature agarose in chemotaxis buffer with or without added attractant (0.1% casamino acids as positive control; 5 or 50 mm organic acids).
Quantitative capillary assays were carried out as described previously (27). Cultures of P. putida KT2440R were grown in MSB medium containing 5 mm succinate, harvested when the A600 was between 0.25 and 0.35, washed once in chemotaxis buffer, and resuspended to an A600 of 0.1.
Bacterial Mutants
The 12 mutants of P. putida KT2440 each deficient in one mcp gene were obtained from the Granada mutant collection. Mutants were generated by random mutagenesis of P. putida with mini-Tn5-Km as described (28). Further information on mutant generation and additional information of these 12 mcp genes in supplemental Table S1.
Complementation of mcpS Mutant
The mcpS gene was amplified from P. putida KT2440 genomic DNA using primers 4520-BamHI forward (5′-GTAGTAGGATCCTGGAGAGCGTGCATGAACAGC-3′) and 4520-SacI reverse (5′-GATGTAGAGCTCCCAGGTTCCAAAGGTCAGACG-3′), where the restriction sites used for cloning into pRK415 (29) are underlined. The cloned gene was sequenced to confirm that no PCR errors had occurred, and pRK415-mcpS was introduced into the mcpS mutant by mating from E. coli S17-1 (30).
Construction of Expression Plasmid for McpS-LBD
The DNA fragment of mcpS encoding amino acids Gly47–Ser283 was amplified with primers that contain restriction sites for NdeI and BamHI. The resulting PCR product was then cloned into pET-28b (Novagen) using the same restriction enzymes that gave rise to pETMcpS. The resulting protein contains an N-terminal His tag.
Overexpression and Purification of McpS-LBD
E. coli BL21(DE3) containing pETMcpS was grown at 30 °C until the culture reached an A600 of 0.6. Isopropyl 1-thio-β-d-galactopyranoside was then added to a final concentration of 0.1 mm and the culture was maintained at 16 °C overnight. Cells were harvested and frozen at −80 °C. Cells derived from a 0.5-liter culture were resuspended in 50 ml of buffer A (20 mm Tris/HCl, 0.2 m NaCl, 1 mm EDTA, 10 mm β-mercaptoethanol, 10 mm imidazol, 5% glycerol, pH 8.0) containing CompleteTM protease inhibitor (Roche) and benzonase (Sigma). Cells were disrupted using French Press, which was followed by centrifugation (45 min at 20,000 × g). The supernatant was loaded onto a 5-ml HisTrapHP column (GE Healthcare) previously equilibrated in buffer A. Elution was performed in a single step with buffer A containing 300 mm imidazol. Protein-containing fractions were pooled, concentrated to 5 ml, dialyzed against 50 mm Tris/HCl, 0.5 m NaCl, pH 8.0, and loaded onto a HiPrepTM 26/60 SephacrylTM S200 gel filtration column (GE Healthcare). Protein was eluted isocratically (1 ml/min). Coomassie-stained SDS-PAGE gels containing 30 μg of McpS-LBD did not show any additional bands.
Buffer System for Biophysical Studies
All subsequent experiments were carried out in polybuffer, pH 6.0. This buffer contains 5 mm Tris, 5 mm MES, 5 mm PIPES adjusted to pH 6.0 with HCl. The study of the pH optimum for binding was carried out in polybuffer adjusted to pH 5.0–8.0.
Isothermal Titration Calorimetry (ITC)
Measurements were done on a VP-microcalorimeter (MicroCal, Amherst, MA) at 20 °C. McpS-LBD was dialyzed overnight against polybuffer, pH 6. For binding studies, protein at 34–38 μm was introduced into the sample cell and titrated with aliquots of ligand solution. The ligand solutions were prepared by dissolving the compounds in dialysis buffer. For dissociation studies, 90 μm McpS-LBD was injected into buffer. Data analysis was carried out using the “one binding site” model of the MicroCal version of ORIGIN, leaving all parameters floating.
Analytical Ultracentrifugation
An Optima XL-I analytical ultracentrifuge (Beckman-Coulter, Palo Alto, CA) was used. The detection was carried out using an UV-visible absorbance detection system. Experiments were conducted at 20 °C using an AnTi50 rotor and absorbance scans were taken at 230 or 280 nm. Sedimentation velocity was performed using epon-charcoal standard double sector centerpieces (12-mm optical path) at a speed of 48,000 rpm. Sedimentation coefficient distributions were determined by direct linear least-squares boundary fitting of the sedimentation velocity profiles using SEDFIT software (31). SEDNTERP software (32) was used for the correction of S values to standard conditions (20 °C and water). Short-column (85 μl) equilibrium runs were carried out at multiple speeds (12,000, 14,000, and 18,000 rpm) and the corresponding scans were measured at 230 and 280 nm. After the equilibrium scans, a high-speed centrifugation run (40,000 rpm) was done to estimate the corresponding baseline offsets. Whole cell apparent weight-average buoyant molecular weight of the protein samples were determined by fitting a single species model to the experimental data using either a MATLAB program based on the conservation of signal algorithm (33) or the Hetero-Analysis program (34). The corresponding protein molecular weights were calculated from the experimental buoyant mass, using 0.725 cm3/g as the protein partial specific volume. Several self-association models at sedimentation equilibrium were globally fitted to multiple experimental data using the Hetero-Analysis program (34). In parallel, a monomer/m-ner/n-mer self-association scheme was also fitted to secondary data (apparent weight-average molecular weight versus protein concentration) using a non-linear least-squares procedure with MATLAB scripts kindly provided by Dr. Allen Minton (NIH).
Differential Scanning Calorimetry (DSC)
Thermal denaturation experiments were carried out with a VP-DSC, capillary-cell microcalorimeter from MicroCal (Northampton, MA) at a scan rate of 120 °C/h. A scan rate of 30 °C/h was also used to determine the effect of scan rate on thermal denaturation. McpS-LBD solutions were prepared by dialysis against polybuffer, pH 6.0, and each ligand compound was subsequently added to the protein samples and buffer. Before each experiment several buffer-buffer baselines were obtained to equilibrate the instrument. The experimental thermograms were baseline-subtracted, corrected from the response of the instrument and normalized by the protein concentration. Reheating runs were systematically carried out to determine the calorimetric reversibility of the denaturation profiles. The calorimetric enthalpies were obtained by integration of the transition peaks.
RESULTS
PP4658 (McpS) Is the Chemoreceptor for Succinate
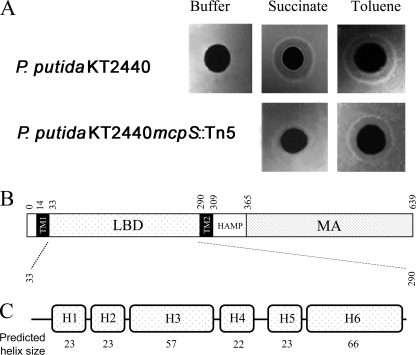
The chemotactic behavior of wild-type P. putida KT2440 and 12 mutants deficient in a single MCP (Table S1) was analyzed using agarose-plug assays. This assay involves placing a solidified agarose-plug containing chemoattractant in contact with a cell suspension, and allowing the formation of rings that are indicative of a chemotactic response. No chemotaxis of P. putida KT2440 (Fig. 1A) was observed for immobilized chemotaxis buffer, but ring formation, indicative of a chemotactic response, was observed for succinate and toluene, which are both known to be chemoattractants for P. putida (10). The screening of the 12 mutants revealed that the strain with a knock-out in the PP4658 open reading frame failed to respond to succinate (Fig. 1A) while maintaining chemotaxis toward toluene. The pp4658 gene is well separated from flanking genes, which do not encode proteins related to chemotaxis or motility (supplemental Fig. S1), and appears to form a single transcription unit. PP4658 was named McpS (succinate).
FIGURE 1.
McpS is the chemoreceptor for succinate. A, agarose-plug assays of P. putida KT2440 and a mutant deficient in pp4658 (mcpS) with succinate and toluene (positive control). B, domain prediction of McpS according to SMART (14): TM, transmembrane region; HAMP, linker domain; MA, methyl-accepting domain. C, consensus secondary structure prediction of McpS-LBD using NPSA (37).
McpS Contains an Unusually Large Sensor Domain
Analysis of the protein sequence of McpS by SMART (14) and DAS (35) allowed us to identify a periplasmic LBD, which spans 257 residues and is flanked by two transmembrane regions (Fig. 1B). As such, the LBD of McpS is ∼100 amino acids larger than the LBD of the 4 MCPs of E. coli. Furthermore, these 4 LBDs all belong to the TarH family (InterPro signature IPR003122), whereas the LBD of McpS remains un-annotated in InterPro (36). The three-dimensional structure of the TarH-LBD shows a 4-helix up-down-up-down bundle arrangement (20). Consensus secondary structure predictions (37) clearly indicate that McpS-LBD consists of six helices separated by short loops (Fig. 1C). Interestingly, of these 6 helices, there are two pairs of helices that span 22–23 amino acids, and one pair of helices that span ∼60 amino acids. Subsequent studies were aimed at establishing to what extent this 6-helix domain is present in chemoreceptors of other bacterial species. We selected randomly 100 chemoreceptor sequences from SMART using the presence of a methyl-accepting domain (IPR004089) and two transmembrane regions as criteria. Using DAS, the sequence limits of the LBD domains were identified and subsequently the secondary structure was predicted as detailed above. Interestingly, from these 100 sequences 11 were found to have a LBD in the range of 240–280 amino acids and a secondary structure consisting of 6 helices. Such sensing domains were found in different species of the genus Pseudomonas (Uniprot, A4XQ69 and Q9HUB1) but also in Rhodopseudomonas (Q07HJ2), Aeromonas hydrophila (Uniprot: A0KGQ7), Marinobacter aquaeolei (A1U770), Deinococcus radiodurans (Q9RYG4), Oceanobacillus iheyensis (Q8EST3), or Bradyrhizobium (A5EAD2). In all cases the LBD of these proteins was found to be un-annotated in SMART. It remains to be established whether this domain corresponds to a novel bacterial sensor domain.
McpS-LBD Interacts Directly with Succinate
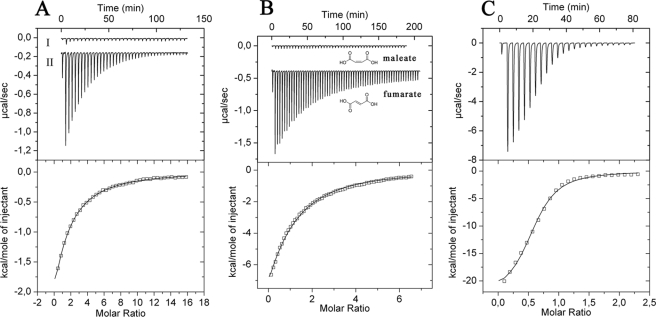
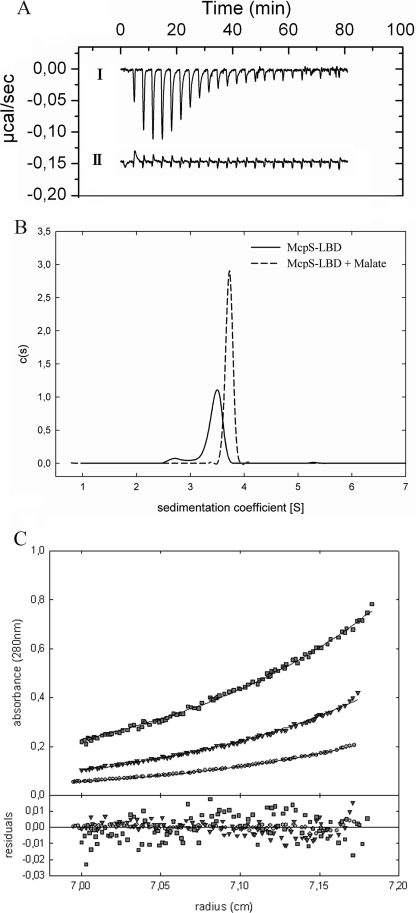
To our knowledge no chemoreceptor that mediates chemotaxis toward succinate has been characterized, and the mode of receptor interaction, either directly or via a binding protein, is unknown. To address this question McpS-LBD was overexpressed, purified, and subjected to ITC studies (26). This technique allows the determination of the thermodynamic forces that drive the interaction, as well as binding constants and stoichiometry. As shown in Fig. 2A titration of buffer with 1 mm succinate resulted in small and uniform peaks representing heats of dilution. In contrast, titration of McpS-LBD with succinate (Fig. 2A) gave rise to large exothermic heat signals due to binding. Data analysis revealed that binding was driven by favorable enthalpy changes that were counterbalanced by unfavorable entropy changes (Fig. 3). To determine the optimum pH for this interaction, the experiment was repeated in polybuffer over a pH range of 5 to 8. The highest binding affinity was measured at pH 6.0 (KD = 82 μm). The binding constants at pH 5.0, 7.0, and 8.0 were 250, 330, and 570 μm, respectively. All subsequent in vitro experiments with McpS-LBD were carried out in polybuffer, at pH 6.0.
FIGURE 2.
Isothermal titration calorimetry studies for the binding of different dicarboxylic acids to McpS-LBD. A, titration of dialysis buffer (I) and 36 μm McpS-LBD (II) with 6.4-μl aliquots of 1 mm succinate. B, titration of 36 μm McpS-LBD with 1 mm maleate and fumarate. C, titration of 100 μm McpS-LBD with 1 mm malate. Upper panels contain the titration raw data and the lower panels show the integrated and dilution-corrected peak areas of the raw data. Data were fitted with the “one binding site model” of the MicroCal version of ORIGIN (Amherst, MA).
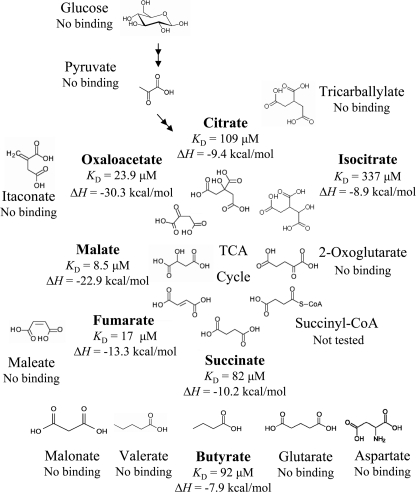
FIGURE 3.
Summary of microcalorimetric titrations of McpS-LBD with a range of different compounds. The full range of compounds analyzed is provided under “supplemental Fig. S2. Shown are the thermodynamic parameters for the 7 compounds that were found to bind.
McpS-LBD Recognizes Most of the TCA Cycle Intermediates and Butyrate with High Specificity
ITC was subsequently employed to determine the ligand profile of McpS-LBD. All compounds that were used are shown in supplemental Fig. S2. Because succinate was found to be a ligand, initial experiments were done with other dicarboxylic compounds. Titration of McpS-LBD with 1 mm aspartate, oxalate (C2-dicarboxylate), malonate (C3-dicarboxylate), glutarate (C5-dicarboxylate), and pimelate (C7-dicarboxylate) did not produce any binding heats. It was found, however, that butyrate bound with an affinity comparable with that of succinate (Fig. 3). Binding appeared to be specific to C4-carboxylates because lactate or valerate (C5-monocarboxylate) did not bind to McpS-LBD.
Next, the remaining TCA cycle intermediates were studied. Interestingly all TCA cycle intermediates, except 2-oxoglutarate, were found to bind (Fig. 3). The common structural feature of fumarate, malate, oxaloacetate, citrate, and isocitrate is that they contain a C4-dicarboxy moiety (including citrate and isocitrate, which are C2-substituted C4-dicarboxylates). In contrast, 2-oxoglutarate, the only TCA cycle intermediate that did not bind, is a C5-dicarboxylate (Fig. 3). Malate was the tightest binding ligand, followed by fumarate and oxaloacetate (Fig. 3).
To assess the specificity of McpS-LBD for recognition of TCA cycle intermediates, maleate, itaconate, and tricarballylate, close structural homologues of TCA cycle intermediates (Fig. 3) were analyzed. None of these compounds showed binding, including maleate, which only differs from fumarate in the configuration of the double bond, suggesting that McpS-LBD shows high binding specificity toward TCA cycle intermediates. As shown in Fig. 2B, large heats of binding were observed for fumarate, whereas titration signals for maleate were identical to the heats of dilution.
To further consolidate this ligand profile, a series of physiologically relevant and structurally less-related compounds were analyzed including various amino acids, sugars, and organic acids (supplemental Fig. S2). No binding was observed with any of these compounds, leading to the conclusion that the in vitro ligand profile of McpS consists of the 7 compounds as highlighted in Fig. 3.
A McpS-LBD Dimer Recognizes One Malate Molecule
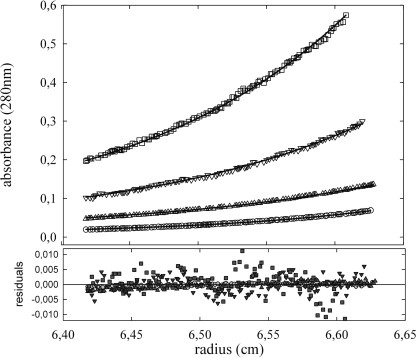
Subsequent studies were aimed at determining the binding stoichiometry between McpS-LBD and its ligands. To this end it is necessary to characterize first the oligomeric state of the protein that was accomplished by analytical ultracentrifugation experiments. Equilibrium sedimentation experiments of McpS-LBD were carried out at protein concentrations between 0.125 and 10 mg/ml (Fig. 4). Data analysis revealed that McpS-LBD exists as a mixture of monomers and dimers in solution, with a monomer-dimer association constant of 164,000 m−1, corresponding to a KD of 6.1 μm. These results are consistent with sedimentation velocity studies reported below.
FIGURE 4.
Monomer-dimer equilibrium of McpS-LBD determined by analytical ultracentrifugation. Experiments were carried out at two sedimentation equilibrium speeds (12,000 and 14,000 rpm) with protein concentrations ranging from 10 to 0.25 mg/ml. For clarity reasons only the experimental radial distribution at 12,000 rpm for the range 1–0.25 mg/ml are shown: 0.125 (○), 0.25 (Δ), 0.5 (▿), and 1 mg/ml (□). The solid lines represent the best-fit curves of the global analysis of the 12 data sets to the monomer-dimer equilibrium model described in the text.
ITC can also be used to determine the binding stoichiometry in cases were binding curves are sigmoid. However, curves that so far had been obtained are hyperbolic (Fig. 2, A and B). ITC curve shape depends on the c value (38), which is a function of affinity and ligand concentration. To generate a sigmoid binding curve, experiments were repeated with the tightest binding ligand, malate, at a high protein concentration (100 μm). In agreement with equilibrium sedimentation studies, McpS-LBD at this concentration was exclusively present as dimer. The resulting sigmoid binding curve is shown in Fig. 2C. Curve fitting gave rise to an n value of 0.56, which is consistent with the binding of a single malate molecule to the McpS-LBD dimer.
Differential Chemotactic Response toward the 7 Ligands
The above studies show that McpS-LBD binds 7 ligands in vitro. Chemotaxis agarose-plug assays were carried out with P. putida KT2440 and its mcpS mutant to evaluate the chemotactic behavior to these 7 ligands. The wild-type strain showed a strong chemotactic response toward succinate, malate, and fumarate (supplemental Fig. S3). This phenotype was characterized by the formation of well defined rings, which started to appear after 4 min. A chemotactic response to oxaloacetate was also observed. However, the intensity of ring formation was weaker and delayed because ring formation started after around 6 min (supplemental Fig. S3). Using this technique no response was observed for citrate, isocitrate, and butyrate.
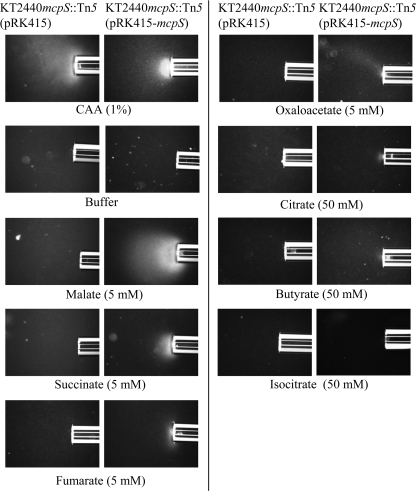
Subsequently, chemotaxis toward these 7 compounds was studied by qualitative and quantitative capillary assays that have a higher sensitivity than the agarose-plug assay. The mcpS mutant strain was complemented with the mcpS gene, which was present in plasmid pRK415-mcpS. The chemotactic behavior of this strain was compared with the mutant strain harboring the empty plasmid pRK415. In contrast to results with the agarose-plug assay, responses to malate, succinate, fumarate, oxaloacetate, citrate, isocitrate, and butyrate were detected (Fig. 5). The responses to citrate, butyrate, and isocitrate were significantly weaker than those of the other four attractants, as a response was only detected with a relatively high attractant concentration. The response to isocitrate was particularly weak. The mutant strain carrying the empty vector responded to the positive control and weakly to butyrate, but not to the other tested organic acids. These results suggest that McpS is the only chemoreceptor for all of the compounds except butyrate, which may be detected by an additional MCP.
FIGURE 5.
Qualitative capillary assays of P. putida toward the 7 McpS-LBD ligands identified in vitro. The mcpS mutant strain carrying pRK415 (empty plasmid) and the complemented mutant strain carrying mcpS on plasmid pRK415 were used for analyses. Photographs were taken after 5 min except those for the isocitrate (after 20 min) and butyrate (after 10 min). In some cases the agarose in the capillary receeded 1–2 mm during the assay and cells were accumulated in the tip of the capillary. Note that citrate, butyrate, and isocitrate were tested at 50 mm to detect a response. CAA, casamino acids.
To determine whether the weak response toward citrate, isocitrate, and butyrate is due to a nonspecific effect leading to cell paralysis, agarose-plug assays to mixtures of toluene with butyrate or isocitrate were carried out (supplemental Fig. S4). Chemotaxis toward toluene is mediated by a different MCP. However, the presence of an equimolar concentration of butyrate and isocitrate did not alter chemotaxis toward toluene indicating that these compounds do not alter cell motility. In addition, microscopic inspection of cells containing citrate, isocitrate, and butyrate did not reveal any inhibition of cell motility. Furthermore, P. putida KT2440 can grow in minimal medium supplemented with any of these 3 compounds.
We hypothesized that there are two groups of ligands: strong attractants (succinate, fumarate, malate, and oxaloacetate) and weak attractants (citrate, isocitrate, and butyrate), which differ significantly in their chemotactic response. It has to be noted that the in vitro binding affinity of the weak attractants butyrate and citrate for McpS-LBD is comparable with that of the strong attractant succinate.
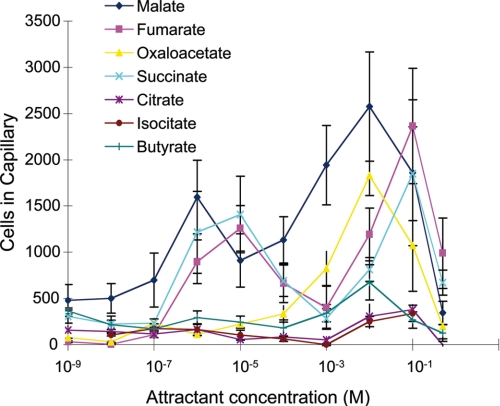
To precisely quantify the differences in the response toward both groups of attractants quantitative capillary assays were conducted (Fig. 6). Confirming the agarose-plug assays malate, fumarate, and succinate showed the strongest response. Interestingly, their response was biphasic with maximum at attractant concentrations of 1–10 μm and 10–100 mm. At all concentrations the response caused by the weak attractants was superior to the buffer control but significantly inferior to the response toward strong attractants.
FIGURE 6.
Concentration dependence of chemotaxis. Quantitative chemotaxis assays of P. putida KT2440 toward the 7 chemoattractants. Data are the means of at least four different capillaries. The average number of cells that swam into capillaries containing buffer only (365 cells/capillary) was subtracted from the data shown.
Both Groups of Attractants Compete in Vitro and in Vivo
To identify the molecular reason for the differential response of weak and strong attractants we wanted to establish whether both groups of compounds share the same binding site at McpS. To this end ITC competition experiments were carried out that involved the titration of McpS-strong attractant complexes with weak attractants and, vice versa. McpS-LBD saturated with succinate or malate (2 mm) was titrated with butyrate and citrate and, vice versa. In all experiments previous saturation with a ligand reduced significantly the binding of the secondary ligand. This is illustrated in Fig. 7, which demonstrates that the presence of malate inhibits citrate binding.
FIGURE 7.
Competition between strong and weak attractants in vitro. To elucidate whether both classes of compounds compete for binding to McpS-LBD, ITC competition experiments were carried out. These experiments involved the titration of McpS-weak attractant complexes with strong attractants and, vice versa. A, microcalorimetric titration of 35 μm McpS-LBD with aliquots of 1 mm citrate. B, microcalorimetric titration of 35 μm McpS-LBD, containing 1 mm malate, with aliquots of 1 mm citrate.
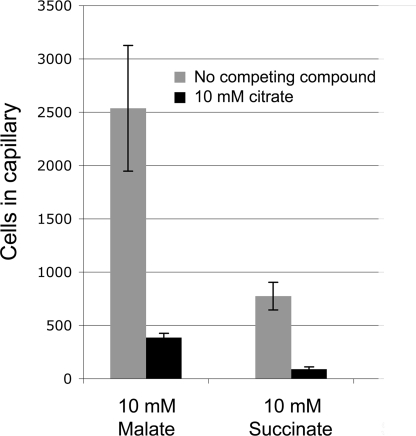
Subsequently quantitative capillary assays were conducted to malate and succinate in the absence and presence of citrate. The competing agent citrate was present in both, the capillary and the bacterial suspension, whereas the attractants succinate and malate were only present in the capillary. Bacteria migrate thus in response to a malate and succinate gradient in the absence or presence of a constant, high concentration of citrate. Under these experimental conditions the citrate concentration in the immediate environment of the receptor LBD is likely to be well above the malate or succinate concentration. In both cases, the presence of citrate reduced significantly the chemotaxis toward succinate and malate, which is exemplified for the latter compound in Fig. 8. These in vitro and in vivo data strongly suggest that weak and strong attractants compete for the same binding site at McpS-LBD.
FIGURE 8.
Competition between strong and weak attractants studies in vivo by capillary chemotaxis assays. The responses of P. putida KT2440 to malate and succinate were tested in the presence and absence of the competing attractant citrate (10 mm), which was added to the cell suspension and the capillary. The data were corrected for background accumulation in capillaries containing buffer or buffer plus citrate (∼38 ± 11 cells for experiments without citrate and ∼20 ± 3 cells for experiments with citrate). The results are averages of at least four capillaries and error bars indicate standard errors.
Strong Attractants Increase the Thermal Stability of McpS-LBD
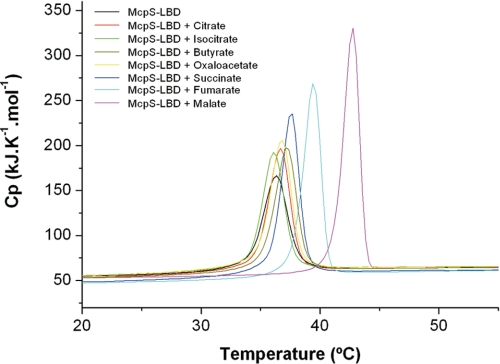
To explain the differential effect of strong and weak attractants in vivo, McpS-LBD in the absence and presence of the 7 ligands were analyzed by DSC. In these experiments McpS-LBD is heated at a constant rate and heat changes caused by the thermal denaturation of the protein was recorded. Samples were scanned against a reference that contains all buffer components but lacks the protein, which implies that the heat measured can be exclusively attributed to protein unfolding. Ligands were added at a concentration that guaranteed the complete saturation of protein. This technique permits the determination of the enthalpy change (ΔH, energy necessary to unfold 1 mol of protein) and the Tm (temperature of the maximum of the thermal denaturation transition) of thermal unfolding (39). The resulting thermograms are shown in Fig. 9 and the derived parameters are listed in Table 1. Re-scans and analyses at different scan rates revealed that thermal denaturation is an irreversible process. Thermal denaturation of protein in the absence of ligand is characterized by a Tm of 36.3 °C and a ΔH of 66 kcal/mol. A group of 4 thermograms (Fig. 9) representing McpS-LBD in complex with citrate, isocitrate, butyrate, and oxaloacetate reveal Tm values similar to that of the protein in the absence of ligand (36.1–37.1 °C), whereas increased ΔH values were observed ranging from 75 to 84 kcal/mol. These compounds are the weak attractants, along with oxaloacetate, which only triggered a modest chemotactic response. Within this group the latter compound was found to have the highest ΔH. On the other hand, Fig. 9 clearly shows that when complexed to the strong attractants malate, succinate, or fumarate, McpS-LBD has a significantly different denaturation profile than when complexed to weak attractants. The thermal denaturation of these 3 complexes is characterized by a significant increase in ΔH (87–106 kcal/mol) and Tm (37.7–42.8 °C). In addition, there is a correlation between ΔH values and the in vivo response: weak attractants have significantly lower enthalpy changes than strong attractants. This is further confirmed by oxaloacetate, which exhibits a weaker chemotactic response than malate, fumarate, and succinate and which has a ΔH inferior to these latter compounds but superior to the weak attractants.
FIGURE 9.
Analysis of McpS-LBD by differential scanning calorimetry in the absence and presence of the ligands. Protein was at a concentration of 36 μm. Ligands were added at a concentration of 1 mm except for isocitrate, for which the concentration was increased to 3 mm to achieve complete saturation of the protein. Derived parameters are given in Table 1.
TABLE 1.
Parameters derived from the analysis of McpS-LBD in the absence and presence of ligands by DSC
Compounds were present at 1 mm, except isocitrate, which was at 3 mm.
| Ligand | ΔH | Tm |
|---|---|---|
| kcal/mol | °C | |
| None | 66 | 36.3 |
| Isocitrate | 75 | 36.1 |
| Citrate | 76 | 36.7 |
| Butyrate | 80 | 37.1 |
| Oxaloacetate | 84 | 36.9 |
| Succinate | 87 | 37.7 |
| Fumarate | 89 | 39.4 |
| Malate | 106 | 42.8 |
Malate Stabilizes the McpS-LBD Dimer
We then hypothesized that ligand binding might cause a stabilization of the dimeric state of McpS-LBD, which might account for the increase in thermal stability as observed by DSC. To address this question a series of biophysical experiments were conducted to investigate the complex between McpS with malate. This latter compound caused the strongest chemotactic response in vivo (Figs. 5 and 6) and had the most pronounced effect on the thermal stability of McpS-LBD in vitro (Fig. 9).
ITC can also be used to study self-association by monitoring heat changes caused by the dilution of a concentrated oligomeric protein into buffer, where heat changes measured represent the transition of a higher to a lower oligomeric state (40). Fig. 10A shows the data for the injection of 90 μm McpS-LBD into dialysis buffer. According to the above analytical ultracentrifugation studies, McpS-LBD at 90 μm is entirely present as a dimer. A series of exothermic peaks are observed for the initial injections, which then diminish in size to reach the level of dilution heats. During the course of this experiment the protein concentration in the ITC cell is increased from 0 to 15.5 μm. In agreement with the equilibrium ultracentrifugation studies (Fig. 4), which revealed a monomer-monomer dissociation constant of 6.1 μm, the heat changes observed for the injection of concentrated McpS-LBD into buffer represent the dissociation of protein dimers into monomers, which is an exothermic process.
FIGURE 10.
Impact of malate binding on the oligomeric state of McpS-LBD. A, ITC data for the injection of: (I) 90 μm McpS-LBD into dialysis buffer; (II) 90 μm McpS-LBD containing 1 mm malate into buffer containing 1 mm malate. B, sedimentation velocity studies of McpS-LBD at 1 mg/ml in the absence of ligand and in the presence of 2 mm malate and butyrate. C, equilibrium sedimentation ultracentrifugation studies of McpS-LBD at 0.25 (○), 0.5 (▿), and 1 mg/ml (□) in the presence of 2 mm malate.
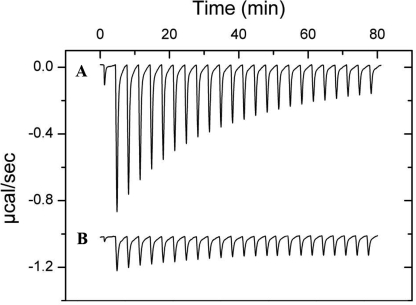
This experiment was repeated in the presence of 2 mm malate, which was added to both, the protein and buffer solutions. Under these conditions the protein is a homogeneous sample entirely saturated with ligand. Under these conditions the heat changes were very small and can be almost entirely attributed to heats of dilution (Fig. 10A), suggesting the absence of significant dimer dissociation.
To verify this hypothesis sedimentation velocity ultracentrifugation studies were conducted. McpS-LBD at 1 mg/ml in the absence and presence of 2 mm malate was submitted to analysis. The sedimentation coefficient distribution obtained for McpS-LBD in the absence of ligand is shown in Fig. 10B and reveals two species with standard S values of 2.7 ± 0.1 S and 3.5 ± 0.2 S, which are compatible with a globular monomer (frictional ratios f/f0 = 1.2) and a dimer that slightly deviates from globular shape (f/f0 = 1.5). The relative abundance of the two species depends upon the protein concentration in a manner that confirms the existence of a monomer-dimer equilibrium (Fig. 4). In the presence of malate, a single peak was observed with an S value of 3.7 ± 0.2 S. This peak is likely to present the protein dimer, whereas the absence of the peak corresponding to the protein monomer is observed.
To verify whether the binding of malate shifts the protein monomer-dimer equilibrium toward the dimer, sedimentation equilibrium studies were conducted in analogy to data reported in Fig. 4 but in the presence of malate. McpS-LBD at different concentrations was incubated with 1 mm malate. The analysis of the resulting data (Fig. 10C) revealed that molecular weights for McpS-LBD did not vary in function of its concentration. At all protein concentrations molecular weights were obtained that are close to the protein dimer. The average molecular weight derived from analyses of different McpS-LBD concentrations was 59,100 ± 2,500, which is close to the expected size of the protein dimer (57,226). At the lowest protein concentration (0.25 mg/ml) a molecular weight of 41,400 ± 200 was determined for McpS in the absence of malate, whereas a value of 59,700 ± 300 was measured in the presence of malate. Taken together, the calorimetric dilution studies, the sedimentation velocity experiments, and the sedimentation equilibrium analyses, it can be concluded that binding malate stabilizes the dimeric form of McpS-LBD.
DISCUSSION
A glimpse at a metabolic map of aerobic microorganisms reveals the central role of the TCA cycle. Apart from its essential role in the generation of NADH, many biosynthetic routes branch off from it and many catabolic pathways funnel products into it. Due to its central metabolic role many bacteria are able to use TCA cycle intermediates as their sole carbon and energy source. Bacteria possessing a complete TCA cycle require only an uptake system for the utilization of these compounds (41, 42) and in addition, several anaerobic routes have been described for the catabolization of TCA cycle intermediates (43). This, combined with the fact that TCA cycle intermediates are abundantly present in a variety of natural habitats (4, 6), explains why bacteria frequently exhibit chemotactic behavior toward these compounds (7–13). However, the molecular mechanism of this type of chemotaxis is still poorly understood as so far only a few receptors have been identified. Those that have been identified are the Tcp receptor in S. typhimurium, which mediates positive and negative chemotaxis to citrate and phenol, respectively (9), and PA2652 of P. aeruginosa, which mediates chemotaxis to malate but not to any of the remaining TCA cycle intermediates (7). P. putida KT2440, the model organism chosen for this study, has a saprophytic lifestyle, is able to efficiently colonize roots and seeds (44), and was found to use organic acids present in root exudates as the primary carbon source during rhizosphere colonization (45).
By screening the chemotactic behavior of bacterial mutants deficient in a single chemoreceptor we have identified the chemoreceptor for succinate, which we term McpS (Fig. 1). In general, the establishment of the complete chemoreceptor ligand profile by analyzing single mutants can be misleading because there are several examples that show that bacteria have multiple receptors for the same compound (3). Therefore, the LBD of McpS was produced as recombinant protein and its ligand profile was established by microcalorimetric screening of compounds (Figs. 2 and 3). The choice of the recombinant LBD for this series of experiments was based on data available on Tar, which demonstrate that the molecular determinants for the recognition of ligands by Tar are exclusively present in the LDB. It was shown that: first, full-length Tar and its recombinant LBD bind aspartate with a similar affinity of ∼3 μm (23, 46, 47); second, both proteins recognize ligands with a stoichiometry of 1 ligand per dimer (23, 48); third, in both cases this binding stoichiometry is caused by strong negative cooperativity between the 2 ligand binding sites per dimer (23, 48); and fourth, in both cases ligand binding causes modest but significant structural alterations (20, 23, 49).
The ligand profile as determined by ITC-based screening of McpS consists of succinate, fumarate, malate, oxaloacetate, citrate, isocitrate, and butyrate. Subsequently, the chemotactic behavior of the McpS mutant and its derivative complemented with the mcpS gene was analyzed. Data show that among the 26 chemoreceptors of P. putida KT2440 McpS is the sole receptor for these 6 TCA cycle intermediates, whereas an additional receptor for butyrate might exist (Fig. 5). The fact that there is a sole receptor distinguishes taxis toward TCA cycle intermediates well from other systems where multiple receptors have been identified for the same compound. This is exemplified by P. aeruginosa, which employs the CtpL and CtpH receptors for taxis toward inorganic phosphate and the PctA, PctB, and PctC chemoreceptors, which have largely overlapping ligand profiles for the 20 L-amino acids (3).
Apart from the determination of the ligand profile, the ITC experiments with McpS-LBD have established the following features of its action. 1) In general, two basic modes in interaction between ligands and chemoreceptors can be distinguished: either by direct binding of the chemoattractant to the receptor, or by the initial formation of a complex between the attractant with an auxiliary protein, which then binds to the receptor, as shown for maltose, galactose, and ribose chemotaxis in E. coli (2). Our ITC experiments demonstrate the direct nature of the interaction of TCA intermediates with McpS, as is the case for citrate binding to Tcp (9), whereas the mode of interaction of PA2652 with malate remains to be elucidated (7).
2) A binding stoichiometry of one ligand molecule per receptor dimer has been observed for Tar (48) and Tsr (50). The currently reported ITC studies show that the same stoichiometry is also observed for the binding of malate to McpS (Fig. 2C). This might indicate that recognition of a single ligand by a receptor dimer is a mode shared by structurally different chemoreceptors.
3) The affinities of chemoattractants for receptors have been determined for Tar and Tsr. Tar binds aspartate with a KD of around 3 μm (46, 47), whereas the KD of serine for Tsr is in the range of 10 to 27 μm (50–52). These studies were all conducted at pH 7.0–7.5 and the pH dependence of ligand binding has not been addressed. Here we show that the affinity is largely dependent on the pH, with the optimum at pH 6. Of the 4 efficient ligands that trigger chemotaxis in vivo, three (malate, fumarate, and oxaloacetate) were recognized by McpS with affinities ranging from 9 to 24 μm (Fig. 3), which are thus comparable with that of serine binding to Tsr. Succinate was recognized with a lower affinity (82 μm).
Our data reveal that the minimal structural requirements of a ligand for binding to McpS are at least 1 carboxylic group present at position 1 of a C4-moiety (citrate and isocitrate can be considered as C2-substituted C4-carboxylic acids, Fig. 3). The only TCA cycle intermediate that is a C5-dicarboxylic acid, oxoglutarate, was devoid of binding. However, most interestingly, isomers or very close derivatives of the 7 ligands identified were devoid of binding. This is illustrated in Fig. 2 for fumarate, a tight binding ligand, and its isomer maleate, which did not bind. Other examples are the absence of binding of aspartate, itaconate, or tricarballylate (Fig. 3). Therefore, structural differences between ligands and non-ligands, for example, between fumarate and maleate, are much smaller than structural differences between ligands, as exemplified by fumarate and citrate. It can therefore be concluded that functional similarities, i.e. belonging to the same metabolic route like the TCA cycle, has dominated over structural similarities in the evolution of the molecular recognition of McpS. These findings provide interesting insight into protein evolution and are in this case clearly related to the evolutionary advantages that arise from chemotaxis toward TCA cycle intermediates. Different studies report that TCA cycle intermediates are present at high concentrations in root exudates and dry plant mass (4, 6). P. putida is a saprophyte and was shown to efficiently colonize plant roots. The physiological relevance of McpS-mediated taxis is thus likely to consist in accessing TCA cycle intermediates from debris of dead plants and root exudates.
However, the magnitude of the chemotactic response toward the 7 attractants varied largely. Malate, succinate, fumarate, and oxaloacetate gave a significant response, whereas citrate, isocitrate, and butyrate gave a response that was close to the detection limit of the chemotaxis assays employed. Several studies have determined the organic acid composition of plant root tissue and plant root exudates (4, 6, 53–55). Citrate is one of the most abundant organic acids in both samples (6, 53, 54), whereas no significant concentrations of isocitrate were detected. In this context the very weak chemotaxis toward 50 mm citrate and isocitrate (Fig. 5) does not appear to be of physiological relevance. Although butyrate is a product of fermentation of anaerobic bacteria such as Clostridia (56) and although this compound is a carbon source for growth of P. putida the weak taxis observed for butyrate does not equally point to a physiological relevance. In summary, among the 7 ligands of McpS, a physiological relevance in the context of taxis toward root exudates and plant debris can only be suggested for the 4 strong attractants succinate, malate, fumarate, and oxaloacetate.
Many chemoreceptors are characterized by a relatively narrow attractant profile. Tcp of S. typhimurium mediates attraction to only citrate (9) and PA2652 of P. aeruginosa to only malate but to no other TCA cycle intermediate (7). This contrasts McpS, which recognizes 6 TCA cycle intermediates and butyrate. Tcp and PA2652 have LBDs of the TarH and Cache-2 (IPR013163) type, respectively, which are both ∼150 residues. McpS-LBD in contrast has a size of 257 amino acids, remains un-annotated in InterPro (36), and shows no significant sequence similarities with the other two LBDs. Therefore, at least three different domains are employed by MCPs to recognize TCA cycle intermediates: the TarH 4-helix bundle, the Cache domain, and the 6-helix structure predicted for McpS.
Protein samples in complex with the 7 ligands were submitted to analyses by differential scanning calorimetry. Interestingly, the 3 ligands that caused the strongest chemotactic response, malate, succinate, and fumarate, induced a significant increase in Tm, whereas the remaining ligands had only a marginal impact on the thermal stability of McpS-LBD (see Fig. 9 and Table 1). Although the affinity of oxaloacetate for McpS-LBD is around three times higher than that for succinate (Fig. 3), the former compound did not significantly increase the protein Tm. The capacity of ligands to increase the Tm (Table 1) appears to correlate better with the final chemotactic response (Fig. 6) than with the binding affinity. This is consistent with the notion that the binding affinity is not the only determinant that defines the magnitude of the final chemotactic response. The determination of the detailed molecular consequences triggered by the binding of strong and weak attractants involves the resolution of the corresponding co-crystal structures, which is an ongoing activity in our laboratory.
In subsequent studies the impact of the binding of malate, the strongest chemoattractant, on the oligomeric state of McpS-LBD was investigated (Fig. 10). Based on equilibrium sedimentation ultracentrifugation studies of protein at different concentrations, a monomer-dimer equilibrium with a KD of 6.1 μm was described in the absence of bound ligand (Fig. 4). When these experiments are repeated in the presence of malate, protein was found to be exclusively present as dimers at all protein concentrations analyzed (Fig. 10C), indicating that malate binding has shifted the monomer-dimer equilibrium toward the dimeric state. This conclusion is supported by microcalorimetric protein dilution studies as well as by sedimentation velocity ultracentrifugation analyses (Fig. 10, A and B).
In this respect clear parallels exist to the Tar chemoreceptor of E. coli. The LBDs of both receptors differ in size (167 amino acids for Tar as compared with 257 amino acids for McpS) and share no significant sequence identity. The Tar-LBD was found to form a 4-helix bundle structure (20). Milliagan and Koshland (23) have studied the effect of aspartate binding on the recombinant LBD of Tar. In the absence of aspartate, a monomer-dimer equilibrium characterized by a KD between 0.5 and 5 μm was reported. Interestingly, in the presence of saturating concentrations of aspartate the KD was at least 2 orders of magnitude lower, indicating a significant stabilization of the LBD dimer (23). The KD for the monomer-dimer equilibrium determined for Tar is thus in agreement with the corresponding value of 6.1 μm as determined for McpS-LBD. In analogy to Milligan and Koshland (23), we establish that malate binding stabilizes the LBD dimer (23). These observations are consistent with the notion that stabilization of the LBD dimer is a molecular consequence of chemoattractant binding common to receptor LBDs, which are unrelated at the sequence level.
The stabilization of the receptor LBD dimer as a consequence of ligand binding is also a feature that has been observed for several eukaryotic receptors such as, for example, the glutamate receptors (57). In analogy to most chemoreceptors this class of receptor has an extracellular LBD and a cytosolic signaling domain that interacts with other proteins. The function of these receptors was found to depend on the presence of Na+ and Cl− that bind, together with glutamate, to the receptor LBD (58). The same authors present electrophysiological measurements that show that increasing NaCl concentrations increase the rate of desensitization of the receptor toward glutamate. In analogy to our work the authors used equilibrium sedimentation experiments to measure the influence of NaCl binding on the monomer-dimer equilibrium of the recombinant LBD. The authors were able to show that NaCl stabilizes the LBD dimer 50-fold as compared with a non-NaCl containing control. It was concluded that the differential stabilization of the LBD dimer by NaCl causes the differences seen in receptor function.
The initial observation that signaling of both eukaryotic and prokaryotic receptors might involve ligand-mediated stabilization of the receptor LBD has been made by Jeff Stock (59). In this article the above mentioned stabilization of the Tar LBD by aspartate was related to the function of several hormone receptors. Data presented here are thus consistent with the initial proposition by Stock. However, further studies are necessary to elucidate to what degree these similarities in the function of prokaryotic and eukaryotic receptors can be generalized.
Supplementary Material
Acknowledgments
We thank Carmen Lorente and M. Mar Fandila for secretarial assistance, Cristina G. Fontana for technical assistance, and Benjamin Pakuts for improving the use of English.
This work was supported by Spanish Ministry of Science and Education Grants CICYT BIO-2006-05668, BFU 2005-0487-C02-02, CSD 2007-00010, and BIO2008-04478-C03-03, Junta de Andalucía Grant CIV344 (to J. L. R.), CIV1912 and P09-RNM-4509 (to T. K.), and the BBVA foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- MCP
- methyl-accepting chemotaxis protein
- DSC
- differential scanning calorimetry
- ITC
- isothermal titration calorimetry
- LBD
- ligand-binding domain
- MES
- 4-morpholineethanesulfonic acid
- PIPES
- 1,4-piperazinediethanesulfonic acid
- TCA
- tricarboxylic acid.
REFERENCES
- 1.Hazelbauer G. L., Falke J. J., Parkinson J. S. (2008) Trends Biochem. Sci. 33, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wadhams G. H., Armitage J. P. (2004) Nat. Rev. Mol. Cell Biol. 5, 1024–1037 [DOI] [PubMed] [Google Scholar]
- 3.Kato J., Kim H. E., Takiguchi N., Kuroda A., Ohtake H. (2008) J. Biosci. Bioeng. 106, 1–7 [DOI] [PubMed] [Google Scholar]
- 4.Kamilova F., Kravchenko L. V., Shaposhnikov A. I., Azarova T., Makarova N., Lugtenberg B. (2006) Mol. Plant Microbe Interact. 19, 250–256 [DOI] [PubMed] [Google Scholar]
- 5.Vílchez S., Molina L., Ramos C., Ramos J. L. (2000) J. Bacteriol. 182, 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas García J. A., Barbas C., Probanza A., Barrientos M. L., Gutierrez Mañero F. J. (2001) Phytochem. Anal. 12, 305–311 [DOI] [PubMed] [Google Scholar]
- 7.Alvarez-Ortega C., Harwood C. S. (2007) Appl. Environ. Microbiol. 73, 7793–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Weert S., Vermeiren H., Mulders I. H., Kuiper I., Hendrickx N., Bloemberg G. V., Vanderleyden J., De Mot R., Lugtenberg B. J. (2002) Mol. Plant Microbe Interact. 15, 1173–1180 [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto K., Imae Y. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parales R. E., Ditty J. L., Harwood C. S. (2000) Appl. Environ. Microbiol. 66, 4098–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson J. B., Bauer W. D. (1993) J. Bacteriol. 175, 2284–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugdahl M. B., Beery J. T., Doyle M. P. (1988) Infect. Immun. 56, 1560–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhold B., Hurek T., Fendrik I. (1985) J. Bacteriol. 162, 190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulrich L. E., Zhulin I. B. (2005) Bioinformatics 21, Suppl. 3, 111, 45–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhulin I. B. (2001) Adv. Microb. Physiol. 45, 157–198 [DOI] [PubMed] [Google Scholar]
- 17.Anantharaman V., Aravind L. (2000) Trends Biochem. Sci. 25, 535–537 [DOI] [PubMed] [Google Scholar]
- 18.Zhulin I. B., Nikolskaya A. N., Galperin M. Y. (2003) J. Bacteriol. 185, 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu E. W., Koshland D. E., Jr. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9517–9520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh J. I., Biemann H. P., Privé G. G., Pandit J., Koshland D. E., Jr., Kim S. H. (1996) J. Mol. Biol. 262, 186–201 [DOI] [PubMed] [Google Scholar]
- 21.Murphy O. J., 3rd, Kovacs F. A., Sicard E. L., Thompson L. K. (2001) Biochemistry 40, 1358–1366 [DOI] [PubMed] [Google Scholar]
- 22.Ottemann K. M., Xiao W., Shin Y. K., Koshland D. E., Jr. (1999) Science 285, 1751–1754 [DOI] [PubMed] [Google Scholar]
- 23.Milligan D. L., Koshland D. E., Jr. (1993) J. Biol. Chem. 268, 19991–19997 [PubMed] [Google Scholar]
- 24.Busch A., Lacal J., Martos A., Ramos J. L., Krell T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13774–13779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guazzaroni M. E., Gallegos M. T., Ramos J. L., Krell T. (2007) J. Biol. Chem. 282, 16308–16316 [DOI] [PubMed] [Google Scholar]
- 26.Krell T. (2008) Microb. Biotechnol. 1, 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X., Wood P. L., Parales J. V., Parales R. E. (2009) J. Bacteriol. 191, 2909–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duque E., Molina-Henares A. J., de la Torre J., Molina-Henares M. A., del Castillo T., Lam J., Ramos J. L. (2007) Pseudomonas, Vol. V, Springer, Dordrecht, The Netherlands [Google Scholar]
- 29.Keen N. T., Tamaki S., Kobayashi D., Trollinger D. (1988) Gene 70, 191–197 [DOI] [PubMed] [Google Scholar]
- 30.Simon R., Priefer U., Pühler A. (1983) Bio/Technology 1, 784–791 [Google Scholar]
- 31.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) Analytical Ultracentrifugation in Biochemistry and Polymer Science, Ed. (Harding S. E., ed) pp. 90–125, The Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 33.Minton A. P. (1996) in Modern Analytical Ultracentrifugation (Schuster T. M., Laue T. M. eds) pp. 81–93, Birkhauser, Boston, MA [Google Scholar]
- 34.Cole J. L. (2004) Methods Enzymol. 384, 212–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cserzö M., Wallin E., Simon I., von Heijne G., Elofsson A. (1997) Protein Eng. 10, 673–676 [DOI] [PubMed] [Google Scholar]
- 36.Hunter S., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., Finn R. D., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Laugraud A., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., Mistry J., Mitchell A., Mulder N., Natale D., Orengo C., Quinn A. F., Selengut J. D., Sigrist C. J., Thimma M., Thomas P. D., Valentin F., Wilson D., Wu C. H., Yeast C. (2009) Nucleic Acids Res. 37, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deléage G., Blanchet C., Geourjon C. (1997) Biochimie 79, 681–686 [DOI] [PubMed] [Google Scholar]
- 38.Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 39.Spink C. H. (2008) Methods Cell Biol. 84, 115–141 [DOI] [PubMed] [Google Scholar]
- 40.Velazquez-Campoy A., Leavitt S. A., Freire E. (2004) Methods Mol. Biol. 261, 35–54 [DOI] [PubMed] [Google Scholar]
- 41.Warner J. B., Lolkema J. S. (2002) Microbiology 148, 3405–3412 [DOI] [PubMed] [Google Scholar]
- 42.Yurgel S. N., Kahn M. L. (2004) FEMS Microbiol. Rev. 28, 489–501 [DOI] [PubMed] [Google Scholar]
- 43.Bott M. (1997) Arch. Microbiol. 167, 78–88 [PubMed] [Google Scholar]
- 44.Molina L., Ramos C., Duque E., Ronchel M. C., García J. M., Wyke L., Ramos J. L. (2000) Soil Biol. Biochem. 32, 315–321 [Google Scholar]
- 45.Lugtenberg B. J., Dekkers L., Bloemberg G. V. (2001) Annu. Rev. Phytopathol. 39, 461–490 [DOI] [PubMed] [Google Scholar]
- 46.Björkman A. M., Dunten P., Sandgren M. O., Dwarakanath V. N., Mowbray S. L. (2001) J. Biol. Chem. 276, 2808–2815 [DOI] [PubMed] [Google Scholar]
- 47.Borkovich K. A., Alex L. A., Simon M. I. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6756–6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biemann H. P., Koshland D. E., Jr. (1994) Biochemistry 33, 629–634 [DOI] [PubMed] [Google Scholar]
- 49.Ottemann K. M., Thorgeirsson T. E., Kolodziej A. F., Shin Y. K., Koshland D. E., Jr. (1998) Biochemistry 37, 7062–7069 [DOI] [PubMed] [Google Scholar]
- 50.Lin L. N., Li J., Brandts J. F., Weis R. M. (1994) Biochemistry 33, 6564–6570 [DOI] [PubMed] [Google Scholar]
- 51.Levit M. N., Stock J. B. (2002) J. Biol. Chem. 277, 36760–36765 [DOI] [PubMed] [Google Scholar]
- 52.Vaknin A., Berg H. C. (2008) J. Mol. Biol. 382, 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipton D. S., Blanchar R. W., Blevins D. G. (1987) Plant Physiol. 85, 315–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson J. F., Allan D. L., Vance C. P. (1994) Plant Physiol. 85, 315–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao H., Wan H., Shaff J., Wang X., Yan X., Kochian L. V. (2006) Plant Physiol. 141, 674–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seedorf H., Fricke W. F., Veith B., Brüggemann H., Liesegang H., Strittmatter A., Miethke M., Buckel W., Hinderberger J., Li F., Hagemeier C., Thauer R. K., Gottschalk G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2128–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tikhonov D. B., Magazanik L. G. (2009) Neurosci. Behav. Physiol. 39, 763–773 [DOI] [PubMed] [Google Scholar]
- 58.Chaudhry C., Plested A. J., Schuck P., Mayer M. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12329–12334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stock J. (1996) Curr. Biol. 6, 825–827 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.