Abstract
The correct localization of integral membrane proteins to subcellular compartments is important for their functions. Synaptotagmin contains a single transmembrane domain that functions as a type I signal-anchor sequence in its N terminus and two calcium-binding domains (C2A and C2B) in its C terminus. Here, we demonstrate that the localization of an Arabidopsis synaptotagmin homolog, SYT1, to the plasma membrane (PM) is modulated by tandem C2 domains. An analysis of the roots of a transformant-expressing green fluorescent protein-tagged SYT1 driven by native SYT1 promoter suggested that SYT1 is synthesized in the endoplasmic reticulum, and then delivered to the PM via the exocytotic pathway. We transiently expressed a series of truncated proteins in protoplasts, and determined that tandem C2A-C2B domains were necessary for the localization of SYT1 to the PM. The PM localization of SYT1 was greatly reduced following mutation of the calcium-binding motifs of the C2B domain, based on sequence comparisons with other homologs, such as endomembrane-localized SYT5. The localization of SYT1 to the PM may have been required for the functional divergence that occurred in the molecular evolution of plant synaptotagmins.
Keywords: Arabidopsis, Membrane Biogenesis, Membrane Proteins, Plant, Plasma Membrane, Protein Targeting, C2 Domain, Synaptotagmin
Introduction
The localization of integral membrane proteins to the appropriate intracellular compartment(s) is important for their functioning. Secretory proteins are transported to the apoplast through the default secretion pathway, in which the proteins move from the endoplasmic reticulum (ER)4 to the Golgi and trans-Golgi network (TGN), and then finally to the plasma membrane (PM). However, the default pathway and sorting mechanism of integral membrane proteins are still unclear.
Synaptotagmin (Syt) family proteins are calcium sensors that regulate exocytosis in mammalian cells (1). In the presynaptic membrane of neuron cells, Syt I is known to regulate the calcium-dependent exocytosis of synaptic vesicles containing neurotransmitter by detecting calcium influx via voltage-dependent calcium channels after activation of neuron cells (2). Some isoforms of Syt are also related to the other calcium-dependent secretions, such as insulin release in pancreatic cell and maintenance of bone mass in bone cells (3, 4). Syt homologs occur throughout the plant kingdom (5). The Arabidopsis genome has five genes encoding Syt homologs (SYT1, -2, -4, and -5; full-length SYT3 cDNA has a stop codon at a functional region). Recently, we demonstrated that Arabidopsis SYT1 is involved in calcium-dependent freezing tolerance, which is related to the membrane-resealing system (6). The membrane-resealing system was initially identified in animal cells as an emergency response system against disruption of the PM, and a mammalian Syt homolog, Syt VII, functions in membrane resealing as a sensor to detect the calcium influx from outside of the cells through the ruptured site of the PM (7–10). In addition, Arabidopsis SYT1 is required for the maintenance of PM integrity and viability of root cells when exposed to severe salt stress, suggesting the involvement of the membrane-resealing system in such harsh conditions (11).
The Syt family has one conserved transmembrane (TM) domain in the N terminus and two conserved calcium-binding domains (C2A and C2B) in the C terminus, and C2A and C2B of mammalian Syt I are actually bound to three and two calcium ions, respectively (1, 2, 7, 12). Note that the membrane proteins with a single TM domain are classified according to protein structure and topogenesis: type I membrane proteins possess an N-terminal cleavable signal sequence and a subsequent stop-transfer sequence and are anchored with the Nexo/Ccyt topology; type II membrane proteins (also called SA-II proteins) possess an uncleaved type II signal-anchor (SA-II) sequence and are anchored with the Ncyt/Cexo topology; type I signal-anchor (SA-I) proteins (also called type III membrane proteins) possess an uncleaved SA-I sequence and are anchored with the Nexo/Ccyt topology; tail-anchored (TA) membrane proteins are anchored by a C-terminal TM domain with the Ncyt/Cexo topology (13, 14). Based on their structure and topology, the Syt family proteins are classified as SA-I proteins (15).
The structure of plant Syts differs from that of conventional mammalian Syts in several ways. The predicted TM domain of plant Syts consists of 23 or 22 amino acid residues, which is almost the same number of amino acid residues reported for mammalian Syts. However, plant Syts lack an N-terminal extension in the extracellular region, but have an SMP (plant synaptotagmin-like, mitochondria proteins) domain just beside the C-terminal side of the TM domain (5, 16). The SMP domain has also been identified in yeast Tcb and mammalian E-Syt families (mentioned later), although the function of the SMP domain in these proteins is unknown (5, 16). Our previous examination indicated that the topology of SYT1 is the same as that of mammalian Syt, i.e. the Nexo/Ccyt topology (6). In addition, like mammalian Syts, the C2A domain of SYT1 has the conserved calcium-binding motif and exhibits the calcium-dependent interaction with lipid membranes; however, the C2B domain of SYT1 exhibits a calcium-independent interaction with the lipid membrane (11).
Mammalian genomes contain thirteen Syt homologs, some of which are known to contain mRNA splicing variants. Mammalian Syt II, which is the most similar protein to Syt I, contains the SA-I sequence that integrates the translated proteins in the ER membrane and orientates C2A and C2B to the cytosol and the N-terminal extension to the extracellular space. It was established that the mammalian Syts examined in several cell types (for instance, neuron cells, endocrine β-cells, sperm cells, and mammalian cultured cells) are located in endomembrane systems (i.e. secretory vesicles, Golgi/TGN, lysosomes, acrosomes, etc.), but not in the PM (17–29). Interestingly, a novel mammalian Syt-related protein family, E-Syt (E-Syt1/KIAA0747/FAM62A, E-Syt2/FAM62B, and E-Syt3/FAM62C), contains five C2 domains (E-Syt1) or three C2 domains (E-Syt2 or E-Syt3) in the C terminus (5, 30, 31). E-Syt1 is localized to some endomembrane systems, but E-Syt2 and E-Syt3 are localized to the PM, indicating that the C2 domain is also involved in subcellular localization (30). In addition, three yeast Syt homologs (Tcb1, Tcb2, and Tcb3) contain five or six C2 domains in their C termini, and Tcb2 seems to be localized to the PM (16, 32). An Arabidopsis synaptotagmin, SYT1, is also localized to the PM (6).
In this study, we analyzed the mechanism by which plant SYT1 is delivered to the PM. Whereas mammalian Syts localize to endomembranes, including synaptic vesicles, the TGN, and endosomes and lysosomes, but not to the PM, Arabidopsis SYT1 localizes to the PM. We were thus interested in establishing whether there is a specific mechanism that facilitates the differential localization of mammalian and plant Syts. We found that SYT1 may be synthesized on rough ER and then delivered to the PM through the Golgi. Moreover, the tandem C2A-C2B domains were found to be necessary for the delivery of SYT1 to the PM. A motif in the C2B domain was found to be required for localization to the PM, which may have been modified during the course of evolution to permit a novel function of plant Syts. To our knowledge, this is the first report demonstrating the localization mechanism of a plant SA-I protein.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Seeds of wild-type Arabidopsis thaliana (ecotype Columbia; A. thaliana Col0) and SYT1-EmGFP-expressing plants, which were used for the observation of leaf cells, were planted and grown for 3 or 4 weeks as reported earlier (6), with a slight modification of light conditions (16-h photoperiod at 125 μmol/m2/s at soil level). For observation of root cells, SYT1-EmGFP-expressing plants were planted on one-half Murashige and Skoog medium with 0.8% agar and grown for 5 days.
cDNA
Plasmid clones of full-length cDNA of SYT1 (MIPS code At2g20990; cDNA clone RAFL05-21-F01) and SYT5 (MIPS code At1g05500; cDNA clone RAFL09-84-A22) were obtained from RIKEN Bioriken BioResource Center (33, 34).
Construction of Plasmid Vectors
To prepare the promoter of SYT1 (SYT1p), the nucleotide sequences of the region upstream of SYT1 were obtained from the TAIR genome data base, and primer sets were designed to amplify SYT1p. An 815-bp SYT1p fragment, which is upstream of the start codon of translation, was amplified and cloned into the pENTR vector (Invitrogene) and sequenced. The fragment containing the promoter region was transferred into the pCAMBIA3300 vector containing a BASTA chemical herbicide resistance gene (bar). The coding sequence of SYT1 fused to a 5×Gly linker was subcloned into the pENTR vector, and then transferred to the 5′-end of a EmGFP gene encoding an enhanced green fluorescence protein EmGFP (Invitrogen) at a SalI/NcoI site in a modified gfp transient expression vector containing a CaMV35S promoter in the pUC18 vector (6, 35). The SYT1-EmGFP chimera gene was transferred into pCMABIA3300-SYT1p. Truncated forms of SYT1 fused to EmGFP and full-length forms of SYT5 fused to EmGFP at C terminus via a 5×Gly linker were cloned into the pUC18-CaMV35S::EmGFP vector using the same method as for full-length SYT1-EmGFP described above. Mutations of SYT1-EmGFP at the C2B domain were performed using an Inverse PCR-based Mutagenesis Kit (TOYOBO). The list of primers and constructs used is presented in supplemental Table S1.
Transformation of Arabidopsis
The constructs were introduced into the A. thaliana Col0 genome using Agrobacterium tumefaciens (GV3101). Seeds of T1 plants were planted on Murashige and Skoog medium with 0.8% agar containing 0.01% BASTA, and EmGFP- or SYT1-EmGFP-expressing plants were selected. T2 plants were used for experiments.
Membrane Isolation and Biochemical Analysis
A crude membrane fraction was prepared according to a previously described method (36). For fractionation of the organelle membrane vesicles, the crude membrane fractions isolated from 5 g of seedlings were loaded onto 15–50% (w/w) sucrose density linear gradient buffer, and ultracentrifuged at 141,000 × g for 20 h at 4 °C (6). For purification of the plasma membrane fraction, a previously described method was used (6, 37). For the protease protection assay, fresh plasma membrane fractions containing 2 μg of proteins in 100 μl of reaction solution were treated with thermolysin (final concentrations of thermolysin were 0, 0.01, 0.1, and 1 μg ml−1) with or without 1% (w/v) Triton-X 100 for 1 h at 4 °C. The reaction was stopped by adding 1 μl of 0.5 m EGTA. For the alkaline assay, the crude membrane fraction containing 20 μg of proteins in 200 μl of reaction solution was treated with 100 mm Na2CO3, and after ultracentrifugation, the supernatant and pellet were separated. The pellet was washed once and then re-suspended in 200 μl of buffer. The proteins in the sample were separated by standard SDS-polyacrylamide electrophoresis on a 12% polyacrylamide gel. A standard silver staining method was used. Immunoblot analyses were performed according to standard procedures. The antigens on polyvinylidene difluoride membranes were visualized using ECL Plus (BD Healthcare). The origin of antibodies used in this study and their dilution ratios are as follows: anti-SYT1, as made previously (6), 1:2,000; anti-GFP, commercial product of TaKaRa Bio, 1:5,000; anti-PAQs (38), 1:20,000; and anti-PMA2, 1:5,000 (recognizes cytosolic region of plasma membrane type H+-ATPase) (39). Vanadate-sensitive ATPase activity was measured in the plasma membrane fractions as described previously (36).
Transient GFP Fusion Protein Expression Assay
Protoplasts were enzymatically isolated from 4-week-old A. thaliana (Col0) leaves (6, 37) or from transgenic Arabidopsis plants expressing CFP-labeled Golgi marker proteins (40). Using a standard polyethylene glycol transfection method (41). 10 μg of plasmid DNA of each construct was transfected into 2 × 104 protoplasts in 100 μl of transfection buffer, and then GFP expression was observed after overnight incubation at room temperature. In the colocalization analysis, the protoplasts were stained with 1 μm ER-Tracker Blue-White DPX (Molecular Probes) for 30 min at room temperature (42, 43).
BFA Treatment
For brefeldin A (BFA) treatment, 5-day-old plants grown on one-half Murashige and Skoog medium with 0.8% agar were used. The seedlings were soaked for 30 min in liquid one-half Murashige and Skoog medium containing 10 μm BFA and 16 μm FM4–64 at room temperature, and then observed with microscopy.
Microscopy
A confocal fluorescent microscope BX-61 with a disc-scanning unit (Olympus) was used. All observations were performed at room temperature. For observations of leaf cells, young leaves were mounted in a piece of 4% low melting agar gel and sliced at a thickness of 75–80 μm with a vibrating blade (HM 650 V, MICROM International) in a buffer containing 1 mm MES/KOH (pH 5.7). For protoplast observation, protoplasts suspended in suspension buffer were attached to a coverslip coated with 10 mg/ml polylysine. EmGFP fluorescence and FM4–64 fluorescence or autofluorescence of chlorophyll in chloroplasts were detected using the U-MGFPHQ and U-MWIG2 filter sets (Olympus), respectively. In the colocalization analysis of truncated SYT1-EmGFP and CFP-labeled Golgi marker protein or ER-Tracker, the following filter sets were used: for EmGFP, HQ500/20× EX (excitation filter), Q515LP BS (dichroic miller), and HQ535/30m EM (emission filter; Chroma); for CFP, the U-MCFPHQ filter set (Olympus); for ER-Tracker, 400DF15 (excitation filter), 415DRLP (dichroic miller), and 450DF65 (emission filter, Omega). The following objective lenses (Olympus) were used: UPlanApo 20×/0.70 numerical aperture (NA) (supplemental Figs. S1 and S2) and UPlanApo 40×/0.85 NA (Figs. 3, 4, 5, and 7). The fluorescence in optical sections in each 2 μm was captured using a cooling monochrome charge-coupled device camera CoolSNAP HQ (Photometrics) and imaged with imaging software (SlideBook, Intelligent Imaging Innovations). All captured images were deconvoluted by a command, and the images presented in Figs. 3C, 4, 5, and 7 were processed to generate projections using a command in SlideBook. The colocalization of EmGFP-labeled proteins and CFP-labeled proteins or ER-Tracker-labeled membranes was evaluated using the Pearson's correlation in SlideBook, which estimates the correlation between the intensity in one channel and the intensity in another channel. The values are calculated as r-values; 0.0 signifies no matching trend in intensities, 1.0 means complete correlation, and −1.0 implies anti-correlation.
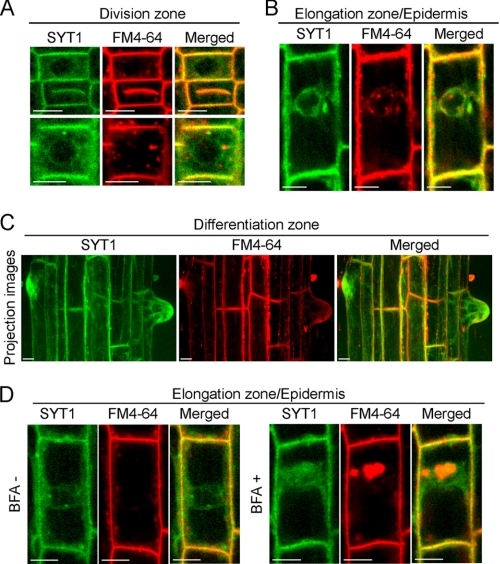
FIGURE 3.
Localization of SYT1-EmGFP in root cells. GFP (green) and FM4–64 (red) emissions were observed in root cells using confocal fluorescence microscopy. Fluorescence images of dividing cells (A) and elongating cells (B). Projection images from optical sections of cells in differentiation zone (C). Root cells in the elongation zone were observed with or without 10 μm BFA (D). Bars indicate 10 μm.
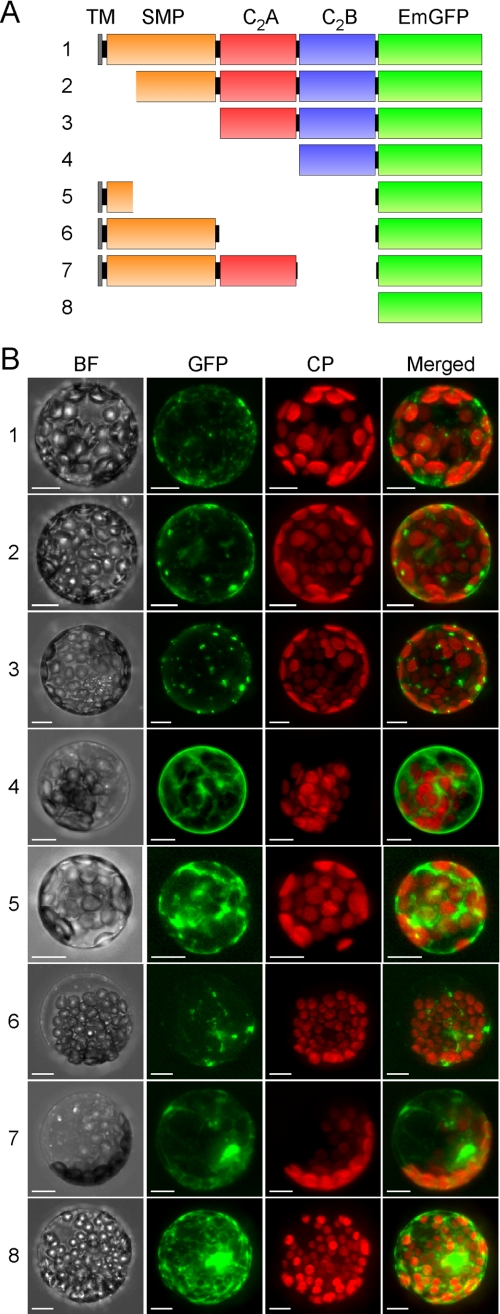
FIGURE 4.
Localization of transiently expressed truncated SYT1-EmGFP proteins in protoplasts. A, schematics of truncated SYT1 with EmGFP (green box) in the C terminus. The TM, SMP, C2A, and C2B domains are represented as yellow, orange, red, and blue boxes, respectively. B, projection images generated from optical sections of EmGFP florescence in protoplasts expressed each truncated SYT1-EmGFP protein. Protoplasts transfected with the plasmid constructs containing the truncated SYT1-EmGFP described in panel A were observed using confocal fluorescence microscopy. The obtained images of each optical section of the protoplasts were reconstructed as projection images. BF, bright field; GFP, GFP fluorescence; CP, fluorescence of chlorophyll; Merged, merged image of GFP with CP. Bars indicate 10 μm.
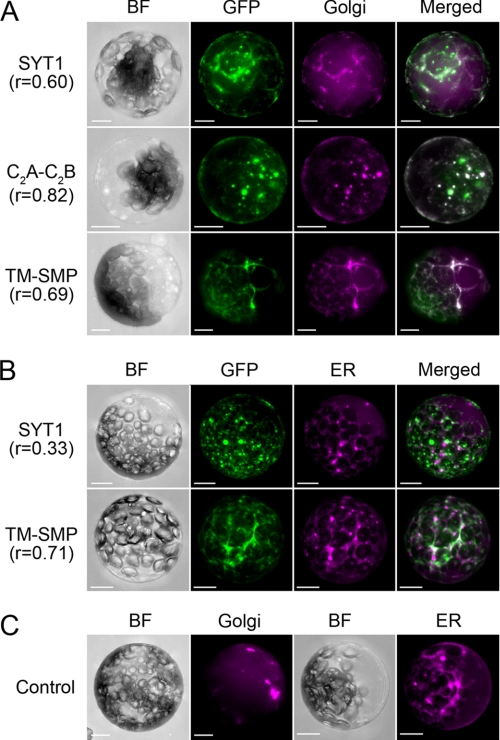
FIGURE 5.
Localization of transiently expressed truncated SYT1-EmGFP proteins in ER- or Golgi-labeled protoplasts. A, SYT1-EmGFP and truncated SYT1-EmGFP proteins were transfected into protoplasts isolated from leaves of a transgenic plant expressing CFP-tagged Golgi marker proteins. B, co-localization analysis of SYT1-EmGFP or TM-SMP with ER-Tracker. C, CFP fluorescence in a transgenic plant expressing CFP-tagged Golgi marker and wild-type protoplasts stained with ER-Tracker. Projection images generated from optical sections of EmGFP florescence and CFP or ER-Tracker in protoplasts expressing full-length or truncated SYT1-EmGFP proteins. Emissions of EmGFP and CFP or ER-Tracker were observed using a confocal fluorescence microscope. The fluorescence images obtained from individual optical sections were reconstructed as projection images. SYT1, full-length SYT1; C2A-C2B, tandem C2A and C2B domains; TM-SMP, TM with SMP, BF, bright field; GFP, GFP fluorescence; Golgi, CFP fluorescence; ER, fluorescence of ER-Tracker; Merged, merged image of GFP with CFP or ER-Tracker. GFP fluorescence was pseudocolored green, and CFP or ER-Tracker fluorescence was pseudocolored magenta. Bars indicate 10 μm.
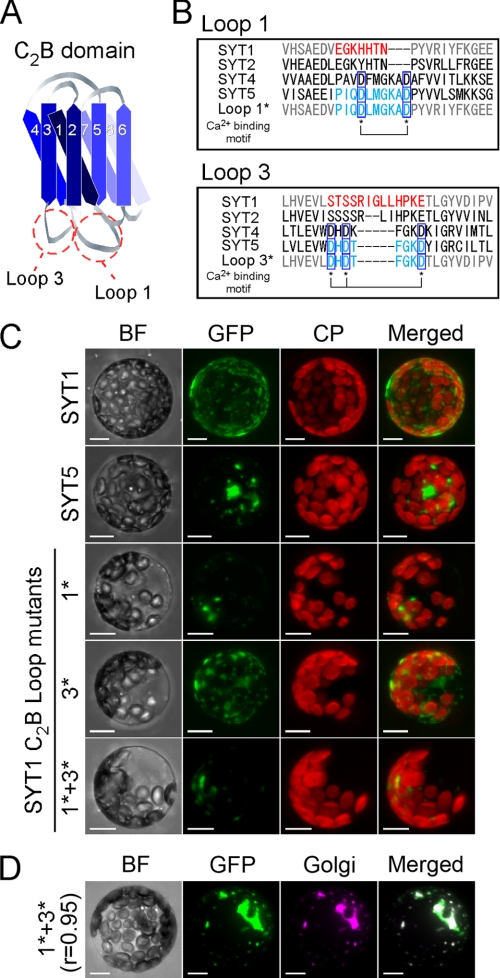
FIGURE 7.
Localization of transiently expressed EmGFP-tagged SYT1, SYT5, and SYT1 proteins mutated at the C2B calcium-binding site in protoplasts. A, structural model of the SYT1 C2B domain. Diagrammatic representation of the C2B domain of SYT1 based on the prediction of putative β-strands in the C2 domains of eukaryotic synaptotagmin-related proteins by Jiménez and Davletov (16). Graduated-blue colored strips indicate putative β-strands. The strands are labeled in numerical order from the N terminus to the C terminus. B, comparison of the calcium-binding sites at Loop 1 and Loop 3. Amino acid sequences of SYT1, SYT2, SYT4, SYT5, and SYT1 mutants are aligned. Asterisks indicate amino acid residues of the conserved calcium-binding motifs. C, projections of optical sections of protoplasts. Transfected protoplasts were observed with confocal fluorescence microscopy. The optical sections were reconstructed as a projection image. GFP and autofluorescence of chlorophyll are green and red, respectively. D, colocalization analysis of SYT1 mutants containing both Loop 1* and Loop 3* in protoplasts isolated from the leaves of Golgi-labeled transgenic plants. GFP and CFP are green and magenta, respectively. BF, bright field; GFP, GFP fluorescence; CP, autofluorescence of chlorophyll; Golgi, CFP fluorescence of CFP-labeled Golgi marker proteins; Merged, merged image of GFP with CP or CFP. Bars indicate 10 μm.
Phylogenetic Analysis and Gene Structure
Nucleotide sequences of SYT-like genes in A. thaliana were retrieved from the genomic database (TAIR, www.arabidopsis.org). To identify SYT-like genes in Oryza sativa, Sorghum bicolor, Medicago truncatula, Populus trichocarpa, Carica papaya, and Vitis vinifera, we performed TBLASTN searches against the available genomic databases using amino acid sequences encoded by Syt genes in A. thaliana as queries. The following databases were used: Rice Annotation Project Data base (rapdb.dna.affrc.go.jp) for O. sativa; JGI Sorghum bicolor v1.0 (genome.jgi-psf.org/Sorbi1/Sorbi1.home.html) for S. bicolor; mips Medicago truncatula Project (mips.gsf.de/proj/plant/jsf/medi/index.jsp) for M. truncatula; JGI Populus trichocarpa v1.1 (genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) for P. trichocarpa; Papaya Genome Project v0.4 in CoGe (synteny.cnr.berkeley.edu/CoGe/) for C. papaya; and Grape Genome Browser (www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/) for V. vinifera. The nucleotide sequences encoding the SMP, C2A, and C2B domains were concatenated, and the amino acid sequences were then aligned using the ClustalW program built into phylogenetic analysis software MEGA4 (www.megasoftware.net/index.html) (44). After excluding the nucleotides at the third position in each codon, the evolutionary distances were calculated using the Tamura-Nei method by using MEGA4 (45). Phylogenetic trees were reconstructed using the neighbor-joining method (46). The bootstrap values were calculated with 1000 replications.
For analysis of gene structures, nucleotide sequences of genomic DNA and full-length cDNA, and annotated gene structure data encoding SYT genes in the Arabidopsis genome were obtained from TAIR. Based on these data, the positions of each exon and intron were compared manually, and exon length to intron length was maintained at a constant ratio.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBankTM/EMBL databases under the following accession numbers: SYT1 (At2g20990), RAFL05–21-F01; SYT2 (At1g20080); SYT3 (At5g04220), AY059741 and BX830716; SYT4 (At5g11100); and SYT5 (At1g05500), RAFL09–84-A22. The accession numbers used in the phylogenetic tree are presented in supplemental Table S2.
RESULTS
SYT1-EmGFP Is Localized to the Plasma Membrane of Leaf Cells
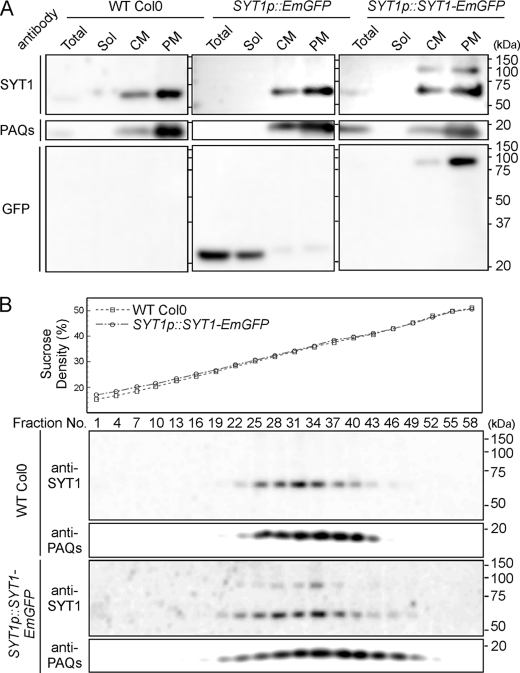
We generated transgenic Arabidopsis plants expressing a fusion protein of SYT1 with EmGFP (Emerald GFP) from the SYT1 native promoter. First, we investigated the biochemical property of SYT1-EmGFP using the crude membrane fraction and the highly purified PM fraction, which were prepared from leaves by means of aqueous two-phase partitioning. In wild-type plants and control plants expressing only EmGFP, anti-SYT1 antibody recognized only 62-kDa proteins, which appeared to be more abundant in the PM fraction than in the crude membrane fraction. In plants expressing SYT1-EmGFP, 89-kDa proteins were detected in addition to the 62-kDa proteins in both the crude membrane and PM fractions (Fig. 1A). In all cases, high levels of the plasma-membrane-type aquaporin (PAQs) were detected in the PM fractions. In addition, the anti-GFP antibody recognized 89-kDa proteins in the PM fraction of plants expressing SYT1-EmGFP and 27-kDa proteins in both the total and soluble protein fractions of control plants expressing EmGFP. The experimental molecular masses of detected SYT1 and SYT1-EmGFP correspond to the calculated molecular masses of these proteins, which are about 62 kDa and 89 kDa, respectively, indicating that SYT1-EmGFP fusion proteins are correctly delivered to the PM in transgenic plants.
FIGURE 1.
Immunoblot analysis of Arabidopsis plants expressing SYT1-EmGFP. A, purification of crude membrane vesicles. Seedlings of wild-type plants (Col0) and transgenic plants expressing EmGFP or SYT1-EmGFP under the SYT1 promoter (i.e. SYT1p::EmGFP or SYT1p::SYT1-EmGFP) were homogenized, and after centrifugation, supernatants were fractionated (Total) into soluble proteins and crude membrane vesicles. After ultracentrifugation, supernatants containing soluble proteins (Sol) and crude membrane vesicles (CM) were fractionated. From the CM fraction, plasma membrane vesicles (PM) were fractionated by the aqueous two-phase partition system. These fractions were separated by SDS-PAGE, and immunoblot analysis was performed using the anti-SYT1, which recognizes the C2A domain, anti-GFP and anti-PAQs, which recognizes total plasma membrane aquaporin, antibodies. Molecular mass is indicated to the right of the blot. B, separation of crude membrane vesicles from wild-type plants and the two transgenic plant seedlings using linear sucrose density gradient centrifugation. Sucrose densities of each fraction were measured (upper panel), and aliquots of each fraction were separated by SDS-PAGE and analyzed by immunoblotting with anti-SYT1 and anti-PAQ antibodies.
To further confirm the localization of SYT1-EmGFP, crude membrane fractions were fractionated using the sucrose density gradient centrifugation method (Fig. 1B). SYT1 signals were detected in the 28–41% sucrose fractions of wild-type plants, with maximum signal intensity approximately at the 35% sucrose fraction, in which signals of PAQs were detected. Also, in the transgenic lines, fractionation patterns of SYT1, SYT1-EmGFP, and PAQ signals were almost identical to those of wild-type plants. These data indicate that both SYT1 and SYT1-EmGFP are localized only to the PM in leaf cells. In addition, observation of leaf cells expressing SYT1-EmGFP shows that GFP signals are detected mainly in the epidermis and palisade layers, are strong in the vascular bundle, and are localized to the edge of cells (supplemental Figs. S1A and S1B). Interestingly, in guard cells, the SYT1-EmGFP signal was localized to the PM on the outer edges of stomata (supplemental Fig. S1C).
Topology of SYT1 and SYT1-EmGFP in Plasma Membrane Vesicles
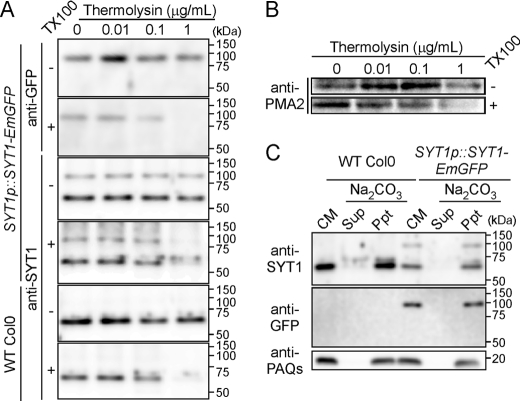
A previous experiment showed that the C2A domain of SYT1 is located in the cytosolic region (6). In the current experiment, because EmGFP was fused to the C terminus of SYT1, the topology of SYT1-EmGFP needed to be confirmed. We used the aqueous two-phase partitioning system to isolate plasma membrane vesicles (6, 36, 37). The ATPase activities in the fresh PM fractions were evaluated by using the specific plasma membrane-type ATPase inhibitor vanadate in the presence and absence of Triton X-100 (supplemental Table S2). The fresh PM fractions prepared in our system contained 78.5% right-side-out PM vesicles. Fresh PM fractions prepared from wild-type plants or the transgenic lines were treated with thermolysin in the presence or absence of Triton X-100. The greater the amount of thermolysin applied with 1% Triton X-100, the weaker the signal of SYT1 and SYT1-EmGFP detected (Fig. 2A). However, the application of thermolysin without Triton X-100 did not affect the intensity of SYT1 and SYT1-EmGFP signals. The signal pattern resulting from thermolysin digestion in the presence or absence of Triton X-100 was similar to that of plasma membrane-type H+-ATPases, which were recognized by anti-PMA2 antibody raised against the C-terminal cytosolic region (Fig. 2B). These results indicate that the topology of SYT1-EmGFP is the same as that of SYT1, and that the EmGFP tag was located in the cytosolic region of the cell. This is because the fresh PM fraction purified by aqueous two-phase partitioning includes much of the right-side-out PM vesicle (47) and, consequently, the portions of proteins located at the inside of vesicles are protected from protease degradation. In addition, SYT1 and SYT1-EmGFP were not solubilized when the crude membrane fraction was subjected to alkaline treatment (Fig. 2C). These results collectively suggest that both SYT1 and SYT1-EmGFP are integrated in the PM and that two C2 domains are located in the cytoplasm of Arabidopsis leaf cells.
FIGURE 2.
Topology and membrane integration of SYT1 and SYT1-EmGFP proteins. A, protease protection assay. The right-side-out plasma membrane vesicles were treated with various concentrations of a low specificity protease thermolysin. After digestion of the plasma membrane proteins, each sample was analyzed by immunoblotting with anti-SYT1 and anti-GFP antibodies. B, degradation pattern of plasma membrane-type H+-ATPase. After receiving the same treatment as the right-side-out plasma membrane vesicles in A, the degradation of the plasma membrane-type H+-ATPase was detected with anti-PMA2 antibody. C, membrane integration of SYT1 and SYT1-EmGFP. Two micrograms of crude membrane vesicle proteins isolated from seedlings of wild-type plants and transgenic plants expressing SYT1-EmGFP were treated with Na2CO3. After ultracentrifugation, the supernatant (Sup) and precipitate (Ppt) of each membrane sample were analyzed by immunoblotting with anti-SYT1, anti-GFP, and anti-PAQs antibodies. The separated proteins were also silver-stained.
Subcellular Localization of SYT1-EmGFP in Root Cells
In roots, the fluorescence signal was strong in the root tip, especially in the quiescent center, columella and epidermis, and vascular bundle, as well as in the differential zone of the tips of root hairs (supplemental Fig. S2). Then, we observed the subcellular localization of SYT1-EmGFP in detail. In division and elongation zones, the fluorescence of SYT1-EmGFP was localized not only to the PM, but also to structures, such as the rough ER, that surrounded the nucleus and extended to the cell surface (Fig. 3, A and B). To visualize the PM, root cells were stained with an impermeable lipid membrane-specific fluorescence dye, FM4–64, which is only taken up into cells via endocytosis and labels the early endosomes (EE) and TGN in plants (48, 49). Although the pattern of SYT1-EmGFP fluorescence was almost identical to that of FM4–64-labeled PM, vesicles labeled with SYT1-EmGFP did not overlap with FM4–64-labeled endosomes (Fig. 3, A and B). In dividing cells, SYT1-EmGFP fluorescence was localized to the cell plate (Fig. 3A). In the differentiation zone, SYT1-EmGFP fluorescence in the epidermis showed that SYT1-EmGFP was not uniformly distributed on the PM and seemed to be associated with some microfibers and accumulated in the tips of root hairs (Fig. 3C).
After roots were treated with BFA, which is an inhibitor of the endocytotic pathway in plant cells (50), several endosomes labeled with FM4–64 in the epidermal cells of the elongation zone resulted in large vesicular structures, which are referred to as the BFA compartment (Fig. 3D). These structures only partially overlapped with vesicles labeled with SYT1-EmGFP. These results suggest that SYT1 is targeted to the PM through the exocytosis pathway but tends to be retained in the PM without being removed via the endocytotic recycling pathway.
Localization of Truncated SYT1-EmGFP Proteins
Next, we tested the contribution of the domain structures of SYT1 for its subcellular localization. Plant Syt proteins consist of four domains: a single TM domain following a few amino acid residues in the N terminus, one SMP domain, which is conserved in plants (51), and two calcium-binding domains, C2A and C2B, in the C terminus. Interestingly, the SMP domain is also conserved in the novel mammal Syt family proteins of E-Syts (16). On the basis of this domain information, seven constructs expressing truncated proteins of SYT1 with EmGFP in the C terminus were designed and introduced into protoplasts isolated from leaves (Fig. 4A).
Full-length SYT1 was found to be mainly, but not uniformly, located in the PM of transformed protoplasts (Fig. 4B, row 1). In addition, in this protoplast system, some of the full-length SYT1 proteins seemed to be localized to vesicles, such as the Golgi or the TGN. The localization pattern of truncated SYT1 proteins containing tandem C2A-C2B and SMP domains was very similar to that of full-length SYT1 (Fig. 4B, row 2). In contrast, the truncated proteins containing only the tandem C2A-C2B domains were localized to vesicles such as the Golgi or TGN, but not to the PM (Fig. 4B, row 3). In addition, the proteins containing only the C2B domain seemed to disperse into the cytoplasm (Fig. 4B, row 4). These results suggest that the appropriate localization of SYT1 to the PM is determined by the tandem C2A-C2B and SMP domains. The truncated proteins containing only the putative TM domain were observed in the cytoplasm, but not in any membrane system (Fig. 4B, row 5). However, the truncated proteins containing both the TM domain and the SMP domain (TM-SMP) were localized to vesicles, but not to the PM (Fig. 4B, row 6). Furthermore, the truncated proteins containing TM, SMP, and C2A domains localized to several compartments, but not to the PM (Fig. 4B, row 7).
Next, to analyze the localization of EmGFP-labeled truncated SYT1 proteins containing TM-SMP or C2A-C2B domains, each construct was transfected into protoplasts isolated from the wild-type plant or a transgenic plant expressing CFP-labeled Golgi marker proteins, and the former protoplasts were stained with ER-Tracker to visualize the ER membrane (Fig. 5). The colocalization of EmGFP-labeled proteins was evaluated using Pearson's correlation (see “Experimental Procedures”). Full-length SYT1 proteins tagged with EmGFP partially colocalized with CFP-tagged Golgi marker proteins (r = 0.60) (Fig. 5A, row 1). In contrast, the localization of truncated proteins containing tandem C2A-C2B domains was strongly correlated with that of Golgi marker proteins (r = 0.82) (Fig. 5A, row 2). The truncated proteins containing TM-SMP domains partially colocalized with Golgi marker proteins (r = 0.69) (Fig. 5A, row 3). In the case of ER-Tracker, fluorescence was observed not only on network-like structures, but also on vesicles (Fig. 5C). EmGFP fluorescence associated with full-length SYT1 slightly overlapped with ER-Tracker fluorescence (r = 0.33) (Fig. 5B, row 1). In contrast, EmGFP fluorescence associated with TM-SMP partially overlapped with network-like structures or vesicles stained with ER-tracker (r = 0.71) (Fig. 5B, row 2).
Phylogenetic Relationship of Plant Syts
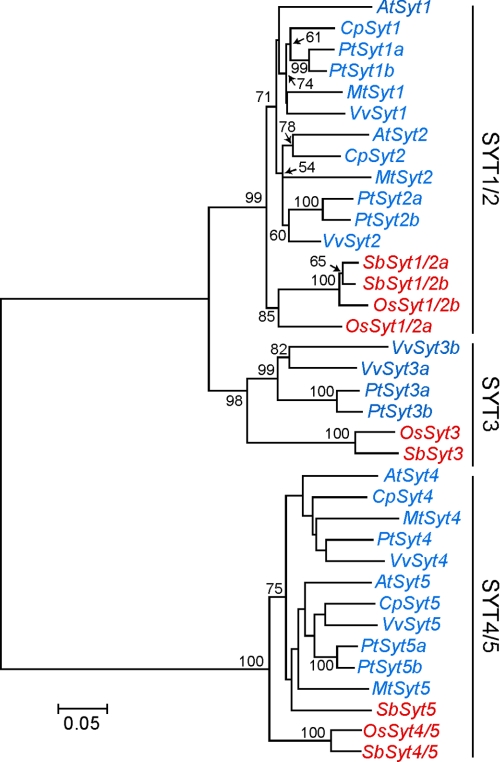
A transient expression assay indicated the importance of the C2A-C2B domain in the PM localization of SYT1. In the Arabidopsis data base (i.e. TAIR), there are five Syt genes that contain only two C2 domains, and these genes may share a common ancestral gene, because the exon-intron structures are highly conserved among the Syt genes (supplemental Fig. S3). From this result, we concluded that the PM localization of SYT1 was responsible for the functional divergence among Arabidopsis Syts. To deduce their evolutionary relationship, we next identified Syt genes containing two C2 domains from the available genomic databases of seven angiosperm plants (monocots: Oryza sativa and Sorghum bicolor; eudicots: A. thaliana, Medicago truncatula, Populus trichocarpa, Carica papaya, and Vitis vinifera). Although Arabidopsis Syt genes have been termed “SYT” (6, 11) in the phylogenetic experiments, they were practically renamed AtSyt, and, similarly, the two-letter species indicators were used to distinguish between the other plant Syt genes. At least four copies of the Syt genes have been retained in each of the plant genomes examined (supplemental Table S3). We did not use AtSyt3 to perform phylogenetic analyses, because the nucleotide sequence of AtSyt3 includes a stop codon in its coding sequence (6) (see also accession numbers AY059741 and BX830716).
In plant Syt genes, the four domain structures described above were conserved, and the most highly conserved region was found in the region of the tandem C2A-C2B domains (supplemental Fig. S4). The amino acid sequences surrounding the C2A and C2B domains and the SMP domain were aligned with the ClustalW program, and then corresponding nucleotide sequences were used to reconstruct a phylogenetic tree based on the neighbor-joining method (Fig. 6). The constructed phylogenetic tree indicates that angiosperm Syt genes had clearly diverged into three clades (the SYT1/2, SYT3, and SYT4/5) and that the SYT3 clade was more closely related to the SYT1/2 clade than to the SYT4/5 clade. Each clade contains Syt genes of both monocots and eudicots. Although the clade of SYT1/2 contained three sister groups and two closely related groups of these were eudicots, the other one comprised monocots. The clade of SYT3 contained a monocotyledonous group and a eudicotyledonous group. Similarly to SYT1/2 clade, SYT4/5 clade also contained two closely related groups of which were eudicots, but the other one comprised monocots. The four Arabidopsis Syt genes (i.e. AtSyt1, AtSyt2, AtSyt4, and AtSyt5) separated into two clades, AtSyt1 and AtSyt2, and AtSyt4 and AtSyt5 were classified into the SYT1/2 and SYT4/5 clades, respectively.
FIGURE 6.
Phylogenetic tree Syts. Phylogenetic relationship between plant Syts was calculated using the neighbor-joining algorithm. Bootstrap support values on the tree branch are indicated. Based on the phylogenetic tree, three clades (i.e. SYT1/2, SYT3, and SYT4/5) were defined. Genes from monocots and eudicots are in red and blue, respectively. The bar shows the rate of nucleotide substitution.
Localization of SYT1 Mutants in the C2B Domain
Next, we compared the amino acid sequences of the C2 domains encoded by the four Syt genes. Fig. 7A is a diagrammatic representation of the C2B domain of SYT1 according to the prediction of β-strands and loops of synaptotagmin-related proteins, including plant Syts by Jiménez and Davletov (16). Two loop structures between the first and second β-strands, and between the third and fourth β-strands, are referred to as Loops 1 and 3, respectively. Multiple alignments among plant Syts show that the C2B domains in the SYT1/2 and SYT3 clades lack conserved motifs in Loops 1 and 3 (supplemental Fig. S4), which are required for the direct interaction with calcium and membrane lipids (52). Although Loop 1 of the C2 domain of Syt generally has a conserved motif motif (D1XXXXXD2), in the C2B domain of Arabidopsis SYT1, the first aspartic acid of D1 is substituted with histidine, and three amino acid residues, including D2, are missing. Although Loop 3 of the C2 domain also generally contains a conserved motif (D′1XD′2XXXXXD′3), the first two aspartic acid residues (D′1 and D′2) and the last aspartic acid residue (D′3) are substituted with serine and glutamic acid, respectively, in the C2B domain of Arabidopsis SYT1 (Fig. 7B). Therefore, the C2B domains of Arabidopsis SYT1 and of the other plant Syts in the SYT1/2 clade completely lacked the calcium-binding motifs. Phylogenetic analysis of plant Syt genes and comparison of the amino acid residues of the C2A or C2B domains suggest that the evolutionary change in the calcium-binding motifs of the C2B domain of the SYT1/2 clade may have occurred after the differentiation of Syt genes into the three clades, and may have resulted in the localization of SYT1 to the plasma membrane in plants.
To test the hypothesis that the localization of SYT1 is affected by the loss of the conserved motifs of Loop 1 and Loop 3 in the C2B domain, we performed a transient expression assay. The localization of SYT1-EmGFP was compared with that of SYT5-EmGFP, which possessed the conserved motifs in Loop 1 and Loop 3 (Fig. 7B), or with the localization of the SYT1-EmGFP mutants, in which the amino acid residues of Loop 1 and/or Loop 3 were exchanged to those of SYT5 (Loop 1* or Loop 3* in Fig. 7B). Although SYT1-EmGFP was localized to the PM and to some endomembranes, SYT5-EmGFP was localized only to endomembranes (Fig. 7C). Interestingly, the SYT1-EmGFP mutants, which has Loop 1* or both Loop 1* and Loop 3*, were localized almost completely to endomembranes, strongly resembling the SYT5-EmGFP localization, but small amounts also localized to the PM (Fig. 7C). However, there was no difference between SYT1-EmGFP and the mutant that only contains Loop 3* (Fig. 7C). Moreover, almost all of the SYT1-EmGFP mutants containing both Loop 1* and Loop 3* colocalized with CFP-labeled Golgi marker proteins (r = 0.93) (Fig. 7D). These results suggest that the localization of SYT1 is determined by the biochemical property of the C2B domain.
DISCUSSION
Syt proteins are classified as SA-I proteins and occur throughout eukaryotes. In animal cells, Syt functions as a calcium sensor to promote the membrane fusion in calcium-dependent events, such as membrane resealing (8). In plant cells, we have recently demonstrated that the Arabidopsis Syt homolog, SYT1, is involved in calcium-dependent membrane resealing, which remarkably enhances freezing tolerance (6). In addition, the C2A domain of SYT1 exhibits calcium-dependent binding to phospholipid membrane (11). Therefore, plant SYT1 may, like mammalian Syt proteins, act as a calcium sensor during membrane-membrane fusion events that are stimulated by calcium ions. However, the localization of plant SYT1 to the PM is different from that of mammalian Syts, because mammalian Syts have ubiquitously been found in endomembranes. Our current study suggests that the evolutionary modification of the C2B domain may be necessary for the delivery of SYT1 to the PM via the exocytotic pathway.
Synthesis and Delivery of SYT1 to the Plasma Membrane
Our biochemical analyses of SYT1-EmGFP driven by the SYT1 native promoter showed that GFP tagging at the C terminus of SYT1 has no effect on its localization and orientation (Figs. 1 and 2). In addition, the biochemical localization of SYT1 to the PM in seedlings agreed with the localization based on GFP fluorescence in leaf epidermal and mesophyll cells (supplemental Fig. S1). In roots, the localization pattern of SYT1-EmGFP differed at the various developmental stages (Fig. 3). This is thought to result from the expression level of the SYT1 gene in root tissue, because the fluorescence pattern of EmGFP driven by the SYT1 promoter in roots corresponded to that of SYT1-EmGFP (supplemental Fig. S2).
Furthermore, we observed the subcellular localization pattern of SYT1-EmGFP in different types of root cells expressing different levels of its transcription. In division and elongation zones, SYT1-EmGFP was localized not only to the PM, but also to the rough ER (Fig. 3, A and D). SYT1 may not be retained in the ER, because a chimeric GFP containing an ER retention signal, HEDL, is localized only to rough ER and cortical ER, but not to the PM (53). In some dividing cells, SYT1-EmGFP was localized to the cell plate (Fig. 3A). The plant cell plate is generated not only by delivery of newly synthesized materials from the ER via the Golgi, but also by endocytic recycling, which provides materials from the PM (54). Recent work based on short term labeling of the PM with FM4–64 shows that vesicles derived from the PM via the endocytotic pathway are localized to the EE and TGN but not to the Golgi (48). In our current study, vesicles containing SYT1-EmGFP did not colocalize completely with vesicles stained with FM4–64 (Fig. 3, A and B), suggesting that SYT1 is localized to the Golgi after synthesis on the ER, and less SYT1 is cycled from the PM back to the cytoplasm through the endocytotic pathway than typical materials incorporated in the PM. This is also supported by the incomplete localization of SYT1-EmGFP to BFA compartments (Fig. 3D), because the BFA compartment is speculated to be composed of TGN and EE membranes, but Golgi vesicles do not fuse to the BFA compartment (48, 50). In addition, colocalization analysis of SYT1-EmGFP driven by the CaMV35S promoter and CFP-labeled Golgi marker showed that SYT1-EmGFP is also partially localized to Golgi vesicles (Fig. 5A). These results suggest that, after SYT1 is synthesized on the rough ER membrane, SYT1 is delivered to the PM by the exocytotic pathway via Golgi, and then tends to be retained in the PM. However, it is unclear whether SYT1 moves directly to the PM or via the TGN.
Interestingly, SYT1-EmGFP was not uniformly localized to the PM and appeared to be located along some microfibers (Fig. 3C). We recently showed that SYT1 accumulates in a fraction of detergent-resistant plasma membrane regions, which are related to microdomains in the membrane, together with actins (55). These results led us to speculate that, after the transport of SYT1 to the PM, SYT1 is redistributed in the PM via a specific mechanism. The localization of RHD2 NADPH oxidase, which is similar to that of SYT1, is affected by treatment with the microfilament-disrupting drug cytochalasin D, but not by the microtubule-disrupting drugs oryzalin or taxol (56). Although both microfilaments and microtubules are localized in the root hair tip, microtubules are aligned with the short axis of cortical cells, whereas microfilaments are not (57). It is possible that several factors facilitate the interaction between SYT1 and microfilaments.
Localization of SYT1 to the PM Is Determined by Tandem C2A-C2B Domains
Plant Syts contain four domains (namely TM, transmembrane; SMP, synaptotagmin-like mitochondria membrane protein; and C2A and C2B, calcium-binding domains) (refer to Fig. 4A, row 1). The TM domain combined with a partial SMP domain was insufficient for anchoring of SYT1 to the membrane, but the TM domain combined with a complete SMP domain may be sufficient for localization to ER (Fig. 4B, rows 5 and 6; Fig. 5B, row 2), suggesting that the SMP domain is required for the integration of Syt into the rough ER membrane. This result also suggests that tandem C2A-C2B domains recruit SYT1 and then helps to transport it to the PM through the exocytotic pathway, because full-length SYT1 is localized to the PM. On the other hand, only tandem C2A-C2B domains are hard to be localized to the PM without SMP domain (Fig. 4B, row 2; Fig. 5A, row 2). In addition, less localization of tandem TM-SMP-C2A domains or of the C2B domain to the PM (Fig. 4B, rows 4 and 7) indicated that SMP-C2A domains or a single C2B domain may not be sufficient to localize SYT1 to the PM. Recently, transient expression analysis using a tobacco protoplast system showed that TM-SMP-C2A of SYT1 is localized to the ER (58). We also confirmed that the TM-SMP-C2A of SYT1 was partially localized to ER (supplemental Fig. S5). This result suggests that tandem C2 domains require the SMP domain to interact with the PM.
We clearly showed that, like mammalian Syts, SYT1 had the Nexo/Ccyt topology (Fig. 2), demonstrating that SYT1 is a SA-I protein. Based on animal and fungal models, the localization mechanism of plant integral membrane proteins has been described in terms of a membrane trafficking pathway. In tobacco cells, modification of the TM domain of human lysosome-associated protein LAMP1 or pea vacuolar sorting receptor VSR/BP80, which are type I membrane proteins with the Nexo/Ccyt topology, shows that the length of TM domain determines the localization of the proteins, with a short TM domain (17 amino acid residues) resulting in the membrane proteins being transported to the ER, and a long TM domain (22 or 23 amino acid residues) resulting in them being transported to the PM (59). In mammalian Syts, the TM length also affects the localization of tail-anchored proteins that possess a single TM domain at the C terminus with an Ncyt/Cexo topology, such as cytochrome b5 or syntaxin (60, 61). For example, the localization of syntaxin, a component of membrane-membrane fusion, to the PM is determined by the long length of its TM domain (≥23 amino acid residues). A prediction program for the presence of TM domain, SOSUI (bp.nuap.nagoya-u.ac.jp/sosui), calculated that the length of the putative TM domain of SYT1 is 23 amino acid residues long, and that the corresponding regions in the other Arabidopsis SYT1 homologous proteins are of the same length. In the case of SYT1, however, the long TM domain does not determine localization to the PM, because truncated SYT1 without tandem C2A-C2B domains cannot localize to the PM (Fig. 4B, lane 6).
Evolutionary Modulation of Plant Synaptotagmin Localization
The phylogenetic tree in Fig. 6 showed that plant Syt genes, which contain two calcium-binding domains in their C termini, were clearly separated into three clades (the SYT1/2, SYT3, and SYT4/5). Each clade contained Syt genes of monocotyledonous and eudicotyledonous plants, suggesting that the three clades diverged before the speciation of monocots and eudicots in the early stages of the angiosperm evolutionary process (Fig. 6A). In addition, the conservation of exon-intron structures among AtSyt1, AtSyt2, AtSyt4, and AtSyt5 implies that the angiosperm Syt genes share a common ancestral gene of SYT1/2, SYT3, and SYT4/5 (Fig. 6B). The most significant difference between the SYT1/2 and SYT3 clades and the SYT4/5 clade is the lack of the calcium-binding motifs in Loop 1 and Loop 3 of the C2B domain (supplemental Fig. S4). It is unclear whether genes in the SYT1/2 and SYT3 clades lost their calcium-binding motifs after duplication of the genes of SYT1/2/3 and SYT4/5 clades. However, a common ancestor of the Syt genes may have retained the calcium-binding motifs both in the C2A domain and in the C2B domain, because the calcium-binding motifs were conserved in both C2 domains in mammalian Syts.
The loss of the calcium-binding motifs in the C2B domain during evolution may result in alterations in the localization of plant Syts in the SYT1/2/3 clades. In fact, SYT1 is localized mainly to the PM and partially to endomembranes, such as Golgi, but SYT5 is localized only to endomembranes (Fig. 7C). Moreover, the SYT1 mutant, which contains the calcium-binding motif in Loop 1 (but not in Loop 3) of the C2B domain by referring SYT5 sequences, was localized mainly to endomembranes. Loop 1 of the C2A domain of mammalian Syt1 bound to calcium was shown to penetrate the lipid bilayer, and NMR data show that Loop 2 and Loop 3 of the C2A domain also bind to phospholipids with calcium (62, 63). Because the C2B domain of SYT1 interacts with liposomes in a calcium-independent manner in vitro (11), the lack of calcium-binding motifs may permit the calcium-independent interaction between SYT1 and the membrane. In addition, the evolutionarily modified amino acid residues of the C2 domains in SYT1, except for arginine and lysine, seem to be more bulky and hydrophobic than those of the SYT4/5 clade (Fig. 7B), raising the possibility that the calcium-independent penetration of Loop 1 into the lipid bilayer occurs and imparts some degree of specificity for the binding to the PM. On the other hand, as described above, the C2B domain alone may not contribute to the localization of SYT1 to the PM, for which tandem C2A-C2B domains are needed.
Reconstructed fragments of the C2A or C2B domain in mammalian Syt I demonstrate that the C2B domain, but not the C2A domain, triggers the membrane-membrane fusion mediated by the SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor) complex in a calcium-dependent manner (64). In addition, the C2B domain interacts in a calcium-independent manner with a complex of SNAP-25/syntaxin known to target SNARE proteins to the PM of neuron cells, and the calcium-independent interaction is imparted by a polybasic region of the C2B domain (65). This calcium-independent interaction of the C2B domain with lipid membrane accelerates the calcium-triggered SNARE-mediated fusion (66). Because the polybasic region is conserved in the C2B domain of plant Syts (see supplemental Fig. S4), plant Syts may interact in a calcium-independent manner with the SNAP-25-syntaxin complex. Therefore, to further understand the evolutionary effect of the absence of calcium-binding motifs in the C2B domain on plant Syts of SYT1/2/3 clades, it is necessary to investigate the interaction between plant SNARE proteins and Arabidopsis SYT1, and to assess their contribution to the localization of SYT1.
Default Pathway and Sorting Mechanism of Plant Syts
Mechanisms or rules that determine the sorting of plant integral membrane proteins to the appropriate compartments after synthesis on rough ER have been proposed, such as the “length rule” described above. However, the length rule is not sufficient to interpret the sorting mechanism of the type I membrane proteins, because signals in the cytoplasmic tail also affect the export from the ER via the Golgi to the prevacuole compartment (67, 68). The localization of plant N-glycan-processing enzymes, which are classified as type II membrane proteins, is dependent not only on the TM domain length but also on signals in the N-terminal cytosolic tail (69). In addition, for the export from the ER to Golgi, both type II and multispanning membrane proteins are influenced by the diacidic motif (D/E)X(D/E) in the cytosolic region, which acts as a signal (70). The cytosolic region of a multispanning protein, plasma membrane H+-ATPase, determines the localization to the PM, because the deletion of the cytosolic region of the protein changes the localization from the PM to the ER (71).
These observations have led to the concept of the default pathway in which the “actual” localization of integral membrane proteins results from changes in the “original” localization (72). At present, after synthesis on rough ER membrane, the most probable destination of the plant default pathway is the tonoplast (59) or PM (73). On the other hand, our results imply that the default destination of plant Syts, which are classified as SA-I proteins, may not be the PM, because the truncated form of SYT1, which lacks the tandem C2 domains, and the form of SYT1 bearing mutations in the calcium-binding motifs of the C2B domain localized to endomembranes, which are mainly Golgi membranes (Figs. 4B, 7C, and 7D).
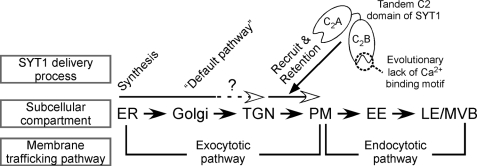
In summary, we propose a delivery model of plant Syts (Fig. 8). First, plant Syts are synthesized on the rough ER membrane, where they have the topology of an SA-I protein. Second, plant Syts are transported from the ER to the default destination of the Golgi through the exocytotic pathway. Third, the tandem C2A-C2B domains of plant Syts in SYT1/2/3 clades mediate the translocation to the PM, depending on the property of Loop 1 in the C2B domain. Fourth, plant Syts in SYT1/2/3 clades tend to be retained in the PM. Although this retention mechanism is unknown, it is possible that the interaction between tandem C2 domains and the PM tethers SYT1 to the PM, and thereby prevents the uptake of SYT1 to the early endosome, the gate to the endocytotic pathway through the late endosome/multivesicular body. Our findings provide new insights not only into the functional divergence in the molecular evolution of the plant Syts, but also into the localization mechanism of plant SA-I proteins.
FIGURE 8.
Model of SYT1 delivery to the plasma membrane. SYT1 is synthesized on ER and delivered to PM through the exocytotic pathway. The default pathway of SYT1 is thought to be Golgi or TGN. Two tandem calcium-binding domains function as a determinant of PM localization and also the prevention of up-taking SYT1 to the endocytotic pathway. PM, plasma membrane; Golgi, Golgi apparatus; TGN, trans-Golgi network; ER, endoplasmic reticulum; EE, early endosome; LE, late endosome; MVB, multivesicular body.
Supplementary Material
Acknowledgments
We thank Prof. M. Maeshima (Nagoya University, Japan), Prof. M. Boutrykindly (Université Catholique de Louvain, Belgium), and A. Nebenführ (University of Tennessee) for kindly providing the antibodies to PAQs, the antibodies to plasma membrane-type H+-ATPase, and transgenic plants expressing CFP-labeled Golgi marker, respectively. We also thank N. Yokota in our laboratory for her technical assistance.
This work was supported by a grant from the 21st Century COE Program to Iwate University (K-03) and the Tohoku University Ecosystem Adaptive Global COE Program (J03) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Grants-in-Aid for Scientific Research (18780242 and 22780288 to Y. K., 20780229 to T. Y., and 17380062 to M. U.), a grant from the Japan Society for the Promotion of Science (19.9498 to N. T.), and the President Fund of Iwate University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S5.
- ER
- endoplasmic reticulum
- Syt
- synaptotagmin
- PM
- plasma membrane
- TGN
- trans-Golgi network
- EE
- early endosome
- SA
- signal anchor
- BFA
- brefeldin A
- GFP
- green fluorescent protein
- EmGFP
- emerald GFP
- CFP
- cyan fluorescent protein
- MES
- 4-morpholineethanesulfonic acid
- SNARE
- soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptors
- SMP
- plant synaptotagmin-like, mitochondria protein
- PAQ
- plasma-membrane-type aquaporin.
REFERENCES
- 1.Südhof T. C. (2002) J. Biol. Chem. 277, 7629–7632 [DOI] [PubMed] [Google Scholar]
- 2.Bai J., Chapman E. R. (2004) Trends Biochem. Sci 29, 143–151 [DOI] [PubMed] [Google Scholar]
- 3.Gustavsson N., Lao Y., Maximov A., Chuang J. C., Kostromina E., Repa J. J., Li C., Radda G. K., Südhof T. C., Han W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3992–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao H., Ito Y., Chappel J., Andrews N. W., Teitelbaum S. L., Ross F. P. (2008) Dev. Cell 14, 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craxton M. (2004) BMC Genomics 5, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamazaki T., Kawamura Y., Minami A., Uemura M. (2008) Plant Cell 20, 3389–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi G. Q., Alderton J. M., Steinhardt R. A. (1995) J. Cell Biol. 131, 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy A., Caler E. V., Andrews N. W. (2001) Cell 106, 157–169 [DOI] [PubMed] [Google Scholar]
- 9.Steinhardt R. A., Bi G., Alderton J. M. (1994) Science 263, 390–393 [DOI] [PubMed] [Google Scholar]
- 10.Terasaki M., Miyake K., McNeil P. L. (1997) J. Cell Biol. 139, 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schapire A. L., Voigt B., Jasik J., Rosado A., Lopez-Cobollo R., Menzel D., Salinas J., Mancuso S., Valpuesta V., Baluska F., Botella M. A. (2008) Plant Cell 20, 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jahn R., Lang T., Südhof T. C. (2003) Cell 112, 519–533 [DOI] [PubMed] [Google Scholar]
- 13.Goder V., Spiess M. (2001) FEBS Lett. 504, 87–93 [DOI] [PubMed] [Google Scholar]
- 14.High S., Flint N., Dobberstein B. (1991) J. Cell Biol. 113, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kida Y., Sakaguchi M., Fukuda M., Mikoshiba K., Mihara K. (2000) J. Cell Biol. 150, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiménez J. L., Davletov B. (2007) Proteins 68, 770–778 [DOI] [PubMed] [Google Scholar]
- 17.Berton F., Cornet V., Iborra C., Garrido J., Dargent B., Fukuda M., Seagar M., Marquèze B. (2000) Eur. J. Neurosci. 12, 1294–1302 [DOI] [PubMed] [Google Scholar]
- 18.Czibener C., Sherer N. M., Becker S. M., Pypaert M., Hui E., Chapman E. R., Mothes W., Andrews N. W. (2006) J. Cell Biol. 174, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuda M., Kanno E., Satoh M., Saegusa C., Yamamoto A. (2004) J. Biol. Chem. 279, 52677–52684 [DOI] [PubMed] [Google Scholar]
- 20.Grimberg E., Peng Z., Hammel I., Sagi-Eisenberg R. (2003) J. Cell Sci. 116, 145–154 [DOI] [PubMed] [Google Scholar]
- 21.Grise F., Taib N., Monterrat C., Lagrée V., Lang J. (2007) Biochem. J. 403, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutt D. M., Cardullo R. A., Baltz J. M., Ngsee J. K. (2002) Biol. Reprod. 66, 50–56 [DOI] [PubMed] [Google Scholar]
- 23.Iezzi M., Kouri G., Fukuda M., Wollheim C. B. (2004) J. Cell Sci. 117, 3119–3127 [DOI] [PubMed] [Google Scholar]
- 24.Levius O., Feinstein N., Linial M. (1997) Eur. J. Cell Biol. 73, 81–92 [PubMed] [Google Scholar]
- 25.Michaut M., De Blas G., Tomes C. N., Yunes R., Fukuda M., Mayorga L. S. (2001) Dev. Biol. 235, 521–529 [DOI] [PubMed] [Google Scholar]
- 26.Monterrat C., Grise F., Benassy M. N., Hémar A., Lang J. (2007) Histochem. Cell Biol. 127, 625–632 [DOI] [PubMed] [Google Scholar]
- 27.Osborne S. L., Herreros J., Bastiaens P. I., Schiavo G. (1999) J. Biol. Chem. 274, 59–66 [DOI] [PubMed] [Google Scholar]
- 28.Perin M. S., Fried V. A., Mignery G. A., Jahn R., Südhof T. C. (1990) Nature 345, 260–263 [DOI] [PubMed] [Google Scholar]
- 29.Ting J. T., Kelley B. G., Sullivan J. M. (2006) J. Neurosci. 26, 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min S. W., Chang W. P., Südhof T. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3823–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagase T., Ishikawa K., Suyama M., Kikuno R., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. (1998) DNA Res. 5, 277–286 [DOI] [PubMed] [Google Scholar]
- 32.Creutz C. E., Snyder S. L., Schulz T. A. (2004) Cell Mol. Life Sci. 61, 1208–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seki M., Carninci P., Nishiyama Y., Hayashizaki Y., Shinozaki K. (1998) Plant J. 15, 707–720 [DOI] [PubMed] [Google Scholar]
- 34.Seki M., Narusaka M., Kamiya A., Ishida J., Satou M., Sakurai T., Nakajima M., Enju A., Akiyama K., Oono Y., Muramatsu M., Hayashizaki Y., Kawai J., Carninci P., Itoh M., Ishii Y., Arakawa T., Shibata K., Shinagawa A., Shinozaki K. (2002) Science 296, 141–145 [DOI] [PubMed] [Google Scholar]
- 35.Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. (1999) Plant J. 18, 455–463 [DOI] [PubMed] [Google Scholar]
- 36.Uemura M., Joseph R. A., Steponkus P. L. (1995) Plant Physiol. 109, 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawamura Y., Uemura M. (2003) Plant J. 36, 141–154 [DOI] [PubMed] [Google Scholar]
- 38.Ohshima Y., Iwasaki I., Suga S., Murakami M., Inoue K., Maeshima M. (2001) Plant Cell Physiol. 42, 1119–1129 [DOI] [PubMed] [Google Scholar]
- 39.Morsomme P., Dambly S., Maudoux O., Boutry M. (1998) J. Biol. Chem. 273, 34837–34842 [DOI] [PubMed] [Google Scholar]
- 40.Nelson B. K., Cai X., Nebenführ A. (2007) Plant J. 51, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 41.Sheen J. (2001) Plant Physiol. 127, 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J. X., Srivastava R., Che P., Howell S. H. (2007) Plant Cell 19, 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wirth J., Chopin F., Santoni V., Viennois G., Tillard P., Krapp A., Lejay L., Daniel-Vedele F., Gojon A. (2007) J. Biol. Chem. 282, 23541–23552 [DOI] [PubMed] [Google Scholar]
- 44.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 45.Tamura K., Nei M. (1993) Mol. Biol. Evol. 10, 512–526 [DOI] [PubMed] [Google Scholar]
- 46.Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 47.Palmgren M. G., Askerlund P., Fredrikson K., Widell S., Sommarin M., Larsson C. (1990) Plant Physiol. 92, 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolte S., Talbot C., Boutte Y., Catrice O., Read N. D., Satiat-Jeunemaitre B. (2004) J. Microsc. 214, 159–173 [DOI] [PubMed] [Google Scholar]
- 49.Lam S. K., Cai Y., Tse Y. C., Wang J., Law A. H., Pimpl P., Chan H. Y., Xia J., Jiang L. (2009) Plant J. 60, 865–881 [DOI] [PubMed] [Google Scholar]
- 50.Robinson D. G., Langhans M., Saint-Jore-Dupas C., Hawes C. (2008) Trends Plant Sci. 13, 405–408 [DOI] [PubMed] [Google Scholar]
- 51.Lee I., Hong W. (2006) FASEB J. 20, 202–206 [DOI] [PubMed] [Google Scholar]
- 52.Fernández-Chacón R., Königstorfer A., Gerber S. H., García J., Matos M. F., Stevens C. F., Brose N., Rizo J., Rosenmund C., Südhof T. C. (2001) Nature 410, 41–49 [DOI] [PubMed] [Google Scholar]
- 53.Ridge R. W., Uozumi Y., Plazinski J., Hurley U. A., Williamson R. E. (1999) Plant Cell Physiol. 40, 1253–1261 [DOI] [PubMed] [Google Scholar]
- 54.Dhonukshe P., Baluska F., Schlicht M., Hlavacka A., Šamaj J., Friml J., Gadella T. W., Jr. (2006) Dev. Cell 10, 137–150 [DOI] [PubMed] [Google Scholar]
- 55.Minami A., Fujiwara M., Furuto A., Fukao Y., Yamashita T., Kamo M., Kawamura Y., Uemura M. (2009) Plant Cell Physiol. 50, 341–359 [DOI] [PubMed] [Google Scholar]
- 56.Takeda S., Gapper C., Kaya H., Bell E., Kuchitsu K., Dolan L. (2008) Science 319, 1241–1244 [DOI] [PubMed] [Google Scholar]
- 57.Baluska F., Salaj J., Mathur J., Braun M., Jasper F., Samaj J., Chua N. H., Barlow P. W., Volkmann D. (2000) Dev. Biol. 227, 618–632 [DOI] [PubMed] [Google Scholar]
- 58.Lewis J. D., Lazarowitz S. G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brandizzi F., Frangne N., Marc-Martin S., Hawes C., Neuhaus J. M., Paris N. (2002) Plant Cell 14, 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedrazzini E., Villa A., Borgese N. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4207–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watson R. T., Pessin J. E. (2001) Am. J. Physiol. Cell Physiol. 281, C215–223 [DOI] [PubMed] [Google Scholar]
- 62.Chae Y. K., Abildgaard F., Chapman E. R., Markley J. L. (1998) J. Biol. Chem. 273, 25659–25663 [DOI] [PubMed] [Google Scholar]
- 63.Chapman E. R., Davis A. F. (1998) J. Biol. Chem. 273, 13995–14001 [DOI] [PubMed] [Google Scholar]
- 64.Gaffaney J. D., Dunning F. M., Wang Z., Hui E., Chapman E. R. (2008) J. Biol. Chem. 283, 31763–31775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rickman C., Archer D. A., Meunier F. A., Craxton M., Fukuda M., Burgoyne R. D., Davletov B. (2004) J. Biol. Chem. 279, 12574–12579 [DOI] [PubMed] [Google Scholar]
- 66.Loewen C. A., Lee S. M., Shin Y. K., Reist N. E. (2006) Mol. Biol. Cell 17, 5211–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.daSilva L. L., Foresti O., Denecke J. (2006) Plant Cell 18, 1477–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.daSilva L. L., Taylor J. P., Hadlington J. L., Hanton S. L., Snowden C. J., Fox S. J., Foresti O., Brandizzi F., Denecke J. (2005) Plant Cell 17, 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saint-Jore-Dupas C., Nebenführ A., Boulaflous A., Follet-Gueye M. L., Plasson C., Hawes C., Driouich A., Faye L., Gomord V. (2006) Plant Cell 18, 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanton S. L., Renna L., Bortolotti L. E., Chatre L., Stefano G., Brandizzi F. (2005) Plant Cell 17, 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lefebvre B., Batoko H., Duby G., Boutry M. (2004) Plant Cell 16, 1772–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rojo E., Denecke J. (2008) Plant Physiol. 147, 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benghezal M., Wasteneys G. O., Jones D. A. (2000) Plant Cell 12, 1179–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.