Abstract
The molecular mechanisms regulating smooth muscle-specific gene expression during smooth muscle development are poorly understood. Myocardin is an extraordinarily powerful cofactor of serum response factor (SRF) that stimulates expression of smooth muscle-specific genes. In an effort to search for proteins that regulate myocardin function, we identified a novel HMG box-containing protein HMG2L1 (high mobility group 2 like 1). We found that HMG2L1 expression is correlated with the smooth muscle cell (SMC) synthetic phenotype. Overexpression of HMG2L1 in SMCs down-regulated smooth muscle marker expression. Conversely, depletion of endogenous HMG2L1 in SMCs increases smooth muscle-specific gene expression. Furthermore, we found HMG2L1 specifically abrogates myocardin-induced activation of smooth muscle-specific genes. By GST pulldown assays, the interaction domains between HMG2L1 and myocardin were mapped to the N termini of each of the proteins. Finally, we demonstrated that HMG2L1 abrogates myocardin function through disrupting its binding to SRF and abolishing SRF-myocardin complex binding to the promoters of smooth muscle-specific genes. This study provides the first evidence of this novel HMG2L1 molecule playing an important role in attenuating smooth muscle differentiation.
Keywords: Differentiation, Gene Transcription, Protein DNA-Interaction, Protein-Protein Interactions, Smooth Muscle, Tissue-Specific Transcription Factors, Transcription Coactivators, Transcription Regulation
Introduction
Differentiated smooth muscle is characterized by the presence of unique isoforms of contractile proteins that are important to maintain the contractile function of cardiovascular, respiratory, genitourinary, and digestive systems. Although the molecular mechanisms regulating smooth muscle-specific gene expression during smooth muscle development are poorly understood, SRF2 has been shown to play a central role in the expression of many different smooth muscle-specific genes including the smooth muscle myosin heavy chain (SM MHC), smooth muscle α- and γ-actin, SM22α, calponin, and telokin genes (1). SRF is an evolutionarily conserved MADS (MDM1, agamous, deficiens, SRF) domain-containing protein that not only binds a highly conserved cis-regulatory element CC(A/T)6GG, termed a CArG box, but also provides a docking surface within the conserved MADS domain for interaction with a wide variety of accessory cofactors.
Of the SRF-associated proteins identified, myocardin has been demonstrated to be an extraordinarily powerful SRF cofactor for stimulating expression of smooth muscle-specific genes (2). In an effort to search for proteins that regulate myocardin function, we identified a novel HMG box-containing protein HMG2L1. HMG proteins are a diverse family of nonhistone chromosomal proteins. The HMG proteins contain DNA binding domains that allow the molecules to produce specific conformational changes in DNA structure, such as bending and twisting (3). HMG box proteins can be divided into two general groups, those that exhibit sequence specific DNA binding and those that bind to DNA nonspecifically. The former group is exemplified by the SOX proteins, a group of transcription factors that play important roles in development (4–6). The latter group is referred to as the canonical HMG proteins and is comprised of three subfamilies, HMG-B, HMG-N, and HMG-A (7). Several studies have demonstrated that HMG proteins may play important roles in smooth muscle gene transcription during phenotypic modulation of SMCs. The HMGA proteins HMG-I/Y have been shown to interact with SRF and enhance SRF-dependent activation of SM22α (8). Conversely, these proteins also have been shown to be up-regulated in proliferating vascular SMCs at a time when SRF-dependent smooth muscle-specific genes are inhibited (9). Similarly, increased expression of SOX18 has been associated with proliferating SMCs (10). The HMG protein SSRP1, which is part of the facilitates chromatin transcription (FACT) chromatin remodeling complex, also has been shown to interact with SRF and synergistically enhances SRF trans-activation on multiple smooth muscle-specific promoters (11).
HMG2L1 originally was identified to negatively regulate Wnt/β-catenin signaling in Xenopus (12), but its function in mammals thus far is unknown. In the current study, our data demonstrate that HMG2L1 is a potent repressor for smooth muscle differentiation through its ability to disrupt the interaction of myocardin with SRF and abolish myocardin-SRF complex binding to the promoters of smooth muscle-specific genes. This study provides the first evidence of this novel HMG2L1 molecule playing an important role in attenuating smooth muscle differentiation.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screen
A two-hybrid screen was carried out by using N-terminal myocardin (1–585 amino acids) essentially as described previously (13). One clone spanning the C-terminal (380–594 amino acids) of HMG2L1 (NM_178017, also known as HMGB2L1 or HMGXB4) was isolated and characterized further.
Cell Culture
10T1/2, COS7, A7r5, and A10 cells were maintained in 10-cm plates containing Dulbecco's modified Eagle's medium with a supplement of 10% fetal bovine serum and antibiotics. For culture of primary human coronary artery SMCs (Cell Applications, Inc.), cells (passages 3–8) were plated in SMC growth medium (MCDB131 containing 5% fetal calf serum, 2 ng/ml human basic fibroblast growth factor, 5 mg/ml human insulin, and 0.5 ng/ml human epidermal growth factor) for 24 h as described in our previous report (13). Cells were incubated in serum-free MCDB131 medium for 0, 2, and 4 days as indicated. Subsequently, total RNA was extracted at the time points indicated using TRIzol reagent (Invitrogen) for qRT-PCR to measure smooth muscle gene and HMG2L1 expression.
Mammalian Expression Constructs, Transfection, Reporter Assay, and Adenoviral Infection
The coding region of mouse HMG2L1 cDNA (encoding amino acids 1–594) was amplified from the positive yeast clone and an IMAGE clone (clone 5347132, Invitrogen) by PCR and ligated to pcDNA3.1/Myc-His vector (Invitrogen), resulting in the expression of a fusion protein with C-terminal His6 and Myc epitope tags. The full-length coding sequence of HMG2L1 was also cloned into a modified pShuttle vector (Clontech) encoding an N-terminal HA tag to generate HMG2L1 adenovirus as reported previously (13, 14). The mammalian expression plasmids for HMG2L1 truncation mutants were generated in pcDNA3.1/Myc-His vectors by the standard PCR cloning strategy. The coding region of mouse HMG-I, HMG-Y, and rat HMGB1 was cloned to pcDNA3.1 His vector (Invitrogen). The Myc or HA-tagged mammalian expression plasmids for myocardin, myocardin-related transcription factor-A, MRTF-B, SRF, thyrotroph embryonic factor β, and adenovirus encoding myocardin were reported in our previous studies (13, 15). An expression plasmid encoding a myocardin leucine zipper mutant was described previously (14). Expression plasmids for fusion proteins of GAL4 DNA binding domain-SRF (pBind-SRF) and VP16 activation domain-myocardin (pAct-myocardin) were generated in our previous study (16). Transient transfection and reporter assays were carried out with FuGENE 6 transfection reagent (Roche Applied Science) as described previously (13, 15). For adenoviral transduction, mouse aortic primary SMCs were isolated from 4–6-week-old mice as described previously (17) and plated in 12-well plates at a density of 7 × 104 cells/well. The next day, cells were transduced with adenovirus encoding HA-tagged nuclear-localized yellow fluorescent protein or HA-tagged HMG2L1 in 10% growth medium. 72 h following transduction, total RNA or protein were extracted from the transduced cells with TRIzol or radioimmune precipitation assay buffer, respectively.
Immunocytochemistry
The coding region of mouse HMG2L1 cDNA was cloned into pTag-RFP-N vector (Evrogen), resulting in the expression of a red fluorescent protein (RFP) fused to C-terminal of HMG2L1. Immunocytochemistry was carried out as described previously (13).
Quantitative Real-time RT-PCR (qRT-PCR) Analysis
Total RNA was isolated with TRIzol reagent, and qRT-PCR was carried out with respective gene-specific primers as reported previously (13). Primers sets for mouse HMG2L1 were designed as sense (5′-GAGAAGCGGCACTCCCGGACCAAG-3′) and antisense (5′-CGGTACTCCTTACAGAATACCTGGTAG-3′). Primers sets for human HMG2L1 were designed as sense (5′-GTATGGTGGCTGTGTCTGGCAGTTTG-3′) and antisense (5′-AGTCCCGGCATGATGTAAGCAATGTTG-3′). All samples were amplified in duplicate, and every experiment was repeated independently two times. Relative gene expression was converted using the 2−ΔΔCt method against an internal control rplp0 (acidic ribosomal phosphoprotein P0) housekeeping gene.
siRNA
Control siRNA or siRNA against HMG2L1 was designed and purchased from Dharmacon. The siRNA sequence for targeting endogenous mouse and rat HMG2L1 was 5′-GCAAGGTGCTTTACTCCTA-3′. For testing the effects of smooth muscle gene expression by depletion of endogenous HMG2L1, control siRNA, or HMG2L1 siRNA was transfected into A10 cells for 48 h, and total RNA was harvested for qRT-PCR as described above.
Coimmunoprecipitation and Western Blotting
A7r5 SMCs were transduced with adenovirus encoding myocardin (no tag) and HA-tagged MRTF-A and MRTF-B, as indicated in Fig. 5. 48 h after transduction, nuclear protein was harvested for coimmunoprecipitation assays with anti-HMG2L1, myocardin, or HA antibodies as described in our previous report (13). Western blot analysis was carried out essentially as described previously (14, 15, 18). Antibodies used in this study were against HA tag (Covance, 1:3,000), HMG2L1 (Sigma, 1:2,000), Myc (Invitrogen, 1:5,000), myocardin (Santa Cruz Biotechnology, M16, 1:2,000), Omni (Invitrogen, 1:3,000), SRF (Santa Cruz Biotechnology, G20X, 1:10,000), T7 (1:10,000, Novagen), and vinculin (Santa Cruz Biotechnology, 1:5,000).
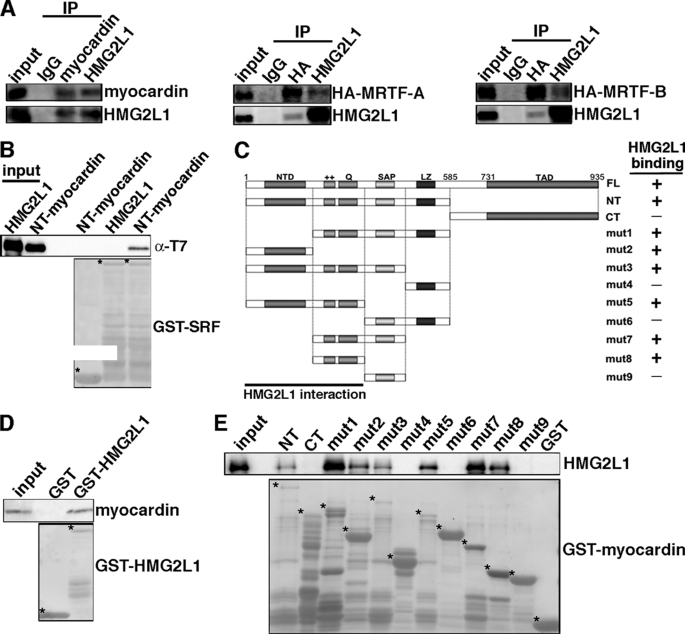
FIGURE 5.
HMG2L1 interacts with myocardin in vivo and in vitro. A, HMG2L1 binds to myocardin family proteins in vivo. Myocardin or HA-tagged MRTF-A/B adenovirus were transduced into A7r5 SMCs. Subsequently, nuclear extract was harvested, and proteins were immunoprecipitated with anti-myocardin, HA, HMG2L1, or control IgG antibodies. The immunoprecipitated proteins were detected by Western blotting using anti-myocardin, anti-HA, and anti-HMG2L1 antibodies, as indicated at the right of the blot. 10% total extract was loaded as input. B, HMG2L1 does not bind to SRF. GST fused to SRF or GST alone was expressed in bacteria, conjugated to glutathione-Sepharose beads, and incubated with bacterially expressed T7 tagged amino-terminal-myocardin (amino acids 1–585) or full-length HMG2L1 as indicated. Bound proteins were identified by Western blotting with an anti-T7 antibody (upper panel). The lower panel showed the expression of the GST-SRF fusion protein or GST alone (marked by an asterisk to the top left of each protein). The interaction between myocardin and SRF served as a positive control. C, schematic representation of myocardin indicating the GST-fusion proteins analyzed and summary of myocardin domains mapped for interacting with HMG2L1 from D and E. NTD, N-terminal domain; ++, basic domain; Q, poly Q domain; LZ, leuzine zipper domain; TAD, transcriptional activation domain; mut: mutant. D, HMG2L1 binds to the full-length myocardin. Bacterially expressed full-length myocardin was incubated with the GST-HMG2L1 fusion protein. Western blotting was performed to detect the myocardin fusion protein that bound to HMG2L1 (upper panel). The lower panel in D indicates the expression of the GST fusion protein of full-length HMG2L1 as detected by a Ponceau S staining. E, HMG2L1 binds to the myocardin N-terminal domain, basic, and poly Q domain. Bacterially expressed HMG2L1 was incubated with the series of GST-myocardin fusion proteins indicated in C. Western blotting was performed to detect the GST-myocardin fusion proteins that bound HMG2L1 (upper panel). The lower panel in E indicates the expression of the myocardin GST fusion proteins as detected by a Ponceau S staining. *, GST fusion protein.
GST Pulldown Assays
Fragments of mouse myocardin, SRF, and HMG2L1 cDNAs were cloned into pGEX-4T vectors (Stratagene) to generate GST fusion proteins or cloned into pET28 vectors (Novagen) to generate T7 fusion proteins and GST pulldown assays were performed as described in our previous reports (13, 19).
Quantitative Chromatin Immunoprecipitation (ChIP) Assays
10T1/2 cells were cultured in 10% growth media and infected with adenovirus encoding Omni-tagged myocardin and HA-tagged YFP or HMG2L1 for 36 h. After cells fixed with formaldehyde ChIP was performed by using anti-Omni epitope tag (myocardin) or anti-SRF antibody essentially as described by the manufacturer (Upstate) and in our previous reports (13). For measuring endogenous HMG2L1 binding to smooth muscle-specific gene promoters, partially differentiated A10 SMCs were harvested for ChIP assay by using anti-HMG2L1 antibody (Sigma).
RESULTS
HMG2L1 Is a Novel Transcription Factor Whose Expression Is Correlated with SMC Synthetic Phenotype
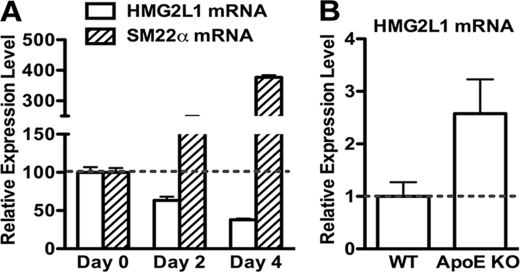
In an effort to identify myocardin regulatory proteins, we discovered a previously uncharacterized HMG box-containing protein HMG2L1 (HMG2-like 1) from a yeast two-hybrid screen using myocardin as bait. The mouse hmg2l1 gene contains 11 exons located on chromosome 8. By using anti-HMG2L1 antibody to detect HMG2L1 expression in adult mouse tissues, we found HMG2L1 is widely expressed in many tissues including smooth muscle-rich tissues such as bladder, colon, and stomach (supplemental Fig. 1A). We next examined the subcellular localization of RFP-fused HMG2L1 or epitope-tagged HMG2L1 in transfected 10T1/2 fibroblast or A10 SMCs and found it to be exclusively nuclear (supplemental Fig. 1B). Previous studies have demonstrated induction of HMG box-containing protein, including SOX18 and HMG-I, can be found in proliferating vascular SMCs (9, 10). To examine the relationship between HMG2L1 and SMC differentiation and proliferation, proliferating human coronary arterial SMCs were cultured in serum-free medium for 2 or 4 days to promote differentiation. Total RNA from these cells were harvested at each time point and then analyzed by qRT-PCR as indicated with gene-specific primers (Fig. 1A). These results show that following serum withdrawal, expression of HMG2L1 mRNA is down-regulated, whereas SM22α mRNA expression is increased, suggesting expression of HMG2L1 is inversely correlated with expression of smooth muscle-specific genes. We further examined HMG2L1 expression in apoE knock-out mice, a mouse model that spontaneously develops arterial atherosclerotic lesions in which SMCs are synthetic and dedifferentiated. We harvested aortic artery tissues from 5-month-old apoE knock-out mice and age-matched control mice to extract RNA. Subsequently, HMG2L1 mRNA was determined by qRT-PCR. Data from this experiment reveal that HMG2L1 expression is increased 2.5-fold in apoE knock-out arteries (Fig. 1B).
FIGURE 1.
HMG2L1 expression during SMC differentiation and in arteries of apoE-null mouse. A, proliferating human coronary SMCs were cultured in serum-free medium for 0, 2, or 4 days as indicated. Total RNA was harvested at each time point from these cells, and gene expression of SM22α and HMG2L1 are examined by qRT-PCR. The gene expression of HMG2L1 and SM22α on day 0 was set to 100 (n = 3). Serum withdrawal results in inducing expression of the smooth muscle gene, whereas it down-regulates expression of HMG2L1. B, aortic artery tissues were harvested from 5-month-old apoE knock-out mice or control age-matched wild-type mice, and HMG2L1 expression was determined by qRT-PCR. The expression level of HMG2L1 in control vessels was set to 1 (n = 6).
Overexpression of HMG2L1 Down-regulates Smooth Muscle-specific Gene Expression
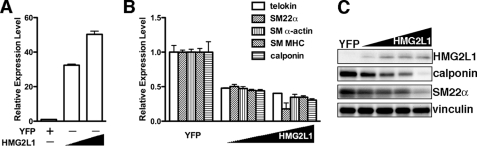
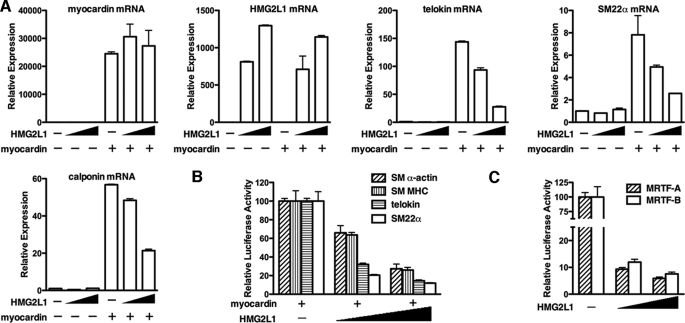
Data presented above demonstrate that HMG2L1 is down-regulated during smooth muscle differentiation and up-regulated in synthetic SMCs. To explore the role of HMG2L1 in the expression of endogenous smooth muscle-specific genes, we transduced HMG2L1 adenovirus into mouse aortic primary SMCs, and RNA or protein was harvested to measure smooth muscle gene expression by qRT-PCR or Western blotting, respectively. All smooth muscle-specific genes examined, including telokin, SM22α, SM α-actin, calponin, and SM MHC were significantly down-regulated 50–75% by HMG2L1 at both mRNA and protein levels (Fig. 2, A–C).
FIGURE 2.
Overexpression of HMG2L1 down-regulates smooth muscle-specific gene expression in primary SMCs. A and B, primary SMCs were prepared from the aortic arteries of 4–6-week-old mice and transduced with adenovirus encoding HMG2L1 or YFP as indicated. 72 h following transduction, mRNA was isolated and analyzed by qRT-PCR to measure HMG2L1 (A) and smooth muscle gene expression (B), respectively. C, YFP control virus or increasing amount of HMG2L1 adenovirus was transduced into primary SMCs as described in A and B for 72 h, and then protein was harvested for Western blotting. Overexpression of HMG2L1 represses smooth muscle-specific gene expression at both mRNA and protein levels in a dose-dependent manner.
Depletion of Endogenous HMG2L1 in Smooth Muscle Cells Results in Up-regulation Smooth Muscle-specific Gene Expression
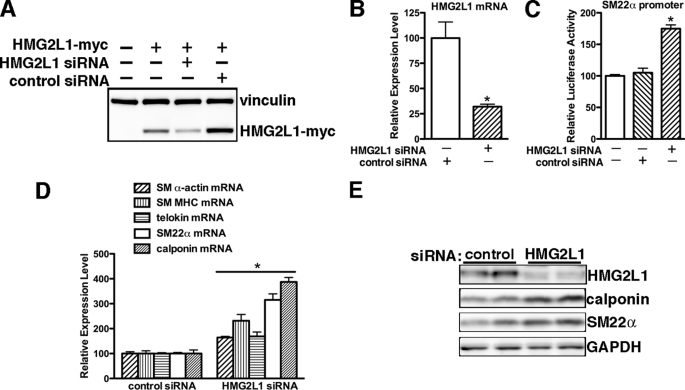
To further determine the role of endogenous HMG2L1 in regulating SM-specific gene expression in partially dedifferentiated vascular SMCs, endogenous HMG2L1 levels were depleted using siRNA. siRNA specific for HMG2L1 was able to significantly knock down expression of exogenous HMG2L1 in Cos7 cells (Fig. 3A) and to decrease endogenous HMG2L1 by 75% (Fig. 3B) in A10 smooth muscle cells. Depletion of HMG2L1 from A10 SMCs resulted in significant increase SM22α promoter activity (Fig. 3C) and mRNA levels of smooth muscle markers of telokin, SM22α, SM α-actin, calponin, and SM MHC mRNA expression to 1.5–4-fold (Fig. 3D). Consistent with this, silencing endogenous HMG2L1 also lead to an increase in calponin and SM22α proteins (Fig. 3E).
FIGURE 3.
Knocking down endogenous HMG2L1 results in increased smooth muscle-specific gene expression in A10 SMCs. A, COS7 cells were transfected with Myc-tagged HMG2L1 expression plasmid followed by either 100 pmol of an RNA duplex directed against HMG2L1 (sequence targets rat and mouse; Dharmacon) or scrambled control RNA duplex (Dharmacon). Western blotting was used to detect exogenous HMG2L1 expression with anti-Myc antibody. Vinculin served as a loading control. B, A10 cells were transfected with either control siRNA or HMG2L1 siRNA duplex. 48 h later, total RNA was harvested, and endogenous HMG2L1 gene expression was analyzed by qRT-PCR. Data are represented as mean ± S.E. of six samples from two independent experiments. *, p < 0.05. Transfection of silencing duplex against HMG2L1 results in an significant decreasing expression of endogenous HMG2L1. C, A10 cells were transfected with either control siRNA or HMG2L1 siRNA for 12 h and then were transfected with SM22α gene luciferase reporter. 24 h later, promoter activity was measured by dual luciferase assay. Reporter activity is normalized to a Renilla luciferase internal control and expressed relative to control transfections (set to 100). Data were presented as mean ± S.E. of six samples from two independent experiments. Silencing endogenous HMG2L1 significantly increases SM22α promoter activity. *, p < 0.05. D and E, A10 cells were transfected with either control siRNA or HMG2L1 siRNA duplex for 48 h as described in B. To measure smooth muscle-specific gene expression, mRNA was harvested for qRT-PCR (D), or protein was harvested for Western blotting (E), respectively. Silencing endogenous HMG2L1 significantly up-regulates smooth muscle-specific gene expression at mRNA and protein levels.
HMG2L1 Abrogates Myocardin-induced Expression of Smooth Muscle-specific Genes and Abolishes MRTF-induced trans-Activation of Smooth Muscle-specific Promoters
Data presented above show that HMG2L1 is a strong repressor for smooth muscle-specific gene expression. Given that HMG2L1 was identified as a myocardin associated protein and myocardin is a potent activator for smooth muscle gene expression, we next sought to determine whether HMG2L1 attenuates myocardin function. We first determined the effects of HMG2L1 on the induction of endogenous smooth muscle-specific genes by myocardin in fibroblast cells. Myocardin or empty pcDNA plasmids were transfected into 10T1/2 cells together HMG2L1 expression plasmids, and 24 h after transduction, RNA was harvested from these cells and subjected to quantitative RT-PCR by using gene-specific primers as indicated (Fig. 4A). Data from this experiment reveals that co-transfection of HMG2L1 with myocardin attenuated the myocardin-induced expression of telokin, calponin, and SM22α in a dose-dependent manner without affecting ectopic myocardin expression (Fig. 4A). We next determined whether HMG2L1 suppressed myocardin or MRTF-A and MRTF-B-induced trans-activation on smooth muscle-specific promoters. Myocardin family members were co-transfected together with telokin, SM22α, SM α-actin, and SM MHC promoter-luciferase reporter genes with or without HMG2L1 expression plasmid into 10T1/2 cells, and the subsequent effects on promoter activity were measured by luciferase assay. Data from these experiments revealed that HMG2L1 repressed the myocardin-mediated activation of each of these promoters in a dose-dependent manner (Fig. 4B). HMG2L1 also attenuated transactivation induced by a dimerization-defective myocardin, indicating that HMG2L1 is unlikely to inhibit myocardin through interfering with its dimerization (supplemental Fig. 2). Furthermore, HMG2L1 attenuated the transactivation of the SM22α promoter induced by myocardin-related transcription factors, MRTF-A and MRTF-B (Fig. 4C). The inhibitory effects of HMG2L1 on myocardin and the MRTFs are specific, as HMG2L1 was not able to affect SRF or TEFβ-induced trans-activation of the telokin promoter (supplemental Fig. 3). On the another hand, myocardin-mediated activation of smooth muscle genes was not significantly attenuated by other HMG box-containing proteins including HMG-I, HMG-Y, or HMGB1, demonstrating the inhibitory effect of HMG2L1 on the transcriptional activity of myocardin was specific to HMG2L1 (supplemental Fig. 4).
FIGURE 4.
HMG2L1 suppresses the induction of endogenous smooth muscle-specific genes by myocardin and abrogates the myocardin and MRTF-induced activation of smooth muscle-specific promoters. A, mouse myocardin expression vector, together with mouse HMG2L1 or empty plasmid pcDNA3.1 were transfected into 10T1/2 cells. 24 h post-transfection, total RNA was harvested from cells, and qRT-PCR was performed to examine expression of endogenous smooth muscle-specific genes as indicated. The ectopic expression of myocardin and HMG2L1 also was measured. Transcript levels first was normalized to acidic ribosomal phosphoprotein (rplp0) internal loading control and then normalized to their respective vector transfected control group. The ΔΔCt method was used to calculate the relative quantity values of gene expression levels. Relative Expression = 2−ΔΔCt and ΔΔCt = (Ct experimental − Ct RPLP0) − (Ct control − Ct RPLP0). Data presented are the mean ± S.E. of four samples obtained from two independent experiments. B, luciferase reporter genes and myocardin expression plasmids were transfected into 10T1/2 cells with or without HMG2L1 expression plasmids. Values are presented as relative luciferase activity compared with myocardin-induced activation (set to 100) and are the mean ± S.E. of six samples from two independent experiments. Co-transfection with HMG2L1 significantly abrogates myocardin-induced transactivation of each of the smooth muscle gene reporters examined in a dose-dependent manner. C, an SM22α promoter reporter gene was transfected into 10T1/2 cells together with MRTF-A or MRTF-B in the presence of increasing amounts of HMG2L1 expression plasmids. All data are normalized to the activation produced by MRTF-A or MRTF-B alone (set to 100).
Myocardin Binds to HMG2L1 in Vivo and in Vitro
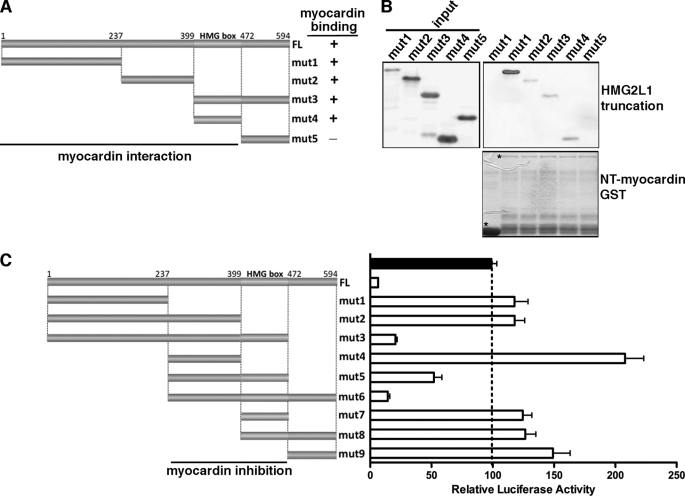
Data described above demonstrated that HMG2L1 negatively regulates myocardin and its family protein MRTF-A and MRTF-B function. We next determined whether HMG2L1 physically binds to myocardin family proteins in vivo by coimmunoprecipitation assays. As there are no antibodies available that detect endogenous myocardin, MRTF-A, and MRTF-B in SMCs, we transduced myocardin, MRTF-A, and MRTF-B adenovirus into A7r5 SMCs and myocardin (for immunoprecipitation of ectopic myocardin), HA (for immunoprecipitation of ectopic MRTF-A or MRTF-B), or HMG2L1 antibodies (for immunoprecipitation of endogenous HMG2L1) were used to immunoprecipitate reciprocally myocardin family protein-HMG2L1 complexes. Western blotting of the precipitated complexes revealed that HMG2L1 complexes contained myocardin, MRTF-A, and MRTF-B, and myocardin family protein complexes contained HMG2L1 (Fig. 5A). GST pulldown assays further confirmed that HMG2L1 binds directly to myocardin but does not bind directly to SRF (Fig. 5B). To understand the mechanism by which HMG2L1 represses myocardin function, we performed GST pulldown assays to map the myocardin domains that bind to HMG2L1. As summarized in Fig. 5C, HMG2L1 binds to the full-length myocardin (Fig. 5D) and binds specifically to two portions of myocardin, the amino-terminal domain and the region that includes the basic domain and poly Q domain (Fig. 5E) that are required for SRF interaction. Conversely, a large fragment of HMG2L1 spanning amino-acids 1 to 472 binds to myocardin (Fig. 6, A and B). To further map HMG2L1 functional domains that inhibit myocardin, we generated a variety of HMG2L1 truncation mutants and co-transfected these together with myocardin and an SM22α reporter gene into 10T1/2 cells. The effects of these HMG2L1 mutants on myocardin transactivation subsequently were measured by luciferase assays (Fig. 6C). Data from this reporter assay revealed that residues between 237 and 472 of HMG2L1, which includes the HMG box domain, are sufficient to inhibit myocardin transcriptional activity.
FIGURE 6.
Characterization of HMG2L1 domains that physically and functionally bind to myocardin. A, schematic illustration of the domain structures of HMG2L1 indicating the positions of the truncation mutants used for mapping studies. mut, mutant. B, bacterially expressed HMG2L1 truncation mutants indicated in A were incubated with the N-terminal myocardin (amino acids 1–585) GST fusion protein (lower panel). Western blotting was performed to detect the HMG2L1 bound to amino-terminal-myocardin GST fusion protein (upper panel). The lower panel in B indicates the expression of the GST or GST-fused amino-terminal-myocardin protein as detected by a Ponceau S staining. Myocardin was found to bind to the HMG2L1 N terminus (amino acids 1–472) as summarized in A. C, HMG box of HMG2L1 is required but not sufficient to abrogate the transactivation of myocardin on the SM22α promoter. An SM22α promoter reporter gene was transfected into 10T1/2 cells together with myocardin in the presence of expression plasmids for full-length of HMG2L1 or a variety of HMG2L1 truncation mutants as indicated at the left panel. All data are normalized to the activation produced by myocardin alone (black bar, set to 100).
HMG2L1 Disrupts the Interaction of Myocardin with SRF and Abolishes Myocardin-SRF Complex Formation on the Promoters of Smooth Muscle-specific Genes
We next determined the mechanism by which HMG2L1 inhibits myocardin. As myocardin has been shown to be able to bind both histone acetyltransferase and histone deacetylases, and another HMG box molecule, such as UBF, has been shown to recruit HDAC (20, 21), we first determined whether the inhibitory activity of HMG2L1 was dependent on its ability to recruit HDAC or disrupt myocardin interaction with its known co-activator such as P300 (20). In luciferase experiments, we found that treating cells with the HDAC inhibitor Trichostatin A augmented the activity of myocardin; however, this did not affect the ability of HMG2L1 to block the activity of myocardin (supplemental Fig. 5). Moreover, overexpression of p300 in 10T1/2 cells was not able to rescue the inhibitory effect of HMG2L1 on the SM22α promoter (supplemental Fig. 6). Because the ability of myocardin to activate smooth muscle-specific genes is dependent strictly on its interaction with SRF, we next tested the possibility that HMG2L1 blocks this interaction. In coimmunoprecipitation assays, the binding between myocardin and SRF was decreased significantly in the presence of HMG2L1 (Fig. 7A). Using a mammalian two-hybrid reporter assay, we also found that HMG2L1 significantly attenuated reporter activity produced by the interaction of SRF and myocardin (Fig. 7B). In partially differentiated A10 SMCs, ChIP assay revealed that endogenous HMG2L1 can bind to the smooth muscle gene CArG region where SRF/myocardin binding (supplemental Fig. 7). Furthermore, data from ChIP assays demonstrated that myocardin binding to the endogenous telokin promoter was remarkably inhibited by exogenous HMG2L1 (Fig. 7C). Taken together, these data demonstrated that HMG2L1 disrupts SRF-myocardin complex formation in vivo. Previous studies have shown that HERP1 and KLF4 can abrogate myocardin-induced activation of smooth muscle gene expression by interfering with SRF binding to the CArG box (22, 23). Because HMG box proteins can bend and distort DNA, it is also possible that the binding of HMG2L1 to myocardin may induce conformational changes in DNA that prevents the formation of stable myocardin-SRF complexes on promoters. We therefore performed ChIP assays from cells transduced with myocardin adenovirus with or without co-transduction of HMG2L1 and then used anti-SRF antibodies to precipitate chromatin complexes as shown in Fig. 7D. Results from this analysis showed that HMG2L1 significantly attenuated myocardin-induced SRF binding to the smooth muscle-specific telokin, SM22α, and SM MHC promoters (Fig. 7D). In sum, these data suggest that the HMG2L1 inhibitory effects on the promyogenetic function of myocardin occurs through disrupting SRF/myocardin interactions, which subsequently decreases both myocardin and SRF binding to smooth muscle-specific genes, thereby attenuating their expression.
FIGURE 7.
HMG2L1 abrogates myocardin and myocardin-mediated SRF binding to the promoters of smooth muscle-specific genes. A, Adenoviruses encoding SRF, myocardin of HMG2L1 were transfected into COS7 cells as indicated. SRF was immunoprecipitated from nuclear extracts of the transduced cells and co-precipitating myocardin was detected by Western blotting. B, 10T1/2 were transfected with pG5Luc reporter plasmid and expression plasmids for fusion proteins of GAL4 DNA binding domain-SRF (pBind-SRF), VP16 activation domain-myocardin (pAct-myocardin) in the presence or absence of HMG2L1. Increased luciferase activity, indicative of protein/protein interactions, was seen when pBind-SRF and pAct-myocardin were co-transfected. In the presence of HMG2L1, the SRF/myocardin interaction-induced reporter activity was significantly abolished (n = 6). *, p < 0.05. C, HMG2L1 represses myocardin binding to smooth muscle-specific gene promoters. ChIP assays using 10T1/2 cells transduced with adenovirus encoding Omni-tagged myocardin, HA-tagged YFP, or HMG2L1. Cross-linked chromatin was immunoprecipitated with anti-Omni antibody (myocardin), and the precipitated DNA was amplified by real-time PCR with telokin gene-specific primers spanning the GArG box region. The myocardin binding is indicated relative to YFP control-transduced cells (set to 1). These data were calculated and normalized to input levels as follows: relative myocardin binding = 2−ΔΔCt, with ΔΔCt = (Ct myocardin ± HMG2L1 − Ct input) − (Ct YFP − Ct input). Data shown are the mean ± S.E. of four samples obtained from two independent experiments. *, significantly different (p < 0.05). D, the myocardin-induced SRF binding to smooth muscle-specific gene promoters is inhibited significantly by the presence of HMG2L1. 10T1/2 cells were transduced with myocardin, HMG2L1, or YFP control adenovirus as indicated. After 36 h, cells were fixed and harvested for chromatin immunoprecipitation assays using an antibody against SRF. The precipitated genomic DNA was purified, and the enrichment of the promoters of SRF-dependent smooth muscle genes was measured by real-time PCR using gene-specific primers as indicated. The effects of myocardin or/and HMG2L1 on SRF binding to smooth muscle-specific genes were indicated relative to those transduced with YFP (set to 1). These data were calculated and normalized to input levels as follows: relative SRF binding = 2−ΔΔCt, with ΔΔCt = (Ct experiment − Ct input) − (Ct YFP − Ct input). Data shown are the mean ± S.E. of four samples obtained from two independent experiments. p < 0.05.
DISCUSSION
HMG transcription factors are the most abundant nonhistone proteins in the nucleus. In the current study, we have identified a novel HMG-box containing protein HMG2L1 as a potent repressor for smooth muscle differentiation that attenuates the myogenic activity of myocardin. Previous studies have shown HMGB family factors are involved in regulating many transcriptional events by physical and functional interaction with multiple transcription factors such as HOX, OCT, steroid hormone receptors, RAG1 and -2, REL, and P53 (24–26). The consequence of these interactions either activates or represses transcription depending on the factors with which they associate, suggesting HMGB proteins have broad functions in distinct cell or tissue contexts. Several studies have demonstrated that HMG proteins may play important roles in smooth muscle gene transcription during phenotypic modulation of smooth muscle cells. For example, the HMG protein SSRP1 and HMGI(Y) have been shown to interact with SRF and enhance SRF-dependent activation of smooth muscle-specific promoters (8, 11). Although the HMG2L1 contains a highly conserved HMG motif similar to SSRP1 and HMGI(Y) proteins, our data demonstrated that the physical and functional interactions of HMG2L1 with myocardin are highly specific. In addition, although HMGI/Y directly binds to SRF, we found HMG2L1 neither directly binds to SRF (Fig. 5B) nor directly affects SRF-induced activation of a smooth muscle-specific promoter (supplemental Fig. 3). Moreover, HMG2L1 does not affect the activation of smooth muscle-specific genes by other transcription factor such as TEFβ (supplemental Fig. 3). These data suggest that it is unlikely that HMG2L1 attenuates expression of smooth muscle-specific genes through global changes in the physical structure of these genes. Rather, our data suggest that HMG2L1 functions to block the binding of SRF and myocardin to attenuate expression of smooth muscle-specific genes. Although the molecular mechanisms underlying the specificity for HMG2L1 remains elusive, our data indicate that the inhibitory effects and high affinity binding to myocardin requires the HMG domain and a region amino-terminal of this domain. As shown in Fig. 6, the HMG box of HMG2L1 is required but not sufficient to convey the HMG2L1 inhibitory effects on myocardin-mediated activation of the smooth muscle gene. As this additional amino-terminal region is not highly conserved among HMG family members, this could account for the specificity of HMG2L1 action on myocardin.
In this study, we show that HMG2L1 can suppress smooth muscle-specific gene expression by preventing myocardin from binding to SRF and attenuating myocardin-SRF complex binding to the promoters of smooth muscle-specific genes (Fig. 7). Our data revealed that HMG2L1 inhibition of myocardin activation on the smooth muscle gene promoter likely is independent of HDAC or P300 recruitment, as neither treatment with an HDAC inhibitor nor exogenous expression of P300 were able to rescue HMG2L1-mediated inhibition of myocardin activity (supplemental Figs. 5 and 6). We also found that HMG2L1 is able to attenuate the activity of a dimerization-defective mutant of myocardin (supplemental Fig. 2), suggesting that inhibition of myocardin dimerization is not a likely inhibitory mechanism. In contrast to previous studies, which showed that KLF4 inhibits smooth muscle differentiation in part by diminishing myocardin expression (23), overexpression or silencing of HMG2L1 did not affect myocardin expression (data not shown). Rather, by coimmunoprecipitation and mammalian two-hybrid assays, we found HMG2L1 can disrupt myocardin binding to SRF (Fig. 7). This can be explained simply by the competitive binding of HMG2L1 and SRF to myocardin, as HMG2L1 and SRF share an overlapping binding site in the N-terminal region of myocardin, which includes basic and glutamine rich domain (Fig. 5C), although we cannot totally rule out other unknown mechanisms. Although HMG2L1 N-terminal fragment (1–236) is able to bind to myocardin with strong affinity, this fragment failed to inhibit myocardin-mediated transactivation (Fig. 6). We speculate that this fragment may interact with the amino-terminal domain of myocardin, whereas the fragment between amino acids 237 and 472 interact with myocardin basic and poly Q domains, which are required for SRF interaction to allow myocardin to activate the smooth muscle gene. As myocardin function is strictly dependent on SRF interaction, binding to these SRF interaction domains of myocardin by HMG2L1 will disrupt the myocardin/SRF interaction, thereby resulting in abolishing myocardin function.
HMG2L1 is an HMG box protein identified initially as a negative regulator of Wnt/β-catenin signaling in Xenopus (12). The functions of HMG2L1 are only beginning to be elucidated, however. Previous studies have demonstrated that HMG proteins HMGI/Y and SOX18 were up-regulated in proliferating vascular SMCs in vitro and in vivo (9, 10). Antisense SOX18 inhibited DNA synthesis and vascular cell growth (10). Furthermore, antisense SOX18 also significantly reduced vascular SMC regrowth after injury in an in vitro model of wound repair (10). These data suggest that SOX18 plays an important role in promoting smooth muscle proliferation following vascular injury. Similarly, we found that HMG2L1 expression is up-regulated in apoE-null mice, which have elevated smooth muscle proliferation. Conversely, HMG2L1 is down-regulated during smooth muscle differentiation in vitro (Fig. 1, A and B). Together, these data suggest that HMG family members may coordinate the complex cellular changes that occur following vascular injury resulting in dedifferentiation and proliferation of vascular SMCs. Although we have demonstrated a role for HMG2L1 in attenuating expression of smooth muscle differentiation genes, this does not preclude possible additional roles in regulating SMC proliferation. Previous studies have shown HMGB1 regulates P53-mediated transcription (27) and HMG2L1 is a potential P53 binding protein (28). Given that P53 is a critical transcription factor involved in cell cycle arrest and activation of apoptotic program (29), it will be very important to assess whether HMG2L1 is also involved in apoptosis or cell cycle regulation in SMCs. Furthermore, the evolutionary conservation from plants to humans of these HMG box-containing proteins suggests a role in fundamental cellular processes (3). Single nucleotide polymorphisms have recently been identified in the HMG2L1 gene that is linked in both bipolar affective disorder and schizophrenia, suggesting that HMG2L1 also plays important roles in neurons (30). In summary, our data demonstrate that HMG2L1 can repress differentiation of vascular smooth muscle cells by interfering with the binding of myocardin to SRF. In future studies, it will be exciting to determine the role of HMG2L1 in smooth muscle and neuron development and their related diseases in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Paul Herring for critical reading the manuscript. We also thank Drs. Eric Olson, Gary Owens, and Michael Parmacek for reagents.
This work was supported by funding of Albany Medical College start-up and a Scientist Development Grant from American Heart Association (to J. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7.
- SRF
- serum response factor
- SMC
- smooth muscle cell
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- RFP
- red fluorescence protein
- qRT
- quantitative real-time
- siRNA
- small interfering RNA
- ChIP
- chromatin immunoprecipitation
- HMG
- high mobility group
- Luc
- luciferase
- SM MHC
- smooth muscle myosin heavy chain
- HDAC
- histone deacytelase.
REFERENCES
- 1.Miano J. M. (2003) J. Mol. Cell Cardiol. 35, 577–593 [DOI] [PubMed] [Google Scholar]
- 2.Pipes G. C., Creemers E. E., Olson E. N. (2006) Genes Dev. 20, 1545–1556 [DOI] [PubMed] [Google Scholar]
- 3.Thomas J. O., Travers A. A. (2001) Trends Biochem. Sci. 26, 167–174 [DOI] [PubMed] [Google Scholar]
- 4.Hong C. S., Saint-Jeannet J. P. (2005) Semin Cell Dev. Biol. 16, 694–703 [DOI] [PubMed] [Google Scholar]
- 5.Wegner M., Stolt C. C. (2005) Trends Neurosci. 28, 583–588 [DOI] [PubMed] [Google Scholar]
- 6.Koopman P. (2005) Trends Genet. 21, 367–370 [DOI] [PubMed] [Google Scholar]
- 7.Bustin M. (2001) Trends Biochem. Sci. 26, 152–153 [DOI] [PubMed] [Google Scholar]
- 8.Chin M. T., Pellacani A., Wang H., Lin S. S., Jain M. K., Perrella M. A., Lee M. E. (1998) J. Biol. Chem. 273, 9755–9760 [DOI] [PubMed] [Google Scholar]
- 9.Chin M. T., Pellacani A., Hsieh C. M., Lin S. S., Jain M. K., Patel A., Huggins G. S., Reeves R., Perrella M. A., Lee M. E. (1999) J. Mol. Cell Cardiol. 31, 2199–2205 [DOI] [PubMed] [Google Scholar]
- 10.García-Ramírez M., Martínez-González J., Juan-Babot J. O., Rodríguez C., Badimon L. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 2398–2403 [DOI] [PubMed] [Google Scholar]
- 11.Spencer J. A., Baron M. H., Olson E. N. (1999) J. Biol. Chem. 274, 15686–15693 [DOI] [PubMed] [Google Scholar]
- 12.Yamada M., Ohkawara B., Ichimura N., Hyodo-Miura J., Urushiyama S., Shirakabe K., Shibuya H. (2003) Genes Cells 8, 677–684 [DOI] [PubMed] [Google Scholar]
- 13.Zhou J., Blue E. K., Hu G., Herring B. P. (2008) J. Biol. Chem. 283, 35383–35392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J., Herring B. P. (2005) J. Biol. Chem. 280, 10861–10869 [DOI] [PubMed] [Google Scholar]
- 15.Zhou J., Hoggatt A. M., Herring B. P. (2004) J. Biol. Chem. 279, 15929–15937 [DOI] [PubMed] [Google Scholar]
- 16.Yin F., Herring B. P. (2005) J. Biol. Chem. 280, 4745–4752 [DOI] [PubMed] [Google Scholar]
- 17.Zhang M., Fang H., Zhou J., Herring B. P. (2007) J. Biol. Chem. 282, 25708–25716 [DOI] [PubMed] [Google Scholar]
- 18.Zhou J., Hu G., Herring B. P. (2005) Mol. Cell. Biol. 25, 9874–9885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J., Zhang M., Fang H., El-Mounayri O., Rodenberg J. M., Imbalzano A. N., Herring B. P. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao D., Wang Z., Zhang C. L., Oh J., Xing W., Li S., Richardson J. A., Wang D. Z., Olson E. N. (2005) Mol. Cell. Biol. 25, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelletier G., Stefanovsky V. Y., Faubladier M., Hirschler-Laszkiewicz I., Savard J., Rothblum L. I., Côté J., Moss T. (2000) Mol. Cell 6, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 22.Doi H., Iso T., Yamazaki M., Akiyama H., Kanai H., Sato H., Kawai-Kowase K., Tanaka T., Maeno T., Okamoto E., Arai M., Kedes L., Kurabayashi M. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 2328–2334 [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., Sinha S., McDonald O. G., Shang Y., Hoofnagle M. H., Owens G. K. (2005) J. Biol. Chem. 280, 9719–9727 [DOI] [PubMed] [Google Scholar]
- 24.Aidinis V., Bonaldi T., Beltrame M., Santagata S., Bianchi M. E., Spanopoulou E. (1999) Mol. Cell. Biol. 19, 6532–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zwilling S., König H., Wirth T. (1995) EMBO J. 14, 1198–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zappavigna V., Falciola L., Helmer-Citterich M., Mavilio F., Bianchi M. E. (1996) EMBO J. 15, 4981–4991 [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee S., Kundu T. K. (2003) Nucleic Acids Res. 31, 3236–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nery F. C., Rui E., Kuniyoshi T. M., Kobarg J. (2006) Biochem. Biophys. Res. Commun. 341, 847–855 [DOI] [PubMed] [Google Scholar]
- 29.Oren M. (1997) Cell 90, 829–832 [DOI] [PubMed] [Google Scholar]
- 30.Potash J. B., Buervenich S., Cox N. J., Zandi P. P., Akula N., Steele J., Rathe J. A., Avramopoulos D., Detera-Wadleigh S. D., Gershon E. S., DePaulo J. R., Jr., Feinberg A. P., McMahon F. J. (2008) Am. J. Med. Genet B. Neuropsychiatr. Genet. 147B, 59–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.