Abstract
Mutations in the amyloid β-protein (Aβ) precursor gene cause autosomal dominant Alzheimer disease in a number of kindreds. In two such kindreds, the English and the Tottori, the mutations produce amyloid β-proteins containing amino acid substitutions, H6R and D7N, respectively, at the peptide N terminus. To elucidate the structural and biological effects of the mutations, we began by examining monomer conformational dynamics and oligomerization. Relative to their wild type homologues, and in both the Aβ40 and Aβ42 systems, the English and Tottori substitutions accelerated the kinetics of secondary structure change from statistical coil → α/β → β and produced oligomer size distributions skewed to higher order. This skewing was reflected in increases in average oligomer size, as measured using electron microscopy and atomic force microscopy. Stabilization of peptide oligomers using in situ chemical cross-linking allowed detailed study of their properties. Each substitution produced an oligomer that displayed substantial β-strand (H6R) or α/β (D7N) structure, in contrast to the predominately statistical coil structure of wild type Aβ oligomers. Mutant oligomers functioned as fibril seeds, and with efficiencies significantly higher than those of their wild type homologues. Importantly, the mutant forms of both native and chemically stabilized oligomers were significantly more toxic in assays of cell physiology and death. The results show that the English and Tottori mutations alter Aβ assembly at its earliest stages, monomer folding and oligomerization, and produce oligomers that are more toxic to cultured neuronal cells than are wild type oligomers.
Keywords: Amyloid, Neurodegeneration, Protein Assembly, Protein Folding, Protein Self-assembly
Introduction
Alzheimer disease (AD)2 is characterized by the accumulation of intraneuronal filaments formed by the microtubule-associated protein Tau and by extracellular parenchymal and vascular amyloid deposits largely comprising the amyloid β-protein (Aβ) (1). Aβ is produced by sequential proteolytic cleavage of the amyloid β-protein precursor (APP) by β- and γ-secretase (2). In some kindreds, AD occurs in an autosomal dominant manner because of mutations in the genes encoding APP or γ-secretase (3). These mutations alter the absolute or relative amounts of Aβ40 or Aβ42 that are produced or they alter peptide primary structure (4–6).
The first intra-Aβ missense mutations that were observed all clustered within the APP gene region encoding amino acids Ala21–Asp23 of Aβ. These mutations included the Flemish (A21G), Dutch (E22Q), Italian (E22K), Arctic (E22G), and Iowa (D23N) (7–11). Each of these mutations alters peptide assembly or metabolism. For example, the Flemish mutation causes a decrease in the fibril extension rate (12). The Dutch, Italian, and Iowa mutations cause disease with fulminant vascular pathology (8, 9, 11). The Arctic mutation causes early onset AD that involves enhanced protofibril formation (13). Recently, a new mutation in the Ala21–Asp23 region, causing a deletion of Glu22 (ΔE22), was reported (14). This deletion causes enhanced peptide oligomerization but no fibrillization.
The only other region of Aβ in which mutations cause amino acid substitutions is the N terminus. Two such mutations, the English (H6R) (15) and Tottori (D7N) (16), cause early onset familial AD. The probands in both of the families were diagnosed clinically as “probable AD.” The proband in the English family was diagnosed at 55 years of age (15). The ages of onset of two affected sisters in the Tottori kindred were 60 and 65 years (16). Initial studies of the effects of the English and Tottori mutations revealed that they do not affect Aβ production, but that they accelerate fibril assembly in the absence of increased protofibril formation (17).
A prominent hypothesis of AD causation posits that Aβ oligomers are the proximate effectors of neurodegeneration (6, 18). Aβ oligomers inhibit hippocampal long term potentiation in mice and rats injected intracerebrally with Aβ oligomers (19–22). The inhibitory effect can be blocked by post hoc injection of Aβ-specific antibodies (23). Townsend et al. (22) found that Aβ trimers fully inhibit long term potentiation, whereas dimers and tetramers have an intermediate potency. Dimers and trimers from the conditioned medium of APP-expressing Chinese hamster ovary cells have been found to cause progressive loss of synapses in organotypic rat hippocampal slices (24). Aβ oligomers extracted from AD brains disrupt synapse structure and function (25). Dimers were the smallest active agents in this latter study.
Most recently, formal structure-cytotoxicity studies of pure Aβ40 oligomer populations produced the first determinations of oligomer specific activity (26). Dimers, trimers, and tetramers all were significantly more toxic than monomers. Importantly, a non-linear dependence of cytotoxicity on oligomer order was observed. Oligomers of higher order were disproportionately more toxic (tetramers ≫ trimers ≫ dimers ≫ monomers). These relationships also were observed in assays of fibril nucleation activity (26).
Here, we sought to elucidate the effects of the English and Tottori mutations on Aβ monomer conformation, oligomerization, oligomer structure, fibril nucleation activity, and cytotoxicity. Our results show that the English and Tottori mutations produce Aβ peptides displaying accelerated statistical coil → α/β → β secondary structure transitions and an increased propensity to form relatively large oligomers. These oligomers are more structured than those of wild type Aβ, are more efficient nucleators of fibril formation, and are significantly more cytotoxic.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Chemicals were obtained from Sigma or Fisher Scientific and were of the highest purity available. Water was produced using a Milli-Q system (Millipore Corp., Bedford, MA).
Peptides
Aβ40 and Aβ42 peptides, and analogues containing the H6R or D7N amino acid substitutions, were synthesized, purified, and characterized essentially as described (27). Briefly, synthesis was performed on an automated peptide synthesizer (model 433A, Applied Biosystems, Foster City, CA) using 9-fluorenylmethoxycarbonyl (Fmoc)-based methods on pre-loaded Wang resins. Peptides were purified using reverse phase-high performance liquid chromatography. Quantitative amino acid analysis and mass spectrometry yielded the expected compositions and molecular weights, respectively, for each peptide. Purified peptides were stored as lyophilizates at −20 °C.
To prepare peptides for study, Aβ peptide lyophilizates were dissolved at a nominal concentration of 25 μm in 10% (v/v) 60 mm NaOH and 90% (v/v) 10 mm phosphate buffer, pH 7.4. After sonication for 1 min in a bath sonicator (model 1510R-DTH, Branson Ultrasonics, Danbury, CT), the peptide solution was centrifuged for 10 min at 16,000 × g.
Photo-Induced Cross-linking of Unmodified Proteins (PICUP) and SDS-PAGE
Cross-linking of Aβ was performed essentially as described (28, 29). Briefly, for Aβ40, the general method was to mix 18 μl of peptide, at a nominal concentration of 25 μm, with 1 μl of 20 mm tris(2,2′-bipyridyl)dichlororuthenium(II) hexahydrate (Ru(bpy)) and 1 μl of 1 mm ammonium persulfate, both dissolved in 10 mm phosphate, pH 7.4. For Aβ42 cross-linking, Ru(bpy) and ammonium persulfate were used at concentrations of 40 and 2 mm, respectively. For some experiments in which higher concentrations of peptides were needed, the molar ratios of Ru(bpy) and ammonium persulfate were adjusted proportionately to the Aβ concentration so that the stoichiometry remained constant. The mixtures then were irradiated for 1 s with visible light and the reaction was quenched with 1 μl of 1 m dithiothreitol (Fisher Scientific) in water. To determine the oligomer size frequency distribution, 18 μl of each cross-linked sample was electrophoresed on a 1.0-mm thick, 10–20% Tris-Tricine gradient gel (Invitrogen). Following electrophoresis, the gels were visualized by silver staining (SilverXpress, Invitrogen). Densitometry was then performed and One-Dscan software (version 2.2.2; BD Biosciences) was used to determine peak areas of baseline-corrected data. Un-cross-linked samples were used as controls in each experiment.
Size Exclusion Chromatography
PICUP reagents were removed from cross-linked samples by size exclusion chromatography. To do so, 1.5-cm diameter cylindrical columns were packed manually with 2 g of Bio-Gel P2 Fine (Bio-Rad), which produced a 6-ml column volume. The column was first washed twice with 25 ml of 50 mm NH4HCO3, pH 8.5. Two-hundred sixteen μl of 50–100 μm cross-linked sample was then loaded. The column was eluted with the same buffer at a flow rate of ≈0.15 ml/min. The first 1 ml of eluate was collected. The fractionation range of the Bio-Gel P2 column is 100–1800 Da. Aβ peptides thus eluted in the void volume, whereas Ru(bpy) (Mr = 748.6), ammonium persulfate (Mr = 228.2), and dithiothreitol (Mr = 154.2) enter the column matrix and are separated from Aβ.
Experiments on Aβ40 and Aβ42 confirmed that the 1-ml void volume fraction contained Aβ oligomers (supplemental Fig. S1). This was shown by examining 18-μl aliquots of the void volume fraction and the subsequent 1-ml fraction using SDS-PAGE and silver staining on 1.0-mm thick, 10–20% Tris-Tricine gradient gels (Invitrogen). Fractions were lyophilized immediately after collection. Reconstitution of the lyophilizates to a nominal concentration of 50 μm in 10 mm PBS, pH 7.4, followed by a repetition of the SDS-PAGE analysis, showed that reagent removal, lyophilization, and reconstitution did not alter the oligomer composition of any of the peptide populations under study (supplemental Fig. S2).
Characterization of Cross-linked Aβ Oligomers
In cytotoxicity experiments, which require relatively long incubation times, it is important to determine whether peptide assembly occurs. Such assembly would complicate the determination of structure-activity correlations. For this reason, we evaluated the assembly state of each oligomer population by incubating the oligomers in 10 mm PBS, pH 7.4, at 37 °C and monitoring fluorescence intensity periodically using ThT. All the peptides, except un-cross-linked Aβ40 and Aβ42, displayed low initial fluorescence intensities that did not change significantly during 2 days of incubation (supplemental Fig. S3). In contrast, the un-cross-linked peptides each exhibited large time-dependent increases in fluorescence intensity (supplemental Fig. S3) that indicated fibril-assembly dependent β-sheet formation (30). The cross-linked peptides thus were stable, as defined by a lack of de novo β-sheet formation.
To address the stability question in the opposite manner, namely whether cross-linked oligomers dissociate over time, we incubated the oligomers in 10 mm PBS, pH 7.4, at 37 °C for 2 days and then analyzed the oligomer distributions by SDS-PAGE, silver staining, and densitometry (supplemental Fig. S4). Immediately after preparation, oligomers comprised ≈84–86% of the species present in each Aβ40 population (Aβ40, 83.5%; H6R, 85.4%; D7N, 84.7%). This range was identical when determined after 2 days of incubation (Aβ40, 84.0%; H6R, 86.4%; D7N, 85.0%). For Aβ42, oligomers comprised ≈72–77% (Aβ42, 71.6%; H6R, 76.6%; D7N, 76.9%) of the populations prior to incubation. Following incubation, this range was 73–77% (Aβ42, 72.8%; H6R, 75.0%; D7N, 77.3%). These data show, within experimental error, that insignificant oligomer dissociation occurs over a 2-day period with any of the six peptides.
Circular Dichroism Spectroscopy (CD)
CD measurements were made by placing 200 μl of sample into a 1-mm path-length CD cuvette (Hellma, Forest Hills, NY) and acquiring spectra using a J-810 spectropolarimeter (JASCO, Tokyo, Japan). The CD cuvettes were maintained on ice prior to introduction into the spectrometer. Following temperature equilibration (≈20 min), spectra were recorded at 22 °C from ≈190 to 260 nm at 0.2-nm resolution with a scan rate of 100 nm/min. Ten scans were acquired and averaged for each sample. Raw data were manipulated by smoothing and subtraction of buffer spectra according to the manufacturer's instructions. The half-time (t½) for the assembly dependent conformational change was determined numerically from the formula: ([θ]t − [θ]min)/([θ]max − [θ]min) = 1/2, where molar ellipticity [θ] always is measured at 198 nm, [θ]t is molar ellipticity at time t, [θ]max is maximal ellipticity, and [θ]min is minimum ellipticity.
Nucleation Activity of Cross-linked Aβ Oligomers
Aβ peptides were dissolved in 10 mm PBS, pH 7.4, to produce final concentrations of 25 μm. After sonication for 1 min using a bath sonicator, the peptide solutions were centrifuged for 10 min at 16,000 × g. Cross-linked Aβ40, Aβ42, of their H6R and D7N analogues, were also prepared at a concentration of 25 μm in 10 mm PBS, pH 7.4. The cross-linked peptides then were added to the un-cross-linked peptides at a 10% (v/v) ratio. The mixtures were incubated at 37 °C for 0–6 days without agitation.
Thioflavin T (ThT) Fluorescence
Ten μl of each peptide sample was added to 190 μl of 20 μm ThT dissolved in 10 mm phosphate buffer, pH 7.4, and then the mixture was vortexed briefly. Fluorescence was determined using an Hitachi F-4500 fluorometer. Excitation and emission wavelength/slit width were 450/5 and 482/10 nm, respectively. The fluorescence intensity was measured three times in succession at time intervals of 10 s and then the three readings were averaged and the average intensity of a ThT blank was subtracted.
Electron Microscopy (EM)
A 10-μl aliquot of each sample was spotted onto glow-discharged, carbon-coated Formvar grids (Electron Microscopy Sciences, Hatfield, PA) and incubated for 20 min. The droplet then was displaced with an equal volume of 2.5% (v/v) glutaraldehyde in water and incubated for an additional 5 min. Finally, the peptide was stained with 8 μl of 1% (v/v) filtered (0.2 μm) uranyl acetate in water (Electron Microscopy Sciences, Hatfield, PA). This solution was wicked off and then the grid was air-dried. Samples were examined using a JEOL CX100 transmission electron microscopy. To determine the distribution of assembly sizes, each electron micrograph was divided into quarters. Two horizontal lines were drawn within each quarter so that each quarter was trisected. The diameters of globular or thread-like structures touching each of these lines were then measured using a ×10 magnifier eyepiece containing a graticule (Electron Microscopy Sciences).
Atomic Force Microscopy (AFM)
Peptide solutions were characterized using a Nanoscope V Dimension 5000 scanning probe microscope (Veeco Digital Instruments, Santa Barbara, CA). All measurements were carried out in the tapping mode under ambient conditions using single-beam silicon cantilever probes. A 10-μl aliquot of each lyophilized peptide, reconstituted to a concentration of 25 μm in 10 mm PBS, pH 7.4, was spotted onto freshly cleaved mica (Ted Pella, Inc., Redding, CA), incubated at room temperature for 5 min, rinsed with water, and then blown dry with air. The sample then was examined and the distribution of assembly sizes was determined as described under “Elecron microscopy (EM)”.
Cell Culture
Rat pheochromocytoma PC12 cells were cultured in 75-cm2 flasks (number 430641, Corning Inc., Corning, NY) at 37 °C in an atmosphere of 5% (v/v) CO2 in air. The medium was F-12K (ATCC) containing 15% (v/v) horse serum, 2.5% (v/v) fetal bovine serum, 100 units/ml of penicillin, 0.1 mg/ml of streptomycin, and 25 μg/ml of amphotericin B. To prepare cells for assay, the medium was removed and the cells were gently washed once with F-12K medium, containing 0.5% (v/v) fetal bovine serum, 100 units/ml of penicillin, 0.1 mg/ml of streptomycin, and 25 μg/ml of amphotericin B. A cell suspension then was prepared by addition of this latter medium, containing 100 μg/ml of nerve growth factor (Invitrogen), to the flasks, followed by agitation. Cell concentration was determined using trypan blue staining, after which cells were plated at a density of 30,000 cells/well (90 μl total volume/well) in 96-well assay plates (Costar number 3610, Corning Inc.). The nerve growth factor-induced differentiation of the cells was allowed to proceed for 48 h, at which time toxicity assays were done.
MTT Assay
Peptide samples were prepared at concentrations ranging from 2.5 to 500 μm, in F-12K medium containing 0.5% (v/v) fetal bovine serum, 100 units/ml of penicillin, 0.1 mg/ml of streptomycin, and 25 μg/ml of amphotericin B. Aliquots of 10 μl were added to differentiated PC12 cells to yield final Aβ concentrations of 0.25, 0.5, 1, 5, 10, 25, and 50 μm. The cells then were incubated for 24 h at 37 °C in a 5% (v/v) CO2 incubator. Fifteen μl of Dye Solution (Promega, Madison, WI) was added to each well of the microtiter plate and then the plate was incubated for an additional 3.5 h. The cells then were lysed by the addition of 100 μl of solubilization solution (Promega, Madison, WI) followed by overnight incubation. MTT signal was determined by measuring absorption at 570 nm (corrected for background absorbance at 630 nm) using a BioTek Synergy HT microplate reader (Bio-Tek Instruments, Winooski, VT). Six replicates were done for each treatment group and the data from three independent experiments were combined and reported as mean ± S.E. Percent toxicity was determined according to the formula: T = ((AAβ − Amedium/(Astaurosporine − Amedium)) × 100, where AAβ, Amedium, and Astaurosporine were absorbance values from Aβ-containing samples, medium alone, or staurosporine alone, respectively. Effective concentration (EC50) was defined as the concentration of each Aβ assembly that produced T = 50%. EC50 was calculated by sigmoidal curve fitting, using GraphPad Prism software (version 4.0a, GraphPad Software, Inc., San Diego, CA).
LDH Assay
LDH activity was determined using the CytoTox-ONE Homogeneous Membrane Integrity assay (Promega, Madison, WI). To do so, peptide solutions of 25 μm concentration were prepared as described for MTT assays and then incubated with nerve growth factor-differentiated PC12 cells for 48 h. One-hundred μl of LDH reagent was then added to each well and the plate was incubated at room temperature on the benchtop in the dark without agitation for 10 min, after which 50 μl of Stop Solution was added and the fluorescence was measured using a BioTek Synergy HT microplate reader set to an excitation wavelength of 560 nm and an emission wavelength of 590 nm. Six replicates were done for each treatment group and the data from three independent experiments were combined and reported as mean ± S.E. Percent toxicity was calculated according to the formula used in the MTT assays, except that the term Astaurosporine was replaced with Alysis.
Statistical Analysis
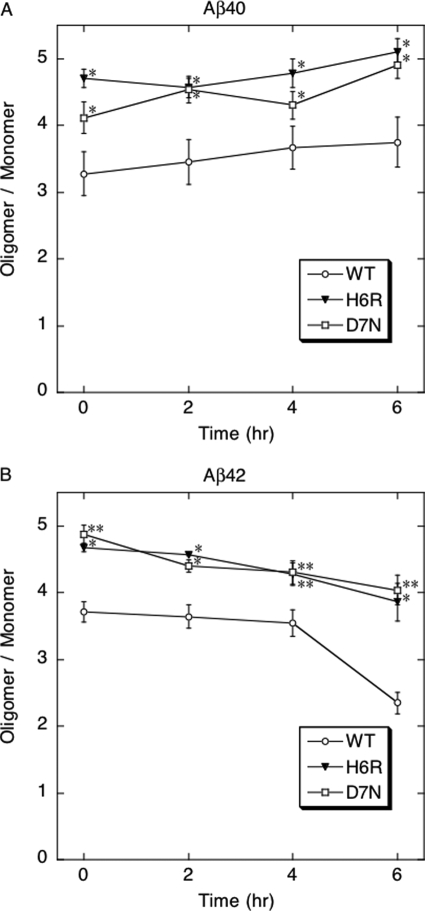
One-way factorial analysis of variance followed by Bonferroni post hoc comparisons were used to determine statistical significance among data sets. For calculation of significance among oligomer/monomer ratios (Fig. 3), paired t tests were done. These tests were implemented within GraphPad Prism software (version 4.0a, GraphPad Software, Inc.). Significance was defined as p < 0.05.
FIGURE 3.
Densitometric analysis of Aβ oligomerization. To produce intensity profiles and calculate the relative amounts of each oligomer type of Aβ40 (A) or Aβ42 (B), One-Dscan software (version 2.2.2; BD Biosciences Bioimaging, Rockville, MD) was used. The metric of oligomer/monomer ratio = (total lane intensity-monomer intensity)/monomer intensity of wild type (WT) (○), H6R (▾), or D7N (□) is expressed as mean ratio ± S.E. Statistical significance of oligomer/monomer differences between each mutant peptide and wild type peptide is indicated by: *, p < 0.05; or **, p < 0.01. Each figure comprises data obtained in three independent experiments.
RESULTS
Secondary Structure Dynamics of Wild Type and Mutant Aβ Peptides
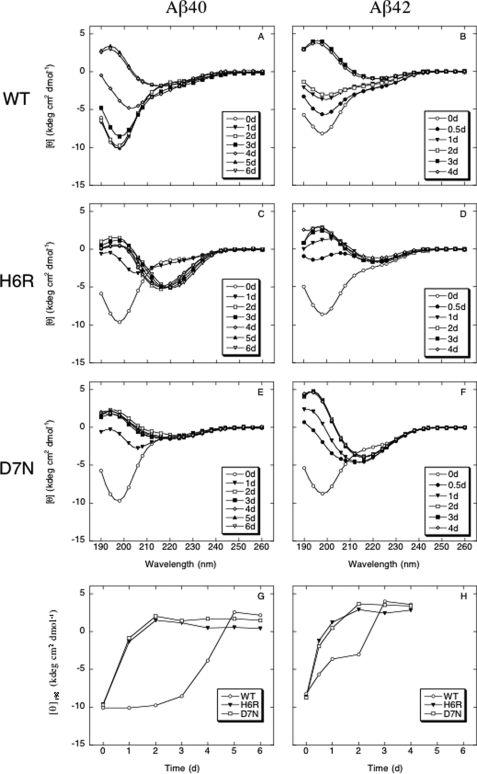
To determine and compare the secondary structure dynamics of wild type Aβ isoforms and N-terminal mutants3 thereof, we used CD to monitor peptide folding and assembly systems during 6 days of incubation at 37 °C. All six peptides initially produced spectra consistent with primarily statistical coil (SC) secondary structure (Fig. 1, A–F). The major feature of these spectra was a large magnitude minimum centered at ≈198 nm. To allow comparisons of the kinetics among the six peptides, the half-time (t½) for the entire transition process was determined using [θ]198, which correlates with SC, as a metric (see “Experimental Procedures”).
FIGURE 1.
Aβ secondary structure dynamics. 25 μm Aβ40 (A, C, and E) or Aβ42 (B, D, and F) of wild type (WT) (A and B), H6R (C and D), or D7N (E and F) were incubated at 37 °C for 7 days in 10 mm phosphate, pH 7.4. Spectra were acquired immediately at the start of the incubation period, day 0 (○), and after days 0.5 (●), 1 (▾), 2 (□), 3 (■), 4 (◇), 5 (▴), and 6 (▿). The spectra presented at each time are representative of those obtained during each of three independent experiments. G and H, molar ellipticity [θ]198 versus time for Aβ40 (G) or Aβ42 (H).
Aβ40 displayed substantial secondary structure changes between days 3 and 5 that were consistent with previously reported SC → α/β → β transitions associated with monomer → protofibril → fibril assembly (31) (Fig. 1A). For Aβ40, t½ ≈ 4 days (Fig. 1G). The English (H6R) and Tottori (D7N) Aβ40 mutants displayed substantially accelerated kinetics, with t½ ≈ 0.6 days for each (Fig. 1, C, E, and G). Conformational changes in the mutant peptides were complete by day 2 and the spectra from days 3–6 were very similar. Visual inspection of the data shows that maximal values of d[θ]/dt were similar among the three peptides. However, no lag period was observed with the mutant peptides, compared with a lag period of ≈2 days with Aβ40 itself. Shorter observation intervals might reveal a lag time for the mutants, but consideration of the observed maximum d[θ]/dt metric in both systems and the delay intervals before the second CD measurements were made (0.5 or 1 days), suggests that this lag period would be very small (≪1 day).
The Aβ42 system displayed similar structural changes to those observed in the Aβ40 system. However, in all cases, the kinetics was accelerated even more and no lag phase was observed for any peptide (subject to the same qualification as noted above for the Aβ40 system) (Fig. 1H). Aβ42 exhibited a t½ < 2 days, approximately 1/2 that of Aβ40. The kinetics for the mutant peptides was faster, with both the English and Tottori Aβ42 peptides exhibiting t½ ≈ 0.4 days.
Peptide Oligomerization
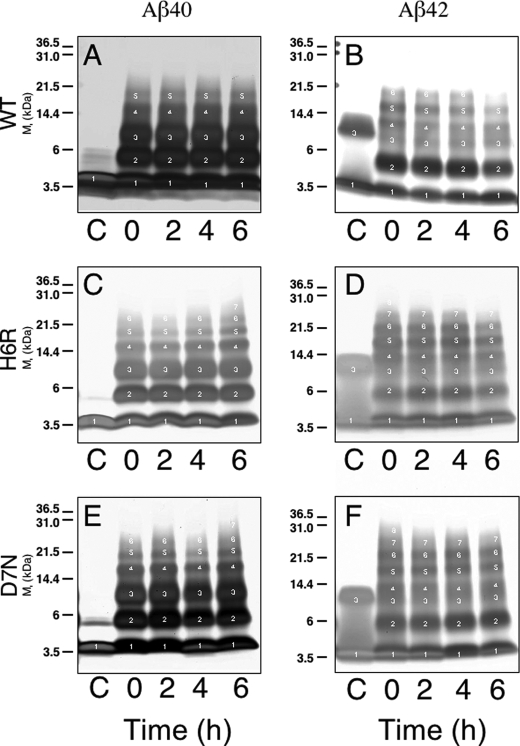
To determine whether the English or Tottori amino acid substitutions affected peptide oligomerization, we incubated the six peptides at a concentration of 25 μm in PBS, pH 7.4, at 37 °C for 0–6 h without agitation. At 0, 2, 4, and 6 h, aliquots were chemically cross-linked using the technique of PICUP (28) and the oligomer size frequency distributions were determined by SDS-PAGE, silver staining, and densitometry. In the un-cross-linked controls, Aβ40 displayed primarily monomers, although weak bands were observed with mobilities consistent with that of dimers (Fig. 2A, lane C). Similar patterns were observed for the homologous English and Tottori peptides (Fig. 2, C and E, lanes C). When the oligomers were stabilized by cross-linking, both mutant peptides displayed distributions comprising monomers through hexamers or heptamers, whereas Aβ40 displayed predominately monomers through pentamers. The distributions were similar at all time points, but a time-dependent trend toward higher orders was observed for both mutant peptides (Fig. 3A). To evaluate the statistical significance of these trends, we determined p values for the differences between wild type peptide and each of the mutant peptides at each time point of the assay. The oligomer/monomer ratios of both mutant peptides, at all time points, were significantly (p < 0.05) higher than those of the wild type peptide (Fig. 3A).
FIGURE 2.
Aβ oligomerization. PICUP, followed by SDS-PAGE and silver staining, was used to study oligomerization of 25 μm Aβ40 (A, C, and E) or Aβ42 (B, D, and F) of wild type (WT) (A and B), H6R (C and D), or D7N (E and F). Lanes C, un-cross-linked Aβ; lanes 0, Aβ cross-linked at the beginning of incubation at 37 °C; lanes 2, Aβ cross-linked after 2 h; lanes 4, Aβ cross-linked after 4 h; lanes 6, Aβ cross-linked after 6 h. Oligomer order is indicated by white numerals over the respective gel bands. The gel is representative of each of three independent experiments.
Un-cross-linked Aβ42 displayed monomers and trimers (Fig. 2B, lane C). Cross-linking resulted in a distribution comprising monomers through hexamers, with a node at pentamer/hexamer, consistent with published work (32). The un-cross-linked mutant peptides produced distributions similar to that of un-cross-linked Aβ42 (cf. Fig. 2, B, D, and F, lanes C). As in the Aβ40 system, the mutant peptides produced distributions exhibiting higher order species, including heptamers and octamers (Fig. 2, D and F, lanes 0, 2, 4, and 6), than observed in wild type Aβ42 distributions. In addition, the oligomer/monomer ratios of both mutant peptides, at all time points, were significantly (p < 0.05 or 0.01) higher than those of wild type peptide (Fig. 3B). However, in contrast to the Aβ40 system, there was a time-dependent trend toward lower orders for all peptides (Fig. 3B).
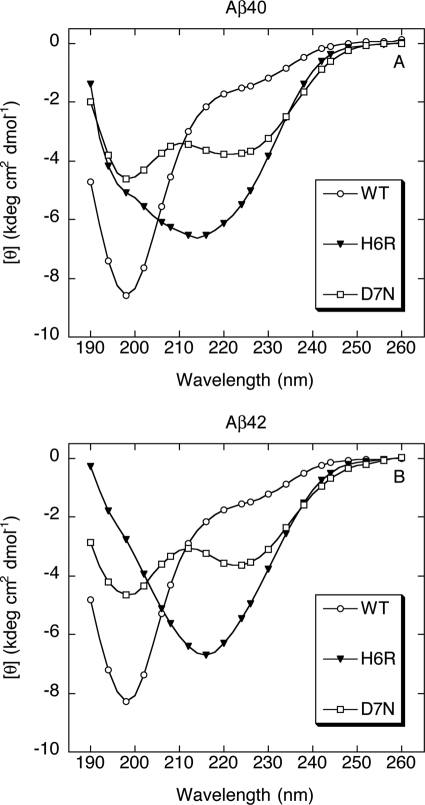
Secondary Structure of Aβ Oligomers
Aβ has been shown to exist predominately as a statistical coil if prepared under conditions designed to prevent its folding and self-assembly (33, 34). However, as shown above, peptide folding and assembly is a dynamic process involving an overall SC → α-helix → β-sheet transition. To determine the conformational states of the stable oligomers formed by the six study peptides, CD was performed following cross-linking, reagent removal, lyophilization, and resolubilization to a concentration of 25 μm in 10 mm PBS, pH 7.4 (Fig. 4). Aβ40 and Aβ42 displayed spectra consistent with the SC state. The major features of these spectra were a substantial minimum centered at ≈198 nm and an inflection point at ≈225 nm. In contrast, the major spectral feature displayed by cross-linked oligomers of the English mutant Aβ40 and Aβ42 peptides was a substantial minimum centered at ≈216 nm, indicative of β-sheet structure. The spectra of the Tottori mutant Aβ40 and Aβ42 oligomers were distinct from those of the wild type or English peptides. The Tottori spectra were “saddle-like,” in that minima at ≈198 and ≈222 nm were observed. The latter minimum is consistent with the presence of the α-helix structure.
FIGURE 4.
Secondary structure of Aβ oligomers. CD spectroscopy was performed after cross-linking of 25 μm Aβ40 (A) or Aβ42 (B) of wild type (WT) (○), H6R (▾), or D7N (□). Each figure is representative of data obtained in each of three independent experiments.
Nucleation of Aβ Fibril Assembly
Aβ fibril assembly has characteristics of a nucleation-dependent polymerization process (35–37). To study the abilities of oligomers of wild type and mutant Aβ40 and Aβ42 to nucleate fibril formation, we monitored the time dependence of ThT fluorescence in seeded fibril formation experiments (Fig. 5). ThT fluorescence does not measure fibril concentration per se (some fibrils do not possess the β-sheet structures to which ThT binds) (38), but fluorescence intensities do correlate with Aβ fibril content (30, 39, 40).
FIGURE 5.
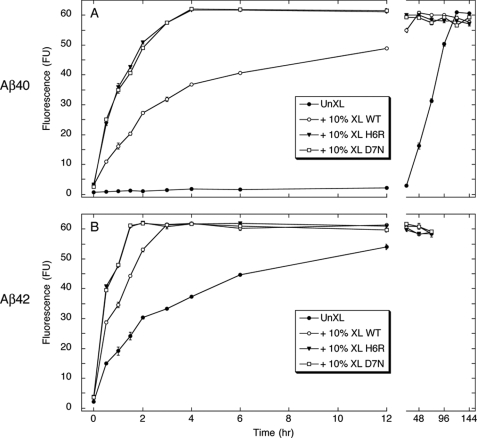
Nucleation of Aβ fibril assembly. Zero % (v/v) (●) or 10% (v/v) cross-linked (XL) WT (○), H6R (▾), or D7N (□) oligomers of Aβ40 (A) or Aβ42 (B) were added to un-cross-linked (UnXL) WT Aβ40 and Aβ42, which then were incubated for 3 or 7 days at 37 °C in 10 mm phosphate-buffered saline, pH 7.4. Aliquots were assayed periodically using ThT. Binding is expressed as mean fluorescence (in arbitrary fluorescence units (FU)) ± S.E. Each figure comprises data obtained in three independent experiments.
Un-seeded Aβ40 displayed a quasi-sigmoidal process curve characterized by an ≈24-h lag time, an ≈96-h period of increasing ThT binding, and a binding plateau occurring after ≈120 h (Fig. 5A). Results were consistent with a nucleation-dependent polymerization process. The unseeded reaction displayed no initial fluorescence increase, within experimental error. Adding 10% (w/w) cross-linked Aβ40 oligomers eliminated the lag period and produced a quasi-hyperbolic increase in fluorescence that reached maximal levels at ≈48 h. Stable wild type oligomers thus functioned as fibril nuclei. Nucleation activity was displayed by the H6R and D7N oligomers as well. In these cases, the initial rates of increase in ThT signal were the same (≈50 fluorescence units/h), within experimental error, and these rates were significantly (p < 0.001) greater than that produced by wild type seeds (≈22 fluorescence units/h). Maximal fluorescence was observed by ≈4 h using mutant seeds, compared with 48 h using wild type seeds, a 12-fold rate increase.
Similar results were obtained in the Aβ42 system (Fig. 5B). The relative initial rates of increase in ThT fluorescence followed the same rank order, namely H6R = D7N (≈80 fluorescence units/h) > wild type (≈58 fluorescence units/h) > un-seeded (≈30 fluorescence units/h), and the rate differences displayed between the mutant and wild type peptide seeds was significant (p < 0.001). The rates for each species were substantially higher than those observed for their respective Aβ40 analogues. Maximal ThT levels for the mutant Aβ42 oligomer-seeded reactions were reached in ≈½ the time as observed for the mutant Aβ40 oligomers (2 versus 4 h). Wild type Aβ42 oligomers produced maximal ThT levels ≈16-fold faster (3 versus 48 h) than did wild type Aβ40 oligomers, and un-seeded Aβ42 assembled ≈5-fold faster than did Aβ40 (1 versus 5 days).
Aβ Assembly Morphology
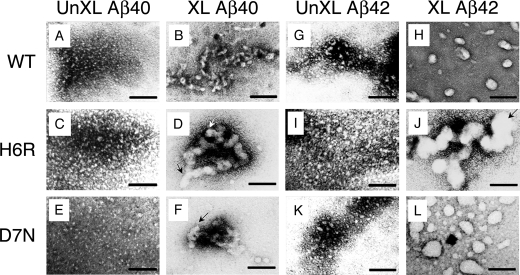
To determine the morphologies of the oligomers formed immediately following peptide dissolution, EM (Fig. 6 and Table 1) and AFM (Fig. 7 and Table 1) were performed on samples following removal of reactants, lyophilization, and resolubilization. In the Aβ40 system, un-cross-linked Aβ produced irregular, globular structures that often had thread-like components. The average diameters (d̄) of the globular components were 1.44 nm (Fig. 6A and Table 1). Un-cross-linked H6R and D7N peptides produced similar globular structures, but with d̄ = 2.34 and 2.15 nm, respectively (Fig. 6, C and E, and Table 1). Analysis of cross-linked oligomers revealed populations of much larger d̄. The Aβ40, H6R, and D7N peptides produced d̄ = 10.3, 15.8, and 13.5, and nm, respectively (Table 1). The structures were quasi-spherical (Fig. 6D, white arrow) or were more complex, including those that appeared to be composed of globular subunits attached to each other in twisted, rope-like structures (Fig. 6, D and F, black arrows).
FIGURE 6.
EM analysis of Aβ40 or Aβ42 assemblies. EM was performed on 25 μm un-cross-linked (A, C, E, G, I, and K) and cross-linked (B, D, F, H, J, and L) Aβ40 (A-F) and Aβ42 (G-L) of wild type (WT) (A, B, G, and H), H6R (C, D, I, and J), or D7N (E, F, K, and L) peptides. Scale bars are 100 nm.
TABLE 1.
Morphological analysis of Aβ assemblies
| Assembly | Diametera | Heightb |
|---|---|---|
| Un-cross-linked WT Aβ40 | 1.44 ± 0.17 (86) | 0.19 ± 0.03 (44) |
| Un-cross-linked H6R Aβ40 | 2.34 ± 0.31 (84) | 0.31 ± 0.04 (96) |
| Un-cross-linked D7N Aβ40 | 2.15 ± 0.31 (66) | 0.30 ± 0.03 (95) |
| Cross-linked WT Aβ40 | 10.30 ± 2.01 (30) | 0.89 ± 0.10 (93) |
| Cross-linked H6R Aβ40 | 15.80 ± 3.61 (24) | 1.71 ± 0.24 (86) |
| Cross-linked D7N Aβ40 | 13.52 ± 2.99 (25) | 1.37 ± 0.15 (92) |
| Un-cross-linked WT Aβ42 | 2.17 ± 0.32 (78) | 0.29 ± 0.05 (52) |
| Un-cross-linked H6R Aβ42 | 2.45 ± 0.27 (131) | 0.34 ± 0.05 (53) |
| Un-cross-linked D7N Aβ42 | 2.46 ± 0.39 (79) | 0.35 ± 0.04 (99) |
| Cross-linked WT Aβ42 | 20.49 ± 4.70 (23) | 1.36 ± 0.31 (43) |
| Cross-linked H6R Aβ42 | 30.06 ± 7.04 (24) | 2.30 ± 0.38 (54) |
| Cross-linked D7N Aβ42 | 25.94 ± 5.33 (32) | 1.72 ± 0.25 (73) |
a Mean diameter ± S.E., in nm, is listed for (n) Aβ assemblies visualized by EM.
b Mean height ± S.E., in nm, is listed for (n) Aβ assemblies visualized by AFM.
FIGURE 7.
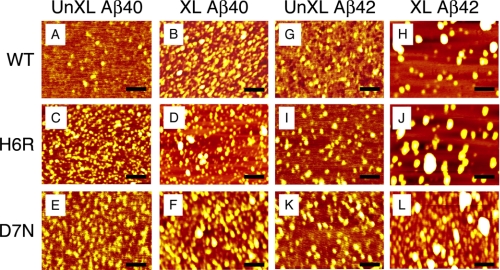
AFM analysis of Aβ40 or Aβ42 assemblies. AFM was performed on 25 μm un-cross-linked (A, C, E, G, I, and K) and cross-linked (B, D, F, H, J, and L) Aβ40 (A–F) and Aβ42 (G–L) of WT (A, B, G, and H), H6R (C, D, I, and J), or D7N (E, F, K, and L) peptides. Scale bars are 100 nm.
In the Aβ42 system, un-cross-linked Aβ42 also displayed the irregular, globular and thread-like structures seen with Aβ40, but these structures were larger (d̄ = 2.17 nm; Fig. 6G and Table 1). The structures observed in the H6R and D7N samples also were larger than those seen in the Aβ40 system (d̄ = 2.45 and 2.46 nm, respectively; Fig. 6, I and K, and Table 1). Cross-linked Aβ42 and D7N peptides displayed predominately quasi-spherical or ellipsoidal assemblies (Fig. 6, H and L). The H6R peptide produced the largest globules, and unlike Aβ42 or the D7N peptide, these globules were often associated into oligoglobular assemblies (Fig. 6J, black arrow). Relative to Aβ40, Aβ42 produced little if any thread-like structures, nor did the mutant peptides. As in the Aβ40 system, cross-linking revealed structures of size substantially larger than found in the un-cross-linked state. The average diameters of the Aβ42, H6R, and D7N oligomers were 20.5, 30.1, and 25.9 nm, respectively (Table 1).
We next studied oligomer morphology using AFM (Fig. 7 and Table 1). The rank order of heights (z axis values) was identical to that determined by EM. For un-cross-linked Aβ40, H6R, and D7N, d̄ = 0.19, 0.31, and 0.30 nm, respectively (Fig. 7, A, C, and E, and Table 1). Larger structures were observed after cross-linking, with d̄ = 0.89, 1.71, and 1.37 nm, respectively, for Aβ42, H6R, and D7N (Fig. 7, B, D, and E, and Table 1).
Qualitatively similar relationships between un-cross-linked and cross-linked samples were observed in the Aβ42 system. Average heights for un-cross-linked Aβ42, H6R, and D7N were 0.29, 0.34, and 0.35 nm, respectively (Fig. 7, G, I, and K, and Table 1), whereas average heights of the cross-linked samples were substantially larger (1.36, 2.30, and 1.72 nm, respectively; Fig. 7, H, J, and L, and Table 1).
We note that the most accurate parameter in AFM studies of proteins is the z axis value (41). For this reason, it can be misleading to consider x-y geometries and to compare these to geometries obtained by EM. Even with z axis values, careful attention must be paid to the effects of sample compression by the AFM probe, which can lead to underestimation of heights (41). Nevertheless, in prior studies of Aβ peptide structure, we have shown that results obtained by the two methods correlated at the r2 > 0.95 level (26).
Relative Biological Activities of Aβ Assemblies
To examine the relative biological activity of each type of oligomer, we assayed MTT metabolism using nerve growth factor-differentiated PC12 cells (42, 43). MTT assays are a rapid and sensitive method for determination of gross Aβ toxicity in cultures of dissociated cells (44). Our experimental goal here was not an elucidation of mechanisms of toxicity, but rather to take advantage of the sensitivity of the assay to determine the concentration dependence of Aβ-mediated toxicity (26). (Peptide-mediated cell death activity, specifically, is discussed in the next section.)
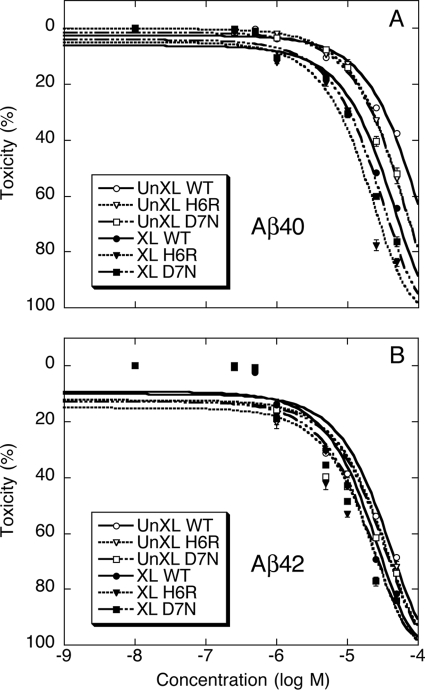
In the Aβ40 system, un-cross-linked wild type, English, and Tottori peptides displayed EC50 values of 95.9 ± 2.26, 47.3 ± 2.0, and 50.6 ± 4.68 μm (mean ± S.E.), respectively (Fig. 8A and Table 2). The difference in toxicity between the wild type and mutant peptides was significant (p < 0.01). Stable oligomers were significantly more toxic. Cross-linked wild type, H6R, and D7N peptides displayed EC50 values of 25.0 ± 0.26, 14.3 ± 0.38, and 19.1 ± 0.30 μm (mean ± S.E.), respectively (Fig. 8A and Table 2). As with the un-cross-linked samples, the mutant cross-linked peptides were significantly more toxic than the cross-linked wild type peptide (p < 0.01).
FIGURE 8.
MTT metabolism. MTT assays were performed on differentiated PC12 cells incubated for 24 h with un-cross-linked (UnXL) wild type (WT) (○), H6R (▿), D7N (□) or cross-linked (XL) WT (●), H6R (▾), D7N (■) of Aβ40 (A) and Aβ42 (B). Each figure is representative of results obtained in each of three independent experiments. Data are expressed as mean percent toxicity ± S.E.
TABLE 2.
Toxicity (EC50) of Aβ assemblies
EC50 values (μm) were determined by sigmoidal curve fitting of the data shown in Fig. 8 by using GraphPad Prism software (version 4.0a, GraphPad Software).
| Assembly | Aβ40 | Aβ42 |
|---|---|---|
| Un-cross-linked WT | 95.9 ± 2.26 | 18.0 ± 0.32 |
| Un-cross-linked H6R | 47.3 ± 2.00 | 13.8 ± 0.63 |
| Un-cross-linked D7N | 50.6 ± 4.68 | 12.4 ± 0.22 |
| Cross-linked WT | 25.0 ± 0.26 | 10.4 ± 0.65 |
| Cross-linked H6R | 14.3 ± 0.38 | 8.0 ± 0.43 |
| Cross-linked D7N | 19.1 ± 0.30 | 8.9 ± 0.18 |
In the Aβ42 system, all peptides were more toxic than their Aβ40 analogues. For the un-cross-linked peptides, this increase in toxicity varied from 3- to 5-fold (Table 2). As in the Aβ40 system, each mutant peptide was equally toxic, within experimental error, and this level of toxicity was significantly (p < 0.01) greater than that of the wild type peptide. Wild type, H6R, and D7N peptides displayed EC50 values of 18.0 ± 0.32, 13.8 ± 0.63, and 12.4 ± 0.22 μm (mean ± S.E.), respectively (Fig. 8B and Table 2).
Cross-linking the Aβ42 peptides increased their toxicity significantly (Table 2), but the relative increase (<2-fold) was less than that observed in the Aβ40 system. Wild type, H6R, and D7N displayed EC50 values of 10.4 ± 0.65, 8.0 ± 0.43, and 8.9 ± 0.18 μm (mean ± S.E.), respectively (Table 2). The mutant peptides were significantly more toxic than wild type Aβ42 (p < 0.05).
Relative Cytolytic Activities of Aβ Assemblies
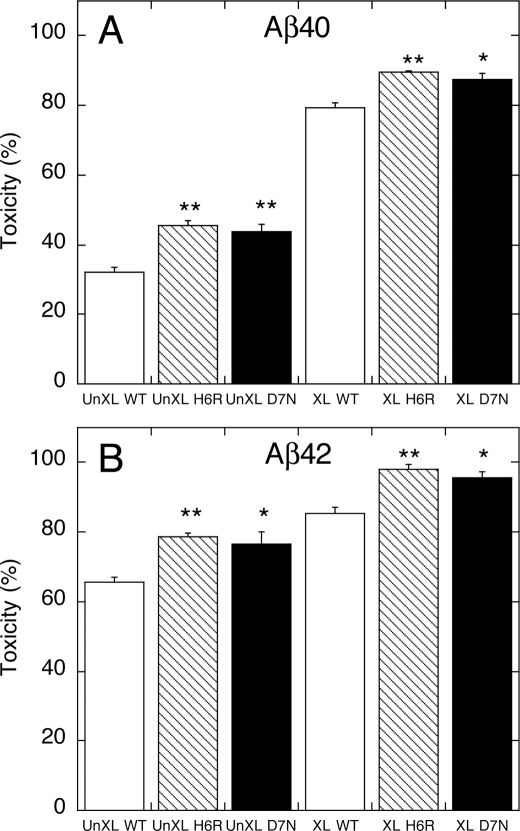
We next determined cytolytic activity by measuring LDH release from peptide-treated, nerve growth factor-differentiated PC12 cells (Fig. 9). The peptide concentration was 25 μm for all peptides. This concentration was chosen because it is within, and toward the lower end, of the effective concentration range (≈10–100 μm) determined in the MTT assays. Examination of the data from the Aβ40 and Aβ42 systems (Fig. 9, A and B) reveals that stabilized oligomers were more toxic than unstabilized oligomers (cf. the three left-most bars with the three right-most bars in each system). Furthermore, the mutant peptides were more toxic than were the wild type homologues (beginning from the left of each panel, cf. bars 2 and 3 with 1, and 5 and 6 with 4).
FIGURE 9.
LDH activity. LDH release was measured after a 48-h incubation of differentiated PC12 cells with un-cross-linked (UnXL) or cross-linked (XL) Aβ40 (A) and Aβ42 (B) of WT, H6R, or D7N at final nominal concentration of 25 μm. Each figure is representative of data obtained in each of three independent experiments. Bars are mean LDH activity ± S.E. Significance was determined using one-way fractional analysis of variance and multiple comparison tests (*, p < 0.05; **, p < 0.01).
DISCUSSION
The etiology of AD is complex, but it is clear that Aβ is a seminal pathogenetic agent (6, 18, 45). In a small percentage of AD cases, missense mutations in the Aβ coding region of the APP gene cause AD or cerebral amyloid angiopathy (3). The study of these mutations offers the possibility of elucidating a biophysical basis for disease causation. In earlier work (17), we showed that the H6R (English) and D7N (Tottori) mutations produce Aβ peptides that display accelerated fibril elongation in the absence of accelerated protofibril formation. Here, we focused primarily on the monomer and oligomer states, the latter of which is hypothesized to be the most important in AD pathobiology (6, 45, 46).
We found, relative to peptides containing the wild type Aβ N-terminal sequence, that both mutations stabilized ordered secondary structure elements within monomers, facilitated oligomerization, and produced larger oligomeric assemblies. Stable oligomers of the mutant peptides were more potent nucleators of higher-order assembly and more cytotoxic than wild type oligomers. Distinct differences in behavior between the English and Tottori mutants also were observed.
Our initial studies were of the conformational dynamics of the Aβ40 and Aβ42 systems. Examination of temporal changes in the secondary structure revealed that both mutations accelerated the rate of the SC → α-helix → β-sheet transition. The acceleration was ≈10-fold in the Aβ40 system and ≈5-fold in the Aβ42 system, as determined by the t½ for the transition. The most significant factor contributing to the acceleration of Aβ40 assembly was diminution in lag time, as the maximal observed transition rate from SC to ordered secondary structures was equivalent among the three different Aβ40 peptides. This suggests that the Aβ mutations cause substantial and immediate increases in peptide structural order4 that facilitate monomer folding and are necessary for higher order assembly, including oligomerization and fibril nucleation and elongation. The effects of the mutations in the Aβ42 system were restricted to transition rate increases because no lag phases were observed. These effects were consistent with experimental and computational studies that demonstrated that Aβ42 displays greater initial structural order, which correlates with its distinct oligomerization behavior relative to Aβ40 (32, 34, 47–49).
The predictions derived from analysis of the conformational dynamics were borne out in the subsequent examination of peptide oligomerization. In both the Aβ40 and Aβ42 systems, the mutant peptides produced oligomer distributions in which the highest observed oligomer order exceeded that of the wild type peptides. In addition, although subtle, two distinct time-dependent trends were observed in the Aβ40 and Aβ42 systems. In the former system, skewing of the oligomer distribution toward higher order was observed, whereas in the latter system, the opposite was seen. The oligomer size frequency distribution is controlled by the proximity, chemical reactivity, and chemical accessibility of amino acid side chains, predominately the phenol group of Tyr (29, 50, 51). Previous studies have shown that Aβ40 undergoes substantial conformational reorganization during its initial folding and assembly (52). This reorganization may facilitate both exposure of reactive groups and oligomerization, which would result in a time-dependent increase in higher-order oligomers. In contrast, Aβ42 exists initially in a relatively more ordered state characterized by the presence of higher-order oligomers. This state facilitates protofibril and fibril formation, producing structures in which reactive groups are less accessible. Hence, over time, the node of the Aβ42 oligomer distribution moves toward lower order.
The increase in structural order caused by the English and Totorri mutations was particularly evident from the secondary structure determination of stabilized oligomers. Neither Aβ40 nor Aβ42 exhibited a substantial content of ordered secondary structure elements (α-helix, β-strand). In contrast, in both systems, the mutant peptides produced oligomers displaying substantial β-strand character. The primary secondary structure element of the English mutant oligomers was β-strand. The Tottori oligomers possessed mixed α-helix/β-strand character. These results emphasize the rapid and substantial acquisition of order in the mutant peptides relative to their wild type homologues.
Increased order was accompanied by increased size, as determined by EM (width) and AFM (z axis value) analyses. In both systems, assemblies formed by the mutant peptides were larger than those formed by their wild type homologues. These data were consistent with our determination of the oligomer size frequency distributions, which showed skewing toward higher order with the mutant peptides. The data also are consistent with recent studies demonstrating a direct correlation between oligomer order and size (26). Examination of the stabilized oligomers in each system showed a consistent rank order of size, English > Tottori > wild type. This rank order was identical to that of β-strand content in the oligomers.
The increase in structural order per se is of fundamental interest with respect to the Aβ system. However, the effects of the mutations on the Aβ assembly process and the biological activities of the assemblies thus produced are particularly relevant to elucidation of disease mechanism(s). For this reason, we examined the relative abilities of stabilized oligomers to nucleate the assembly of ThT-positive structures. We found that Aβ40 exhibited a substantial lag period (≈24 h) that was eliminated by addition of wild type and mutant oligomers. The rank order of the rates of propagation of ThT-positive structure (dFU/dt) was H6R = D7N > wild type. In the Aβ42 system, no lag period was observed for any peptide, but the rank order of dFU/dt was identical. The mutations thus not only facilitate a structural organization of Aβ, but the structure(s) thus formed are potent nucleators of the assembly of ThT-positive Aβ structures.
We completed our studies by assessing the biological activities of the mutant peptides relative to their wild type homologues. In both MTT and LDH assay systems, and for the un-cross-linked and cross-linked states in both the Aβ40 and the Aβ42 systems, the mutant peptides were significantly more toxic than were their wild type homologues. In the MTT assays, the English mutant Aβ40 oligomers had a significantly lower EC50 value than did the Tottori oligomers. For Aβ42, the same trend existed, but the difference was not significant. LDH assays produced identical trends. The rank order of toxicity of the English and Tottori oligomers correlates with the rank orders of β-strand secondary structure and oligomer size. The English mutation thus appears to have a greater affect on peptide structure and activity than does the Tottori.
The consistency among the biophysical and biological data supports the conclusions that: 1) stabilization of the oligomer state significantly increases peptide toxicity; and 2) the English and Tottori mutations are capable of further increasing this toxicity through their effects on the kinetics of peptide assembly and the structures of the assemblies thus formed.
What types of structural effects might be caused by the English and Tottori mutations? Two classes of effects exist, local and global. By definition, the local effects include alteration in peptide chain structure at or immediately adjacent to the sites of the amino acid substitutions. It is not clear whether local folding motifs may be affected. What is clear is that oligomerization and higher order assembly processes are affected. Again, by definition, this means that intermolecular interactions must be altered. Three mechanisms of alteration may operate: 1) the same monomer fold necessary to mediate oligomerization of wild type peptides has a higher occurrence frequency (is more stable) and the same residues mediating the intermolecular interactions are involved (but do not include Arg6 or Asn7); 2) intermolecular interactions involve Arg6 or Asn7 and these interactions are more stable than those involving the wild type His6 or Asp7; or 3) a different monomer fold forms, one that has a higher propensity for self-association but which may or may not require direct intermolecular interactions involving His6 or Asn7.
A relative dearth of knowledge exists about Aβ N-terminal structural dynamics. Mechanisms other than those proposed above are possible. Recent structure determinations of co-crystals of Aβ-(1–7) and various antibody (Fab) fragments reveal that antibodies with nominal specificity for the Aβ N terminus actually bind both extended and helical conformers (53). These data emphasize the structural plasticity of the N terminus. We do note, however, that turns located in the N-terminal half of Aβ have been observed in experimental (for a comprehensive review, see Ref. 54) and computational (34, 55) studies. A turn at Ser8–Gly9 might be particularly important because it can bring the N-terminal quarter of the peptide into contact with the central hydrophobic cluster region. Maji et al. (56) proposed that competition between the N and C termini to form a stable complex with the central hydrophobic cluster underlay these effects. For example, a single amino acid substitution, Asp1 → Tyr, alters substantially the Aβ assembly kinetics and oligomer size frequency distribution (56). The proximity of His6 and Asp7 to the putative Ser8–Gly9 turn suggests that the English and Tottori mutations may affect this structural element, and through this effect, alter overall peptide assembly. It is intriguing, in this respect, that the N terminus of the yeast Sup35 prion, which is not involved in forming the amyloid core of the prion, has been found to play an essential role in prion formation and fibril assembly (57).
Supplementary Material
Acknowledgments
We thank Drs. Mohammed Inayathullah and Robin Roychaudhuri for advice and critical comments.
This work was supported, in whole or in part, by National Institutes of Health Grant AG027818 and the Japan Human Science Foundation, a Pergolide Fellowship (Eli Lilly Japan), and the Mochida Memorial Foundation for Medical and Pharmaceutical Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
We refer to the English (H6R) and Tottori (D7N) peptides as “mutants” only for ease in distinguishing them from wild type Aβ40 and Aβ42. This designation does not refer directly to the corresponding DNA sequences.
Structural order in this context refers to the relative lack of SC elements in secondary structure space.
- AD
- Alzheimer disease
- Aβ
- amyloid β-protein
- AFM
- atomic force microscopy
- APP
- amyloid β-protein precursor
- LDH
- lactate dehydrogenase
- MTT
- 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- PICUP
- photo-induced cross-linking of unmodified proteins
- Ru(bpy)
- tris(2,2′-bipyridyl)dichlororuthenium(II) hexahydrate
- ThT
- Thioflavin T
- PBS
- phosphate-buffered saline
- SC
- statistical coil
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Teplow D. B. (1998) Amyloid 5, 121–142 [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D. J. (2004) Ann. Intern. Med. 140, 627–638 [DOI] [PubMed] [Google Scholar]
- 3.Bertram L., Tanzi R. E. (2008) Nat. Rev. Neurosci. 9, 768–778 [DOI] [PubMed] [Google Scholar]
- 4.Borchelt D. R., Thinakaran G., Eckman C. B., Lee M. K., Davenport F., Ratovitsky T., Prada C. M., Kim G., Seekins S., Yager D., Slunt H. H., Wang R., Seeger M., Levey A. I., Gandy S. E., Copeland N. G., Jenkins N. A., Price D. L., Younkin S. G., Sisodia S. S. (1996) Neuron 17, 1005–1013 [DOI] [PubMed] [Google Scholar]
- 5.Tomita T., Maruyama K., Saido T. C., Kume H., Shinozaki K., Tokuhiro S., Capell A., Walter J., Grünberg J., Haass C., Iwatsubo T., Obata K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roychaudhuri R., Yang M., Hoshi M. M., Teplow D. B. (2009) J. Biol. Chem. 284, 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendriks L., van Duijn C. M., Cras P., Cruts M., Van Hul W., van Harskamp F., Warren A., McInnis M. G., Antonarakis S. E., Martin J. J. (1992) Nat. Genet. 1, 218–221 [DOI] [PubMed] [Google Scholar]
- 8.Levy E., Carman M. D., Fernandez-Madrid I. J., Power M. D., Lieberburg I., van Duinen S. G., Bots G. T., Luyendijk W., Frangione B. (1990) Science 248, 1124–1126 [DOI] [PubMed] [Google Scholar]
- 9.Bugiani O., Padovani A., Magoni M., Andora G., Sgarzi M., Savoìardo M., Bizzi A., Giaccone G., Rossi G., Tagliavini F. (1998) Neurobiol. Aging 19, S238 [Google Scholar]
- 10.Kamino K., Orr H. T., Payami H., Wijsman E. M., Alonso M. E., Pulst S. M., Anderson L., O'Dahl S., Nemens E., White J. A. (1992) Am. J. Hum. Genet. 51, 998–1014 [PMC free article] [PubMed] [Google Scholar]
- 11.Grabowski T. J., Cho H. S., Vonsattel J. P., Rebeck G. W., Greenberg S. M. (2001) Ann. Neurol. 49, 697–705 [DOI] [PubMed] [Google Scholar]
- 12.Betts V., Leissring M. A., Dolios G., Wang R., Selkoe D. J., Walsh D. M. (2008) Neurobiol. Dis. 31, 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilsberth C., Westlind-Danielsson A., Eckman C. B., Condron M. M., Axelman K., Forsell C., Stenh C., Luthman J., Teplow D. B., Younkin S. G., Näslund J., Lannfelt L. (2001) Nat. Neurosci. 4, 887–893 [DOI] [PubMed] [Google Scholar]
- 14.Tomiyama T., Nagata T., Shimada H., Teraoka R., Fukushima A., Kanemitsu H., Takuma H., Kuwano R., Imagawa M., Ataka S., Wada Y., Yoshioka E., Nishizaki T., Watanabe Y., Mori H. (2008) Ann. Neurol. 63, 377–387 [DOI] [PubMed] [Google Scholar]
- 15.Janssen J. C., Beck J. A., Campbell T. A., Dickinson A., Fox N. C., Harvey R. J., Houlden H., Rossor M. N., Collinge J. (2003) Neurology 60, 235–239 [DOI] [PubMed] [Google Scholar]
- 16.Wakutani Y., Watanabe K., Adachi Y., Wada-Isoe K., Urakami K., Ninomiya H., Saido T. C., Hashimoto T., Iwatsubo T., Nakashima K. (2004) J. Neurol. Neurosurg. Psychiatry 75, 1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hori Y., Hashimoto T., Wakutani Y., Urakami K., Nakashima K., Condron M. M., Tsubuki S., Saido T. C., Teplow D. B., Iwatsubo T. (2007) J. Biol. Chem. 282, 4916–4923 [DOI] [PubMed] [Google Scholar]
- 18.Walsh D. M., Selkoe D. J. (2007) J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 19.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 20.Klyubin I., Walsh D. M., Cullen W. K., Fadeeva J. V., Anwyl R., Selkoe D. J., Rowan M. J. (2004) Eur. J. Neurosci. 19, 2839–2846 [DOI] [PubMed] [Google Scholar]
- 21.Wang H. W., Pasternak J. F., Kuo H., Ristic H., Lambert M. P., Chromy B., Viola K. L., Klein W. L., Stine W. B., Krafft G. A., Trommer B. L. (2002) Brain Res. 924, 133–140 [DOI] [PubMed] [Google Scholar]
- 22.Townsend M., Shankar G. M., Mehta T., Walsh D. M., Selkoe D. J. (2006) J. Physiol. 572, 477–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klyubin I., Walsh D. M., Lemere C. A., Cullen W. K., Shankar G. M., Betts V., Spooner E. T., Jiang L., Anwyl R., Selkoe D. J., Rowan M. J. (2005) Nat. Med. 11, 556–561 [DOI] [PubMed] [Google Scholar]
- 24.Shankar G. M., Bloodgood B. L., Townsend M., Walsh D. M., Selkoe D. J., Sabatini B. L. (2007) J. Neurosci. 27, 2866–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., Brett F. M., Farrell M. A., Rowan M. J., Lemere C. A., Regan C. M., Walsh D. M., Sabatini B. L., Selkoe D. J. (2008) Nat. Med. 14, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono K., Condron M. M., Teplow D. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14745–14750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh D. M., Lomakin A., Benedek G. B., Condron M. M., Teplow D. B. (1997) J. Biol. Chem. 272, 22364–22372 [DOI] [PubMed] [Google Scholar]
- 28.Bitan G., Lomakin A., Teplow D. B. (2001) J. Biol. Chem. 276, 35176–35184 [DOI] [PubMed] [Google Scholar]
- 29.Bitan G., Teplow D. B. (2004) Acc. Chem. Res. 37, 357–364 [DOI] [PubMed] [Google Scholar]
- 30.LeVine H., 3rd (1999) Methods Enzymol. 309, 274–284 [DOI] [PubMed] [Google Scholar]
- 31.Kirkitadze M. D., Condron M. M., Teplow D. B. (2001) J. Mol. Biol. 312, 1103–1119 [DOI] [PubMed] [Google Scholar]
- 32.Bitan G., Kirkitadze M. D., Lomakin A., Vollers S. S., Benedek G. B., Teplow D. B. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teplow D. B. (2006) Methods Enzymol. 413, 20–33 [DOI] [PubMed] [Google Scholar]
- 34.Yang M., Teplow D. B. (2008) J. Mol. Biol. 384, 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomakin A., Chung D. S., Benedek G. B., Kirschner D. A., Teplow D. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1125–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomakin A., Teplow D. B., Kirschner D. A., Benedek G. B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7942–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarrett J. T., Lansbury P. T., Jr. (1993) Cell 73, 1055–1058 [DOI] [PubMed] [Google Scholar]
- 38.Groenning M. (2010) J. Chem. Biol. 3, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeVine H., 3rd (1993) Protein Sci. 2, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naiki H., Nakakuki K. (1996) Lab. Invest. 74, 374–383 [PubMed] [Google Scholar]
- 41.Khurana R., Ionescu-Zanetti C., Pope M., Li J., Nielson L., Ramírez-Alvarado M., Regan L., Fink A. L., Carter S. A. (2003) Biophys. J. 85, 1135–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 43.Abe K., Saito H. (1998) Neurosci. Res. 31, 295–305 [DOI] [PubMed] [Google Scholar]
- 44.Rönicke R., Klemm A., Meinhardt J., Schröder U. H., Fändrich M., Reymann K. G. (2008) PLoS ONE 3, e3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haass C., Selkoe D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 101–112 [DOI] [PubMed] [Google Scholar]
- 46.Kirkitadze M. D., Bitan G., Teplow D. B. (2002) J. Neurosci. Res. 69, 567–577 [DOI] [PubMed] [Google Scholar]
- 47.Grant M. A., Lazo N. D., Lomakin A., Condron M. M., Arai H., Yamin G., Rigby A. C., Teplow D. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16522–16527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lazo N. D., Grant M. A., Condron M. C., Rigby A. C., Teplow D. B. (2005) Protein Sci. 14, 1581–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teplow D. B., Lazo N. D., Bitan G., Bernstein S., Wyttenbach T., Bowers M. T., Baumketner A., Shea J. E., Urbanc B., Cruz L., Borreguero J., Stanley H. E. (2006) Acc. Chem. Res. 39, 635–645 [DOI] [PubMed] [Google Scholar]
- 50.Fancy D. A., Kodadek T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6020–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fancy D. A., Denison C., Kim K., Xie Y., Holdeman T., Amini F., Kodadek T. (2000) Chem. Biol. 7, 697–708 [DOI] [PubMed] [Google Scholar]
- 52.Maji S. K., Amsden J. J., Rothschild K. J., Condron M. M., Teplow D. B. (2005) Biochemistry 44, 13365–13376 [DOI] [PubMed] [Google Scholar]
- 53.Gardberg A., Dice L., Pridgen K., Ko J., Patterson P., Ou S., Wetzel R., Dealwis C. (2009) Biochemistry 48, 5210–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lazo N. D., Maji S. K., Fradinger E. A., Bitan G., Teplow D. B. (2005) in Amyloid Proteins-The Beta Sheet Conformation and Disease (Sipe J. C. ed) pp. 385–492, Wiley-VCH, Weinheim [Google Scholar]
- 55.Sgourakis N. G., Yan Y., McCallum S. A., Wang C., Garcia A. E. (2007) J. Mol. Biol. 368, 1448–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maji S. K., Ogorzalek Loo R. R., Inayathullah M., Spring S. M., Vollers S. S., Condron M. M., Bitan G., Loo J. A., Teplow D. B. (2009) J. Biol. Chem. 284, 23580–23591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnan R., Lindquist S. L. (2005) Nature 435, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.