Abstract
Double strand breaks (DSBs) can be repaired by homology independent nonhomologous end joining (NHEJ) pathways involving proteins such as Ku70/80, DNAPKcs, Xrcc4/Ligase 4, and the Mre11/Rad50/Nbs1 (MRN) complex. DSBs can also be repaired by homology-dependent pathways (HDR), in which the MRN and CtIP nucleases produce single strand ends that engage homologous sequences either by strand invasion or strand annealing. The entry of ends into HDR pathways underlies protocols for genomic manipulation that combine site-specific DSBs with appropriate informational donors. Most strategies utilize long duplex donors that participate by strand invasion. Work in yeast indicates that single strand oligonucleotide (SSO) donors are also active, over considerable distance, via a single strand annealing pathway. We examined the activity of SSO donors in mammalian cells at DSBs induced either by a restriction nuclease or by a targeted interstrand cross-link. SSO donors were effective immediately adjacent to the break, but activity declined sharply beyond ∼100 nucleotides. Overexpression of the resection nuclease CtIP increased the frequency of SSO-mediated sequence modulation distal to the break site, but had no effect on the activity of an SSO donor adjacent to the break. Genetic and in vivo competition experiments showed that sequence conversion by SSOs in the immediate vicinity of the break was not by strand invasion or strand annealing pathways. Instead these donors competed for ends that would have otherwise entered NHEJ pathways.
Keywords: DNA Recombination, DNA Repair, Gene Knockout, Gene Therapy, Nucleic Acid, Oligonucleotide, Triple Helix
Introduction
Non-homologous end joining (NHEJ)3 repair is un-templated, as there is no requirement, or even apparent use, for an informational reference sequence acting in trans. NHEJ might be regarded as a collection of operations (end processing by nucleases, limited single strand exposure of the ends, fill in by polymerases, ligation), without requirement for a specific order of events (1). In mammalian cells the best characterized version of NHEJ, often termed “classical” (C-NHEJ) (2, 3), utilizes several factors, including Ku70/80, Artemis, XLF, DNAPKcs, Xrcc4/DNA ligase 4, polλ, polμ, and XLF/Cernunnos (4–8). Although precise joining of restriction enzyme cleaved ends can occur, the pathway is frequently mutagenic, resulting in the introduction of short deletions at the break site (9–11).
Deletions at the DSB are sometimes bounded by short homology elements located on either side of the break. These have been described as the product of a microhomology mediated end joining (MMEJ) pathway (5, 12). Deletions without microhomologies are also recovered and recent work suggests that the MRN complex plays an important role in both versions of deletional NHEJ (2, 13–15). A backup pathway involving poly(ADP-ribose) polymerase 1 and ligase III has been described as well (16, 17). It should be noted that whether end joining in the absence of one of the canonical factors is evidence of different discrete pathways, or simply a variation on a fundamental scheme, is a matter for discussion (7). Nonetheless, NHEJ is the dominant mode of DSB repair (not associated with replication) in mammalian cells, and is operative throughout the cell cycle (18). In yeast, much DSB repair is via HDR pathways. However, NHEJ is fully functional and all mammalian proteins, with the exception of DNAKcs and Artemis have yeast homologs (21).
HDR pathways are active during the S and G2 phase of the cell cycle (18, 19), and compete with NHEJ (20, 21). Transfected duplex donor sequences can be the source of homology, providing an opportunity to introduce a novel sequence into the genome. Consequently there has been considerable effort directed toward developing strategies for genomic modulation based on targeted DSBs and duplex donors (22–26). Entry of a DSB into HDR pathways requires 5′-3′ strand resection exposing 3′-end single strand tails (27, 28). Resection requires the MRN complex in conjunction with Brca1 and the resection nuclease CtIP, which interacts with MRN and promotes endonucleolytic activity of the complex in a cell cycle-regulated fashion (29–32). In yeast, which lack Brca1, resection can extend over thousands of bases (33–37). However, in mammalian cells, proteins such as Rad51, Rad51C, Xrcc3, and NBS1 associate with DNA extending only 1–2 kb from the break, indicating a more limited resection (38, 39).
The resected ends can hybridize to homologous sequences by Rad51/Brca2-mediated strand invasion (required for homologous recombinational repair) (40, 41). The Rad51 paralogue proteins, Rad51B, -C, -D, Xrcc2, and Xrcc3, support Rad51 activity (39, 42–44). Fanconi anemia (FA) proteins are also thought to contribute to HDR pathways at broken replication forks (45). Recently, a FancG-dependent complex of Brca2/FancD2/Xrcc3 was described and implicated in replication associated HDR (46).
Homology-dependent interactions can also occur between elements on either side of a break via the SSA pathway (47). Repeated sequences in the extended single strand tails can anneal with each other, forming a structure that is resolved by deletion of the intervening sequence and one copy of the repeat. In mammalian cells Rad52 and Ercc1/Xpf make important contributions to this pathway (48–52).
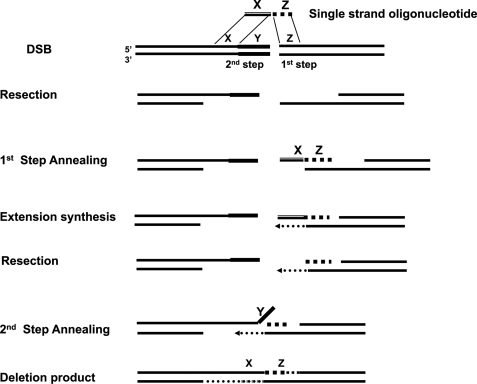
Most strategies for targeted gene modification use double-stranded donors, which enter classical Rad51/Brca2-dependent homologous recombinational repair pathways (53–55). However, important work in yeast from the Resnick (56) laboratory has shown that single-strand oligonucleotides (SSOs) can modify the sequence around a DSB. They demonstrated that SSOs could be used to manipulate sequences thousands of bases from the break site. They proposed that the SSOs enter an SSA pathway with two cycles of annealing of the donor oligonucleotide to resected ends on either side of the DSB (Fig. 1) (56). Their model explains SSO-mediated sequence modulation in yeast, and provides a conceptual basis for analysis of related pathways in mammalian cells.
FIGURE 1.
The two-step annealing model for sequence modulation by an SSO donor at a double strand break, based on the schematic of Storici et al. (56). In this illustration the SSO contains homology elements X and Z that are contiguous in the oligonucleotide, but separated by Y in the genome. After introduction of a double strand break at the target site the ends are resected to reveal extended 3′ terminal single strands. The 1st step side (Z) of the SSO donor anneals with the complementary single strand region, and the remainder of the oligonucleotide serves as a template for extension synthesis to generate a new double-stranded end. Additional resection (or oligonucleotide displacement) would reveal an extended 3′ terminal single strand with homology to region X on the other (2nd step) side of the break. Hybridization of the complementary X regions in the 2nd annealing step forms an intermediate that is resolved by elimination of the Y region sequence, fill in, and ligation to yield a precise deletion at the rejoined break.
Sequence conversion in mammalian cells by SSO donors at targeted DSBs has been described (57, 58). The DSB was introduced by the well known homing endonuclease I-SceI in stably integrated marker genes. DSBs can also be generated as a result of the cellular response to DNA damage. We have shown that targeted psoralen cross-links induce small deletions typical of NHEJ repair of DSBs (59, 60). Consistent with the induction of DSBs, the site-specific cross-links can also stimulate SSO-mediated sequence conversion of the target region (61).
The activity of SSO donors at sites of DSBs raises questions about the applicability of the yeast model to the mammalian cell system. In the experiments described in this report we have tested, in mammalian cells, two postulates derived from the yeast two-step annealing model: efficient sequence modulation can occur distal to the break, and SSO donors enter a SSA pathway. Our results argue that, in mammalian cells, SSO-directed information transfer is restricted to the immediate vicinity of the DSB. Furthermore, the SSO donors compete for ends that enter an NHEJ, rather than an SSA, pathway. Because NHEJ operates throughout the cell cycle this opens the possibility that directed modulation of genomic sequences could be realized at targeted DSBs in nonproliferating cells.
MATERIALS AND METHODS
Cells, Culture, and Synchronization
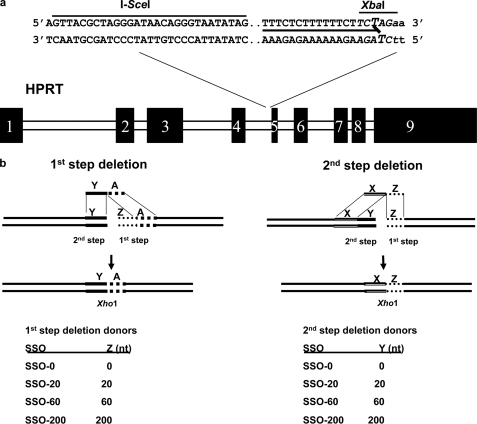
Chinese hamster ovary (CHO) cells, wild type, and repair-deficient derivatives were grown in minimal Eagle's medium (Invitrogen) supplemented with penicillin/streptomycin and 10% fetal bovine serum. CHO cells expressing a dominant-negative form of Rad51 (62) (the kind gift of Dr B. Lopez), Xrcc3-deficient irs1SF cells (63), the FancG knock-out line KO40 (64), and the Brca2-deficient line V-C8 (65) have all been described. V-C8 cells complemented by expression of wild type Brca2 were the generous gift of Drs. S. Kowalczykowski and R. Jensen. The construction of the AA8-I-SceI cell line, by targeted knock-in, was described in a previous publication (61). This cell line is repair proficient and contains an I-SceI site in intron 4 immediately 5′ of the polypurine:polypyrimidine element that is part of the splice junction and the triplex target site (Fig. 2a).
FIGURE 2.
The 1st and 2nd step SSO deletion donors. a, the pso-TFO and I-SceI target sites in the Chinese hamster Hprt gene in intron 4 adjacent to exon 5. The triplex target site is denoted by a solid line and the cross-linked thymidines are in enlarged font. The first 2 nt of exon 5 are in lowercase. An XbaI site is located immediately adjacent to exon 5. b, schematic of the two kinds of SSO deletion donors. 1st step deletion donors are designed to introduce precise deletions in the region involved in the first annealing step. Conversely 2nd step deletion donors introduce deletions in the region engaged in the 2nd annealing step. The size of the deletion is denoted by the identification of the donor, SSO-60 will introduce a 60-nt deletion, etc. The donors also introduce an XhoI site and eliminate the XbaI site as shown a. In the SSO-0 donor the homology elements immediately adjacent to either side of the break are contiguous to one another, and the conversion product has no deletion.
The AA8-SSA cell line was constructed by targeted knock-in into intron 5 of a 270-nt sequence present in intron 4 of the hamster Hprt gene. AM-12 CHO cells contain a variant triplex psoralen target site adjacent to a mutated exon 5, and are Hprt deficient. A duplex donor construct (containing, in the 5′ to 3′ order, 1000 nt from intron 4, the I-SceI recognition sequence, the wild type triplex-psoralen target site, a wild type exon 5, an insertion of the 270-nt intron 4 element, and 1000 nt of intron 5) was co-electroporated with the AM-12 variant pso-TFO into AM-12 cells. After photoactivation, the cells were cultured for 72 h and then incubated in hypoxanthine/aminopterin/thymidine (10−4 m hypoxanthine, 5 × 10−6 m aminopterin, 10−5 m thymidine) medium to select for colonies with a wild type Hprt gene. Candidate clones were examined by PCR, restriction enzyme digestion, and sequence analysis to confirm the construction.
Prior to experiments, cells were cultured in medium containing hypoxanthine/aminopterin/thymidine to remove pre-existing Hprt-deficient cells. CHO were synchronized in G0/G1 by a variation of the method described by Sawai et al. (66). Briefly, cells were plated at subconfluent levels and the next day the medium changed to α-minimal Eagle's medium with 2% fetal bovine serum and 2% Me2SO. After 48 h the cells (85–88% G0/G1) were washed and incubated with complete medium containing 100 μm mimosine for 16 h to block them in early S phase (∼90% early S phase cells). After 16 h the cells were released from the mimosine block by feeding with α-minimal Eagle's medium, 10% fetal bovine serum.
Oligonucleotides, Transfection Hprt Mutation Assay
The 17-nt TFO, AE-07 (referred to in the text as pso-TFO), against the hamster Hprt target, was synthesized and purified as described previously (67). The sequence of the SSO-0 100-nt single strand oligonucleotide donor, which introduces inactivating mutations into exon 5 as well as a novel XhoI site, was discussed in a prior publication (61). This donor was homologous to the region immediately adjacent to either side of the double strand break generated by the targeted cross-link site, or the I-SceI homing endonuclease, at the intron 4/exon 5 junction of the CHO Hprt gene. The 100-nt deletion donor oligonucleotides contained 50 nt homologous to the region immediately adjacent to one or the other side of the DSB target site. The remaining 50 nt were homologous to sequences located at the indicated distance from the DSB site. Oligonucleotides and/or plasmids (the CtIP-GFP expression plasmid was the kind gift of Dr. Stephen Jackson) were introduced into cells by electroporation using an Amaxa nucleoporator (Lonza). Transfection of the pso-TFO was followed by incubation for 3 h and then exposure in the Rayonet chamber to UVA light for 3 min at 1.8 J/cm2. The cells were cultured for 7 days and then plated in medium containing 20 μm 6-thioguanine. Cells were also plated in medium without 6-thioguanine to determine plating efficiency. After 7–10 days, colonies were counted and the mutation frequencies calculated as the ratio of 6-thioguanine-resistant colonies/total colony forming cells. In experiments with MIRIN (Tocris), following electroporation and photoactivation, cells were incubated with 60 μm compound for 20 h, after which the medium was replaced with standard medium and the cells processed as described.
Analysis of Hprt Target Region in 6-Thioguanine-resistant Colonies
Individual colonies were picked and expanded in 96-well dishes. DNA was extracted and the exon 5 target region was amplified using forward primer 5′-CTAGTTTGAGGCCAGCTTTGGC and reverse primer 5′-GGGATTCCAGGCATGCCTTACTG, which yield a 750-bp fragment with DNA from wild type cells. Digestion of the PCR product with the appropriate restriction enzyme was used to identify clones in which sequence conversion had occurred. Conversion was confirmed by sequence analysis.
RESULTS
Activity of SSO Donors
In earlier publications we described the development of biologically active triple helix forming oligonucleotides (TFOs) linked to psoralen, a photoactive cross-linking agent (67, 68). Some experiments presented in this report employed a psoralen-linked TFO (pso-TFO) designed to introduce a cross-link into a site immediately adjacent to exon 5 in the Chinese hamster Hprt gene. Electroporation of the pso-TFO was followed by a brief exposure to long wave ultraviolet light (UVA) to activate the psoralen. In other experiments we used I-SceI to introduce the break (see Fig. 2a for a schematic of the target site) (61). I-SceI is continuously active for the lifetime of the encoding plasmid and transcription/translation product, thus there are many cleavage events during the course of an experiment. In contrast, the psoralen cross-link is introduced once, during a single exposure to UVA. The requirement for photoactivation permits control of the time of cross-linking and subsequent processing, in contrast to the uncontrolled expression of I-SceI. Based on the yeast two-step model we anticipated that the activity of the SSO donors would be dependent on SSA. Consequently we introduced the targeted cross-link into cells synchronized in S phase (“Materials and Methods”).
In our previous study we demonstrated the sequence conversion activity of an SSO donor (here referred to as SSO-0) designed to interact with regions immediately adjacent to either side of a DSB. The oligonucleotide was 100 nt long and contained 50 nt homologous to the sequences adjacent to one side of the break site, and 50-nt homologous to the other side. A few nucleotides in the SSO-0 donor differed from the wild type sequence such that conversion created a new restriction site and also inactivated the Hprt gene (61). The activity of the SSO-0 donor implied sufficient single strand exposure at the break to support stable hybridization with the oligonucleotide in both the first and second step, as proposed in the two-step model (56, 69) (Fig. 1). Prompted by the success of the SSO-0 donor we attempted to introduce a defined deletion of 400 bases. We co-electroporated a plasmid encoding I-SceI together with a 100-mer SSO donor (SSO-400) containing 50 nt of homology to a region 200 nt upstream, and 50 nt of homology to a region 200 nt downstream, of the break site. In a parallel experiment, the SSO-400 donor was co-electroporated with the pso-TFO followed by UVA treatment. Cells in which the 400-nt deletion occurred would be deficient in Hprt by loss of exon 5. Of course cells could also be Hprt deficient as the result of NHEJ-mediated deletions, in which the SSO-400 donor played no part. We examined 384 Hprt-deficient colonies from the experiment with the pso-TFO/UVA (frequency of Hprt deficient colonies = 0.5%), and 192 colonies from the experiment with I-SceI (1% Hprt-deficient colonies). We recovered a single clone with the precise deletion from each set, for an absolute frequency of ∼0.002%. This contrasted with the 200-fold greater activity of the SSO-0 donor (61).
Sequence Conversion by SSO Deletion Donors
The minimal activity of the SSO-400 donor was unexpected given the demonstration of end resection extending to ∼2 kb from the break site (38, 39) and the success of deletion donors in the yeast system. Consequently we wanted to assess the distance from the DSB over which the SSO donors would be active, in both steps in the conversion pathway. We designed two series of 100-mer SSO donors that contained 50 nt homologous to the region immediately adjacent to one or the other side of the break (Fig. 2b). In the 1st step deletion donors the 50-nt portion involved in the first annealing step was homologous to sequences progressively further from the break on the 1st step side. The other 50-nt portion of the oligonucleotide was homologous to the sequences immediately adjacent to the break on the 2nd step side. Conversion by these donors would produce clones with deletions on the side of the break engaged in the 1st annealing step.
With the 2nd step deletion donors the SSO would hybridize, in the first step of the pathway, to the 50-nt region immediately adjacent to the break on the 1st step side. The other half of the SSO was designed such that its complement, synthesized by extension, and revealed by subsequent processing, would hybridize in the 2nd step with regions of homology progressively further away from the break on the 2nd step side. Conversion by these donors would produce cell clones with defined deletions in the 2nd step direction. The stepwise scheme for the 2nd step deletion donor is diagrammed in Fig. 1. The activity of the different donors would reflect the efficiency of sequence conversion as a function of distance from the break. However, in contrast to the SSO-400 donor, in each of these deletion donors, half of the sequence was identical to the corresponding half of SSO-0, whose activity was well established (61).
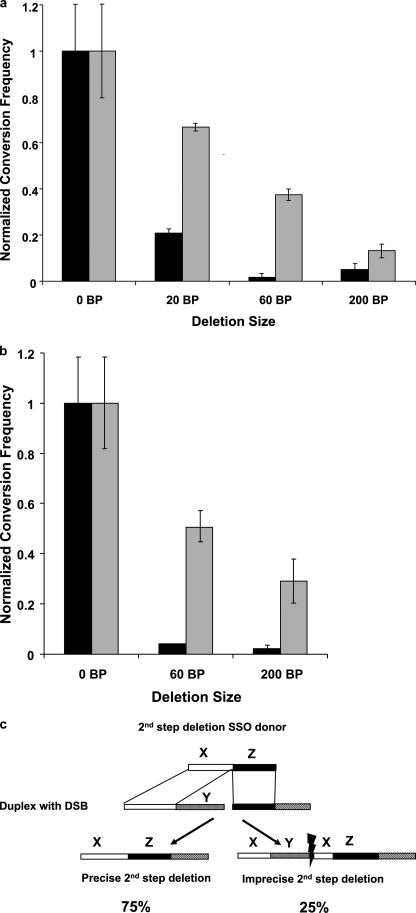
In the first series of experiments we measured donor activity in cells treated in S phase with the pso-TFO/UVA. The SSO donors were designed to introduce deletions of 0 (SSO-0), 20, 60, and 200 bases in the first and second step directions. Additionally, all donors introduced the same new restriction site as SSO-0. After co-electroporation of the pso-TFO and an SSO donor, followed by photoactivation and passage, colonies with mutant Hprt genes were isolated. The target region from these clones was amplified and examined by restriction digestion to identify the products of sequence conversion by the SSO donors. The results demonstrated a pronounced fall in activity as the deletion distance increased, such that the 200-nt 1st step deletion donor was 25-fold less active than the SSO-0 (Fig. 3a). Although the activity of the 2nd step donors also declined as a function of distance from the break, they were more active than their 1st step counterparts.
FIGURE 3.
Activity of 1st and 2nd step deletion donors at targeted DSBs in repair proficient cells. a, frequency (and standard error) of deletion size for 1st step (solid) and 2nd step (gray) deletion SSO donors at a DSB provoked by a targeted cross-link. b, activity of 1st and 2nd step deletion SSO donors at a DSB introduced by I-SceI. c, clones with imprecise products containing partial duplications and deletions of the X and Y sequence elements, as well as clones with precise deletions of the Y region, were recovered.
We repeated the experiment in cells in which the I-SceI expression plasmid was introduced with the deletion donors. The results were quite similar to those with the pso-TFO/UVA. There was a marked decline in activity relative to SSO-0, with the 200-bp 1st step deletion donor only 2% as active. Again the 2nd step deletion donor was more active than the 1st step donor (about 13-fold for the 200-nt deletion donor) (Fig. 3b). These results indicated that the donor activity was sharply reduced as a function of distance from the DSB, independent of the agent that induced the break. They also demonstrated an asymmetry in the activity, with the 2nd step deletion donors always more active than the 1st step donors.
Precise and Imprecise Conversion Products
The variance in activity of the 1st and 2nd step deletion donors was unexpected. Some insight into the basis for this was derived from a sequence analysis of the region adjacent to the break site in the clones in which conversion had occurred.
The PCR products of the target region from clones from experiments with the SSO-0 donor contained the sequence expected from this donor (61). We then examined the sequences of clones derived from the 200-nt deletion donors. The sequences from the 1st step donors were in accord with expectation indicating faithful sequence conversion. However, the analysis of clones derived from the experiment with the 2nd step donor revealed two kinds of products. Approximately 75% were precise deletions, exactly as expected from the design of the donor. However, the remaining clones contained small deletions, as well as repeats, of sequence found in the region of the SSO donor that would be engaged during the second annealing step (diagrammed in Fig. 3c). These products were consistent with a pathway involving annealing in the first step as predicted by the two-step model. In the second step repair was completed by NHEJ, rather than another cycle of annealing.
Activity of the 2nd Step Deletion Donor in Cells Deficient in DSB and HDR Repair
We were curious about the influence of repair functions in SSO-mediated sequence conversion. Genes of particular interest were components of the FA/BRCA pathway, which have been implicated in both cross-link (70, 71) and DSB repair, including NHEJ (72, 73) and HDR pathways (74). Thus we examined the activity of the SSO-0 and the 200-nt 2nd step deletion donor in cells with mutant or impaired FA and HDR proteins.
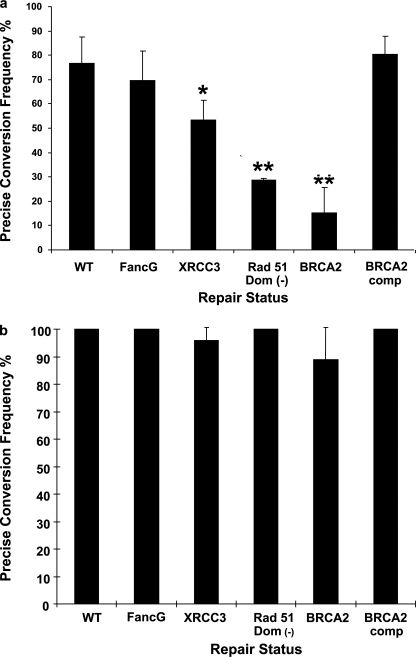
The pso-TFO was introduced into cells deficient in FancG, Xrcc3, Rad51 (75), or Brca2. In the first series of experiments we co-electroporated the 200-nt 2nd step deletion donor. The influence of the repair deficiency on the frequency of the precise deletion product is shown in Fig. 4a. Although the results in the FancG-deficient cells were indistinguishable from wild type (75% precise conversion), there was a decline in the frequency of precise deletions in cells with defects in HDR. In the Xrcc3-deficient cells the frequency of precise products was 50%. There were further reductions in cells expressing the dominant-negative Rad51 and in the Brca2-deficient cells, which had 15% precise deletion products. Complementation of Brca2-deficient cells with the wild type BRCA2 gene restored the frequency of the precise deletion product to wild type levels. These results argued that strand invasion functions were required for the majority of precise conversion events by the SSO 2nd step deletion donor (see supplemental Fig. S1 for a schematic and explanation).
FIGURE 4.
Activity of SSO donors in cells with deficiencies in FA and HDR genes. a, cells with deficiencies as indicated were transfected with the pso-TFO and the 200-nt 2nd step deletion donor SSO. Hprt-deficient colonies were recovered and the frequency (and standard error) of colonies with precise 200-nt deletions was determined. The statistical significance of the difference between an individual cell line and the wild type cells was p < 0.05 (*) and p < 0.001 (**). b, activity of the SSO-0 donor in cells with deficiencies in FA and HDR genes.
The relationship between the 2nd step deletion donor and the HDR status of the cells contrasted with the results with the SSO-0 donor. There was no linkage between the frequency of conversion products and the status of strand invasion functions. The results in all the host cells were similar to those recovered from wild type cells (Fig. 4b).
Overexpression of CtIP Increases the Activity of the 1st Step Deletion Donor
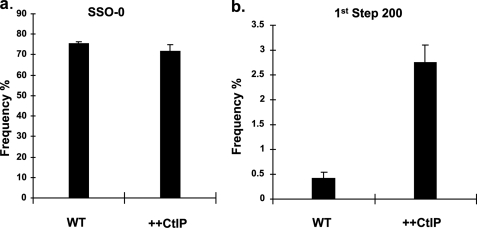
In the light of the very low activity of the 1st step deletion donor, it was of interest to ask if the opportunities for productive interaction by this donor could be increased, by increasing the resection activity at the ends. CtIP is an important modulator of resection in mammalian cells, whose activity is stimulated by phosphorylation during S phase (29, 31, 32, 76, 77). Consequently, we asked if overexpression of CtIP protein could enhance the activity of the oligonucleotides. Cells were co-electroporated with the pso-TFO, the expression plasmid encoding a CtIP-GFP fusion, and either the SSO-0 donor, or the 200-nt 1st step deletion donor. Expression of the CtIP-GFP fusion was verified by fluorescence analysis of the transfected cells. Examination of the Hprt-deficient clones revealed that there was no effect of CtIP overexpression on the activity of the SSO-0 donor (Fig. 5a). However, when we examined the product distribution from the experiment with the 200-nt 1st step deletion donor we observed a 6-fold increase in activity (Fig. 5b). This experiment could be most simply interpreted as indicating that enhanced resection activity, mediated by CtIP, generated a higher frequency of substrates available for annealing with the 200-nt 1st step deletion donor. On the other hand, this activity did not generate additional substrates for the SSO-0 donor.
FIGURE 5.
Overexpression of CtIP increases the activity of the 200-nt 1st step deletion donor. Cells were co-electroporated with the CtIP expression plasmid, pso-TFO, and either: a, the SSO-0 donor or, b, the 200-nt 1st step deletion donor. After recovery of Hprt-deficient colonies the frequency of the SSO donor (and standard error) directed product was determined.
Lack of Competition between SSO-0 and SSA
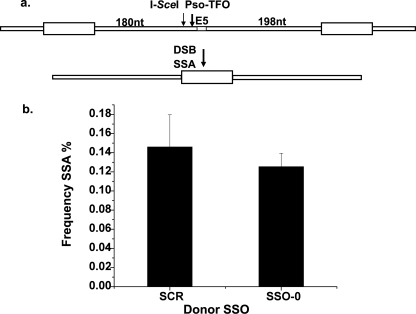
The design of our experiments was influenced by the studies in yeast in which the SSO donors were shown to enter an SSA pathway (56). However, the decidedly low activity of the 200-nt deletion donors (particularly the 1st step donor) suggested that either the SSA pathway was relatively inactive in the cells employed in our experiments, or the SSO donors did not actually enter this pathway. To distinguish these possibilities we constructed a new cell line (AA8-SSA) by targeted knock-in (“Materials and Methods”) (Fig. 6a). We introduced into intron 5 a 270-nt sequence also found in intron 4, as well as the I-SceI recognition sequence and the pso-triplex target sequence adjacent to exon 5. Intron 5 proved to be tolerant of the 270-nt insertion and the Hprt gene had wild type activity. The 270-nt direct repeats were positioned ∼200 nt on either side of exon 5, the same region that had been targeted by the 200-nt deletion donor SSOs. Repair by SSA of a DSB induced by I-SceI or the targeted psoralen cross-link would remove exon 5, one copy of the repeat, and the sequence between them. In preliminary experiments breaks were induced in the AA8-SSA cells either by I-SceI or the pso-TFO/UVA, Hprt-deficient colonies were recovered, and the amplification products from the exon 5 region were characterized. The analysis showed efficient SSA activity with both agents, with absolute frequencies in the 0.5–1% range (not shown). These results demonstrated the capacity of the cells to support SSA between sequence elements that had been the target of the ineffective 200-nt deletion donors.
FIGURE 6.
Competition between the SSO-0 donor and the SSA pathway. a, schematic of the genomic region containing the SSA construct. The cell line AA8-SSA was constructed by pso-TFO targeted knock-in (“Materials and Methods”) such that a 270-nt sequence in intron 4 was placed in intron 5 as a direct repeat. Resolution by SSA of a DSB introduced by I-SceI, or the targeted cross-link (at arrows), results in a precise deletion of one copy of the repeat, and the sequence between the repeats. This removes exon 5 and inactivates the Hprt gene. b, the AA8-SSA cells were transfected with the pso-TFO and either the SSO-0 or a scrambled (SCR) donor oligonucleotide. The frequency of SSA events was determined by analysis of the target region in Hprt-deficient clones. The standard error for each determination is indicated.
We then performed an SSO competition experiment with the pso-TFO/UVA as the inducing agent. In one set of samples an oligonucleotide with no homology to the target (scrambled, SCR) was co-electroporated. In another set, the SSO-0 donor was co-introduced. Hprt-deficient colonies were recovered and the target region in individual clones characterized to measure the frequency of SSA. SSA events were identified at equivalent levels in both sample sets (Fig. 6b). Consequently the frequency of SSA was unaffected by the presence of the SSO-0 donor. The amplification products from the clones from the samples in which the SSO donor had been present, but SSA had not occurred, were examined further. SSO-0 mediated sequence conversion was evident in about half of those colonies. Thus the SSO-0 donor was active although it failed to compete with the SSA pathway.
The SSO-0 donor employed in the preceding experiment engaged the sequences immediately adjacent to the break site, not in the repeated elements. Because the repeated elements had clearly participated in SSA we asked if an oligonucleotide with homology to them would interfere with their entry into the pathway. However, the introduction of a 100-mer oligonucleotide, identical in sequence to 100 nt in the repeat element, had no effect on the frequency of SSA. This was a further indication that the SSO donors were not able to compete for ends that entered the SSA pathway.
Competition between SSO-0 and NHEJ
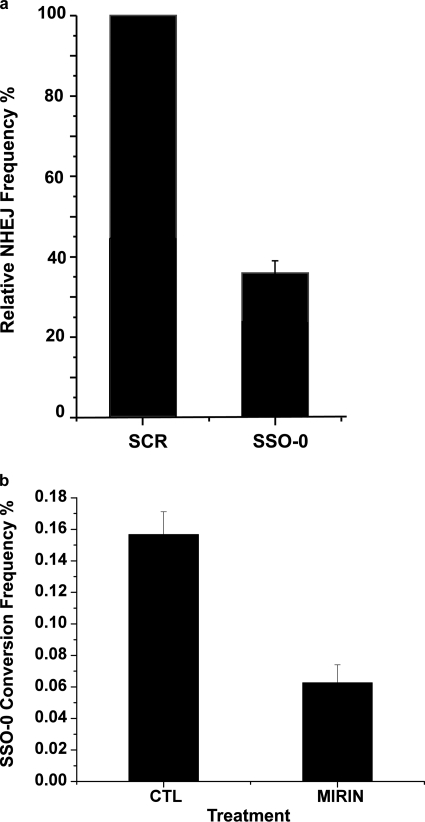
The results of the experiment with the AA8-SSA cells could be summarized as follows. In the absence of the SSO-0 donor the introduction of a targeted DSB in the AA8-SSA cells resulted in precise deletions derived from SSA, and deletions with a range of sizes due to NHEJ. In the presence of the SSO-0 donor the frequency of SSA events was unaffected, whereas the frequency of NHEJ events was reduced. This suggested that sequence conversion by the SSO-0 donor occurred at the expense of the products of NHEJ. To address this possibility directly, we measured the frequency of NHEJ events, induced by the pso-TFO/UVA, in the presence of the scrambled oligonucleotide donor or SSO-0. The results showed that the frequency of NHEJ events was suppressed by the SSO-0 donor oligonucleotide as compared with the scrambled oligonucleotide (Fig. 7a). These data favored the conclusion that SSO-mediated sequence conversion at DSBs occurred via the NHEJ pathway.
FIGURE 7.
Competition between the SSO-0 donor and the NHEJ pathway. a, cells were co-electroporated with the pso-TFO and either the SSO-0 or a scrambled (SCR) oligonucleotide sequence donor. The frequency of NHEJ was determined in the Hprt-deficient clones. b, cells were co-electroporated with the pso-TFO and SSO-0 donors. Following photoactivation, one aliquot of cells was placed in standard medium containing the same amount of Me2SO as used in the MIRIN-treated sample (CTL). Another was incubated in medium supplemented with 60 μm MIRIN for 20 h, after which the cells were returned to standard medium. The frequency (and standard error) of SSO-0 conversion events was determined.
NHEJ has variant versions, including MMEJ, in which interactions between microhomologies are thought to stabilize end:end intermediates prior to formation of an intact, rejoined, duplex (12). Sequence analysis of NHEJ products formed at the Hprt target site, in the absence of the SSO-0 donor, indicated a slight bias against MMEJ products compared with simple NHEJ events (MMEJ:NHEJ, 1:1.3). The analysis of the residual NHEJ products in the experiments with the SSO-0 donor demonstrated a shift that favored MMEJ events (MMEJ:NHEJ, 2:1). This suggested that ends that entered the MMEJ pathway were somewhat less available for engagement by the SSO donor than ends that were joined without microhomology involvement.
The classical version of NHEJ is dependent on the Ku80/70 complex as well as DNAPKcs and the XLF/Xrcc4/Ligase IV complex (7). In our previous study we found that the frequency of SSO-0 sequence conversion was unaffected by deficiency in DNAPKcs (61). We asked if the frequency of conversion would be affected in cells deficient in Ku80 or Xrcc4. However, we again found no distinction between these hosts and wild type cells (not shown). Consequently it appeared that the SSO-0 activity was not dependent on factors required for the classical pathway.
Recent work has shown a requirement for Mre11 activity in NHEJ (13). In that study it was shown that that the frequency of end joining could be suppressed by incubation of human and hamster cells with MIRIN, an inhibitor of MRN (78, 79). We introduced the pso-TFO and SSO-0 donor into cells and, following photoactivation, incubated them with or without MIRIN for 20 h. They were then fed MIRIN-free medium and the cells were processed as before. The cells were recovered from MIRIN treatment and there was no toxicity. The exposure to the drug reduced the recovery of colonies with SSO-0 conversion events by 2.2-fold (Fig. 7b). The reduction in activity was similar to the MIRIN-induced decline in end joining events following DSB formation by I-SceI cleavage, reported recently (13). Thus a treatment that reduced NHEJ activity also reduced activity of the SSO donor.
DISCUSSION
Investigator initiated sequence transfer to the genome can be regarded as the exploitation of a DNA repair pathway, defined by the initiating lesion and informational donor. To develop more efficient protocols it is important to identify the pathway(s) taken by combinations of lesion and donor. In the work reported here we have inquired as to the identity of the DSB repair pathway engaged by an SSO donor at the site of a DSB in mammalian cells. Our experiments addressed some of the tenets of the two-step annealing model from the yeast work: 1) oligonucleotide donors are active at distances from a DSB; 2) activity is independent of strand invasion functions; and 3) SSO donors participate in SSA.
Activity of SSO Donors Distal to a DSB
SSO donors designed to introduce deletions of a few hundred nucleotides were much less active than donors that engaged the region immediately adjacent to the break. This was most apparent with the oligonucleotides that we termed 1st step deletion donors. On the other hand the 2nd step deletion donors were more effective in generating precise deletions. Product analysis in repair-deficient cells indicated that the greater activity of these donors in wild type cells resulted from entry into multiple DSB repair pathways in the 2nd step. Multiple pathway involvement in DSB repair has been described previously (20, 80, 81). However, the underlying conclusion from these experiments was that, in contrast to the situation in yeast, the opportunities for simple annealing 200 nt from either side of the DSB were quite limited.
SSO Donors Do Not Compete with Ends That Enter SSA Pathways
SSA has a pronounced dependence on the ERCC1/XPF complex in mammalian cells (49, 50, 52). In our previous study we found that the activity of the ERCC1/XPF complex was not required for the activity of the SSO-0 donor (61). This, and the failure of the SSO-0 donor to influence the frequency of SSA events in the AA8-SSA cells, argues that the oligonucleotides do not compete for ends that enter the SSA pathway. Thus, resected ends that enter the SSA pathways are largely refractory to engagement by the oligonucleotides. Consequently, yeast and mammalian cells are quite different in this regard.
One explanation for the difference in SSO donor activity in yeast and mammalian cells could be the greater extent of resection in yeast (82). However, the 200-nt deletion donors were designed to generate deletions well within the range of resection in mammalian cells (38, 39). Furthermore, the SSA events in the AA8-SSA cells required annealing interactions between homology elements located 200 nt at either side of the break site. Consequently it was unlikely that the extent of resection was the limiting factor for the deletion donors. It seems more likely that the differences between yeast and mammalian cells are due to DSB repair proteins found in the latter (DNAPKcs, Artemis, Brca1, and Brca2, and some of the Rad51 paralogues (21)) and absent in the former. Presumably the additional proteins are important for maintaining the stability of mammalian genomes with greater complexity and repeated sequence content than in yeast.
It was possible to increase the activity of the 200-nt 1st step deletion donor, which can engage ends only by annealing, by overexpression of CtIP. We interpret this as indicating that the additional CtIP activity generated some resected ends that were free of the restriction on annealing with the deletion donor oligonucleotide. The increase in CtIP activity did not influence activity of the SSO-0 donor, suggesting that activity of this protein is not rate-limiting for the activity of a donor that engages sequences immediately adjacent to the break. It should be noted that whereas CtIP is important for SSA and homologous recombinational repair, it has been shown that total levels of NHEJ events at a break are independent of CtIP activity (83).
The SSO-0 Donor Enters an NHEJ Pathway
Sequence conversion at the DSB site by the SSO-0 donor was accompanied by a reduction in the frequency of NHEJ events. Thus this donor engaged ends that would have been repaired by an NHEJ pathway. Although NHEJ has no requirement for a reference sequence with homology to the region of the break, our results argue that information transfer to the site of a DSB can occur via this pathway when a homologous oligonucleotide is present. Thus some of the intermediates generated during NHEJ processing (7) are targets for annealing by the SSOs, which would enter a scheme similar to that elaborated by the Resnick group, but differing in the distance of activity (from the break) and the enabling pathway.
What factors underlie SSO-0 donor activity? SSO-0 activity was unaffected by deficiencies in Ku80, DNAPKcs, and Xrcc4, all associated with C-NHEJ. However, the results of experiments with MIRIN indicate that the MRN complex is important for activity of the SSO-0 donor. This is consistent with recent work on the role of MRN in NHEJ, and argues that the oligonucleotide engages ends that would otherwise enter one or another of the deletional NHEJ pathways (13, 14). The interpretation is complicated by the multiple roles played by the MRN complex in DSB repair (15, 84). It is the sensor that detects damage and initiates the recruitment of a cascade of repair factors (85). It is also involved in strand resection, an obligate step in both HR and SSA pathways (86, 87). Thus it would seem that the oligonucleotide must engage an intermediate that appears subsequent to the initial MRN-bound DSB, an intermediate that is not a precursor to the strand resection pathways (see supplemental Fig. S2 for a schematic). There are many different gene functions in NHEJ and it will be important to identify those required for the activity of the SSO donors.
Although obviously limited in the amount of information relative to duplex donors, the SSOs would appear to offer certain advantages. They can be synthesized quickly and be introduced into cells at much greater molar equivalents than plasmid- or phage-derived duplexes. Additionally, they are amenable to chemical modifications that improve nuclease resistance and hybrid stability, while retaining template function (88). The results presented here suggest another feature that might be useful. NHEJ is operative throughout the cell cycle, including in G0 and G1 (89, 90). Furthermore, it functions in terminally differentiated cells (91, 92) and in stem cells (93, 94). If SSO sequence conversion donors are active at DSBs in noncycling cells, then it might be possible to develop protocols for sequence conversion at targeted breaks without requirement for cells in S phase, during which time the genome is most vulnerable to rearrangement.
Supplementary Material
Acknowledgments
We thank Drs. S. Jackson, R. Jensen, S. Kowalczykowski, and B. Lopez for cell lines and plasmids.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program, NIA, and by the Fanconi Anemia Research Foundation, through a R&D contract with MedStar Research Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- NHEJ
- non-homologous end joining
- DSB
- double strand break
- HDR
- homology-dependent repair
- MRN
- mre11-RAD50-NBS1
- MMEJ
- microhomology-mediated end joining
- SSA
- single strand annealing
- FA
- Fanconi anemia
- SSO
- single strand oligonucleotide
- Hprt
- hypoxanthine phosphoribosyltransferase
- pso-TFO
- psoralen-linked triple helix forming oligonucleotide
- CHO
- Chinese hamster ovary
- nt
- nucleotide(s)
- GFP
- green fluorescent protein.
REFERENCES
- 1.Lieber M. R., Lu H., Gu J., Schwarz K. (2008) Cell Res. 18, 125–133 [DOI] [PubMed] [Google Scholar]
- 2.Xie A., Kwok A., Scully R. (2009) Nat. Struct. Mol. Biol. 16, 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zha S., Boboila C., Alt F. W. (2009) Nat. Struct. Mol. Biol. 16, 798–800 [DOI] [PubMed] [Google Scholar]
- 4.Capp J. P., Boudsocq F., Bertrand P., Laroche-Clary A., Pourquier P., Lopez B. S., Cazaux C., Hoffmann J. S., Canitrot Y. (2006) Nucleic Acids Res. 34, 2998–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guirouilh-Barbat J., Rass E., Plo I., Bertrand P., Lopez B. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20902–20907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capp J. P., Boudsocq F., Besnard A. G., Lopez B. S., Cazaux C., Hoffmann J. S., Canitrot Y. (2007) Nucleic Acids Res. 35, 3551–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieber M. R. (2008) J. Biol. Chem. 283, 1–5 [DOI] [PubMed] [Google Scholar]
- 8.Tsai C. J., Kim S. A., Chu G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7851–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hefferin M. L., Tomkinson A. E. (2005) DNA Repair 4, 639–648 [DOI] [PubMed] [Google Scholar]
- 10.Burma S., Chen B. P., Chen D. J. (2006) DNA Repair 5, 1042–1048 [DOI] [PubMed] [Google Scholar]
- 11.Gu J., Lu H., Tippin B., Shimazaki N., Goodman M. F., Lieber M. R. (2007) EMBO J. 26, 1010–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McVey M., Lee S. E. (2008) Trends Genet. 24, 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B. S. (2009) Nat. Struct. Mol. Biol. 16, 819–824 [DOI] [PubMed] [Google Scholar]
- 14.Zhuang J., Jiang G., Willers H., Xia F. (2009) J. Biol. Chem. 284, 30565–30573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinkelmann M., Spehalski E., Stoneham T., Buis J., Wu Y., Sekiguchi J. M., Ferguson D. O. (2009) Nat. Struct. Mol. Biol. 16, 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M., Wu W., Wu W., Rosidi B., Zhang L., Wang H., Iliakis G. (2006) Nucleic Acids Res. 34, 6170–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Audebert M., Salles B., Calsou P. (2004) J. Biol. Chem. 279, 55117–55126 [DOI] [PubMed] [Google Scholar]
- 18.Mao Z., Bozzella M., Seluanov A., Gorbunova V. (2008) Cell Cycle 7, 2902–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleh-Gohari N., Helleday T. (2004) Nucleic Acids Res. 32, 3683–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helleday T., Lo J., van Gent D. C., Engelward B. P. (2007) DNA Repair 6, 923–935 [DOI] [PubMed] [Google Scholar]
- 21.Shrivastav M., De Haro L. P., Nickoloff J. A. (2008) Cell Res. 18, 134–147 [DOI] [PubMed] [Google Scholar]
- 22.Porteus M. (2008) Methods Mol. Biol. 435, 47–61 [DOI] [PubMed] [Google Scholar]
- 23.Jarjour J., West-Foyle H., Certo M. T., Hubert C. G., Doyle L., Getz M. M., Stoddard B. L., Scharenberg A. M. (2009) Nucleic Acids Res. 37, 6871–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eklund J. L., Ulge U. Y., Eastberg J., Monnat R. J., Jr. (2007) Nucleic Acids Res. 35, 5839–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moehle E. A., Rock J. M., Lee Y. L., Jouvenot Y., Dekelver R. C., Gregory P. D., Urnov F. D., Holmes M. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson J. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5653–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helleday T. (2003) Mutat. Res. 532, 103–115 [DOI] [PubMed] [Google Scholar]
- 28.Huertas P. (2010) Nat. Struct. Mol. Biol. 17, 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. (2007) Nature 450, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P. (2008) Nature 455, 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L., Nievera C. J., Lee A. Y., Wu X. (2008) J. Biol. Chem. 283, 7713–7720 [DOI] [PubMed] [Google Scholar]
- 32.Yun M. H., Hiom K. (2009) Nature 459, 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lengsfeld B. M., Rattray A. J., Bhaskara V., Ghirlando R., Paull T. T. (2007) Mol. Cell 28, 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clerici M., Mantiero D., Lucchini G., Longhese M. P. (2005) J. Biol. Chem. 280, 38631–38638 [DOI] [PubMed] [Google Scholar]
- 35.White C. I., Haber J. E. (1990) EMBO J. 9, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugawara N., Wang X., Haber J. E. (2003) Mol. Cell 12, 209–219 [DOI] [PubMed] [Google Scholar]
- 37.Zierhut C., Diffley J. F. (2008) EMBO J. 27, 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berkovich E., Monnat R. J., Jr., Kastan M. B. (2007) Nat. Cell Biol. 9, 683–690 [DOI] [PubMed] [Google Scholar]
- 39.Rodrigue A., Lafrance M., Gauthier M. C., McDonald D., Hendzel M., West S. C., Jasin M., Masson J. Y. (2006) EMBO J. 25, 222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies A. A., Masson J. Y., McIlwraith M. J., Stasiak A. Z., Stasiak A., Venkitaraman A. R., West S. C. (2001) Mol. Cell 7, 273–282 [DOI] [PubMed] [Google Scholar]
- 41.Thorslund T., West S. C. (2007) Oncogene 26, 7720–7730 [DOI] [PubMed] [Google Scholar]
- 42.Lio Y. C., Mazin A. V., Kowalczykowski S. C., Chen D. J. (2003) J. Biol. Chem. 278, 2469–2478 [DOI] [PubMed] [Google Scholar]
- 43.Godthelp B. C., Wiegant W. W., van Duijn-Goedhart A., Schärer O. D., van Buul P. P., Kanaar R., Zdzienicka M. Z. (2002) Nucleic Acids Res. 30, 2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu N., Schild D., Thelen M. P., Thompson L. H. (2002) Nucleic Acids Res. 30, 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson L. H., Hinz J. M. (2009) Mutat. Res. 668, 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson J. B., Yamamoto K., Marriott A. S., Hussain S., Sung P., Hoatlin M. E., Mathew C. G., Takata M., Thompson L. H., Kupfer G. M., Jones N. J. (2008) Oncogene 27, 3641–3652 [DOI] [PubMed] [Google Scholar]
- 47.Lin F. L., Sperle K., Sternberg N. (1984) Cold Spring Harbor Symp. Quant. Biol. 49, 139–149 [DOI] [PubMed] [Google Scholar]
- 48.Van Dyck E., Stasiak A. Z., Stasiak A., West S. C. (2001) EMBO Rep. 2, 905–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark J. M., Pierce A. J., Oh J., Pastink A., Jasin M. (2004) Mol. Cell. Biol. 24, 9305–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Minawi A. Z., Saleh-Gohari N., Helleday T. (2008) Nucleic Acids Res. 36, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugiyama T., New J. H., Kowalczykowski S. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmad A., Robinson A. R., Duensing A., van Drunen E., Beverloo H. B., Weisberg D. B., Hasty P., Hoeijmakers J. H., Niedernhofer L. J. (2008) Mol. Cell. Biol. 28, 5082–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donoho G., Jasin M., Berg P. (1998) Mol. Cell. Biol. 18, 4070–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moynahan M. E., Pierce A. J., Jasin M. (2001) Mol. Cell 7, 263–272 [DOI] [PubMed] [Google Scholar]
- 55.Stark J. M., Hu P., Pierce A. J., Moynahan M. E., Ellis N., Jasin M. (2002) J. Biol. Chem. 277, 20185–20194 [DOI] [PubMed] [Google Scholar]
- 56.Storici F., Snipe J. R., Chan G. K., Gordenin D. A., Resnick M. A. (2006) Mol. Cell. Biol. 26, 7645–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radecke F., Peter I., Radecke S., Gellhaus K., Schwarz K., Cathomen T. (2006) Mol. Ther. 14, 798–808 [DOI] [PubMed] [Google Scholar]
- 58.Wang Z., Zhou Z. J., Liu D. P., Huang J. D. (2008) Oligonucleotides. 18, 21–32 [DOI] [PubMed] [Google Scholar]
- 59.Majumdar A., Khorlin A., Dyatkina N., Lin F. L., Powell J., Liu J., Fei Z., Khripine Y., Watanabe K. A., George J., Glazer P. M., Seidman M. M. (1998) Nat. Genet. 20, 212–214 [DOI] [PubMed] [Google Scholar]
- 60.Richards S., Liu S. T., Majumdar A., Liu J. L., Nairn R. S., Bernier M., Maher V., Seidman M. M. (2005) Nucleic Acids Res. 33, 5382–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Majumdar A., Muniandy P. A., Liu J., Liu J. L., Liu S. T., Cuenoud B., Seidman M. M. (2008) J. Biol. Chem. 283, 11244–11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lambert S., Lopez B. S. (2000) EMBO J. 19, 3090–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu N., Lamerdin J. E., Tebbs R. S., Schild D., Tucker J. D., Shen M. R., Brookman K. W., Siciliano M. J., Walter C. A., Fan W., Narayana L. S., Zhou Z. Q., Adamson A. W., Sorensen K. J., Chen D. J., Jones N. J., Thompson L. H. (1998) Mol. Cell 1, 783–793 [DOI] [PubMed] [Google Scholar]
- 64.Tebbs R. S., Hinz J. M., Yamada N. A., Wilson J. B., Salazar E. P., Thomas C. B., Jones I. M., Jones N. J., Thompson L. H. (2005) DNA Repair 4, 11–22 [DOI] [PubMed] [Google Scholar]
- 65.Wiegant W. W., Overmeer R. M., Godthelp B. C., van Buul P. P., Zdzienicka M. Z. (2006) Mutat. Res. 600, 79–88 [DOI] [PubMed] [Google Scholar]
- 66.Sawai M., Takase K., Teraoka H., Tsukada K. (1990) Exp. Cell Res. 187, 4–10 [DOI] [PubMed] [Google Scholar]
- 67.Puri N., Majumdar A., Cuenoud B., Miller P. S., Seidman M. M. (2004) Biochemistry 43, 1343–1351 [DOI] [PubMed] [Google Scholar]
- 68.Puri N., Majumdar A., Cuenoud B., Natt F., Martin P., Boyd A., Miller P. S., Seidman M. M. (2002) Biochemistry 41, 7716–7724 [DOI] [PubMed] [Google Scholar]
- 69.Storici F., Durham C. L., Gordenin D. A., Resnick M. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14994–14999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Niedernhofer L. J., Lalai A. S., Hoeijmakers J. H. (2005) Cell 123, 1191–1198 [DOI] [PubMed] [Google Scholar]
- 71.Mirchandani K. D., D'Andrea A. D. (2006) Exp. Cell Res. 312, 2647–2653 [DOI] [PubMed] [Google Scholar]
- 72.Donahue S. L., Campbell C. (2002) J. Biol. Chem. 277, 46243–46247 [DOI] [PubMed] [Google Scholar]
- 73.Donahue S. L., Lundberg R., Saplis R., Campbell C. (2003) J. Biol. Chem. 278, 29487–29495 [DOI] [PubMed] [Google Scholar]
- 74.Nakanishi K., Yang Y. G., Pierce A. J., Taniguchi T., Digweed M., D'Andrea A. D., Wang Z. Q., Jasin M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lambert S., Lopez B. S. (2002) Oncogene 21, 4065–4069 [DOI] [PubMed] [Google Scholar]
- 76.Takeda S., Nakamura K., Taniguchi Y., Paull T. T. (2007) Mol. Cell 28, 351–352 [DOI] [PubMed] [Google Scholar]
- 77.Huertas P., Jackson S. P. (2009) J. Biol. Chem. 284, 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dupré A., Boyer-Chatenet L., Sattler R. M., Modi A. P., Lee J. H., Nicolette M. L., Kopelovich L., Jasin M., Baer R., Paull T. T., Gautier J. (2008) Nat. Chem. Biol. 4, 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garner K. M., Pletnev A. A., Eastman A. (2009) Nat. Chem. Biol. 5, 129–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richardson C., Jasin M. (2000) Mol. Cell. Biol. 20, 9068–9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hartlerode A. J., Scully R. (2009) Biochem. J. 423, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mimitou E. P., Symington L. S. (2008) Nature 455, 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bennardo N., Cheng A., Huang N., Stark J. M. (2008) PLoS Genet. 4, e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J. M., Chang S., Ferguson D. O. (2008) Cell 135, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuan J., Chen J. (2010) J. Biol. Chem. 285, 1097–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Limbo O., Chahwan C., Yamada Y., de Bruin R. A., Wittenberg C., Russell P. (2007) Mol. Cell 28, 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Williams R. S., Moncalian G., Williams J. S., Yamada Y., Limbo O., Shin D. S., Groocock L. M., Cahill D., Hitomi C., Guenther G., Moiani D., Carney J. P., Russell P., Tainer J. A. (2008) Cell 135, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bell N. M., Micklefield J. (2009) ChemBioChem. 10, 2691–2703 [DOI] [PubMed] [Google Scholar]
- 89.Takata M., Sasaki M. S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S. (1998) EMBO J. 17, 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Delacôte F., Lopez B. S. (2008) Cell Cycle 7, 33–38 [DOI] [PubMed] [Google Scholar]
- 91.Karanjawala Z. E., Murphy N., Hinton D. R., Hsieh C. L., Lieber M. R. (2002) Curr. Biol. 12, 397–402 [DOI] [PubMed] [Google Scholar]
- 92.Meulle A., Salles B., Daviaud D., Valet P., Muller C. (2008) PLoS ONE 3, e3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nijnik A., Dawson S., Crockford T. L., Woodbine L., Visetnoi S., Bennett S., Jones M., Turner G. D., Jeggo P. A., Goodnow C. C., Cornall R. J. (2009) J. Clin. Invest. 119, 1696–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen M. F., Lin C. T., Chen W. C., Yang C. T., Chen C. C., Liao S. K., Liu J. M., Lu C. H., Lee K. D. (2006) Int. J. Radiat. Oncol. Biol. Phys. 66, 244–253 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.