Abstract
Smooth muscle cells (SMCs) retain remarkable plasticity to undergo phenotypic modulation in which the expression of smooth muscle markers is markedly attenuated while conversely expression of extracellular matrix (ECM) is dramatically up-regulated. Myocardin is perhaps the most potent transcription factor for stimulating expression of smooth muscle-specific genes; little is known, however, about whether myocardin can orchestrate ECM expression to act in concert with smooth muscle differentiation program. In this study, we demonstrated myocardin coordinate smooth muscle differentiation by inducing transcription of microRNA-143 (miR-143), which attenuates ECM versican protein expression. Previous studies have shown that versican is a chondroitin sulfate proteoglycan of the ECM that is produced by synthetic SMCs and promotes SMC migration and proliferation. Our data demonstrated that myocardin significantly represses versican expression in multiple cell lines, and this occurs through the induction of miR-143 by myocardin. By a modified reverse transcribed PCR, we found that miR-143 specifically binds to the 3′-untranslated region of versican mRNA. Reporter assays validated that miR-143 targets versican 3′-untranslated region through an evolutionarily conserved miR-143 binding site. Furthermore, overexpression of miR-143 significantly represses versican expression, whereas conversely, depletion of endogenous miR-143 results in up-regulation of versican expression. In addition, we demonstrated that myocardin represses versican through induction of miR-143. Finally, we found that the regulation of versican by miR-143 is involved in platelet-derived growth factor BB-induced SMC migration. This study provides the first evidence that myocardin, in addition to activating smooth muscle-specific genes, regulates ECM expression through induction of microRNAs during smooth muscle differentiation.
Keywords: Extracellular Matrix, Gene Expression, MicroRNA, Tissue-specific Transcription Factors, Transcription Regulation
Introduction
Smooth muscle cells (SMCs)2 are the major contractile component of the vascular, respiratory, genitourinary, and digestive systems. Fully differentiated SMCs are characterized by the presence of a unique repertoire of contractile proteins and exhibit a very low rate of synthesis of extracellular matrix (ECM) components. Under many pathological conditions, including hypertension and atherosclerosis, SMCs undergo phenotypic modulation, in which the expression of smooth muscle markers is markedly attenuated, and conversely, the synthesis and expression of ECM is dramatically up-regulated (1). The mechanisms that result in down-regulation of contractile proteins and up-regulation of ECM during phenotypic modulation of smooth muscle are poorly understood.
Serum response factor (SRF) plays a central role in the transcriptional regulation of many smooth muscle-specific genes (2). SRF is an evolutionarily conserved MADS (MDM1, agamous, deficiens, SRF) domain-containing transcription factor, which binds a highly conserved cis-regulatory element CC(A/T)6GG, termed a CArG box, which can be found in the majority of promoters/enhancers of smooth muscle genes (2). The physical association of SRF with various cell-restricted and/or signal-dependent accessory factors confers co-activator or co-repressor activity via ternary complex formation. Of the SRF-associated proteins identified, myocardin is perhaps the most potent for stimulating expression of all CArG-dependent SMC marker genes (3). Although the function of myocardin in regulating smooth muscle-specific genes has been intensively investigated, little is known about whether myocardin can orchestrate ECM expression to act in concert with the smooth muscle differentiation program.
Recent studies have shown that the vascular ECM plays an important role in the phenotypic modulation of SMCs (4). Healthy arteries contain sparse amount of proteoglycans and glycoproteins. In vascular diseases, however, proteoglycans and collagen predominate in the vascular lesions (5). The increasing expression of proteoglycans and collagen, in turn, promotes smooth muscle phenotypic modulation, leading to increased vascular SMC migration, proliferation, and secretion of ECM (6). Versican is a large chondroitin sulfate proteoglycan of the ECM that can be produced by synthetic smooth muscle cells and is increased after vascular injury and accumulates in advanced atherosclerotic plaques (7). Versican has been implicated in several atherogenic events, including stimulation of vascular SMC growth and migration, retention of lipoproteins, and promotion of thrombogenesis (7). In addition to atherosclerosis and restenosis in the cardiovascular system, elevated expression of versican also can be seen in cancer metastasis (8). Although several lines of evidence showing transcriptional and post-transcriptional regulation of versican expression have been provided (9–11), the mechanisms controlling versican expression during smooth muscle differentiation and phenotypic modulation have not been fully elucidated.
In recent years, emerging evidence has demonstrated that microRNAs (miRNAs) play key roles in the control of smooth muscle proliferation and differentiation (12–16). miRNA is a class of ∼22-nucleotide noncoding RNA that can repress gene expression by annealing with complementary sequences in the 3′-untranslated regions (UTRs) of target mRNAs, leading to mRNA degradation and translational inhibition. The expression of a single coding gene can be regulated by many different miRNAs, and each miRNA might control hundreds of distinct target genes, providing a complex functional network regulation at the post-transcriptional level (17). Recent studies have demonstrated that the miR-143 and miR-145 clusters are co-transcribed and are cardiovascular specific miRNAs (13, 18–20). These miRNAs are critical to modulate smooth muscle phenotype by promoting smooth muscle differentiation and repressing proliferation of SMCs (12, 13, 19). In this study, we demonstrate that myocardin coordinates the smooth muscle differentiation program to suppress ECM versican gene expression by inducing transcription of miR-143.
EXPERIMENTAL PROCEDURES
Cell Culture and PDGF-BB Treatment
Mouse 10T1/2 fibroblast cells, green monkey kidney fibroblast COS7 cells, rat A10 and A7r5 aortic SMCs were purchased from ATCC and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and antibiotics. PAC1 SMCs were gifts of Dr. Li Li (Wayne State University). Mouse primary aortic SMCs were prepared from aorta dissected from 4-week-old mice essentially as described previously (21). For PDGF-BB treatment experiments, rat primary aortic SMCs were prepared by enzymatic dispersion from descending aorta of adult Sprague-Dawley rats. Rat primary or PAC1 SMCs were grown to 80–90% confluence and serum-starved for 2 days and then treated with recombinant rat 20 or 50 ng/ml platelet-derived growth factor BB (PDGF-BB) (Calbiochem), as described in the figure legends. Cells treated with vehicle served as control. Following PDGF-BB or vehicle administration, cells were harvested for total RNA to detect microRNA and gene expression by PCR or harvested for protein to measure gene expression by Western blotting.
Adenoviral Construction and Cell Infection
The full-length protein coding region for myocardin cardiac-enriched isoform was subcloned into a multiple cloning site of a pAdTrack vector (22). This vector contains two independent cytomegalovirus-driven transcription cassettes: one for green fluorescent protein (GFP) and one for myocardin, allowing direct observation of the efficiency of transduction by visualization of GFP expression. The myocardin-pAdTrack vector then was electroporated into BJ5183-competent AdEasy cells (Stratagene), resulting in a recombination of the expression cassettes into the AdEasy adenoviral backbone. The recombinant adenoviral DNA was transfected into HEK293 cells for viral packaging, and the adenovirus was harvested for infection as described previously (22, 23). GFP control virus was generated by same method except no insert in pAdTrack vector.
Quantitative Real-time Reverse Transcription (RT)-PCR (qRT-PCR) Analysis
Total RNA was isolated with TRIzol reagent (Invitrogen). For qRT-PCR, 1 μg of RNA was utilized as a template for RT with random hexamer primers using the Superscript first strand cDNA synthesis kit (Invitrogen). PCR was performed with 200 ng of cDNA and SYBR Green PCR master mix (Applied Biosystems) with respective gene-specific primers as we previously reported (24) or otherwise listed in the supplemental Table 1. Quantification of the reaction product was carried out using the ABI7300 real-time detection system. All samples were amplified in duplicate, and every experiment was repeated independently two times. Relative gene expression was converted using the 2−ΔΔCt method against the internal control (acidic ribosomal phosphoprotein P0) RPLP0 house-keeping gene.
Western Blotting
Western blot analysis was carried out essentially as described previously (23, 25, 26). 30 μg of protein were fractionated on 5 or 15% SDS-polyacrylamide gels, electrophoretically transferred to a nitrocellulose membrane. The membrane was then probed with a series of antibodies. Antibodies used in this study were against glyceraldehyde-3-phosphate dehydrogenase (1:2000; Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), GFP (1:2000; Invitrogen), myocardin (M16, 1:2000; Santa Cruz Biotechnology, Inc.), PARP (1:2000; Cell Signaling), SM α-actin (1:10,000; Sigma), SM22α (1:5000; Sigma), versican (LS-C25140, 1:2000; Lifespan Biosciences), and vinculin (1:5000; Santa Cruz Biotechnology, Inc.).
miRNA qPCR
Total RNA isolated with TRIzol from control or treated cells was used to reverse transcribe and convert small non-coding RNAs into quantifiable cDNAs with the QuantiMir RT kit (System Biosciences). Forward primers specific for microRNAs were used to perform quantitative real-time PCR to detect the corresponding microRNA expression. The signals were first normalized to expression levels of the internal control U6 snRNA transcript, and the ΔΔCt method was used to calculate the relative quantity values of microRNA expression levels. Relative Expression = 2−ΔΔCt and ΔΔCt = (Ct experimental − Ct U6) − (Ct control − Ct U6). The relative microRNA expression in control samples was set to 1.
RT-PCR for Detection miRNA Binding to 3′-UTR
Total RNA was collected from early passages of rat primary aortic SMCs using TRIzol (Invitrogen). Reverse transcription was primed with DNA oligonucleotides corresponding to miR-143 (5′-TGAGATGAAGCACTGTAGCTC-3′) and miR-145 (5′-GTCCAGTTTTCCCAGGAATCCCT-3′). A sample primed with random hexamer was used as a positive control, and an unprimed sample was used as a negative control. After reverse transcription, all samples were treated with RNase H at 37 °C for 1 h. The treated RT product was then amplified by PCR with versican or RPLP0 gene-specific primers as listed in supplemental Table 1. PCR product were resolved in 3% agarose gel and stained with ethidium bromide. Pictures of the staining gel were acquired under UV light.
MicroRNA Expression and Reporter Gene Assays
An approximate 500-bp fragment spanning mir-143, mir-145, mir-133a, or mir-365 genomic region was cloned into pMIF-miR vector (System Biosciences) under the control of the cytomegalovirus promoter to express mature microRNAs. For generating versican 3′-UTR reporter, a 1.9-kbp fragment of versican 3′-UTR containing a putative miR-143 binding site was amplified by PCR from mouse genomic DNA and then inserted to 3′ of the firefly luciferase gene in pMir-report vector (Ambion). Mutation of miR-143 seeding sites in versican 3′-UTR was carried out with the QuikChange site-directed mutagenesis kit (Stratagene). Mouse and human versican gene reporters were constructed by cloning fragments of versican promoter into the pGL2B luciferase vectors (Promega). The mouse versican promoter-luciferase reporter includes nucleotides −676 to +153 of the mouse versican gene, and the human versican gene promoter includes nucleotides −678 to +156 relative to the transcription start site. All plasmids were sequenced to verify the integrity of the insert. Transfection was carried out with Fugene6 transfection reagent (Roche Applied Science) as previously described (23). The level of promoter activity was evaluated by measurement of the firefly luciferase activity relative to the internal control thymidine kinase-Renilla luciferase activity using the Dual Luciferase Assay System essentially as described by the manufacturer (Promega). A minimum of six independent transfections were performed, and all assays were replicated at least twice. Results are reported as the mean ± S.E.
siRNA, microRNA Mimic, and Inhibitor Transfection
Control siRNA or siRNA SMARTpool against rat myocardin and SRF were purchased from Dharmacon. siRNA duplex against versican (which targets mouse, rat, and human versican) was designed as 5′-CTGCAAAGATGGTTTCATT-3′ and purchased from Dharmacon. A10 cells were transfected either with control siRNA or myocardin, SRF, or versican siRNA duplex using Lipofectamine 2000 transfection reagent (Invitrogen) following the manufacturer's protocol. 48 h later, total RNA was harvested for qRT-PCR as described above. Scrambled control or mature microRNA mimics for miR-143, miR-145, and miR-133a were purchased from Qiagen and transfected with Lipofectamine 2000 transfection reagent into cells at a final concentration of 20 μm for 2 days. For knocking down endogenous microRNA, the scrambled miRNA inhibitor control and miR-143 inhibitor were purchased from Qiagen and transfected into cells for 2 days, as same as transfection of miRNA mimics. Cells were harvested for qRT-PCR or Western blot as described above.
SMC Migration Assay
PAC1 SMCs were grown in medium containing 10% fetal bovine serum for 12 h post-transfection with anti-miR-143 RNA duplex and then transfected with silencing versican duplex for an additional 24 h. The two-round transfected cells were trypsinized and seeded into Boyden chambers (PET track-etched, 8-μm pores, 24-well format; BD Biosciences) in serum-free Dulbecco's modified Eagle's medium. Chambers were then immersed in 10% fetal bovine serum medium containing 50 ng/ml PDGF-BB for 5 h. The top sides of the membranes were swabbed to remove cells, and then cells on the bottom surface of the membrane were fixed with 4% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole to visualize nuclei, and counted under fluorescence microscopy. Five identically located fields per membrane were averaged for quantification of migrated cell numbers.
SMC Proliferation Assay
The proliferation of A7r5 SMCs was measured by a cell proliferation WST-1 kit (Roche Applied Science) in a 96-well format. Following silencing of miR-143 and versican, A7r5 cells were maintained in 0.2% fetal bovine serum medium for an additional 48 h to allow growth arrest. Cells were then treated with 50 ng/ml PDGF-BB in 5% medium. After 24 h, the rates of proliferation were determined with incubation of 10 μl/well WST-1 for 4 h, and then the absorbance at 450 nm was measured with a plate reader.
RESULTS
Overexpression of Myocardin Inhibits Versican Expression
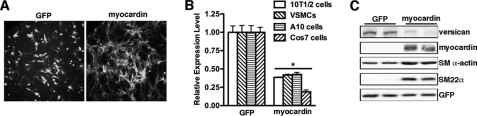
During smooth muscle phenotypic modulation the expression of smooth muscle genes are negatively coupled with expression of a subset of ECM proteins. Because myocardin is a potent transcription factor that is able to activate smooth muscle-specific gene expression in most cell types, we hypothesize that myocardin may also play an important role in coordinating changes in ECM expression. To test this hypothesis, we performed a PCR array screen (SABiosciences) of ECM genes expressed in 10T1/2 cells transduced with myocardin or control GFP adenovirus (Fig. 1A). From this screen, we found that myocardin significantly suppresses expression of a chondroitin sulfate proteoglycan protein, versican (data not shown). Because versican expression is correlated with the synthetic phenotype of SMCs and promotes SMC migration and proliferation (7, 27), we further examined the mechanisms regulating versican expression. In agreement with the role of versican in promoting SMC proliferation and migration, we found that the expression level of versican was significantly elevated in several arterial injury models, including rat carotid artery balloon injury, mouse carotid artery ligation injury, and ApoE knock-out mouse arteries (supplemental Fig. 1). In order to eliminate the possibility that the down-regulation of versican induced by myocardin was cell type-dependent, we next tested the effects of myocardin on versican expression in multiple cell lines. Consistent with a PCR array screen that was performed in mouse 10T1/2 fibroblast cells, overexpression of myocardin significantly suppressed versican mRNA expression by 60–80% in multiple cell lines, including mouse primary vascular SMCs, rat A10 SMCs, and monkey kidney COS7 fibroblast cells (Fig. 1B). At the protein level, myocardin overexpression in 10T1/2 cells significantly elevated expression of SM22α and SM α-actin, whereas versican expression was remarkably down-regulated (Fig. 1C). These results suggest that overexpression of myocardin can suppress versican expression at both the mRNA and protein level.
FIGURE 1.
Effects of overexpression of myocardin on versican gene expression. A, adenovirus encoding GFP or myocardin (myocardin and GFP are encoded on the same adeno-vector by two independent expression cassettes) was transduced into mouse 10T1/2 fibroblast cells for 72 h, and pictures were taken under fluorescence microscope. Note that cell morphology is profoundly altered after overexpression of myocardin. B, adenovirus encoding GFP or myocardin was transduced into mouse 10T1/2 cells, mouse primary aortic vascular SMCs, rat A10 SMCs, and monkey kidney fibroblast COS7 cells, as indicated. After 72 h of infection, cells were harvested with TRIzol for total RNA, and qRT-PCR was conducted to examine endogenous versican expression. *, p < 0.05. C, adenovirus encoding GFP or myocardin was transduced into mouse 10T1/2 cells for 72 h, and then protein lysate was prepared for Western blotting with the indicated antibodies. The equal level of GFP and myocardin transduction is indicated by the expression of GFP immunosignal. Overexpression of myocardin resulted in down-regulation of versican expression at both mRNA and protein levels. Error bars, S.E.
Depletion of Endogenous Myocardin Up-regulates Versican Expression
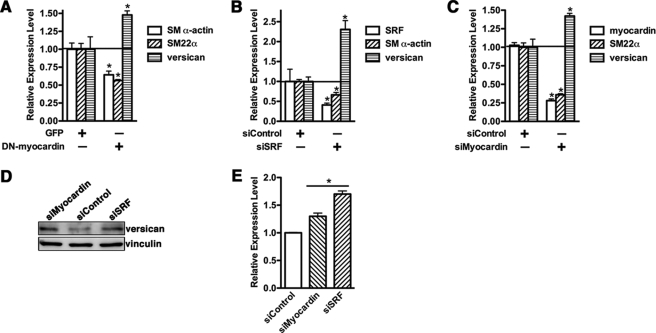
To further assess the role of myocardin in versican expression, we used a carboxyl-terminal deletion mutant of myocardin (amino acids 1–585) that behaves as a dominant negative (DN) (28, 29). A10 SMCs were infected with either GFP control or DN-myocardin adenovirus, and expression of SM α-actin, SM22α, and versican mRNA was analyzed by qRT-PCR. Results from this experiment showed that DN-myocardin suppressed SM α-actin and SM22α mRNA expression significantly, whereas versican expression was increased (Fig. 2A). Because myocardin function is strictly dependent on SRF, we next sought to examine the requirement of SRF on myocardin-mediated versican suppression by knocking down SRF in SMCs. Data from this experiment demonstrate that depletion of endogenous SRF expression to 40% significantly increased versican expression 2-fold while repressing expression of smooth muscle contractile protein genes (Fig. 2B). Furthermore, the role of endogenous myocardin on versican expression was tested by knocking down myocardin expression in SMCs. Consistent with data of SRF depletion, knocking down endogenous myocardin also resulted in a significant increase of versican expression (Fig. 2C). In addition, silencing endogenous SRF or myocardin in A10 SMCs also results in a significant elevation of versican expression at the protein level (Fig. 2, D and E). These results collectively demonstrate that myocardin negatively regulates expression of versican.
FIGURE 2.
Effects of knocking down myocardin level on versican gene expression. A, A10 cells were infected with adenovirus encoding DN myocardin or a GFP control as indicated. 72 h following infection, total RNA was prepared from infected cells and analyzed by qRT-PCR. Overexpression of DN-myocardin significantly inhibits smooth muscle gene expression, whereas it increases versican expression. B, A10 cells were transfected with siRNA duplex against SRF or scrambled siRNA duplex as a control. 72 h following transfection, total RNA was harvested from cells using TRIzol reagent, and RT-PCR was performed to measure endogenous SM α-actin, SRF, and versican mRNA expression. C, siRNA duplex against myocardin or control siRNA duplex was transfected into A10 SMCs, and total RNA was harvested for qRT-PCR as described in B. Changes in expression that were statistically different from the siRNA control (set to 1) are designated by an asterisk (p < 0.05, Student's non-paired t test). D, silencing of myocardin or SRF in A10 smooth muscle cells was performed as described in B and C. 48 h following silencing, duplex transfection cells were harvested for Western blotting with anti-versican antibody. Western blotting data are representative of three independent experiments. E, densitometry of versican or vinculin was measured, and the relative expression of versican was plotted as (silencing of myocardin or SRF/vinculin)/(silencing of control/vinculin). The relative expression of versican in the silencing control group was set to 1. *, p < 0.05. Knockdown of SRF and myocardin resulted in a significant up-regulation of versican expression in SMCs. Error bars, S.E.
Myocardin Induces miR-143 Transcription
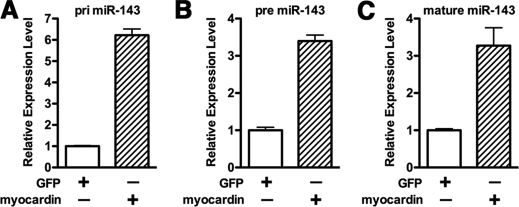
Because the data described above demonstrated that myocardin is a potent suppressor of versican, we next sought to investigate the mechanism by which myocardin suppresses versican expression. First we tested the possibility that myocardin directly affects versican expression at the transcriptional level. We cloned luciferase reporters containing mouse or human versican promoters, which have previously been shown to be predominantly controlled by AP1 and T-cell factor (TCF)/Lymphoid Enhancer Factor transcription factors (30). Data from these reporter assays revealed that myocardin had no effect on versican promoter activity (supplemental Fig. 2) and TCF/LEF-mediated reporter activity (data no shown), whereas the both human and mouse versican promoters can be significantly activated by a constitutively active β-catenin, a known TCF/LEF coactivator. Thus, we reasoned that myocardin-induced suppression of versican is through an indirect mechanism. We hypothesized that myocardin may be acting through inducing microRNA expression. MicroRNA can suppress gene expression through inducing target mRNA degradation and/or translational inhibition. To identify possible microRNA targets, we searched for miRNAs that would be predicted to bind to the versican 3′-UTR and whose promoter region contains a CArG box(es). From this search, miR-143 was identified as a potential candidate. Prior to completion of our studies, several independent reports demonstrated that miR-143/145 are generated from a bicistronic primary RNA that is induced by myocardin in cardiomyocytes (13, 20). We have independently confirmed that overexpression of myocardin markedly induces primary, precursor, and mature miR-143 expression in 10T1/2 cells (Fig. 3, A–C) and other cell types (data not shown), suggesting that this induction by myocardin is at the transcriptional level.
FIGURE 3.
Myocardin induces miR-143 transcription. 10T1/2 cells were transduced with adenovirus encoding myocardin or GFP as a control for 72 h. Subsequently, cells were harvested with TRIzol, and primary (pri) (A), precursor (pre) (B), and mature form of miR-143 (C) were measured by qPCR as indicated. Overexpression of myocardin can significantly induce miR-143 transcription. Error bars, S.E.
miR-143 Binds to the Versican 3′-UTR
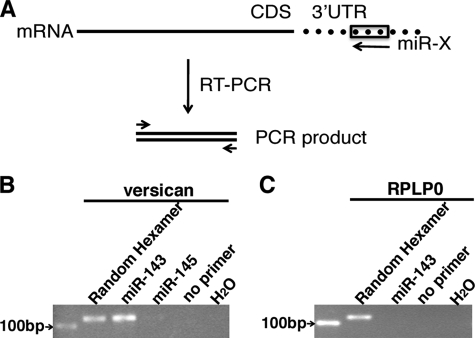
Data described above have shown that transcription of miR-143 can be induced by overexpression of myocardin, and miRBase and miRNA.org prediction algorithms suggest that miR-143 may target versican mRNA. To confirm this latter prediction, we used a miR-143 oligonucleotide as a primer in a reverse transcription reaction to examine whether it would prime the versican mRNA harvested from primary rat aortic SMCs (Fig. 4A). Data from this modified RT-PCR method revealed that an oligonucleotide corresponding to miR-143, but not miR-145, primed first strand synthesis from the versican mRNA (Fig. 4B), whereas a house-keeping gene control RNA, PRLP0, cannot be primed by miR-143 (Fig. 4C), suggesting that a miR-143 binding site exists in the versican 3′-UTR.
FIGURE 4.
miR-143 binds to versican 3′-UTR. A, schematic diagram for the RT-PCR method to determine the interaction of miR-143 with versican 3′-UTR. An oligonucleotide corresponding to miR-X is utilized as a reverse primer to perform the RT reaction, and subsequently the RT product was amplified with a pair of gene-specific primers by PCR. CDS, coding sequence; UTR, untranslated region. B and C, RT-PCR analysis of versican and house-keeping gene RPLP0 using a miR-143 or miR-145 oligonucleotide to prime first-strand synthesis. Following extraction of total RNA from primary rat aortic SMCs, RT was conducted with either a DNA oligonucleotide corresponding to miR-143 or miR-145, a random hexamer (positive control), or no primer (negative control), as indicated. Subsequently, PCR was performed with primers specific to versican, house-keeping gene PRLP0, or water, and the PCR product was separated in agarose gel and visualized with ethidium bromide staining. A 100-bp DNA marker was loaded as a reference. By this modified RT-PCR approach, miR-143 was found to specifically bind to versican 3′-UTR.
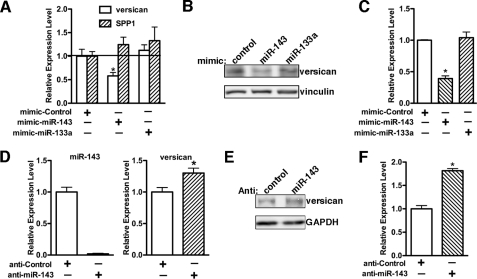
miR-143 Suppresses Versican Expression
To verify that versican is indeed the cognate target of miR-143, we first determined the effects of the overexpression of miR-143 on endogenous expression of versican at the mRNA level by qRT-PCR (Fig. 5A) and at the protein level by Western blot analysis (Fig. 5, B and C). Transfection of mimic miR-143 into 10T1/2 cells produced a significant reduction of versican expression by 50% at the mRNA level and 60% at the protein level without affecting expression of another matrix gene, SPP1 (secreted phosphoprotein 1). In contrast, overexpression of miR-133a had no effect on versican or SPP1 mRNA expression (Fig. 5A). We next sought to assess role of the endogenous miR-143 in versican expression by a loss-of-function assay. Transfection of an anti-miR-143 RNA oligonucleotide dramatically knocked down endogenous miR-143 expression to 5% and resulted in a significant increase in expression of versican mRNA and protein to 1.3- and 1.8-fold, respectively (Fig. 5, D–F), suggesting that miR-143 affects versican mRNA stability and translation.
FIGURE 5.
miR-143 suppresses versican expression. A, mimic RNA oligonucleotides for scrambled control, miR-143, and miR-133a were transfected into 10T1/2 cells for 72 h. Subsequently, cells were harvested with TRIzol, and qRT-PCR was performed to measure versican and SPP1 mRNA expression (A), or cell protein was collected for Western blot to detect versican protein expression (B), and the relative expression of versican from three independent experiments was quantified and plotted in C. *, p < 0.05. Overexpression of miR-143 results in a significant down-regulation of versican expression at both mRNA and protein levels. D, the scrambled miRNA inhibitor control or miR-143 inhibitor was transfected into A10 SMCs for 2 days. Cells were harvested for qPCR to measure miR-143 expression or qRT-PCR to examine versican mRNA expression. Protein lysates were also collected from the transfected cells for Western blot analysis to measure versican protein expression (E). The relative expression of versican from three independent experiments was quantified and plotted in F. *, p < 0.05. Knockdown of endogenous miR-143 led to a significant up-regulation of versican expression, and this enhancement effect can be seen more dramatically at the protein level. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Error bars, S.E.
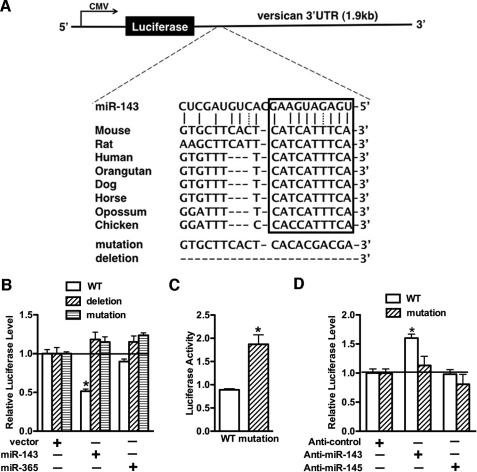
miR-143 Directly Targets Versican 3′-UTR
miRBase and miRNA.org prediction algorithms indicated that the 3′-UTR of the mouse versican gene harbors a putative consensus site for miR-143 binding, and this binding site is evolutionarily conserved among the species of vertebrates (Fig. 6A). To experimentally validate that versican is a miR-143 target gene, transient transfection experiments were performed using the luciferase reporters containing the versican 3′-UTR region, including the putative miR-143 binding site. Data from these experiments revealed that luciferase activity of the wild-type versican 3′-UTR luciferase reporter was significantly decreased by 50% in the presence of miR-143 but not a control miR-365, whereas luciferase activity of constructs in which the miR-143 binding site was mutated or deleted were not affected by miR143 overexpression (Fig. 6B). These data suggest that miR-143 modulates versican expression through the consensus miR-143 binding site in the versican 3′-UTR. Furthermore, mutation of this miR-143 binding site results in a significant increase in reporter activity in A10 SMCs, and knocking down endogenous miR-143 significantly augments the wild type reporter without affecting the mutant reporter in A10 SMCs (Fig. 6, C and D).
FIGURE 6.
miR-143 directly targets versican 3′-UTR. A, schematic diagram of the luciferase reporters containing wild type (WT), deletion, or mutation of the putative miR-143 binding site in the 3′-UTR of versican. The boxed region indicates an evolutionarily conserved “seed” sequence among vertebrate species for miR-143 target recognition. The strategy for generating deletion or mutation of versican 3′-UTR reporter is shown at the bottom. B, 10T1/2 cells were co-transfected with miR-143 or miR-365 expression plasmid and the luciferase reporter constructs harboring the wild type, mutation, or deletion of the predicted miR-143 binding site of versican 3′-UTR, as indicated. Co-transfection of miR-143 significantly inhibits luciferase activity of the reporter harboring wild type versican 3′-UTR, which was abolished when the potential miR-143 target site was mutated or deleted. *, p < 0.05. C, a wild type mouse versican 3′-UTR luciferase construct or a mutant construct containing the mutation of a potential miR-143 binding site and a minimal thymidine kinase-driven Renilla luciferase internal control plasmid were transfected into A10 SMCs, and luciferase activity was measured. The data presented are the luciferase activity normalized with Renilla internal control and mean ± S.E. of six samples. Statistical differences in the activity of the two reporter genes are indicated by an asterisk (p < 0.001). Mutation of the putative miR-143 binding site in the versican 3′-UTR reporter results in increasing luciferase activity. D, A10 cells were transfected with either control miRNA inhibitor (Anti-control), miR-143 inhibitor (Anti-miR-143), or miR-145 inhibitor (Anti-miR-145) for 12 h and then were transfected with luciferase reporter containing wild type or miR-143 binding site mutant of versican 3′-UTR together with a minimal thymidine kinase (TK) promoter-Renilla reporter gene. 24 h later, promoter activity was measured by a dual luciferase assay. Reporter activity was normalized to a Renilla luciferase internal control and expressed relative to control miRNA inhibitor transfections (set to 1). Data are presented as mean ± S.E. of six samples. Silencing endogenous miR-143 augmented wild type versican 3′-UTR reporter activity but had no effect on mutant. *, p < 0.05. Error bars, S.E.
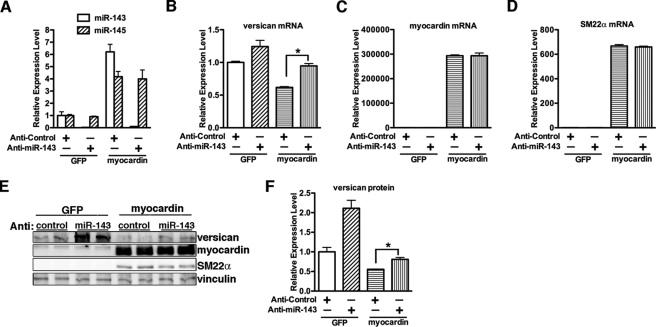
Myocardin-mediated Suppression of Versican Requires miR-143 Expression
To assess if miR-143 is responsible for the myocardin-dependent down-regulation of versican, miR-143 inhibitor was transfected into 10T1/2 cells. Cells were then transduced with myocardin adenovirus, and the expression levels of versican, myocardin, and the SMC contractile protein gene SM22α were examined by qRT-PCR or Western blotting. Overexpression of myocardin significantly induced miR-143 and miR-145 expression in 10T1/2 cells, and transfection of miR-143 inhibitor specifically knocked down miR-143 levels in the presence or absence of myocardin (Fig. 7A). This depletion of miR-143 significantly attenuated myocardin-induced down-regulation of versican at both the mRNA and protein levels (Fig. 7, B, E, and F) without affecting myocardin-mediated induction of SM22α or altering myocardin expression (Fig. 7, C–E). This result suggests that the myocardin-mediated suppression on versican expression is occurring through myocardin-mediated induction of miR-143.
FIGURE 7.
Myocardin represses versican expression through induction of miR-143. A, 10T1/2 cells were transduced with myocardin or control GFP adenovirus overnight, and miRNA inhibitor for scrambled control or against miR-143 was transfected as indicated. 48 h following transfection, cells were harvested with TRIzol for miRNA reverse transcription, and quantitative PCR was conducted to examine the miR-143 and miR-145 expression. Treatment with miR-143 inhibitor can significantly knock down endogenous and myocardin-mediated induction of miR-143 expression. B–D, versican, myocardin, and SM22α mRNA were measured by qRT-PCR in the myocardin-overexpressed 10T1/2 cells with or without transfection of the miR-143 inhibitor. E and F, versican protein expression was determined by Western blotting (E), and its relative expression was quantified in F. *, p < 0.05. Depletion of miR-143 significantly attenuated myocardin-mediated suppression of versican expression but not on smooth muscle-specific gene SM22α. Error bars, S.E.
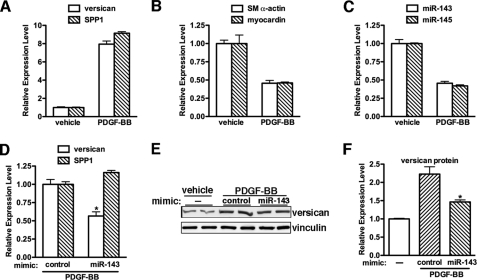
PDGF-BB-induced Expression of Versican Is Partially Dependent on PDGF-BB-mediated Suppression of miR-143
Data described above demonstrated that miR-143 is responsible for myocardin-induced suppression of versican expression in SMC differentiation. We next sought to determine whether changes in miR-143 expression may be involved in up-regulating versican expression during smooth muscle dedifferentiation. PDGF-BB is a potent growth factor known to induce rapid down-regulation of smooth muscle contractile protein genes (31) and promote SMC proliferation and migration (32) while increasing production of matrix proteins, including versican and SPP1 (33, 34). Consistent with previous reports, PDGF-BB treatment on rat primary aortic SMCs increased versican and SPP1 expression, whereas we also found that it decreased SM-specific gene and miR-143 expression following 48-h treatment (Fig. 8, A–C). To explore the role of miR-143 in PDGF-BB-induced versican expression, rat primary aortic SMCs were transfected with miR-143 or control mimics, followed by PDGF-BB treatment for 48 h, and then versican expression was assayed by qRT-PCR or Western blotting. Data from these experiments revealed that exogenous miR-143 significantly attenuated PDGF-BB-mediated up-regulation of versican expression to 50% at both mRNA and protein levels without affecting PDGF-BB-induced SPP1 expression (Fig. 8, D–F). These data demonstrated that PDGF-mediated suppression of miR-143 is involved in up-regulation of versican during smooth muscle dedifferentiation.
FIGURE 8.
PDGF induces versican expression through inhibition of miR-143. Rat primary aortic SMCs were treated with 20 ng/ml PDGF-BB or vehicle for 48 h. Cells were then harvested with TRIzol for total RNA, and qRT-PCRs were performed to measure matrix protein versican and SPP1 (A), smooth muscle gene SM α-actin and myocardin (B), and microRNA miR-143 and miR-145 expression (C). D, qRT-PCR was conducted to examine the versican and SPP1 expression in rat primary SMCs that were PDGF-BB-treated for 48 h following transfection with miR-143 mimic or control mimic overnight. The PDGF-BB treatment-induced relative expression of versican or SPP1 in mimic control transfected cells was set to 1. n = 4. *, p < 0.05. E and F, PDGF-BB-treated rat primary aortic SMCs with or without transfection of mimic miR-143 were harvested for Western blotting with anti-versican antibody. The relative expression of versican was quantified and is plotted in F. The relative expression of versican in vehicle-treated cells was set to 1. *, p < 0.05. Exogenous expression of miR-143 significantly attenuated PDGF-BB-induced versican expression. Error bars, S.E.
Regulation of Versican by miR-143 Is Crucial for PDGF-BB-induced SMC Migration
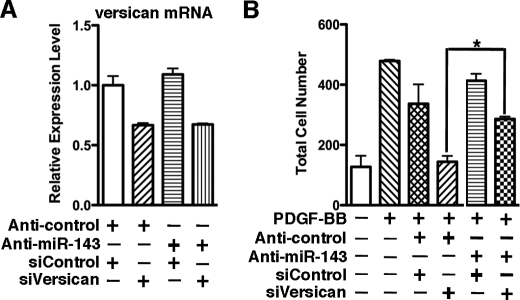
Data described above demonstrated that PDGF-BB-induced suppression of miRNA-143 contributes the up-regulation of versican. We next sought to determine whether this regulatory mechanism affects the SMC phenotype. As determined by a transwell migration assay, transfection of silencing duplex against versican in PAC1 SMCs can successfully knock down 40% of endogenous expression of versican and attenuated PDGF-BB-induced SMC migration (Fig. 9, A and B). However, sequential knockdown of endogenous miR-143 and versican in PDGF-BB-treated SMCs can significantly rescue the depletion of veriscan-mediated inhibition of SMC migration, suggesting that the regulation of versican by miR-143 is essential for PDGF-BB-induced SMC migration. In contrast, this regulatory mechanism is not involved in SMC differentiation, apoptosis, and proliferation (supplemental Fig. 3).
FIGURE 9.
Sequential knockdown of miR-143 and versican in PDGF-BB treated SMCs can significantly rescue the depletion of veriscan-mediated inhibition of SMC migration. A, PAC1 SMCs were sequentially transfected with silencing RNA duplex against versican and/or miR-143, as described under “Experimental Procedures” and harvested for qRT-PCR to measure versican mRNA expression. B, following two-round transfection of silencing versican and/or anti-miR-143 RNA duplex, as indicated, PAC1 SMCs were seeded in a Boyden chamber (8-μm pores; BD Biosciences) in serum-free Dulbecco's modified Eagle's medium and then immersed in 10% fetal bovine serum medium with 50 ng/ml PDGF-BB for 5 h. The migrated cells on the bottom surface of the membrane were fixed with 4% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole (Invitrogen) to visualize nuclei, and counted under fluorescence microscopy. Five identically located fields per membrane were averaged for quantification of migrated cell numbers. p < 0.05. Error bars, S.E.
DISCUSSION
Myocardin is an extremely potent transcription factor that is sufficient and required to induce all CArG-dependent SMC contractile protein genes (3). In this study, we discovered a novel role of myocardin in simultaneously suppressing the ECM protein versican through induction of miR-143. Because high level expression of versican is correlated with SMC dedifferentiation, the decreased expression of versican in response to myocardin suggests that myocardin can orchestrate changes in ECM expression to inhibit proliferation and migration while stimulating the smooth muscle differentiation program. In contrast, during smooth muscle dedifferentiation induced by PDGF-BB treatment, the suppression of miR-143 expression results in up-regulation of versican expression, thereby promoting SMC migration (Fig. 9), while at the same time PDGF-BB causes decreased myocardin and smooth muscle gene expression (Fig. 8).
Versican is a chondroitin sulfate proteoglycan that is present in the extracellular matrix (ECM) of normal blood vessels. Arterial SMCs are the principal source of versican in both arteries and veins, although endothelial cells and myofibroblasts derived from adventitial fibroblasts also have been shown to synthesize versican (27). Versican expression is associated with a synthetic SMC phenotype and increases dramatically in all forms of vascular disease (7). For example, versican is one of the principal genes that are up-regulated in stented and nonstented restenotic lesions (7) and in distinct experimental arteriosclerosis models (supplemental Fig. 1). Interference with versican synthesis by antisense inhibits the proliferation of arterial SMCs, highlighting the importance of the versican component in SMC phenotypic modulation (35). Moreover, versican has been shown to be an important modulator of tumor cell invasion and metastasis (8). Increased expression of the versican is strongly associated with poor outcome for many different cancers, representing one mechanism whereby cancer cells alter their surrounding microenvironment to facilitate malignant growth and invasion (8). Therefore, a greater understanding of the regulation of versican expression may contribute to the development of therapeutic methods not only to cure or prevent neointimal formation in vascular diseases but also for tumor invasion. There is strong evidence that versican expression can be regulated at the transcriptional and post-transcriptional levels (9, 11, 36). Several transcriptional regulatory elements have been identified in the versican proximal promoter, including AP-1, Sp1, AP-2, and two TCF-4 sites. Furthermore, AP-1 and TCF-4 binding sites have been shown to be the main regulatory regions directing versican production during melanoma progression (36), and the synthesis of versican in vascular SMCs was predominantly regulated by a β-catenin·TCF complex (30). Versican expression is also regulated at the post-transcriptional level by a subset of microRNAs. It has been shown that miR-138 and miR-199a* can directly target versican mRNA 3′-UTR, resulting in down-regulation of versican expression (9, 11). In the current study, we found that miR-143 can also bind to the versican 3′-UTR at an evolutionarily conserved miR-143 binding site (Fig. 6) and suppress versican mRNA and protein expression (Fig. 5). Our findings that versican expression is increased in injured vascular smooth muscle tissues (supplemental Fig. 1) are consistent with recent studies showing that overexpression of miR-143 can significantly reduce balloon injury-induced neointimal formation (19). This is also consistent with the recent finding that vascular SMCs from miR-143/145 knock-out mice are locked in a synthetic state (18–20). In addition, our findings that PDGF-BB mediated up-regulation of versican requires concomitant down-regulation of miR-143 suggest that miR-143 is a critical regulator of SMC phenotypic modulation (Figs. 8 and 9). In further support of this, spontaneous arteriosclerosis plagues were seen in miR-143 and miR-145 double mutant mice with evidence of SMC and macrophage cell accumulation in the neointimal area (18). Given that versican expression in these animals was increased and that versican can promote SMC migration (Fig. 9) and proliferation and attract macrophage adhesion (5), we propose that the up-regulation of versican, at least in part, contributes to this process of neointimal formation.
An unbiased quantitative proteomics approach recently revealed that versican was significantly up-regulated 2.7-fold in miR-143/145 double knock-out mice without affecting versican mRNA expression (18). In contrast, in the current study, we found that depletion of endogenous miR-143 significantly increased versican expression at both the mRNA and protein levels, although more dramatic effects were seen at the protein level (Fig. 5). This discrepancy may be explained by different PCR primers for assessing versican expression or measuring versican expression in a cell culture system versus animal tissues. Nevertheless, the greater effect of miR-143 on versican protein as opposed to mRNA that we observed (Fig. 5) suggests that miR-143 can suppress versican expression both through regulating mRNA degradation and inhibiting translation. We did not find any evidence for a direct regulation of versican by miR-145 (Figs. 4B and 6D), although we cannot completely rule out this possibility. In addition, by TargetScan search, we found that other ECM genes, including collagen (type V α2) and fibronectin (type III domain-containing 5), are potential targets of miR-143 (data not shown). Taken together with previous studies, our study suggests that miR-143 will be a potential therapeutic target for smooth muscle-related vascular diseases.
miR-143 is co-transcribed along with miR-145 in a CArG-dependent manner (13, 20). Overexpression of myocardin and the myocardin family protein, MRTF-A, in mouse cardiomyocytes significantly up-regulated miR-143 and miR-145 expression through an evolutionarily conserved CArG box within the miR-143 and miR-145 cluster gene promoter (20). Consistent with this, overexpression of myocardin in 10T1/2 fibroblast cells and other cell lines also activates miR-143 and miR-145 transcription (Figs. 3 and 7A) (data not shown). Given that myocardin is able to suppress versican expression in multiple cell lines, we believe that the mechanism by which myocardin down-regulates versican is the same. In addition, miR-145 has been shown to be necessary for myocardin-induced smooth muscle differentiation in 10T1/2 cells and suppresses multiple repressors of smooth muscle differentiation, including Klf4, Klf5, and CamkII-δ (12, 13, 20). Together with our finding that miR-143 represses ECM versican expression, these studies highlight that myocardin coordinates smooth muscle differentiation while simultaneously repressing ECM expression by inducing miR-143 and miR-145 transcription. In summary, this study reveals a novel role of myocardin orchestrating changes in ECM expression via induction of miRNA transcription to inhibit SMC migration while simultaneously promoting smooth muscle differentiation.
Supplementary Material
Acknowledgment
We thank Dr. Paul Herring for critical reading of the manuscript.
This work was supported by a grant from the Albany Medical College start-up fund and an American Heart Association Scientist Development Grant (to J. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- SMC
- smooth muscle cell
- ECM
- extracellular matrix
- miRNA or miR
- microRNA
- UTR
- untranslated region
- PDGF
- platelet-derived growth factor
- GFP
- green fluorescent protein
- RT
- reverse transcription
- qRT-PCR
- quantitative reverse transcription PCR
- SM
- smooth muscle
- siRNA
- small interfering RNA
- TCF
- T-cell factor
- SRF
- serum response factor.
REFERENCES
- 1.Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 2.Miano J. M. (2003) J. Mol. Cell Cardiol. 35, 577–593 [DOI] [PubMed] [Google Scholar]
- 3.Pipes G. C., Creemers E. E., Olson E. N. (2006) Genes Dev. 20, 1545–1556 [DOI] [PubMed] [Google Scholar]
- 4.Raines E. W. (2000) Int. J. Exp. Pathol. 81, 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wight T. N. (2008) Front. Biosci. 13, 4933–4937 [DOI] [PubMed] [Google Scholar]
- 6.Evanko S. P., Angello J. C., Wight T. N. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 1004–1013 [DOI] [PubMed] [Google Scholar]
- 7.Wight T. N., Merrilees M. J. (2004) Circ. Res. 94, 1158–1167 [DOI] [PubMed] [Google Scholar]
- 8.Ricciardelli C., Sakko A. J., Ween M. P., Russell D. L., Horsfall D. J. (2009) Cancer Metastasis Rev. 28, 233–245 [DOI] [PubMed] [Google Scholar]
- 9.Morton S. U., Scherz P. J., Cordes K. R., Ivey K. N., Stainier D. Y., Srivastava D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17830–17835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahmani M., Wong B. W., Ang L., Cheung C. C., Carthy J. M., Walinski H., McManus B. M. (2006) Can. J. Physiol. Pharmacol. 84, 77–92 [DOI] [PubMed] [Google Scholar]
- 11.Lee D. Y., Shatseva T., Jeyapalan Z., Du W. W., Deng Z., Yang B. B. (2009) PLoS One 4, e4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Y., Liu X., Yang J., Lin Y., Xu D. Z., Lu Q., Deitch E. A., Huo Y., Delphin E. S., Zhang C. (2009) Circ. Res. 105, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordes K. R., Sheehy N. T., White M. P., Berry E. C., Morton S. U., Muth A. N., Lee T. H., Miano J. M., Ivey K. N., Srivastava D. (2009) Nature 460, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis B. N., Hilyard A. C., Nguyen P. H., Lagna G., Hata A. (2009) J. Biol. Chem. 284, 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X., Cheng Y., Zhang S., Lin Y., Yang J., Zhang C. (2009) Circ. Res. 104, 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C. (2008) Clin. Sci. 114, 699–706 [DOI] [PubMed] [Google Scholar]
- 17.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 18.Boettger T., Beetz N., Kostin S., Schneider J., Krüger M., Hein L., Braun T. (2009) J. Clin. Invest. 119, 2634–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elia L., Quintavalle M., Zhang J., Contu R., Cossu L., Latronico M. V., Peterson K. L., Indolfi C., Catalucci D., Chen J., Courtneidge S. A., Condorelli G. (2009) Cell Death Differ. 16, 1590–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin M., Small E. M., Sutherland L. B., Qi X., McAnally J., Plato C. F., Richardson J. A., Bassel-Duby R., Olson E. N. (2009) Genes Dev. 23, 2166–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M., Fang H., Zhou J., Herring B. P. (2007) J. Biol. Chem. 282, 25708–25716 [DOI] [PubMed] [Google Scholar]
- 22.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J., Hoggatt A. M., Herring B. P. (2004) J. Biol. Chem. 279, 15929–15937 [DOI] [PubMed] [Google Scholar]
- 24.Zhou J., Zhang M., Fang H., El-Mounayri O., Rodenberg J. M., Imbalzano A. N., Herring B. P. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J., Hu G., Herring B. P. (2005) Mol. Cell. Biol. 25, 9874–9885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J., Herring B. P. (2005) J. Biol. Chem. 280, 10861–10869 [DOI] [PubMed] [Google Scholar]
- 27.Yao L. Y., Moody C., Schönherr E., Wight T. N., Sandell L. J. (1994) Matrix Biol. 14, 213–225 [DOI] [PubMed] [Google Scholar]
- 28.Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 29.Du K. L., Ip H. S., Li J., Chen M., Dandre F., Yu W., Lu M. M., Owens G. K., Parmacek M. S. (2003) Mol. Cell. Biol. 23, 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahmani M., Read J. T., Carthy J. M., McDonald P. C., Wong B. W., Esfandiarei M., Si X., Luo Z., Luo H., Rennie P. S., McManus B. M. (2005) J. Biol. Chem. 280, 13019–13028 [DOI] [PubMed] [Google Scholar]
- 31.Holycross B. J., Blank R. S., Thompson M. M., Peach M. J., Owens G. K. (1992) Circ. Res. 71, 1525–1532 [DOI] [PubMed] [Google Scholar]
- 32.Jawien A., Bowen-Pope D. F., Lindner V., Schwartz S. M., Clowes A. W. (1992) J. Clin. Invest. 89, 507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan-Albuquerque N., Garat C., Desseva C., Jones P. L., Nemenoff R. A. (2003) J. Biol. Chem. 278, 39830–39838 [DOI] [PubMed] [Google Scholar]
- 34.Wang X., Louden C., Ohlstein E. H., Stadel J. M., Gu J. L., Yue T. L. (1996) Arterioscler. Thromb. Vasc. Biol. 16, 1365–1372 [DOI] [PubMed] [Google Scholar]
- 35.Huang R., Merrilees M. J., Braun K., Beaumont B., Lemire J., Clowes A. W., Hinek A., Wight T. N. (2006) Circ. Res. 98, 370–377 [DOI] [PubMed] [Google Scholar]
- 36.Domenzain-Reyna C., Hernández D., Miquel-Serra L., Docampo M. J., Badenas C., Fabra A., Bassols A. (2009) J. Biol. Chem. 284, 12306–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.