Abstract
Conventional protein kinase C (PKC) isoforms are essential serine/threonine kinases regulating many signaling networks. At cell adhesion sites, PKCα can impact the actin cytoskeleton through its influence on RhoGTPases, but the intermediate steps are not well known. One important regulator of RhoGTPase function is the multifunctional guanine nucleotide dissociation inhibitor RhoGDIα that sequesters several related RhoGTPases in an inactive form, but it may also target them through interactions with actin-associated proteins. Here, it is demonstrated that conventional PKC phosphorylates RhoGDIα on serine 34, resulting in a specific decrease in affinity for RhoA but not Rac1 or Cdc42. The mechanism of RhoGDIα phosphorylation is distinct, requiring the kinase and phosphatidylinositol 4,5-bisphosphate, consistent with recent evidence that the inositide can activate, localize, and orient PKCα in membranes. Phosphospecific antibodies reveal endogenous phosphorylation in several cell types that is sensitive to adhesion events triggered, for example, by hepatocyte growth factor. Phosphorylation is also sensitive to PKC inhibition. Together with fluorescence resonance energy transfer microscopy sensing GTP-RhoA levels, the data reveal a common pathway in cell adhesion linking two essential mediators, conventional PKC and RhoA.
Keywords: Adhesion, Fluorescence Resonance Energy Transfer (FRET), Phosphatidylinositol, Protein Kinase C (PKC), Signal Transduction, Cell Adhesion, Cell Signaling, Phosphospecific Antibody, RhoGDIalpha, RhoGTPase
Introduction
Protein kinase C (PKC)6 isoforms have a wide array of functions in cell adhesion, cytoskeleton, trafficking, and polarity (1–3) to transcription, survival, and differentiation (4). Their activities can intersect with a number of signaling cascades and networks and are stimulated through several distinct receptor families. Broadly, there are three PKC subfamilies, conventional, novel, and atypical, and they operate at different subcellular sites with discrete functions (5, 6). The PKCα conventional isoform may regulate a variety of functions, including redox reactions and cell survival, often upstream of extracellular signal-regulated kinase (ERK) and phosphatidylinositol 3-kinase/AKT pathways and NF-κB synthesis. In addition, this isoenzyme can be localized to sites of cell adhesion and regulate actin cytoskeletal function and migration (7–9). Besides the canonical pathway of PKC activation through phospholipase C-mediated generation of diacylglycerol and inositol trisphosphate, a second method of PKCα activation is known involving uncleaved phosphatidylinositol 4,5-bisphosphate (10–12). Moreover, in vitro experiments reveal that Ca2+ is not required for this activation (13) and is consistent with data showing high PtdIns(4,5)P2 affinity for the C2 domain of PKCα, mediated by a polybasic cluster of amino acids, which leads to persistence of membrane association (14, 15) with decreased Ca2+ requirement (16).
Downstream events from PKCα in fibroblast adhesion to fibronectin, for example, are not known other than an eventual up-regulation of GTP-RhoA levels, with concomitant cytoskeletal reorganization (8, 17). The localization of p190RhoGAP may be intrinsic to the process (17), although a precise role for PKCα activity is unclear. The three mammalian RhoGDI isoforms are cytosolic proteins that sequester RhoGTPases in their GDP-bound form, effectively forming sinks that remove the G proteins from cycling through the guanine exchange factor and GTPase-activating protein-mediated activation/deactivation process (18, 19). In addition, RhoGDI can interact with cytoskeletal proteins such as ezrin-radixin-moesin that could target RhoGTPase activity at a subcellular level (20). Therefore, it is an oversimplification to ascribe only a sequestration function to GDI proteins. Because RhoGDIα, the most abundant isoform, binds several RhoGTPases, including GDP-RhoA, -Rac1, and -Cdc42 (18), regulatory mechanisms that control its function have come under some recent scrutiny (19, 21). RhoGDIα has two domains, an N-terminal domain of around 70 amino acids and an immunoglobulin-like C-terminal domain of ∼130 amino acids. NMR studies have shown that the N-terminal domain of the free protein is flexible in solution but with residues 9–20 and 36–58 forming transient helices (22). This part of the GDI molecule is stabilized by interactions with GTPases and is therefore a potential target for regulation through protein modification, such as phosphorylation. Here, we test the hypothesis that RhoGDIα phosphorylation by PKC may represent a major step in the pathway to RhoA, with onward activation of downstream effectors regulating cell adhesion, cytoskeletal, and other cellular events.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Reagents
Rat embryo fibroblasts were maintained in α-minimal essential medium with 5% fetal calf serum (Labtech International), at 37 °C, 10% CO2 and used between passages 6–20. Wild-type Drosophila Schneider 2 (S2) cells were provided by Dr. Finn Werner (University College London) and were maintained in DES Expression medium (Invitrogen) containing 10% fetal calf serum. PKCα-expressing S2 cells were provided by Dr. Ssang-Taek Lim (University of California, San Diego). For selection and maintenance of stably transfected populations, cells were cultured in DES Expression medium with 10% fetal bovine serum and 300 μg/ml hygromycin-B (Invitrogen). MDCK (type II) cells from ATCC were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum (Hyclone), and human erythroleukemia K562 cells from ATCC were in Dulbecco's modified Eagle's medium/F-12 medium with 10% fetal bovine serum. Polyclonal antibodies against RhoGDI (A20 and K21) and RhoA (119) were from Santa Cruz Biotechnology (Heidelberg, Germany). Mouse monoclonal antibodies against Rac1 (clone 23A8) and Cdc42 (clone 44) were from Upstate Biotechnology and BD Transduction Laboratories. Anti-His tag polyclonal antibody was from Cell Signaling Technology. The phosphospecific antibody against RhoGDI Ser(P)-34 was produced by Quality Controlled Biochemicals (on behalf of BIOSOURCE International) using the peptide sequence CPAQK(pS)IQEI (residues 30–38 with an additional N-terminal cysteine) as an immunogen coupled to keyhole limpet hemocyanin. To purify phospho-specific antibodies, two columns were prepared. The peptides CPAQKSIQEI and CPAQK(pS)IQEI were immobilized through the free cysteine residues to maleimide-activated agarose. Serum was first passed through the unphosphorylated peptide affinity column to remove antibodies recognizing nonphosphorylated RhoGDI. Flow-through fractions from the first column were applied to the second column, with the immobilized phosphopeptide. After washing with phosphate-buffered saline, antibodies recognizing phosphopeptide were eluted with 0.1 m glycine HCl, pH 2.8, and the eluates were neutralized with 1 m Tris, pH 8.0, immediately.
PMA, Gö6976, and GF109203X were from Calbiochem; PtdSer, diolein (DL), histone III-S, ATP, GTPγS, NAD, U73122, and bovine plasma fibronectin were from Sigma. Inositides PtdIns(4,5)P2, PtdIns(3,4,5)P3, and PtdIns were from Biomol or Sigma. [γ- 32P]ATP, GTPγ35S, and [32P]NAD were obtained from Amersham Biosciences. PKC αβγ purified from rabbit brain was from Upstate Biotechnology.
Plasmids, cDNAs, Site-directed Mutagenesis, Recombinant Protein Expression, and Purification
Wild-type human RhoA and Rac1 cDNA cloned in pRk5-Myc expression vector were from Dr. A. Hall (Memorial Sloan-Kettering Cancer Center, New York); pGEX-2T RhoA was from Dr. V. Braga (Imperial College London, UK), and pTAT-C3 was from Dr. J. Bertoglio (INSERM, France). cDNAs encoding wild-type RhoGDIα and an N-terminal truncation mutant lacking the first 66 amino acids (RhoGDI(67–204)) in the pHis-parallel vector (23) were a kind gift from Dr. Z. Derewenda (University of Virginia). All plasmids were transformed in the BL21(pLysS) strain, and proteins were purified on a TALON (Qiagen) or Co2+-charged chelating Sepharose fast flow resin (Amersham Biosciences) according to Sheffield et al. (23). The hexahistidine tag was removed by incubating with recombinant tobacco etch virus (AcTEV, Invitrogen) as described by the manufacturer, and the protease, cleaved tags, and any uncleaved protein were removed by metal-chelating affinity chromatography. For full-length RhoGDI, an extra gel filtration step on a HiPrep 16/60 Sephacryl S-100 high resolution column (Amersham Biosciences) was performed on an AKTA high pressure liquid chromatography system (Amersham Biosciences). Proteins were >95% pure as evidenced by Coomassie stain. Protein concentration was calculated with the BCA assay kit (Pierce). Primers for site-directed mutagenesis are listed in supplemental Table 1. cDNAs were confirmed by sequencing, and bacterially expressed proteins were purified as described above.
Expression and Purification of RhoGDI-Rho Protein Complexes
Purification of Rac1-RhoGDI complexes from induced S2 cells was carried out essentially as described by Read and Nakamoto (24). Briefly, cells were harvested in cold wash buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 10% (v/v) glycerol, 20 μg/ml aprotinin, 20 μg/ml leupeptin, 5 μg/ml pepstatin, 1 μm Pefabloc, and 1 mm phenylmethylsulfonyl fluoride) and lysed by three passes through a French press at 20,000 p.s.i. After a 15-min centrifugation at 14,000 rpm, supernatants were centrifuged at 100,000 × g for 1 h. Supernatants were incubated for 1 h with TALON resin, washed in wash buffer, and eluted in wash buffer containing 150 mm imidazole. The eluate was passed five times over an equilibrated anti-Myc-agarose (9E10) column (Sigma), washed in wash buffer, and bound protein complexes eluted with wash buffer containing 100 μg/ml Myc peptide (Sigma).
Yeast strain SY1 transformed with plasmids encoding for FLAG-RhoGDI and His6-RhoA were gifts from Dr. R. Nakamoto (University of Virginia). Yeast growth and purification of a stoichiometric 1:1 protein complex of RhoA-RhoGDI by sequential passage over TALON and FLAG-agarose (Sigma) columns were performed as described previously, as were protein concentration calculations (24). In phosphorylation experiments, tags from the recombinant RhoA and RhoGDI proteins were removed by recombinant tobacco etch virus protease. Tags, uncleaved proteins, and the protease were removed on respective affinity columns. Purity of the RhoGDI-Rho complexes was confirmed by silver staining. The Myc tag on Rac1 was not removed.
Expression and Purification of Recombinant PKCα
Drosophila S2 cells transfected with plasmid encoding full-length PKCα and methods for protein purification were reported previously (25). Recombinant PKC activity was determined by comparing the incorporation of phosphate in histone III-S or RhoGDI with that of commercial PKC αβγ, and comparable activities were used for all experiments.
Other Recombinant Proteins
Recombinant purified vinculin tail protein was a generous gift from Dr. D. Critchley (University of Leicester). Recombinant GST-RhoA and TAT-C3 proteins were purified as described previously (26, 27). Cell lysates and recombinant proteins were analyzed by SDS-PAGE and Western blotting (28). Western blots were analyzed with NIH Image version 1.61.
Detection of Ser-34 Phosphorylation in Endogenous RhoGDI
MDCK cells were serum-starved for 5–8 h and treated with either 200 nm PMA or vehicle (DMSO) for 10 min (29). To confirm ruffle membrane formation in MDCK cells upon PMA treatment, phase contrast images of cells fixed in 2% glutaraldehyde were recorded on an Olympus IX71 microscope with a DP50 camera and ×20 objective. Images were processed with Viewfinder Lite version 1.0 and Studio Lite version 1.0. In some experiments, cells were pretreated with 1 μm PKC inhibitors Gö6976 or GF109203X for 1 h or 3 μm U73122 for 15 min prior to PMA treatment. K562 cells (0.8 × 106) were suspended in serum-free Dulbecco's modified Eagle's medium/F-12 medium and then treated with 800 nm PMA for 5–30 min at 37 °C. Cells were washed with cold Tris-buffered saline containing phosphatase inhibitors (20 mm β-glycerophosphate, 25 mm NaF, and 1 mm orthovanadate) and then lysed with buffer containing 50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 10 mm MgCl2, protease inhibitor mixture, and phosphatase inhibitors. Cleared lysates were analyzed by Western blotting of 12% SDS-polyacrylamide gels with pan-GDI antibodies (A20) and Ser(P)-34 RhoGDI antibody. Rat embryo fibroblasts (REF) and G361 human melanoma cells were treated as above but were not PMA-treated. Plasmids were transfected in MDCK cells with Lipofectamine LTX (Invitrogen) according to the manufacturer's protocol. Ectopically expressed RhoGDI was detected by anti His tag antibodies followed by fluorochrome-conjugated (Alexa Fluor 488/546) secondary antibodies (Invitrogen).
Pulldown of RhoGTPases with Bacterially Expressed GDIs
REF in growth medium were lysed in 50 mm Tris, pH 7.6, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, protease inhibitor mixture (Roche Applied Science), 20 mm β-glycerophosphate, 25 mm NaF, and 1 mm orthovanadate. Cleared lysates were incubated with bacterially expressed GDIs (wild type or mutant) immobilized onto Talon beads at 4 °C for 1 h. After washing beads with lysis buffer three times, proteins pulled down with GDIs were separated by SDS-PAGE and analyzed by Western blotting with RhoA, Rac1, and Cdc42 antibodies.
Kinase Assay
Protein kinase C assays were performed as described previously (13). Calcium chloride was added to 750 μm when PtdSer/DL were the lipid co-factors and was omitted or included at the indicated concentrations when PtdIns(4,5)P2 was used. Reactions proceeded for 10 min with histone III-S or 30 min for RhoGDI or vinculin substrates at room temperature. Samples were resolved by 15% SDS-PAGE, and phosphorylation was detected by autoradiography on a Fuji-BAS reader and quantified using the TINA-BAS densitometry software. For Western blotting [γ-32P]ATP was omitted from the reaction mixture; 25 ng of phosphorylated protein were resolved by 15% SDS-PAGE and immunoblotted as described above.
For determination of phosphorylation stoichiometry, 10 pmol of RhoGDI protein (free or complexed to RhoA) were phosphorylated for 1 h with 10 ng of PKCαβγ in a total of 20 μl of reaction buffer containing 50 mm HEPES, pH 7.4, 3 mm magnesium acetate, 50 μm PtdIns(4,5)P2, and 50 μm ATP (1 μCi of [γ-32P]ATP; 3 Ci/mmol, 2 mCi/ml). Reactions were stopped by spotting onto P81 phosphocellulose filters (Whatman) and washed in 1% (v/v) orthophosphate buffer. Filters were analyzed by scintillation counting and converted to moles of phosphate using radiolabel specific activity and calibration of scintillation counting efficiency. Radiolabel incorporated into an equimolar amount of the S34A point mutant was considered background and subtracted from assays of wild-type RhoGDI. Results were obtained from triplicate experiments.
Nucleotide Exchange Reaction and RhoA ADP-ribosylation
Nucleotide exchange assays and ADP-ribosylation experiments were performed as described previously (30, 31). Briefly, 10 pmol of GST-RhoA or RhoA complexed to RhoGDI (either nonphosphorylated or phosphorylated) were diluted to a final volume of 50 μl in nucleotide exchange assay buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 10 mm EDTA) containing 45 pmol of GTPγS (0.5 μCi [35S]GTPγS; 1,115 Ci/mmol, 1 mCi/ml). The nucleotide exchange reactions (at 30 °C) were stopped with cold assay buffer (without EDTA), adsorbed on nitrocellulose filters (0.45 μm; Schleicher and Schuell), and assayed (30). Counts were converted to moles of GTPγS as before.
ADP-ribosylation experiments were performed essentially as described previously (31). Briefly, GST-RhoA fusion protein or 20 μl of kinase or a control containing 500 ng of RhoA protein (∼20 pmol) were brought to a volume of 40 μl in a buffer containing 50 mm Tris-HCl, pH 7.4, 2 mm MgCl2, 1 mm EDTA, and 40 pmol of NAD (1 μCi of [32P]NAD; 1,000 mCi/mmol, 10 mCi/ml). Fifty ng (or amounts indicated) of recombinant TAT-C3 protein (27) were added last. The samples were incubated for 5 min at room temperature, stopped by addition of 5 μl of sample buffer, denatured at 100 °C, and resolved by 15% SDS-PAGE for visualization by autoradiography.
Silencing of Rat RhoGDIα and Ectopic Expression of Human RhoGDIα Proteins
Oligonucleotide-based silencing of rat RhoGDIα was by pre-designed sense and antisense oligonucleotides obtained from Qiagen. The target sequence was Rn_Arhgdia_1_HP (bases 393–413). Annealed duplexes were transfected in REF cells with Oligofectamine (Invitrogen). Expression levels of RhoGDIα were determined after 48 h by Western blotting, with a luciferase sequence negative control (28). For expression of human RhoGDIα (wild type and S34A and S34D mutants), the cDNA was amplified by PCR from the pHis parallel vector using the primers F1 and R1 (see supplemental material). The amplified PCR product was digested, ligated into the KpnI/EcoRI site of the pcDNA3.1/HisA vector (Invitrogen), and sequenced. Recombinant RhoGDIα contained a hexahistidine tag to distinguish it from endogenous RhoGDIα. Plasmids were transfected in REF cells as described above. Because human RhoGDIα contains the same sequence as rat RhoGDIα siRNA target sequence, silent mutations of five nucleotides in the target sequence were introduced by overlap extension PCR. Primers F1 and 193:5′ or R1 and 192:5′ were used for first PCR. The second PCR used the first PCR products and primers F1 and R1. The amplified PCR product was subcloned into the KpnI and EcoRI sites of pcDNA3.1/HisA.
Confocal Microscopy and FRET Analysis
REF were transfected with siRNA specific for rat RhoGDI and imaged on a Zeiss Axiovert 200 M inverted microscope equipped with a Zeiss 510 META confocal head. All images were analyzed with Zeiss software. FRET analysis by acceptor photobleaching was carried out as described previously (32) with some modification. Briefly, REF cells co-transfected with pRaichu 1502 (from M. Matsuda (33)) and pcDNA-RhoGDI (wild type) or pcDNA-RhoGDI mutants (S34A or S34D) were plated onto coverslips, then fixed for 5 min in 4%(w/v) formaldehyde, washed with phosphate-buffered saline, and visualized with a C-Apochromat 40X/1.2-W objective lens using a 458 nm argon laser light and Meta detector (462.6–516.1 nm) for CFP excitation and emission and a 514-nm argon laser light and Meta detector (516–591 nm) for YFP. FRET efficiency was measured by acceptor photobleaching; pre-bleach CFP and YFP images were collected simultaneously following excitation at 458 nm for CFP and 514 nm for YFP. A selected region was irradiated with the 514-nm laser line (100% intensity, 60 iterations, using a 458/514-nm dual dichroic mirror) to bleach YFP. Post-bleach CFP and YFP images were collected simultaneously (at 458/514 nm) immediately following photobleaching. FRET was measured as an increase in CFP fluorescence intensity following YFP photobleaching. FRET efficiency was calculated as 100 × ((CFP post-bleach − CFP prebleach)/CFP post-bleach) using the FRET Macro in the Zeiss Aim software, correcting for CFP and YFP background in each channel.
RESULTS
Conventional PKC Phosphorylates RhoGDIα in Vitro on Serine 34
The recombinant RhoGDIα fusion protein was first cleaved with recombinant tobacco etch virus (23) to remove the hexahistidine tag, because the two threonine residues in the tag linker region are PKC substrates (supplemental Fig. 1A). Similarly, we noted, as published previously, that GST is a PKCα substrate (on Ser-93) (34), ruling out fusion proteins based on this system. Recombinant RhoGDIα was phosphorylated by PKC in the presence of PtdIns(4,5)P2 but much less in the presence of the more commonly used phosphatidylserine/diacylglycerol/calcium (PtdSer/DL/Ca2+). Essentially identical results were obtained with recombinant mammalian PKCα purified from Drosophila S2 cells (25) or a mixture of conventional PKCs (PKCαβγ) purified from rabbit brain (Fig. 1A). Both preparations have low intrinsic activity in the absence of lipid activators (25). As a control, histone III-S was efficiently phosphorylated by both PKC preparations in the presence of PtdSer/DL/Ca2+ and to a lesser degree by PtdIns(4,5)P2 (Fig. 1B), consistent with our previous data (13). We and others have shown that PKCα activity stimulated by PtdIns(4,5)P2 in combination with the cytoplasmic domain of syndecan-4 can be independent of divalent cations (13, 35). This was apparent with RhoGDIα phosphorylation, which was independent of Ca2+ and persisted in the presence of EGTA (Fig. 1A). In contrast, a documented physiological substrate for PKC, vinculin tail (36, 37), showed different characteristics to RhoGDIα (Fig. 1B). PtdSer/DL/Ca2+ provided optimal conditions for phosphorylation, whereas PtdIns(4,5)P2 alone was insufficient for effective PKCα-mediated phosphorylation unless Ca2+ was present in low concentrations (Fig. 1B).
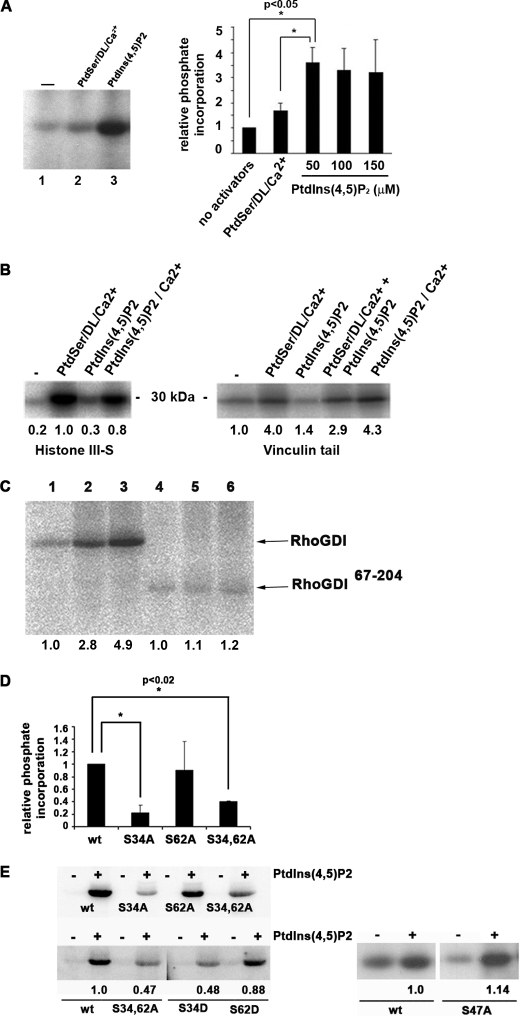
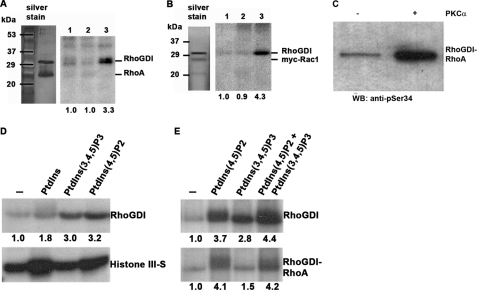
FIGURE 1.
RhoGDIα is a substrate for PKCα on serine 34, in a PtdIns(4,5)P2-dependent manner. A, phosphorylation of RhoGDI by PKCαβγ. Kinase activators were phosphatidylserine/diolein/calcium or phosphatidylinositol 4,5-bisphosphate in the presence of radiolabeled ATP followed by SDS-PAGE and autoradiography. The graph shows quantitation of RhoGDIα phosphorylation by PKCα with increasing inositide levels. The mean ± S.D. of three independent experiments is shown. Phosphorylation in the presence of 50 μm PtdIns(4,5)P2 is significantly higher than that without activators or PtdSer/diolein/Ca2+ (*, p < 0.05). B, phosphorylation of histone IIIS (left) and vinculin tail (right) by recombinant PKCα in the presence of the shown activators and [32P]ATP. Proteins were resolved by SDS-PAGE and autoradiography. Unlike RhoGDIα, both substrates were phosphorylated inefficiently by PKCα in the presence of PtdIns(4,5)P2 unless calcium ions were present (25 μm). Mean values from triplicate experiments are shown. C, although wild-type RhoGDIα was phosphorylated by recombinant PKCα (lanes 1–3), a truncated form consisting of the immunoglobulin domain alone (residues 67–204; lanes 4–6) was not. Lanes 1 and 4, no activators; lanes 2 and 5, PtdSer/diolein/calcium; lanes 3 and 6, PtdIns(4,5)P2. Quantitation is shown, normalized to lanes 1 (for lanes 1–3) and lane 4 (for lanes 4–6). D, phosphorylation of RhoGDIα by PKCαβγ is mostly abrogated by a serine-alanine mutation at residue 34, but a similar mutation of serine 62 has no effect. A representative autoradiogram is shown, together with quantitation by a phosphorimager from triplicate experiments (mean ± S.D., *, p < 0.02 compared with wild type). wt = wild-type RhoGDIα. E, mutation of serine 34 to aspartate also reduced RhoGDIα phosphorylation by PKCαβγ, although an equivalent mutation of serine 62 had no effect. Quantitation from autoradiograms is shown below. S34A/S62A (S34,62A) is a double alanine mutant. Although serine 47 is a potential PKC phosphorylation target, its mutation to alanine had no effect on overall RhoGDIα phosphorylation.
Phosphorylation of RhoGDIα by PKCα was maximal at 50 μm PtdIns(4,5)P2, and under these conditions, calculations revealed between 0.25 and 0.5 mol of phosphate per mol of RhoGDIα, with a mean of 0.3. The necessity for PtdIns(4,5)P2 in RhoGDIα phosphorylation is unusual but consistent with a known site in the C2 region of PKCα that binds this lipid in the absence of Ca2+ and becomes activated (10). The data might also suggest an interaction between PtdIns(4,5)P2 and RhoGDI, but none was detected by plasmon resonance spectroscopy.7
In silico analysis revealed multiple potential phosphorylation sites on RhoGDIα, several of them bearing a strong PKC phosphorylation consensus motif. As RhoGDI proteins are composed of two discrete domains (supplemental Fig. 1D), an N-terminal truncation mutant lacking the first 66 amino acids and comprising only the immunoglobulin domain (RhoGDIα(67–204)) was used as substrate (Fig. 1C). Kinase assays showed that the immunoglobulin-like domain was not phosphorylated by PKCα (Fig. 1C) suggesting that the site(s) were N-terminal or, conceivably, that the N terminus provides a PKCα-docking site for RhoGDIα phosphorylation at a separate site.
Of several serine residue mutations within the N-terminal domain of full-length, wild-type RhoGDIα, that of Ser-34 to Ala had the most marked effect, reducing phosphorylation by recombinant PKCα or PKCαβγ by around 70%, relative to the wild-type protein (Fig. 1D). In contrast, mutation of the consensus site Ser-62 to Ala had no effect on phosphorylation mediated by purified brain PKC (Fig. 1D). Ser-to-Asp substitutions resulted in comparable phosphorylation levels relative to the Ser-to-Ala single point mutants (Fig. 1E), indicating that acquisition of negative charge on residue 62 does not affect phosphorylation of RhoGDIα, therefore excluding co-operativity between residues 34 and 62. A control mutation of Ser-47 was also completed for two reasons. First, it lies in the strongest consensus PKC phosphorylation motif, and second this residue is involved in hydrogen bonding with Thr-37 of Cdc42 that coordinates the Mg2+ ion of the GTPase (38), making it a possible regulatory residue. However, phosphorylation by PKCα was not reduced in a S47A mutant protein (Fig. 1E). Therefore, Ser-34 was identified as the major phosphorylation site.
Serine 34 lies at the base of helix α2, forming part of the helix-loop-helix motif that binds the switch regions of GTPases (helices α2 and α3) and which NMR spectroscopy shows to be unstable or transient when RhoGDI is not complexed to GTPases (22). To further show that this residue could be phosphorylated in the wild-type RhoGDIα protein, a phosphospecific antibody was prepared and characterized. The sequence (C)PAQK(pS)IQEI was used as immunogen, having 100% sequence conservation in mammalian RhoGDIs and may therefore represent a conserved phosphorylation site. The affinity-purified antibody reacted with in vitro PKCα phosphorylated RhoGDIα, with minimal immunoreactivity toward the unphosphorylated protein (Fig. 2A). In addition, virtually no reactivity of the antibody was seen in Western blots of control RhoGDIα lysates from K562 cells (Fig. 2B), whereas reactivity of the antibody against phosphorylated RhoGDIα was detected in phorbol ester-treated K562 cells (Fig. 2B). A general anti-phosphothreonine antibody did not detect the in vitro phosphorylated protein, indicating the absence of threonine phosphorylation on RhoGDI (Fig. 2A). As a positive control for the phosphothreonine antibody, fibroblast lysates from untreated and phosphatase-inhibited cells were analyzed by Western blotting (supplemental Fig. 2). In total, the data show that Ser-34 is a major site in RhoGDIα that can be phosphorylated by PKCα and, as shown below, regardless of whether the GDI is bound to GTPase or not.
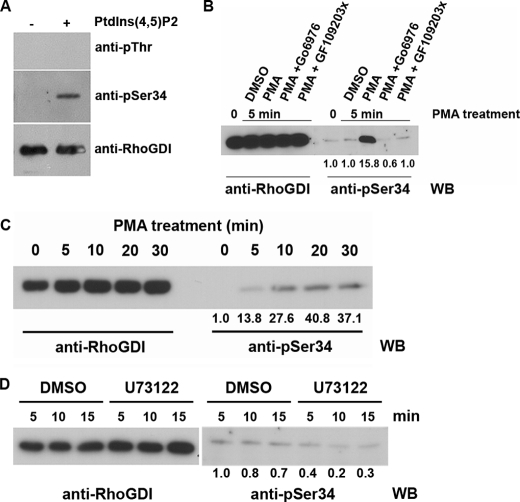
FIGURE 2.
Phosphorylation of RhoGDIα on serine 34 in cells occurs in a PKC-dependent manner. A, 25 ng of in vitro phosphorylated RhoGDIα was immunoblotted with a phosphothreonine antibody, an affinity-purified phosphospecific antibody (pSer-34), and an antibody against total RhoGDIα. B and C, K562 cells undergo rapid phosphorylation of RhoGDIα on serine 34 when treated with 800 nm PMA to promote cell adhesion but not if pretreated with 1 μm PKCα/β inhibitor (Gö69796) or a more general PKC inhibitor (GF109203X). C, phosphorylation is shown to persist for at least 30 min after 800 nm PMA treatment. Total RhoGDIα in the cell lysates is shown on the left as loading controls. Quantitation of the phosphorylation from triplicate Western blots (WB) is shown. D, RhoGDIα phosphorylation in rat embryo fibroblasts is not eliminated by inhibition of phospholipase Cγ. Cells were treated with 3 μm U73122 (or DMSO as vehicle control) for 5, 10, or 15 min, and phosphorylated RhoGDIα was detected by Western blotting with the phosphospecific antibody. Quantitation of triplicates is shown below the blots, and total RhoGDIα is shown in the left 6 lanes.
Serine 34 Phosphorylation of RhoGDIα Is Endogenous and Regulated by Adhesion in a Range of Cell Types
Phosphorylation of endogenous RhoGDIα on Ser-34 in vivo was shown with K562 erythroleukemic cells, epithelial, fibroblastic, and G361 melanoma cells (Figs. 2 and 3 and supplemental Fig. 3). Upon phorbol ester treatment of K562 cells, known to activate integrins and promote cell-matrix adhesion (39), levels of Ser-34, phosphorylated RhoGDIα climbed rapidly from an undetectable level in resting nonadherent cells. Phosphorylation was detectable within 5 min and was maintained through 30 min (Fig. 2, B and C). PKCα is known to be present in K562 cells (40), and treatment of the cells with Gö6976 or GF109203X completely inhibited the rise in RhoGDI phosphorylation (Fig. 2B). The former inhibitor is known for its more specific inhibition of conventional PKC isoforms, particularly PKCα. The GF109203X inhibitor has a wider spectrum of PKC isoform inhibition (41).
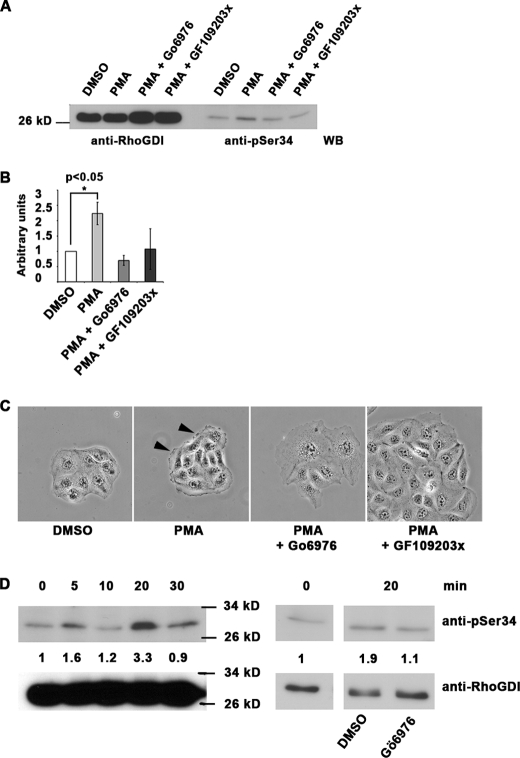
FIGURE 3.
Serine 34 phosphorylation of RhoGDIα is a regulator of PKC-mediated adhesion modulation in MDCK cells. A–C, 10-min PMA treatment (200 nm) of MDCK cells leads to phosphorylation of RhoGDIα on serine 34 that is inhibited by 1 μm Gö6976 PKCαβ inhibitor or a broader spectrum PKC inhibitor (GF109203X). Total RhoGDIα is shown in the four left lanes. Quantitation of triplicates is shown in B, ± S.D. Phase contrast images (C) show the characteristic ruffling induced by 200 nm PMA (arrowheads), but this was not seen where PKC inhibitors (1 μm) were present. Scale bar, 50 μm. D, MDCK cells were treated with 50 ng/ml HGF for 0–30 min, and serine 34 phosphorylation of RhoGDIα was quantitated. Molecular mass marker positions are shown. On the right, sensitivity of this phosphorylation to PKCαβ inhibition at the 20-min time point is shown. Quantitation data for the experiment shown are presented but represent one of three repeated experiments. WB, Western blot.
Rat embryo fibroblasts and G361 melanoma cells adherent to fibronectin-coated surfaces contained Ser-34-phosphorylated RhoGDIα. Some fibroblast cultures were pretreated with the phospholipase C inhibitor, U73122 (Fig. 2D). Inhibitor activity was confirmed by altered distribution of a green fluorescent protein fusion protein with the pleckstrin homology domain of phospholipase Cδ1 that has specificity for PtdIns(4,5)P2 (data not shown) (42). This inhibitor triggered only a partial loss of RhoGDIα phosphorylation (around 50%) in adherent REF cells. This is consistent with in vitro data suggesting that PtdIns(4,5)P2 rather than the products of its degradation by phospholipase C are relevant for this phosphorylation (Fig. 1 and below). Where phosphatase inhibitors were omitted from lysis buffers, markedly decreased reactivity of the phospho-RhoGDIα antibody was seen, again suggesting that the antibody was specific for phosphorylated forms of the GDI (supplemental Fig. 3) and that the phosphorylation is labile.
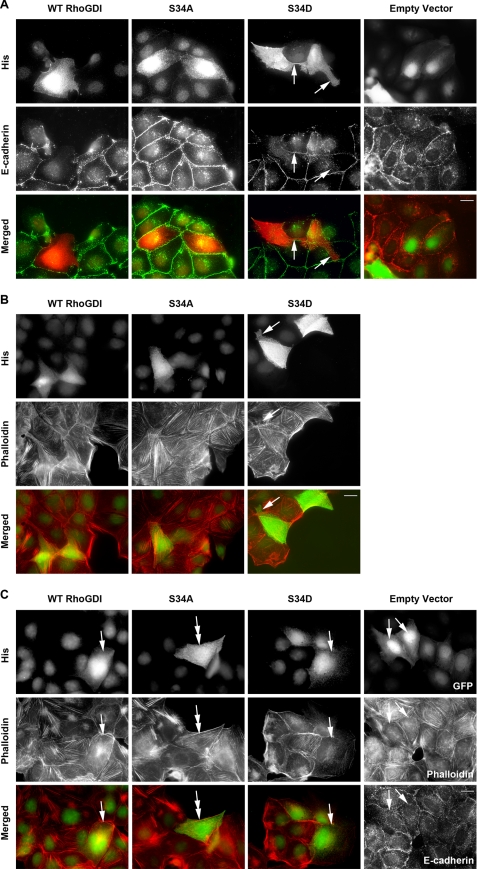
In a well characterized model of PKCα activation and translocation (43), MDCK cells were treated with phorbol ester, which triggers membrane protrusion, and increased GTP-RhoA levels with activation of Rho kinases. This is accompanied by adherens junction loss and F-actin turnover, not microfilament assembly as seen in mesenchymal cells. Western blotting showed that levels of Ser-34-phosphorylated RhoGDIα rapidly increased with phorbol ester treatment, by 2–4-fold, but this was suppressed in parallel cultures by treatment with Gö6976 or GF109203X PKC inhibitors (Fig. 3, A and B). The PKCα and -β isoforms are present in MDCK cells (44). Both PKC inhibitors also suppressed the morphological changes associated with phorbol ester treatment (Fig. 3C). In further experiments, MDCK cells were transfected with empty vector, wild-type RhoGDIα, or mutants with alanine (S34A) that cannot be phosphorylated or aspartic acid (S34D) as a phosphomimetic in place of serine 34. Cultures were untreated or phorbol ester-treated and then stained for E-cadherin or F-actin (Fig. 4) to follow adhesion changes. The most notable effect was that in untreated S34D cells, extensive protrusions were accompanied by poor adherens junction formation, unlike empty vector controls. On PKC activation by phorbol ester, there was an extensive spreading response with loss of F-actin containing microfilament bundles in control (empty vector) or wild-type RhoGDI-expressing cells (Fig. 4C). In phorbol ester-treated S34A cells, however, microfilament bundles persisted, compared with neighboring untransfected cells, with commensurate lack of protrusions. Morphology and adherens junction formation in untreated S34A cells appeared normal. As a whole, the data suggest that RhoGDI (pseudo)phosphorylated at residue 34 has a strong impact on cell adhesion events, resembling the effects of PKC activation by phorbol ester.
FIGURE 4.
RhoGDIα status regulates MDCK cytoskeletal organization. MDCK epithelial cells were transfected with empty vector or RhoGDI cDNAs encoding His-tagged wild-type (WT RhoGDI), S34A, or S34D mutations. Transfected cells were detected by His antibodies or green fluorescent protein co-transfected with the empty vector. Cultures were stained for E-cadherin as a marker of adherens junctions (A and control in C) or phalloidin for F-actin (B and C). Micrographs in C show cultures treated for 10 min with 200 nm phorbol ester, which triggers loss of adherens junctions and reorganization of the actin cytoskeleton, in a Rho-dependent manner. In each case single fluorescence and merged images are shown. A, E-cadherin is abundant in cell-cell contact sites in all cases except S34D, where the cells exhibit protrusions (arrows) and do not have extensive E-cadherin on their surface. Staining for the actin cytoskeleton (B) shows that all transfected and untransfected cells have some F-actin containing microfilament bundles. Protrusions in S34D cells are labeled with arrows. In phorbol ester treated cultures (C), microfilament bundles are lost or are condensed into small aggregates (arrows) in all cases except S34A cells, where bundles persist (double arrows). Increased cell spreading is also characteristic of phorbol ester-treated cells, again not seen in the S34A transfected cells. Scale bar, 10 μm.
Hepatocyte growth factor (HGF) is known to induce morphological and behavioral changes in MDCK cells, with the acquisition of a motile phenotype and cell scattering (45). Roles for GTP-RhoA with adducin as a downstream target have been reported (45, 46). This model was used in further experiments, where PMA is replaced with a physiological ligand. Treatment of cultures with 50 ng/ml HGF led to a rapid but transient increase in RhoGDI phosphorylation on Ser-34, with levels returning to base line in 30 min (Fig. 3D). This phosphorylation was sensitive to Gö6976 PKCα/β inhibitor (Fig. 3D). In total, the data show the widespread nature of RhoGDIα phosphorylation on Ser-34 in a variety of cell types, and its responsiveness to adhesion changes where there are roles for RhoA.
Acquisition of Negative Charge on Serine 34 Decreases RhoGDIα Binding to RhoA and Facilitates RhoA Activation
To characterize the molecular mechanisms regulated by RhoGDIα phosphorylation on Ser-34, RhoGDIα in complexes with Rac1 or RhoA were tested as potential PKCα substrates. For purification of RhoGDIα-protein complexes, human RhoGDI and Rac1 cDNAs were co-expressed recombinantly in Drosophila S2 cells, which can prenylate GTPases, thus producing a functional complex. However, although S2 cells were able to express both RhoGDIα and Rac1 proteins, RhoA expression was minimal, and complexes with RhoGDIα were not obtainable. For this reason, recombinant RhoA-RhoGDIα complex was purified from yeast cells (24). Kinase assays with purified RhoGDIα-RhoA complexes revealed a similar regulation of phosphorylation to monomeric RhoGDIα. Phosphorylation did not occur when PtdSer/DL/Ca2+ were used as activators but was dependent on the presence of PtdIns(4,5)P2 (Fig. 5, A and B). Potentially, by partially opening the protein complex (30), PtdIns(4,5)P2, exposes the phosphoacceptor site thus making it available for phosphorylation. RhoGDI is therefore a substrate for PKCα when complexed to either RhoA or Rac1, whereas the GTPases themselves were not phosphorylated (Fig. 5, A and B). The site of RhoGDIα phosphorylation when complexed to RhoA was confirmed to be Ser-34, as illustrated by Western blotting with the phosphospecific antibody (Fig. 5C). Therefore, bound GTPase does not affect the availability of the Ser-34 residue of RhoGDIα to be phosphorylated by PKCα.
FIGURE 5.
RhoGDIα can be phosphorylated on serine 34 when complexed with RhoA or Rac1. A and B, RhoGDIα was prepared in complex with either RhoA or Rac1 (0.5 μg/assay) and subjected to phosphorylation by 3 ng of PKCα for 10 min. In both figures, a silver-stained SDS-polyacrylamide gel is shown on the left and an autoradiograph on the right. The positions of GDI and GTPase are marked. Lanes 1, no PKC activators; lanes 2, PtdSer/diolein/calcium (250/32/750 μm); and lanes 3, PtdIns(4,5)P2 (50 μm). Neither GTPase is phosphorylated detectably by PKCα. C, RhoGDIα complexed with RhoA (0.5 μg of protein) was mixed with (50 μm) PtdIns(4,5)P2 ± 3 ng of PKCα for 10 min and in a Western blot (WB) by a phosphospecific antibody recognizing Ser(P)-34 RhoGDI. The result shows that the phosphospecific antibody can recognize the RhoGDI even when complexed with RhoA. D and E, three inositides (50 or 25 μm of each where two were mixed) were compared as PKCαβγ activators for RhoGDIα, histone III-S. or RhoA/RhoGDIα complex phosphorylation. For complexed RhoGDIα, PtdIns(4,5)P2 was superior, with no additive effect of PtdIns(3,4,5)P3, although histone III-S phosphorylation was increased by all the inositides. Quantitation is shown below the autoradiograms.
Confirmation of phosphoinositide dependence for RhoGDIα phosphorylation was obtained through the use of alternative inositides, PtdIns and PtdIns(3,4,5)P3. As shown in Fig. 5D, PtdIns(3,4,5)P3 addition resulted in significant phosphorylation of RhoGDIα, whereas PtdIns only minimally induced RhoGDI phosphorylation. On the other hand, PtdIns and PtdIns(4,5)P2 promoted phosphorylation of histone III-S (Fig. 5D), whereas PtdIns(3,4,5)P3 was much less efficient, as reported previously (47). This again demonstrated that the nature of the substrate can influence the activation of the kinase and its subsequent phosphorylation. PtdIns(3,4,5) P3 could act in an additive manner with PtdIns(4,5)P2 for the phosphorylation of monomeric RhoGDIα, but it could not promote the phosphorylation of RhoGDIα complexed with RhoA (Fig. 5E). This suggests PtdIns(4,5)P2 as the physiologically relevant regulator of the association between RhoA and RhoGDI and its ability to promote RhoGDI phosphorylation by PKCα.
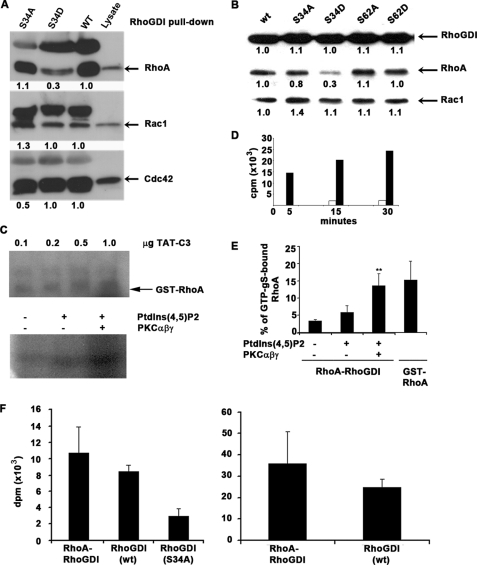
To assess whether Ser-34 phosphorylation regulates complex formation with GTPases, phospho-mimetic or -abolishing mutants of RhoGDIα were used in pulldown assay s from fibroblast lysates (Fig. 6A). When Ser-34 was mutated to Asp, there was significantly reduced RhoGDIα binding to RhoA (by around 75%) but not Rac1 or Cdc42. Mutation to Ala had minimal effect on the binding to any of the three GTPases. In a second assay, mutations of Ser-34 and Ser-62 to either Asp or Ala were compared. Single mutations of Ser-62 to either Asp or Ala had no effect on the binding of either RhoA or Rac1 (Fig. 6B).
FIGURE 6.
Phosphomimetic RhoGDIα at residue 34 leads to decreased retention of RhoA but no change in binding of Rac1 or Cdc42. A, wild type (WT) and RhoGDIα mutated to alanine (S34A) or aspartate (S34D) were compared in GTPase pulldown assays with lysate from rat embryo fibroblasts. In each Western blot, the right lane shows the GTPase detected in the lysate. Quantitation shows that phosphomimetic RhoGDIα has decreased ability to bind RhoA, although binding to Rac1 and Cdc42 is unaffected. Nonspecifically reacting polypeptides are present in the upper portion of each Western blot. B, serine 34 and 62 mutants were compared in their abilities to bind RhoA or Rac1 from rat embryo fibroblast lysates. Although mutation of serine 62 to alanine (S62A) or aspartate (S62D) had no impact on GTPase binding, mutation of serine 34 to aspartate (S34D) reduced RhoA binding specifically. The corresponding alanine mutant at serine 34 (S34A) bound both GTPases similarly to the wild-type (wt) RhoGDIα. Quantitation of the Western blots is shown. C, C3 transferase ADP ribosylates RhoA once the RhoGDIα-RhoA complex has been phosphorylated by PKCαβγ. Varying amounts of TAT-C3 transferase were added to GST-RhoA (upper panel). In the lower panel, 500 ng of RhoA in complex with RhoGDIα were untreated (−) or incubated with 50 μmol of PtdIns(4,5)P2 ± PKCαβγ (lower panel). In both cases, 1 μCi of [32P]NAD was included in a 5-min reaction at room temperature, and autoradiographs are shown from one of four independent experiments. D, exchange of GDP for GTP in RhoA is high in free GTPase but low when complexed to RhoGDIα. Ten pmol of RhoA either alone (black bars) or complexed to RhoGDIα (white bars) was incubated with 30 pmol (0.5 μCi) of GTPγS for the indicated times, and radiolabel incorporation into the GTPase was measured by liquid scintillation spectroscopy. E, phosphorylation of RhoGDIα in complex with RhoA-GDP by PKCαβγ, in the presence of 50μmol PtdIns(4,5)P2, leads to increased nucleotide exchange for GTP. PKC phosphorylation of 10 pmol of RhoGDIα-RhoA complex for 1 h at room temperature was followed by a GTP exchange reaction in the presence of radiolabeled (0.5 μCi) GTPγS. The results are shown as mean ± S.D. from three independent experiments. ** Significantly more nucleotide exchange occurs with RhoGDIα phosphorylation (p < 0.05) than without. F, stoichiometry of RhoGDIα phosphorylation. Left panel, 10 pmol of wild-type RhoGDI complexed with RhoA (left), wild-type RhoGDI (center), or S34A RhoGDI alone (right) were incubated with 10 ng of PKCαβγ and 50 μmol of PtdIns(4,5)P2 for 1 h at room temperature, and radiolabel incorporation was measured. Values are mean ± S.D. from three experiments. Right panel, data from the left panel were recalculated to show stoichiometry of Ser-34 phosphorylation, subtracting background (S34A RhoGDI values) ± S.D. There was no significant difference in phosphorylation whether the RhoGDI was complexed with RhoA or not.
The effect of RhoGDIα phosphorylation on its complex with RhoA was studied using ADP-ribosylation of RhoA by the C3 transferase (Fig. 6C). This approach has been employed before as an indication of partial release of RhoA by RhoGDI following incubation of the RhoA-RhoGDI complex with phosphoinositides (30). The C3 exotransferase is a bacterial toxin that ADP-ribosylates RhoA on Asn-41 (48) and results in RhoA inactivation through enhanced interaction with RhoGDI (49). When purified RhoA-RhoGDI complexes were incubated with TAT-C3, little ADP-ribosylation occurred either in the presence or absence of PtdIns(4,5)P2. In contrast, ADP-ribosylation was evident in samples that had been phosphorylated by PKCα (Fig. 6C). This result indicates that following phosphorylation of RhoGDIα by PKCα, the RhoA switch I region is exposed sufficiently to allow rapid ADP-ribosylation by the C3 exotransferase, suggestive of activation of RhoA. This was further tested by measuring the incorporation of [35S]GTPγS into RhoA complexed to RhoGDIα following magnesium depletion, an in vitro condition that mimics the in vivo effects of guanine exchange factors (26). As shown in Fig. 6D, free RhoA rapidly exchanged its bound nucleotide (GDP) with GTP, and the reaction was saturated after 15 min, with an efficiency of ∼15%. RhoA complexed to RhoGDIα did not undergo nucleotide exchange, as expected, with only 3% incorporating GTPγS. Following phosphorylation of RhoGDIα by PKCαβγ, a mean of 13% of RhoA exchanged to GTP (Fig. 6E), representing a 4-fold increase compared with the untreated control. A significant increase in GTP load could not be observed following incubation with PtdIns(4,5)P2 alone. These combined data indicate that RhoA can be sufficiently released from its inhibitory complex with RhoGDI following phosphorylation of the latter by PKCαβγ, to facilitate its activation.
Under these conditions of RhoGDIα phosphorylation by PKCαβγ, ∼0.3 mol of phosphate were incorporated per mol of RhoGDI molecule (Fig. 6F). As there appears to be only one major phosphorylation site, this would represent 30% of RhoGDIα. Theoretically, 30% of the bound RhoA would therefore be able to exchange its bound nucleotide for GTPγS. Under the experimental conditions employed, the data are consistent with approximately half of this potential pool of RhoA exchanging nucleotide for GTPγS.
RhoGDI Phosphorylation in Fibroblasts Regulates GTP-RhoA Levels
Further experiments with fibroblasts tested the hypothesis that Ser-34 phosphorylation of RhoGDIα leads to elevated GTP-RhoA levels. Primary fibroblasts were transfected with the pRaichu 1502 cDNA, the protein acting as a sensitive indicator of GTP-RhoA levels when analyzed by FRET microscopy (supplemental Fig. 4A) (33). In this system, FRET efficiency is inversely proportional to RhoA-GTP levels. In a base-line characterization of the fibroblast system, stress fiber and focal adhesion formation were manipulated through transfection of syndecan-4 cDNA constructs. In combination with the pRaichu 1502 cDNA, cells were transfected with wild-type (S4W) or cytoplasmically truncated (S4I) syndecan-4 cDNA. Wild-type syndecan-4 promotes stress fiber and focal adhesion formation, although the S4I acts as a dominant negative, because it cannot bind PKCα (11, 50–52). Stress fibers and focal adhesions are depleted in S4I-transfected cells. Stress fiber formation in these fibroblasts is sensitive to inhibition of GTP-RhoA by the C3 transferase or inhibition of Rho kinases by Y27632 (28). As expected, FRET efficiency decreased significantly in S4W cells compared with control transfected or S4I cells, indicative of higher GTP-RhoA levels (supplemental Fig. 4). In S4I cells, FRET efficiency was increased significantly above control levels.
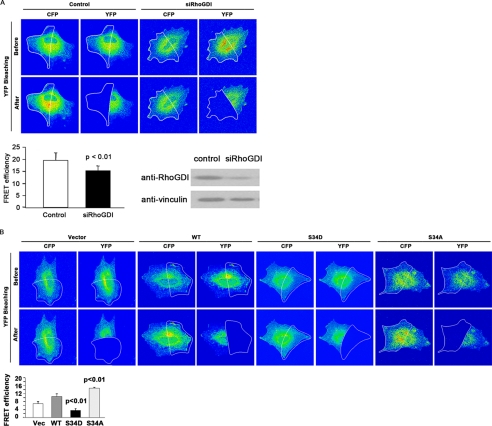
To manipulate RhoGDIα status in rat fibroblasts, endogenous levels were first reduced by siRNA. Reduction of RhoGDIα levels in REF cells by siRNA techniques led to a drop in FRET efficiency, commensurate with a rise in GTP-RhoA levels as seen by FRET microscopy with the pRaichu 1502 probe (Fig. 7A). This was expected, because the GDI protein serves as a sink for GDP-bound Rho family members. In this background, wild-type RhoGDI cDNA or Ser-34 mutants (to alanine or aspartate) were introduced into the cells for further FRET analysis. Compared with an empty vector, expression of wild-type RhoGDIα was accompanied by decreased GTP-RhoA levels. This effect was enhanced by overexpression of the S34A mutant of RhoGDIα (Fig. 7B). Importantly, overexpression of the S34D phosphomimetic mutant had an opposite effect with elevated GTP-RhoA levels (Fig. 7B). These data confirm that Ser-34 can be a key RhoGDIα regulatory site in vivo. Upon phosphorylation, the GDI cannot retain RhoA efficiently, facilitating its activation by nucleotide exchange.
FIGURE 7.
Serine 34 of RhoGDIα in fibroblasts regulates RhoA-GTP levels. A, rat embryo fibroblasts were cotransfected with pRaichu 1502 and either control siRNA or siRNA oligonucleotides for RhoGDIα. After 48 h, cells were lysed and immunoblotted with anti-RhoGDI antibody. Vinculin was used as a loading control. FRET efficiency was measured after acceptor photobleaching and was quantitated (n = 20). Efficiency was reduced after reduction in RhoGDI protein levels, indicative of higher GTP-RhoA levels. B, rat fibroblasts with endogenous levels of RhoGDIα reduced by siRNA were co-transfected with pRaichu1502 and one of His vector (Vec), His-tagged RhoGDI (WT), or His-tagged RhoGDI mutants (S34D or S34A). FRET efficiency was measured after acceptor photobleaching, and quantitation is shown (n = 20). Expression of phosphomimetic RhoGDIα leads to decreased FRET efficiencies and therefore elevated RhoGTP levels. The converse was measured after expression of the nonphosphorylatable S34A mutant. p < 0.01 indicates a FRET signal significantly different to the vector only control (t test).
DISCUSSION
RhoGDI proteins regulate the cycling and distribution of RhoGTPases (18, 19). Co-crystallization data of RhoGDIα bound to Cdc42 or Rac1 (38, 53) reveal that residues 34–57 form an ordered helix-loop-helix motif characterized by hydrophobic interactions. Acquisition of negative charge at Ser-34 may disrupt the stability of the helix-loop-helix motif, interfering with the GTPase interaction and causing release of the GTPase from RhoGDI. In addition, there are extensive interactions between GDI N termini and switch I and II regions of the GTPases. GTPase binding establishes a bridge that brings regions of the N- and C-terminal domains of RhoGDI into proximity. A striking example is Arg-66 (Cdc42), the guanidinium group of which exhibits hydrogen bonding with Asp-185 as well as Pro-30 and Ala-31 of the GDI. Additionally, hydrophobic interactions between the aliphatic portion of the Arg-66 with Gln-32 and Ile-122 of the GDI were observed (38). Because many of the key residues of the RhoGDI N-terminal region lie immediately adjacent to Ser-34, these interactions may be disturbed by phosphorylation. However, Arg-66 of Cdc42 is highly conserved in Rac1 and RhoA, and indeed there is high sequence conservation through the switch II region, consistent with its role in GTP hydrolysis regulation, so the RhoA-specific effect of Ser-34 phosphorylation deserves further analysis.
In vitro studies identifying the site and consequences of RhoGDIα phosphorylation are supported by in vivo studies with a Ser-34-phosphospecific antibody. Four widely differing cell types were investigated, and in each case endogenous phosphorylation of the Ser-34 residue could be detected. In MDCK cells, phorbol ester stimulates cytoskeletal reorganization and PKCα translocation to the membrane (43), although integrin-mediated adhesion is promoted by phorbol ester in K562 cells, also associated with actin cytoskeletal reorganization (39). In both cell types, RhoGDIα phosphorylation on Ser-34 rapidly increased in concert with these adhesion events. Additionally, and consistent with in vitro data, RhoGDIα phosphorylation was blocked by the Gö6976 inhibitor, which has highest activity against PKCα and PKCβ1 (41) The increase in Ser-34 phosphorylation of RhoGDIα in MDCK cells responding to phorbol ester, or HGF, is particularly interesting. Both stimulants activate RhoA rather than Rac (45, 54, 55), although the molecular mechanism is unknown. The result is actin cytoskeletal reorganization and loss of cell-cell junctions. Moreover, two studies with mouse keratinocytes and KB cells showed that motility in response to these same agents was blocked both by C3 transferase and overexpression of wild-type RhoGDI (55, 56). These studies therefore implicated RhoGDI, with which the current data are entirely consistent. A key result is that introduction of the S34D mutant of RhoGDIα into MDCK cells activated the motility responses of MDCK cells without a need for PKC activation. In contrast, such responses were not seen in wild-type or S34A RhoGDI-expressing cells. However, we cannot rule out additional roles for guanine exchange factors and GTPase-activating proteins. Gentile et al. (57) showed, for example, that HGF-promoted invasion is associated with down-regulated expression of the RacGAP, Arhgap12.
Further experiments utilized the Raichu construct as a reporter for GTP-Rho levels (33). This sensitive indicator combined with FRET microscopy enabled the impact of RhoGDIα phosphorylation to be assessed. Expression of the S34D form of RhoGDIα in fibroblasts yielded a FRET decline (i.e. GTP-RhoA levels increased). This provided in vivo evidence that RhoGDIα phosphorylation regulates RhoGTP levels. In contrast, no elevation in GTP-RhoA levels was seen by transfection of wild-type RhoGDIα or an S34A form. In these cases, FRET was increased, suggesting that a form of RhoGDIα that cannot be phosphorylated sequesters GDP-RhoA. Although wild-type RhoGDIα can theoretically be phosphorylated when overexpressed, increased protein levels alter the equilibrium in favor of decreased overall phosphorylation, consistent with Takaishi et al. (55).
Efficient Ser-34 phosphorylation of RhoGDIα by conventional PKC occurs in the presence of PtdIns(4,5)P2, not with the often used combination of phosphatidylserine, diacylglycerol, and calcium. This inositol phospholipid can mediate PKCα activation, and a lysine-rich-binding site in the C2 domain was identified (10). We, and others, showed that syndecan-4-mediated activation of PKCα in the presence of PtdIns(4,5)P2 was calcium-independent (13, 35), and this mechanism of kinase activation corresponds well to the current data showing that the PKCα/PtdIns(4,5)P2 combination efficiently phosphorylated RhoGDIα. The C2 domain of PKCα has a higher affinity for PtdIns(4,5)P2 than phosphatidylserine, so the inositide may be a kinase-targeting site (15, 58). Recent work shows that PKCα C2 domain may adopt a different membrane orientation when bound to PtdIns(4,5)P2 compared with phosphatidylserine (59). In turn, this may modulate kinase activity, and our study provides some evidence that substrate specificity might be engendered this way. Therefore, we predict PKCα activation resulting from phospholipase C activity, i.e. the generation of diacylglycerol, would not trigger RhoGDI phosphorylation and RhoA activation. By the same token, because inositol 1,4,5-trisphosphate and Ca2+ release from the endoplasmic reticulum are consequences of phospholipase C activity, the phosphorylation of RhoGDIα is presumed to be independent of calcium fluxes. Consistent with this, we find that the PtdIns(4,5)P2-driven PKCα activation is indeed Ca2+-independent. Moreover, RhoGDIα phosphorylation persisted in the presence of a characterized phospholipase C inhibitor, U73122 (60).
Our data differ from two reports suggesting that in endothelial cells Ser-96 of RhoGDIα was subject to phosphorylation by PKCα, with a downstream increase in GTP-RhoA levels (61) or release of RhoG (62), a close relative of Rac1 (63). Although the methods and cells are different, we could not demonstrate phosphorylation of Ser-96 in vitro. A truncated form of RhoGDIα(67–204), missing the N-terminal region but containing Ser-96, was not phosphorylated by PKCα. Knezevic et al. (61) utilized N-terminally green fluorescent protein-tagged RhoGDIα that may have affected the ability of PKCα to phosphorylate Ser-34. In contrast to that study, we prepared a phosphospecific antibody that revealed both the widespread occurrence of Ser-34 phosphorylation and regulation in response to PKC activation. Serine 96 lies in a consensus PKC phosphorylation site that is present in the immunoglobulin domain of mammalian RhoGDIα but not conserved across other vertebrates or isoforms. Based on structural data, the Ser-96 residue does not apparently form part of a contact site with GTPase or the hydrophobic pocket that captures the prenyl moiety. In contrast, Ser-34 is conserved across all vertebrate RhoGDIα isoforms and is present in all vertebrate β and γ isoforms for which there are data. Although Ser-34 is a major PKC phosphorylation site, in vitro experiments suggested some residual phosphorylation after GDI mutation of Ser-34 to alanine or aspartate. The amounts varied but could represent another phosphorylation site, presumably also in the N-terminal portion of RhoGDIα.
Because RhoGDIα is an abundant cytoplasmic protein, its role is potentially significant. Our work complements that of DerMardirossian et al. (64) where p21-activated kinase phosphorylation of RhoGDIα on Ser-101 and Ser-174 causes specific release of Rac1. In this case, the key sites are C-terminal and are close to each other and to the prenyl-binding cleft of the GDI (38). It is proposed that the negative charge may destabilize this hydrophobic interaction and cause release of the Rac1. This must be a very specific effect because Rho and Cdc42 are also prenylated yet are unaffected. A second model of RhoGDI regulation involves Tyr-156 phosphorylation by Src, but only when not complexed to a GTPase (21). This not only reduced affinity of the GDI for RhoA, Rac1, and Cdc42, but it also triggered a membrane or cortical redistribution of the protein. The current data show that Ser-34 phosphorylation can occur in free or GTPase-bound RhoGDIα. Phosphorylation on Tyr-156 was only noted in cells overexpressing a constitutively active form of Src (21), perhaps an indicator of its transient nature. Here, Ser-34 phosphorylation was observed in several different untransfected cell types, with levels changing in response to adhesion events.
Focal adhesion and microfilament bundle assembly is promoted by GTP-RhoA (65, 66), with activation of the Rho kinases, with ROCK I playing a major role in primary fibroblasts (28). Rho kinases phosphorylate myosin light chain and the myosin-binding subunit of myosin phosphatase, leading to myosin II-driven contraction. The transmembrane heparan sulfate proteoglycan, syndecan-4, supports focal adhesion assembly through the binding and persistent activation of PKCα, in the presence of PtdIns(4,5)P2, an event upstream of GTP-RhoA (8, 11, 25). In MDCK cells, Rho kinase activity is required both for focal adhesion/stress fiber formation but also lamellipodial ruffle stabilization, possibly through phosphorylation of adducin (29, 67). However, the upstream events that lead to the accumulation and targeting of GTP-RhoA were not known. Here, evidence suggests that a major downstream target of PKCα/PtdIns(4,5)P2 is RhoGDIα with phosphorylation on Ser-34. The result of phosphorylation is release or decreased capture of the GTPase, leading to nucleotide exchange and interaction with downstream effectors.
Supplementary Material
The work was supported in part by Wellcome Trust Programme Grant 065940 (to J. R. C.), Danish National Research Foundation, Haensch Fond, Wilhelm Pedersen Fond, and University of Copenhagen (to J. R. C., H. A. B. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
G. Wuytens and P. Zimmermann, personal communication.
- PKC
- protein kinase C
- PtdIns
- phosphatidylinositol
- Ptd
- phosphatidyl
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- MDCK
- Madin-Darby canine kidney cell
- REF
- rat embryo fibroblast
- FRET
- fluorescence resonance energy transfer
- GDI
- guanine dissociation inhibitor
- DL
- diolein
- PMA
- phorbol 12-myristate 13-acetate
- siRNA
- small interfering RNA
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- HGF
- hepatocyte growth factor.
REFERENCES
- 1.Ivaska J., Kermorgant S., Whelan R., Parsons M., Ng T., Parker P. J. (2003) Biochem. Soc. Trans. 31, 90–93 [DOI] [PubMed] [Google Scholar]
- 2.Larsson C. (2006) Cell. Signal. 18, 276–284 [DOI] [PubMed] [Google Scholar]
- 3.Suzuki A., Ohno S. (2006) J. Cell Sci. 119, 979–987 [DOI] [PubMed] [Google Scholar]
- 4.Reyland M. E. (2009) Front. Biosci. 14, 2386–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mochly-Rosen D., Gordon A. S. (1998) FASEB J. 12, 35–42 [PubMed] [Google Scholar]
- 6.Steinberg S. F. (2008) Physiol. Rev. 88, 1341–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaken S., Leach K., Klauck T. (1989) J. Cell Biol. 109, 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dovas A., Yoneda A., Couchman J. R. (2006) J. Cell Sci. 119, 2837–2846 [DOI] [PubMed] [Google Scholar]
- 9.Bass-Zubek A. E., Hobbs R. P., Amargo E. V., Garcia N. J., Hsieh S. N., Chen X., Wahl J. K., 3rd., Denning M. F., Green K. J. (2008) J. Cell Biol. 181, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbalán-García S., García-García J., Rodríguez-Alfaro J. A., Gómez-Fernández J. C. (2003) J. Biol. Chem. 278, 4972–4980 [DOI] [PubMed] [Google Scholar]
- 11.Keum E., Kim Y., Kim J., Kwon S., Lim Y., Han I., Oh E. S. (2004) Biochem. J. 378, 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Multhaupt H. A., Yoneda A., Whiteford J. R., Oh E. S., Lee W., Couchman J. R. (2009) J. Physiol. Pharmacol. 60, Suppl. 4, 31–38 [PubMed] [Google Scholar]
- 13.Oh E. S., Woods A., Lim S. T., Theibert A. W., Couchman J. R. (1998) J. Biol. Chem. 273, 10624–10629 [DOI] [PubMed] [Google Scholar]
- 14.Manna D., Bhardwaj N., Vora M. S., Stahelin R. V., Lu H., Cho W. (2008) J. Biol. Chem. 283, 26047–26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marín-Vicente C., Nicolás F. E., Gómez-Fernández J. C., Corbalán-García S. (2008) J. Mol. Biol. 377, 1038–1052 [DOI] [PubMed] [Google Scholar]
- 16.Guerrero-Valero M., Marín-Vicente C., Gómez-Fernández J. C., Corbalán-García S. (2007) J. Mol. Biol. 371, 608–621 [DOI] [PubMed] [Google Scholar]
- 17.Bass M. D., Morgan M. R., Roach K. A., Settleman J., Goryachev A. B., Humphries M. J. (2008) J. Cell Biol. 181, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DerMardirossian C., Bokoch G. M. (2005) Trends Cell Biol. 15, 356–363 [DOI] [PubMed] [Google Scholar]
- 19.Dovas A., Couchman J. R. (2005) Biochem. J. 390, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi K., Sasaki T., Mammoto A., Takaishi K., Kameyama T., Tsukita S., Takai Y. (1997) J. Biol. Chem. 272, 23371–23375 [DOI] [PubMed] [Google Scholar]
- 21.DerMardirossian C., Rocklin G., Seo J. Y., Bokoch G. M. (2006) Mol. Biol. Cell 17, 4760–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golovanov A. P., Hawkins D., Barsukov I., Badii R., Bokoch G. M., Lian L. Y., Roberts G. C. (2001) Eur. J. Biochem. 268, 2253–2260 [DOI] [PubMed] [Google Scholar]
- 23.Sheffield P., Garrard S., Derewenda Z. (1999) Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 24.Read P. W., Nakamoto R. K. (2000) Methods Enzymol. 325, 15–25 [DOI] [PubMed] [Google Scholar]
- 25.Lim S. T., Longley R. L., Couchman J. R., Woods A. (2003) J. Biol. Chem. 278, 13795–13802 [DOI] [PubMed] [Google Scholar]
- 26.Self A. J., Hall A. (1995) Methods Enzymol. 256, 3–10 [DOI] [PubMed] [Google Scholar]
- 27.Sebbagh M., Renvoizé C., Hamelin J., Riché N., Bertoglio J., Bréard J. (2001) Nat. Cell Biol. 3, 346–352 [DOI] [PubMed] [Google Scholar]
- 28.Yoneda A., Multhaupt H. A., Couchman J. R. (2005) J. Cell Biol. 170, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukata Y., Kimura K., Oshiro N., Saya H., Matsuura Y., Kaibuchi K. (1998) J. Cell Biol. 141, 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fauré J., Vignais P. V., Dagher M. C. (1999) Eur. J. Biochem. 262, 879–889 [DOI] [PubMed] [Google Scholar]
- 31.Aktories K., Just I. (1995) Methods Enzymol. 256, 184–195 [DOI] [PubMed] [Google Scholar]
- 32.Bastiaens P. I., Majoul I. V., Verveer P. J., Söling H. D., Jovin T. M. (1996) EMBO J. 15, 4246–4253 [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshizaki H., Ohba Y., Kurokawa K., Itoh R. E., Nakamura T., Mochizuki N., Nagashima K., Matsuda M. (2003) J. Cell Biol. 162, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez P., Mitton B., Kranias E. G. (2005) Biotechnol. Lett. 27, 1869–1873 [DOI] [PubMed] [Google Scholar]
- 35.Horowitz A., Simons M. (1998) J. Biol. Chem. 273, 25548–25551 [DOI] [PubMed] [Google Scholar]
- 36.Weekes J., Barry S. T., Critchley D. R. (1996) Biochem. J. 314, 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziegler W. H., Tigges U., Zieseniss A., Jockusch B. M. (2002) J. Biol. Chem. 277, 7396–7404 [DOI] [PubMed] [Google Scholar]
- 38.Hoffman G. R., Nassar N., Cerione R. A. (2000) Cell 100, 345–356 [DOI] [PubMed] [Google Scholar]
- 39.Lub M., van Vliet S. J., Oomen S. P., Pieters R. A., Robinson M., Figdor C. G., van Kooyk Y. (1997) Mol. Biol. Cell 8, 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hocevar B. A., Morrow D. M., Tykocinski M. L., Fields A. P. (1992) J. Cell Sci. 101, 671–679 [DOI] [PubMed] [Google Scholar]
- 41.Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marmé D., Schächtele C. (1993) J. Biol. Chem. 268, 9194–9197 [PubMed] [Google Scholar]
- 42.Fujii M., Ohtsubo M., Ogawa T., Kamata H., Hirata H., Yagisawa H. (1999) Biochem. Biophys. Res. Commun. 254, 284–291 [DOI] [PubMed] [Google Scholar]
- 43.Sciorra V. A., Daniel L. W. (1996) J. Biol. Chem. 271, 14226–14232 [DOI] [PubMed] [Google Scholar]
- 44.Balboa M. E., Insel P. A. (1998) Mol. Pharmacol. 53, 221–227 [DOI] [PubMed] [Google Scholar]
- 45.Kodama A., Matozaki T., Fukuhara A., Kikyo M., Ichihashi M., Takai Y. (2000) Mol. Biol. Cell 11, 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukata Y., Oshiro N., Kinoshita N., Kawano Y., Matsuoka Y., Bennett V., Matsuura Y., Kaibuchi K. (1999) J. Cell Biol. 145, 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kochs G., Hummel R., Fiebich B., Sarre T. F., Marmé D., Hug H. (1993) Biochem. J. 291, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lerm M., Schmidt G., Aktories K. (2000) FEMS Microbiol. Lett. 188, 1–6 [DOI] [PubMed] [Google Scholar]
- 49.Genth H., Gerhard R., Maeda A., Amano M., Kaibuchi K., Aktories K., Just I. (2003) J. Biol. Chem. 278, 28523–28527 [DOI] [PubMed] [Google Scholar]
- 50.Longley R. L., Woods A., Fleetwood A., Cowling G. J., Gallagher J. T., Couchman J. R. (1999) J. Cell Sci. 112, 3421–3431 [DOI] [PubMed] [Google Scholar]
- 51.Couchman J. R. (2003) Nat. Rev. Mol. Cell Biol. 4, 926–937 [DOI] [PubMed] [Google Scholar]
- 52.Mostafavi-Pour Z., Askari J. A., Parkinson S. J., Parker P. J., Ng T. T., Humphries M. J. (2003) J. Cell Biol. 161, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grizot S., Fauré J., Fieschi F., Vignais P. V., Dagher M. C., Pebay-Peyroula E. (2001) Biochemistry 40, 10007–10013 [DOI] [PubMed] [Google Scholar]
- 54.Kitajo H., Shibata T., Nagayasu H., Kawano T., Hamada J., Yamashita T., Arisue M. (2003) Oncol. Res. 10, 1351–1356 [PubMed] [Google Scholar]
- 55.Takaishi K., Sasaki T., Kato M., Yamochi W., Kuroda S., Nakamura T., Takeichi M., Takai Y. (1994) Oncogene 9, 273–279 [PubMed] [Google Scholar]
- 56.Nishiyama T., Sasaki T., Takaishi K., Kato M., Yaku H., Araki K., Matsuura Y., Takai Y. (1994) Mol. Cell. Biol. 14, 2447–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gentile A., D'Alessandro L., Lazzari L., Martinoglio B., Bertotti A., Mira A., Lanzetti L., Comoglio P. M., Medico E. (2008) Oncogene 27, 5590–5598 [DOI] [PubMed] [Google Scholar]
- 58.Son H., Lim Y., Kim J., Park H., Choi S., Han I., Kim W. S., Park S., Bae Y., Oh E. S. (2006) Arch. Biochem. Biophys. 454, 1–6 [DOI] [PubMed] [Google Scholar]
- 59.Landgraf K. E., Malmberg N. J., Falke J. J. (2008) Biochemistry 47, 8301–8316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horowitz L. F., Hirdes W., Suh B. C., Hilgemann D. W., Mackie K., Hille B. (2005) J. Gen. Physiol. 126, 243–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knezevic N., Roy A., Timblin B., Konstantoulaki M., Sharma T., Malik A. B., Mehta D. (2007) Mol. Cell Biol. 27, 6323–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elfenbein A., Rhodes J. M., Meller J., Schwartz M. A., Matsuda M., Simons M. (2009) J. Cell Biol. 186, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Buul J. D., Allingham M. J., Samson T., Meller J., Boulter E., García-Mata R., Burridge K. (2007) J. Cell Biol. 178, 1279–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DerMardirossian C., Schnelzer A., Bokoch G. M. (2004) Mol. Cell 15, 117–127 [DOI] [PubMed] [Google Scholar]
- 65.Ridley A. J., Hall A. (1994) EMBO J. 13, 2600–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Machesky L. M., Hall A. (1997) J. Cell Biol. 138, 913–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Royal I., Lamarche-Vane N., Lamorte L., Kaibuchi K., Park M. (2000) Mol. Biol. Cell 11, 1709–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.