Abstract
The key gluconeogenic enzyme fructose-1,6-bisphosphatase (FBPase) is induced when Saccharomyces cerevisiae are starved of glucose. However, when glucose is added to cells that have been starved for 3 days, FBPase is degraded in the vacuole. FBPase is first imported to Vid (vacuole import and degradation) vesicles, and these vesicles then merge with the endocytic pathway. In this report we show that two additional gluconeogenic enzymes, isocitrate lyase and phosphoenolpyruvate carboxykinase, were also degraded in the vacuole via the Vid pathway. These new cargo proteins and FBPase interacted with the TORC1 complex during glucose starvation. However, Tor1p was dissociated from FBPase after the addition of glucose. FBPase degradation was inhibited in cells overexpressing TOR1, suggesting that excessive Tor1p is inhibitory. Both Tco89p and Tor1p were found in endosomes coming from the plasma membrane as well as in retrograde vesicles forming from the vacuole membrane. When TORC1 was inactivated by rapamycin, FBPase degradation was inhibited. We suggest that TORC1 interacts with multiple cargo proteins destined for the Vid pathway and plays an important role in the degradation of FBPase in the vacuole.
Keywords: Cell/Endocytosis, Cell/Trafficking, Membrane/Trafficking, Metabolism/Gluconeogenesis, Protein/Degradation, Subcellular Organelles/Vesicles
Introduction
The yeast Saccharomyces cerevisiae has been used as a model system to study protein trafficking pathways that target proteins or lipids from donor membranes to acceptor membranes (1–20). The yeast vacuole is homologous to the mammalian lysosome and plays an important role in protein degradation (1–3). A number of protein targeting pathways to the vacuole have been studied. For example, the Vps pathway delivers carboxypeptidase Y from the endoplasmic reticulum to the Golgi and then to the vacuole for maturation (1–6). The Cvt pathway, on the other hand, carries aminopeptidase I from the cytosol to the vacuole for maturation (7–12). Organelles such as peroxisomes can also be delivered to the vacuole for degradation, either by micropexophagy or macropexophagy (13–15). Finally, under starvation conditions, a non-selective macroautophagy pathway targets organelles and proteins to the vacuole (7–12). A unique autophagy pathway that delivers specific cytosolic proteins to the vacuole for degradation has been studied in our laboratory (16–20). The gluconeogenic enzymes fructose-1,6-bisphosphatase (FBPase)2 and malate dehydrogenase (MDH2) are induced when cells are starved of glucose. These enzymes are then degraded when cells are replenished with fresh glucose (21).
The site of FBPase and MDH2 degradation is dependent on the duration of starvation. When cells are starved for a short period of time (1 day), FBPase and MDH2 are degraded in the proteasome (21). By contrast, these proteins are degraded in the vacuole when glucose is added to cells that have been starved of glucose for longer periods of time (3 days) (21). We have identified a number of VID genes involved in the degradation of FBPase in the vacuole. Interestingly, many of the VID genes function not only in the vacuolar-dependent pathway but also in the proteasomal pathway as well (21). Conversely, GID genes were identified for the proteasomal pathway (22, 23), and they are also required for vacuolar-dependent degradation of FBPase and MDH2 (21). Thus, the same set of genes can be utilized for both the proteasomal and the vacuolar degradation pathways.
For the vacuolar pathway, FBPase is associated with Vid (vacuole import and degradation) vesicles (24), which then merge with the endocytic pathway (25). FBPase import into Vid vesicles requires the heat shock protein Ssa2p (26), Vid22p (27), and cyclophilin (28). The biogenesis of Vid vesicles appears to be regulated by the UBC1 gene. For instance, FBPase accumulates in the cytosol in mutants lacking UBC1, suggesting the Vid vesicle formation requires this gene (29). Purified Vid vesicles also contain COPI coatomer proteins (25). COPI vesicles are involved in multiple protein trafficking pathways including the retrograde trafficking from the Golgi to the endoplasmic reticulum, intra-Golgi trafficking, and endosomal trafficking (30–33). In mammalian cells and in yeast coatomer components are found on endosomes and play critical roles in endocytic trafficking and in multivesicular body sorting (34–39). We have shown that coatomer proteins form a large protein complex with Vid24p, a unique Vid vesicle marker (29). In the absence of functional coatomer subunits, Vid24p association with Vid vesicles was reduced, suggesting that coatomer subunits are required to recruit Vid24p to Vid vesicles (25).
We have used the coatomer ϵ subunit Sec28p to follow the Vid vesicle trafficking pathway (25). Sec28p was observed on Vid vesicles as well as in multiple compartments along the endocytic pathway in both wild type strains and in mutant strains that block FBPase degradation. Based on Sec28p-GFP localization studies in various mutants, we proposed that Sec28p resides on Vid vesicles. These vesicles initially merge with the endocytic pathway and then with the vacuole. The merger of the endocytic and Vid pathways was further supported by co-localization experiments using the Δvph1 mutant. FBPase was in the lumen of endocytic compartments at later time points in this strain. Thus, this provides direct evidence that FBPase utilizes the endocytic pathway before being delivered to the vacuole (25).
Although we have shown that the Vid pathway is tightly linked to the endocytic pathway, there are many questions remaining to be answered. For example, how do cells recognize proteins that are destined for degradation? Also, where do Vid vesicles come from? Along these lines, Sec28p-GFP was found in vesicles that appear to bud from the vacuole membrane in the ret2–1 mutant that contained a defective δ subunit (25). Could these retrograde vesicles represent Vid vesicles? Alternatively, might these vesicles traffic back to the plasma membrane? This might occur, for instance, if the plasma membrane were the site of Vid vesicle formation.
To address the above questions, we attempted to identify cellular proteins that interact with FBPase. To identify such proteins, we used affinity chromatography to purify FBPase-interacting proteins under native conditions. We then subjected the bound material to MALDI spec analysis. Via this approach, we identified several candidate proteins including Tco89p, a specific member of the TORC1 complex that also contains Lst8p, Kog1p and Tor1p (40–43). Binding of various components of TORC1 to FBPase was confirmed by in vivo immunoprecipitation. Kinetic studies indicated that Tor1p was released from FBPase in response to glucose. However, in cells over-expressing TOR1, FBPase degradation was inhibited. Likewise, in cells lacking TCO89, FBPase degradation was blocked. Both Tco89p and Tor1p were found in anterograde endocytic compartments. In addition, they were in retrograde transport vesicles that form from the vacuole membrane. When TORC1 was inactivated by rapamycin, FBPase degradation was inhibited. Taken together, our results suggest TORC1 interacts with multiple cargo proteins destined for the Vid pathway, and it plays an essential role in the degradation of cargo proteins in the vacuole.
EXPERIMENTAL PROCEDURES
Cell Culture, Media, and Antibodies
Cells used in this study are listed in Table 1. The deletion strains derived from BY4742 were from Euroscarf (Euroscarf, Germany). Primers used to tag FBPase, Icl1p, Pck1p, Tco89p, and Tor1p with HA or GFP are listed in Table 2. Cells were grown in glucose-starved conditions in 1% yeast extract, 2% peptone, 1% potassium acetate, 0.5% glucose (YPKG) for the indicated times then shifted to media containing 2% glucose for various periods of time points. Tco89-protein A fusion was obtained from Open Biosystems. Cells expressing Tco89p-myc as well as cells expressing Tor1p-HA, Kog1p-HA, or Lst8p-HA were obtained from Dr. T. Powers (University of California, Davis, CA). The Δpep4Δprb1Δprc1 mutant strain was from Dr. A. Kornberg (Stanford University). The Δvid22Δpep4 double mutant was produced by replacing the PEP4 gene with the pep4::URA3 fragment in the Δvid22 strain (Euroscarf). The pep4::URA3 plasmid was linearized by EcoR1 and XhoI digestions and transformed into the Δvid22 strain. Transformants were screened for the presence of the p2 form of carboxypeptidase Y (CPY) by Western blotting with anti-CPY antibodies. Monoclonal anti-HA was purchased from Roche Applied Science. FBPase antibodies were produced as described (17). MDH2 antibodies were from Dr. L. McAlister-Henn (University of Texas, Houston, TX). The enhanced chemiluminescence kit was purchased from PerkinElmer Life Sciences. Rapamycin was purchased from Sigma.
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| BY4742 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| HLY1049 | MATαura3 leu2 his3 ICL1-HA::TRP1 |
| HLY635 | MATαura3–52 LEU2 trp1Δ63 his3Δ200 GAL2 |
| HLY1082 | MATa ade2 his3 trp1 leu2 ura3 pep4Δ::HIS3 prb1ΔhisG prc1ΔhisG ICL1-HA::TRP1 |
| HLY1052 | MATα leu2Δ0 lys2Δ0 ura3Δ0 pep4::kanMX4 ICL1-GFP::HIS3 |
| HLY2802 | MATα leu2Δ0 lys2Δ0 ura3Δ0 vid22::kanMX4 pep4::URA3 ICL1-GFP::HIS3 |
| HLY1032 | MATαura3 leu2 his3 PCK1-HA::TRP1 |
| HLY1081 | MATa ade2 his3 trp1 leu2 ura3 pep4Δ::HIS3 prb1ΔhisG prc1ΔhisG PCK1-HA::TRP1 |
| HLY1037 | MATα leu2Δ0 lys2Δ0 ura3Δ0 pep4::kanMX4 PCK1-GFP::HIS3 |
| HLY2749 | MATα leu2Δ0 lys2Δ0 ura3Δ0 vid22::kanMX4 pep4::URA3 PCK1-GFP::HIS3 |
| HLY227 | MATa his3 trp1 leu2 ura3 vid24::TRP1 |
| HLY1051 | MATa his3 trp1 leu2 ura3 vid24::TRP1 ICL1-HA::HIS3 |
| HLY1034 | MATa his3 trp1 leu2 ura3 vid24::TRP1 PCK1-HA::HIS3 |
| Δtco89 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tco89::kanMX4 |
| HLY2149 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TCO89-Protein A::URA3 |
| HLY2173 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TCO89-V5-His6::URA3PCK1-HA::HIS3 |
| HLY2182 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TCO89-V5-His6::URA3 ICL1-HA::HIS3 |
| PLY122 | MATα leu2 ura3 trp1 ade2 3HA-TOR1::HIS3 |
| PLY336 | MATa leu2-3 ura3 trp1 ade2-1can1-100 3HA-TOR2::HIS3 TCO89-MYC::TRP1 |
| PLY306 | MATahis3 leu2 ura3 ade2 can1-100 KOG1-HA::TRP1 |
| PLY307 | MATαura3 leu2 his3 LST8-HA::TRP1 |
| HLY228 | MATαleu2Δ0 lys2Δ0 ura3Δ0 VID24-HA::HIS3 |
| HLY1023 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vam3::kanMX4 VID24-HA::HIS3 |
| Δtor1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tor1::kanMX4 |
| HLY1927 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TOR12u URA3 |
| HLY2242 | MATα leu2Δ0 lys2Δ0 ura3Δ0 TCO89-GFP::HIS3 |
| HLY2710 | MATα leu2Δ0 lys2Δ0 ura3Δ0 TCO89-GFP::HIS3 TOR1::URA3 2μ |
| HLY2218 | MATα leu2Δ0 lys2Δ0 ura3Δ0 TOR1-GFP::HIS3 |
| HLY1080 | MATα leu2Δ0 lys2Δ0 ura3Δ0 FBPase-GFP::HIS3 |
TABLE 2.
Primers used in this study
Tco89p-protein A and Tco89p-V5-His6 Pulldown Assay
Tco89-protein A and Tco89p-V5-His6 were expressed on multicopy plasmids. Tco89p-protein A was pulled down with IgG beads, and Tco89p-V5-His6 was pulled down with nickel beads. Interaction was determined by Western blotting with anti-V5 or anti-FBPase antibodies.
Immunoprecipitation
For immunoprecipitation experiments, cells expressing HA-tagged Tor1p, Kog1p, and Lst8p were immunoprecipitated with FBPase, MDH2, Icl1p, and Pck1p. Lysates were precipitated, separated into unbound and bound fractions, and blotted with HA antibodies. For FBPase and Tco89p-myc interactions, FBPase was immunoprecipitated, and the bound and unbound fractions were then blotted with FBPase and myc antibodies to detect FBPase and Tco89p-myc, respectively.
Subcellular Fractionation
Cells with various tags were subjected to subcellular fractionation as described (16, 26). Briefly, cells were grown in glucose starvation conditions and then refed with fresh glucose for various periods of time. Lysates were fractionated by differential centrifugation followed by immunoblotting with anti-FBPase antibodies or HA antibodies for cells expressing Vid24p-HA.
Fluorescence Microscopy
Cells expressing FBPase-GFP, Tco89p-GFP, or Tor1p-GFP were grown in glucose starvation conditions. Cells were refed with fresh glucose for various periods of time. For anterograde transport, FM4-64 (FM) was added at the same time with glucose. For retrograde transport, FM was added 16 h before glucose shift to label the vacuole. For rapamycin treatment experiments, cells were transferred to media containing fresh glucose in the presence of rapamycin. The distribution of Tco89p-GFP, Tor1p-GFP, and FBPase-GFP was determined in untreated or treated cells.
RESULTS
Isocitrate Lyase and Phosphoenolpyruvate Carboxykinase Are Degraded in the Vacuole in a VID24-dependent Manner
We have shown that FBPase and MDH2 can be degraded either in the proteasome or the vacuole, depending on the duration of glucose starvation (25). When glucose is added to 1-day-starved cells, these proteins are degraded in the proteasome. By contrast, when glucose is added to 3-day-starved cells, they are degraded in the vacuole. For the vacuole-dependent pathway, both FBPase and MDH2 are degraded via the Vid vesicle-mediated pathway (21).
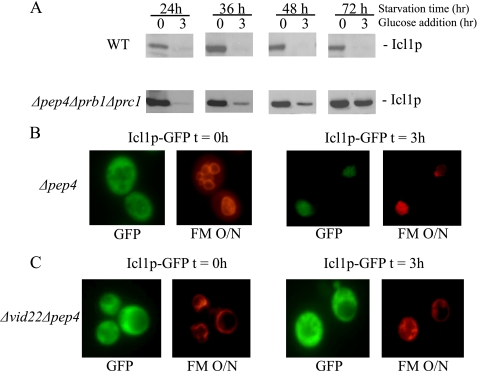
In this study, we asked the question of whether other cytosolic proteins utilized the Vid pathway for degradation in the vacuole. Isocitrate lyase (Icl1p) is a cytosolic protein involved in gluconeogenesis. This protein, as with other gluconeogenic enzymes such as phosphoenolpyruvate carboxykinase (Pck1p), is inactivated in response to glucose (44, 45). Therefore, we first determined whether Icl1p and Pck1p were degraded in the vacuole. For these studies, we used a Δpep4Δprb1Δprc1 mutant strain lacking the three major vacuole proteinases, proteinase A, B, and C. This strain was used previously to show that FBPase and MDH2 are targeted to the vacuole for degradation when 3-day-starved cells are shifted to glucose (21). If Icl1p is degraded in the vacuole, then the degradation of this protein should be retarded in the Δpep4Δprb1Δprc1 strain. By contrast, if Icl1p is not degraded in the vacuole, then this mutant should have little effect on the degradation of this protein. Wild type and Δpep4Δprb1Δprc1 mutants were transformed to express Icl1p-HA, starved for 24–72 h, and then transferred to media containing fresh glucose. In wild type cells, Icl1p was degraded in response to glucose whether cells were starved for 1 or 3 days (Fig. 1A). In this study, unless otherwise indicated, all the tagged proteins were expressed from their own promoters after integration to their chromosomal loci. In the Δpep4Δprb1Δprc1 mutant, however, Icl1p was degraded when glucose was added to 1-day-starved cells, but its degradation was retarded when 3-day-starved cells were refed with fresh glucose (Fig. 1A). Thus, the degradation of Icl1p does not require vacuole proteinases after short term starvation, but vacuole proteinases are required when cells are subjected to 3-day starvation before shifting to glucose.
FIGURE 1.
Icl1p is targeted to the vacuole for degradation in 3-day-starved cells. A, wild type (WT) and Δpep4Δprb1Δprc1 cells expressing Icl1p-HA were starved for 24–72 h and then shifted to glucose. The degradation of Icl1p was examined by Western blotting. B, Icl1p-GFP was expressed in 3-day-starved Δpep4cells that were labeled with FM for 16 h to visualize the vacuole. Cells were then refed with glucose for 0 and 3 h. GFP and FM were visualized by fluorescence microscopy. O/N, overnight. C, Icl1p-GFP was expressed in the Δvid22Δpep4 cells that were starved for 3 days. FM was added to cells for 16 h to label the vacuole before glucose addition. Icl1p-GFP and FM were examined by fluorescence microscopy.
Next, we examined whether Icl1p was targeted to the vacuole when glucose was added to 3-day-starved cells. If this is the case, then this protein should accumulate in the vacuole of cells deficient in vacuole proteolysis. Icl1p was tagged with GFP and transformed into Δpep4 cells. This strain was used because the Δpra1Δprb1Δprc1 mutant displayed a high level of autofluorescence, and as such, it was not suitable for the GFP study. In 3-day-starved Δpep4 cells, Icl1p-GFP was in the cytosol at t = 0 min (Fig. 1B). However, after the addition of glucose for 3 h, Icl1p-GFP was detected in the vacuole. Thus, Icl1p is targeted from the cytosol to the vacuole in response to glucose in 3-day-starved cells.
The VID22 gene is required for FBPase import into Vid vesicles. Therefore, we determined whether Icl1p-GFP was targeted to the vacuole in the Δvid22Δpep4 double mutant strain. Icl1p-GFP was expressed in the Δvid22Δpep4 double mutant, and the distribution of Icl1p-GFP was examined (Fig. 1C). The majority of Icl1p-GFP was outside the vacuole during glucose starvation. This protein remained outside the vacuole after the addition of glucose for 3 h. Thus, Icl1p is not targeted to the vacuole in the Δvid22Δpep4 strain.
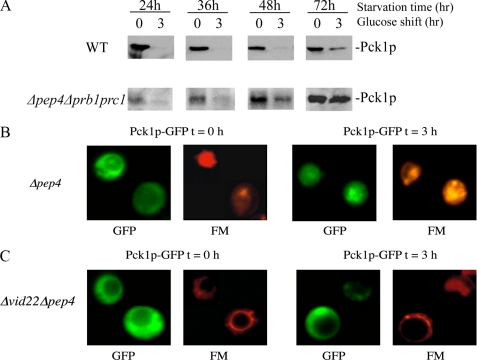
The above strategy was next used to study the degradation of another key gluconeogenic enzyme Pck1p. Glucose addition caused Pck1p degradation in wild type cells that were starved for 24–48 h (Fig. 2A). By contrast, there was a progressive defect in the degradation of Pck1p in Δpep4Δprb1Δprc1 mutant cells that were starved for longer periods of time (Fig. 2A). The level of Pck1p was low in 1-day-starved Δpep4Δprb1Δprc1 strain. Pck1p may not be fully induced in 1-day-starved Δpep4Δprb1Δprc1 mutant. Alternatively, Pck1p may be partially degraded by non-vacuolar or residual vacuolar proteinases in 1-day-starved Δpep4Δprb1Δprc1 cells. Note that the degradation of Pck1p was slightly delayed in 3-day-starved wild type cells. However, this process was significantly retarded in the Δpep4Δprb1Δprc1 cell.
FIGURE 2.
Pck1p is targeted to the vacuole in 3-day-starved Δpep4cells. A, wild type (WT) and Δpep4Δprb1Δprc1 cells expressing Pck1p-HA were glucose-starved for 24–72 h and then refed with glucose for 0 and 3 h. Pck1p degradation in response to glucose was examined. B, Pck1p-GFP was expressed in Δpep4 cells that were starved for 3 days. FM was added to cells for 16 h to label the vacuole. Cells were then transferred to media containing fresh glucose for 3 h, and Pck1p distribution was visualized using fluorescence microscopy. C, Pck1p-GFP was expressed in Δvid22Δpep4 double mutant that was starved for 3 days. FM was added to cells for 16 h to label the vacuole. Cells were then transferred to media containing fresh glucose for 0 and 3 h. Pck1p-GFP and FM were examined by fluorescence microscopy.
To test whether Pck1p was targeted to the vacuole in prolonged starved cells, Pck1p was tagged with GFP, and its distribution was determined in Δpep4 cells. Before glucose shift, most of the protein was outside the vacuole, although some uneven distribution was seen (Fig. 2B). However, by 3 h after the addition of glucose, the majority of Pck1p-GFP was detected in the vacuole in Δpep4 cells. These results indicate that Pck1p is indeed targeted to the vacuole for degradation, when 3-day-starved cells are replenished with fresh glucose.
We next determined whether Pck1p-GFP was targeted to the vacuole in cells lacking the VID22 gene. Pck1p-GFP was expressed in the Δvid22Δpep4 double mutant, and the distribution of Pck1p-GFP was determined (Fig. 2C). During glucose starvation, the majority of Pck1p-GFP was outside the vacuole. Again, uneven distribution of Pck1p-GFP was seen at t = 0 in this double mutant. After the addition of glucose for 3 h, Pck1p-GFP remained outside the vacuole. Because Pck1p-GFP was not targeted to the vacuole after the addition of glucose for 3 h in cells lacking the VID22 gene, our results suggest that this gene plays an important role in the targeting of Pck1p to the vacuole.
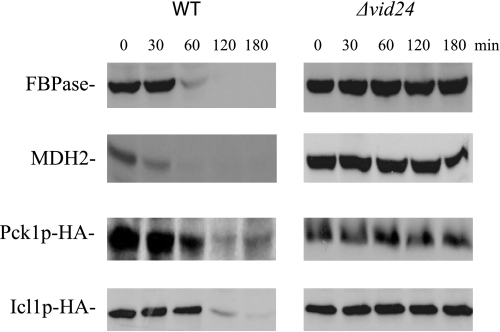
Proteins can be delivered to the vacuole via multiple routes, including the Vid trafficking pathway. Therefore, we tested whether Icl1p and Pck1p were degraded in the vacuole via the Vid pathway. If this is the case, then the degradation of these proteins should be blocked in cells lacking VID genes. Along these lines, the VID24 gene plays a role in FBPase trafficking after the import of FBPase into Vid vesicles (46). Therefore, if Icl1p and Pck1p are degraded in the vacuole via the Vid dependent pathway, their degradation should be affected in cells lacking this gene. In wild type cells, most of the FBPase, MDH2, Pck1p, and Icl1p were degraded in response to glucose (Fig. 3). However, the degradation of these proteins was reduced in a strain lacking VID24. Thus, FBPase, MDH2, Icl1p, and Pck1p are targeted to the vacuole via the Vid pathway in 3-day-starved cells. In this study, we focused on the role that TORC1 played in the vacuole-dependent pathway. Therefore, all subsequent experiments were performed in cells that were starved for 3 days and then transferred to media containing fresh glucose for the indicated time points.
FIGURE 3.
FBPase, MDH2, Pck1p, and Icl1p are degraded in response to glucose in a VID24 dependent manner. Wild type (WT) and Δvid24 mutants were transformed to express Pck1p-HA and Icl1p-HA individually. These cells were transferred to media containing fresh glucose for the indicated times, and their degradation was examined with anti-FBPase, anti-MDH2 antibodies, or with HA antibodies.
TCO89 Is Required for the Degradation of FBPase, MDH2, Icl1p, and Pck1p
Having established that multiple proteins utilize the Vid vacuolar degradation pathway, we asked the question as to how these proteins were recognized for degradation. First, we looked for proteins that may form an interaction with cargo proteins. Potential interacting proteins were isolated using lysates from wild type cells that had been shifted to glucose for 20 min. Total lysates were solubilized with Triton X-100, and FBPase-interacting proteins were captured using an FBPase affinity column. Proteins were eluted from the column and then subjected to MALDI analysis. Via this approach, a number of proteins including Tco89p were identified (Table 3).
TABLE 3.
Putative FBPase-interacting proteins identified by MALDI
| Gene | Functions |
|---|---|
| KCS1 | Inositolhexakisphosphate (IP6) and inositolheptakisphosphate (IP7) kinase |
| YHR097C | Putative protein of unknown function |
| HSC82 | Cytoplasmic chaperone of the Hsp90 family |
| GYP1 | Cis-golgiGTPase-activating protein (GAP) for the Rab family members Ypt1p |
| AKL1 | Ser-Thr protein kinase, member of the Ark kinase family |
| PPZ1 | Serine/threonine protein phosphatase Z |
| ARP5 | Actin-related protein |
| ARP6 | Actin-related protein |
| ARP8 | Actin-related protein |
| MDS3 | Protein with an N-terminal kelch-like domain |
| UBC2 | Ubiquitin-conjugating enzyme (E2) |
| UBP14 | Ubiquitin-specific protease |
| YML050W | Putative protein of unknown function |
| HSV2 | Phosphatidylinositol 3,5-bisphosphate-binding protein |
| TCO89 | Subunit of TORC1 |
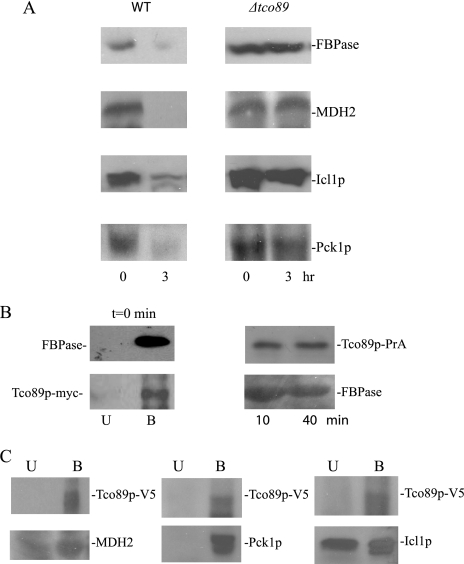
To test the role of Tco89p in the FBPase degradation pathway in the vacuole, we examined FBPase degradation in a Δtco89 strain. FBPase was degraded normally in 3-day-starved wild type cells. However, FBPase degradation was defective in Δtco89 cells that were starved for 3 days (Fig. 4A). Therefore, Tco89p plays an important role in the degradation of FBPase in the vacuole.
FIGURE 4.
Tco89p is required for the degradation of FBPase, MDH2, Icl1p, and Pck1p. A, wild type (WT) cells and cells lacking TCO89 were starved for 3 days. Cells were then transferred to media containing fresh glucose for 0 and 3 h. The degradation of FBPase, MDH2, Icl1p, and Pck1p was examined. B, left panel, wild type strain expressing Tco89p-myc was starved for 3 days. FBPase was immunoprecipitated from total lysates at t = 0 min. The bound and unbound fractions were subjected to immunoblotting with anti-FBPase and anti-myc antibodies. Right panel, Tco89p-protein A fusion proteins were precipitated from wild type cells that were refed with fresh glucose for the indicated times. The bound fractions were blotted with anti-FBPase antibodies. C, Tco89p-V5-His6 fusion proteins were precipitated from wild type cells that expressed Pck1p-HA or Icl1p-HA. The presence of MDH2, Pck1p and Icl1p in the unbound (U) and bound (B) fractions was detected by Western blotting.
Having established that Icl1p and Pck1p utilized the Vid pathway, we next tested whether Tco89p played a similar role in the degradation of these proteins in the vacuole. Wild type and Δtco89 cells were starved for 3 days and shifted to glucose. Under these conditions, wild type cells degraded MDH2, Icl1p, and Pck1p in response to glucose. By contrast, the degradation of these proteins was delayed in cells lacking TCO89 (Fig. 4A). Thus, Tco89p is involved in the vacuolar degradation of multiple proteins selected for the Vid pathway.
TORC1 Binds to Multiple Proteins Destined for the Vid Pathway
As noted above, Tco89p was identified as an FBPase-interacting protein using lysates from 3-day-starved cells. To confirm the results of our MALDI study, we examined the in vivo interaction of Tco89p with FBPase. For this study, we obtained a strain that expressed Tco89p-myc on its own chromosomal locus. This strain was starved for 3 days, and FBPase was precipitated from total lysates at t = 0 min. The bound and unbound fractions were then immunoblotted with myc to detect Tco89p-myc. The majority of FBPase and Tco89p-myc were detected in the bound fractions (Fig. 4B, left panel). We also utilized Tco89p-protein A or Tco89p-V5-His6 fusion proteins that were transformed into wild type cells and expressed on multicopy plasmids for interaction studies. Transformed cells were starved for 3 days and then shifted to glucose for the indicated times. Tco89p was precipitated first, and FBPase in the precipitates was detected by Western blotting. Under these conditions, FBPase was found in the material that co-precipitated with Tco89p-protein A (Fig. 4B, right panel).
We next tested whether other Vid cargo proteins interacted with Tco89p. Tco89p-V5- His6 was precipitated from cells that were starved for 3 days, and the unbound and bound materials were immunoblotted with antibodies directed against MDH2, Icl1p-HA, and Pck1p-HA. As expected, the majority of Tco89p was in the bound fraction (Fig. 4C). Likewise, high levels of MDH2, Icl1p, and Pck1p were also detected in the bound fractions, indicating that Tco89p interacts with these cargo proteins.
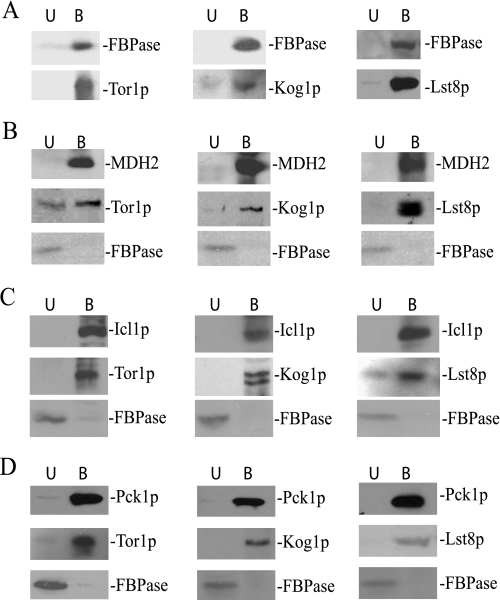
Tco89p is a specific component of the TORC1 complex (40). Therefore, we tested whether other components of TORC1 might bind to our cargo proteins. FBPase was precipitated from total lysates from cells expressing Tor1p-HA, Kog1p-HA, or Lst8p-HA. The presence of TORC1 and FBPase in the bound and unbound fractions was then examined by Western blotting. In these strains the majority of the FBPase was in the bound fractions (Fig. 5A). Significant amounts of Tor1p, Kog1p, and Lst8p were also detected in the bound fraction (Fig. 5A).
FIGURE 5.
The TORC1 complex binds to proteins destined for the Vid pathway. A, FBPase was precipitated from wild type cells expressing Tor1p-HA, Kog1p-HA, or Lst8p-HA. The presence of these HA-tagged proteins was then detected in the unbound (U) and the bound (B) fractions. The tagged cells were used to precipitate MDH2 (B), Icl1p (C), and Pck1p (D) in total lysates. The HA-tagged proteins and FBPase in the unbound and bound fractions were then detected by Western blotting with anti-HA antibodies or anti-FBPase antibodies.
Next, we examined whether TORC1 components bound to MDH2, Icl1p, and Pck1p. For these experiments, MDH2, Icl1p, and Pck1p were precipitated from total lysates from cells expressing Kog1p-HA, Lst8p-HA, or Tor1p-HA. The unbound and bound fractions were then immunoblotted with HA antibodies. As expected, the majority of MDH2 (Fig. 5B), Icl1p (Fig. 5C), and Pck1p (Fig. 5D) were precipitated in the bound fractions. Likewise, significant amounts of Tor1p, Lst8p, and Kog1p were found in the bound fractions. Under these conditions, most of the FBPase was in the unbound fraction when TORC1 components were precipitated with MDH2, Icl1p, or Pck1p (Figs. 5, B–D).
Tor1p Is Released from FBPase after the Addition of Glucose
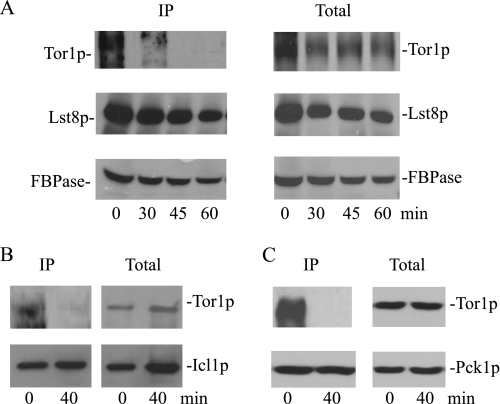
We next determined whether the addition of glucose affects these interactions. Tagged cells were transferred to media containing fresh glucose for various periods of time. FBPase was then precipitated from total lysates, and the bound material was blotted with HA to detect Tor1p and Lst8p. In these experiments, high amounts of FBPase were precipitated at each time point (Fig. 6A). The levels of bound Tor1p, however, changed over time. For example, high levels of Tor1p were bound to FBPase at t = 0 min, but these levels were significantly reduced after the addition of glucose. At these time points, substantial amounts of Tor1p were detected in total lysates. Thus, glucose appears to trigger a rapid dissociation of Tor1p from FBPase. This is in contrast to Lst8p, where a high percentage of this protein remained associated with FBPase after a glucose shift.
FIGURE 6.
Tor1p is released from FBPase following a glucose shift. A, wild type cells expressing Tor1p-HA or Lst8p-HA were glucose-starved and then refed with fresh glucose for the indicated time points. FBPase was precipitated from total lysates, and the presence of Tor1p and Lst8p in the bound fractions was determined by Western blotting. Only one FBPase blot was shown. IP, immunoprecipitated. B and C, Icl1p and Pck1p were precipitated from cells expressing Tor1p-HA that were transferred from low to high glucose for 0 and 40 min. The kinetics of Tor1p-HA interaction with these proteins in the bound fraction was shown.
We next examined whether Tor1p dissociated from Icl1p and Pck1p (Fig. 6, B and C). During glucose starvation, Tor1p was bound to Icl1p and Pck1p. However, after a glucose addition, there was a drastic decrease of Tor1p in the bound fractions. Taken together, these results suggest that glucose triggers a rapid dissociation of Tor1p from these proteins.
TOR1 Overexpression Inhibits FBPase Degradation
Because Tor1p is released from cargo proteins shortly after the addition of glucose, one possible explanation is that Tor1p is inhibitory and the dissociation of Tor1p from cargo proteins might relieve the inhibition, thereby allowing for their degradation. For the following experiments, we used FBPase as a model protein to study the Vid pathway. MDH2, Icl1p, and Pck1p are likely to follow the same pathway, as they are all degraded in the vacuole in a VID24-dependent manner when cells are starved for 3 days. Furthermore, these proteins all bind to the TORC1 complex and require the TCO89 gene for degradation.
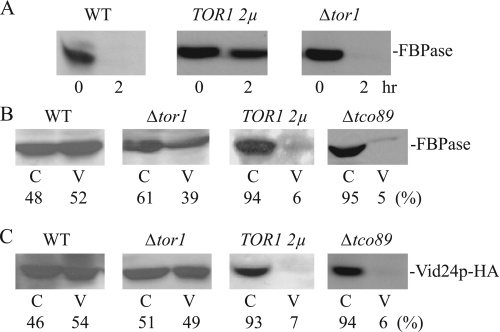
If Tor1p is inhibitory, then overexpression of the TOR1 gene should block FBPase degradation, whereas the deletion of TOR1 would either accelerate degradation or have little effect. To test this idea, wild type cells were transformed to overexpress TOR1, and FBPase degradation was examined. FBPase degradation was delayed in cells overexpressing TOR1 (Fig. 7A). By contrast, the deletion of TOR1 had little effect on FBPase degradation. Thus, excessive Tor1p appears to be inhibitory for FBPase degradation.
FIGURE 7.
TOR1 overexpression inhibits FBPase degradation. A, wild type (WT) cells overexpressing TOR1 on multicopy plasmids and cells lacking this gene were examined for FBPase degradation. B and C, wild type cells, Δtor1cells, cells overexpressing TOR1, and the Δtco89 strain were glucose-starved for 3 days. These cells were transferred to media containing fresh glucose for 20 min. The distribution of FBPase and Vid24p in the cytosolic enriched (C) and the Vid vesicle enriched (V) fractions was examined. Relative ratios of FBPase and Vid24p in these fractions were quantitated by ImageJ software.
We next determined which step of the FBPase degradation pathway was affected by TOR1 overexpression. Wild type cells and wild type cells that overexpressed TOR1 were shifted to glucose for 20 min, and the distribution of FBPase was examined. If overexpression of TOR1 inhibits the targeting of FBPase to Vid vesicles, then FBPase should remain in the cytosol in cells overexpressing this gene. By contrast, if TOR1 overexpressing inhibits a step after the sequestration of FBPase in Vid vesicles, FBPase should be in Vid vesicle-containing fraction in these cells. In wild type cells, a fraction of FBPase was found in the Vid vesicle-enriched fraction (Fig. 7B). In cells lacking the TOR1 gene, a percentage of FBPase was also detected in the Vid vesicle-enriched fraction. By contrast, very little FBPase was detected in the Vid vesicle fraction in TOR1-overexpressing cells. We used the same strategy to study the role of Tco89p in the FBPase degradation pathway. The majority of FBPase was in the cytosolic fraction in cells lacking the TCO89 gene.
We next examined the distribution of Vid24p in cells overexpressing TOR1 as well as in cells lacking this gene. Vid24p is a peripheral protein resides on Vid vesicles. If Vid vesicles are formed, then a fraction of Vid24p should be detected in the Vid vesicle-enriched fraction. By contrast, if Vid vesicles are not formed, then most of the Vid24p should not be in the Vid vesicle-enriched fraction. Vid24p was tagged with HA and expressed in wild type cells, in cells lacking TOR1, and in cells overexpressing TOR1. These cells were transferred to media containing fresh glucose for 20 min, and total lysates were subjected to differential centrifugation. The distribution of Vid24p in the cytosolic versus Vid vesicle-enriched fraction was then examined (Fig. 7C). In wild type cells, a portion of Vid24p was present in the Vid vesicle-enriched fraction. Likewise, in cells lacking TOR1, a percentage of Vid24p was also detected in the Vid vesicle-enriched fraction. By contrast, in cells overexpressing TOR1, low levels of Vid24p were detected in Vid vesicle-enriched fraction. In a similar manner, low amounts of Vid24p were found in the Vid vesicle-enriched fraction in cells lacking the TCO89 gene.
Tco89p and Tor1p Associate with Endosomes and Retrograde Vesicles from the Vacuole Membrane
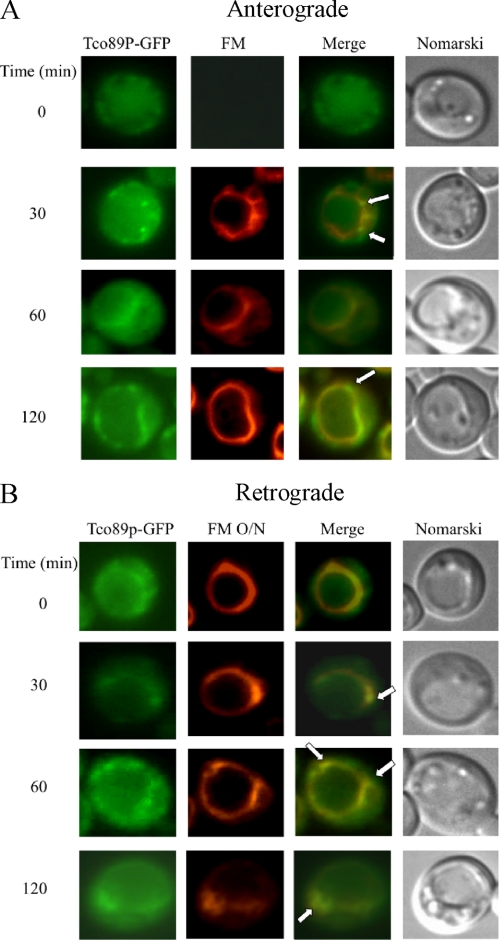
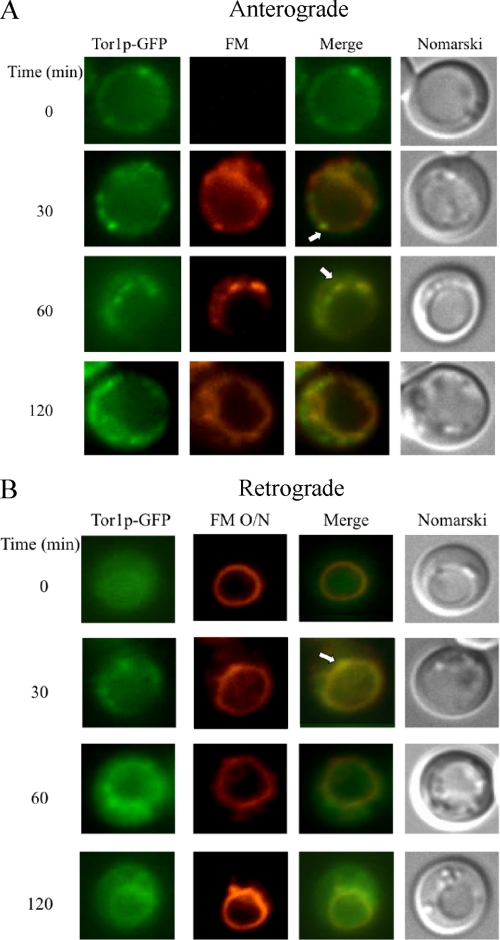
Tco89p localizes to plasma membranes and vacuoles in cells that are grown exponentially (40, 41, 47, 48). However, the distribution of this protein under our growth conditions has not been examined. Furthermore, whether these proteins move in or out of these compartments has not been investigated. We have shown that the Vid pathway merges with the endocytic pathway (25). Therefore, we utilized the FM dye to determine whether Tco89p travels to parts of the endocytic pathway. FM is internalized from the plasma membrane, traffics to endosomes, and reaches the vacuole membrane (49). To examine localization, Tco89p-GFP was expressed in wild type cells that were grown under 3-day glucose starvation conditions and then transferred to media containing fresh glucose for various periods of time. Note that during prolonged starvation, most cells displayed a very large vacuole, which made our interpretation difficult. Therefore, to better visualize the distribution of GFP-tagged proteins, cells with smaller vacuoles or with the vacuole on one side of the cells were selected for presentation purposes. During glucose starvation, Tco89p-GFP appeared to be in the cytosol and in punctate structures (Fig. 8A). However, after the addition of glucose for 30 min, a portion of Tco89p-GFP appeared in punctuate structures (Fig. 8A, arrows), some of which co-localized with FM-labeled dots located around the vacuole membrane. At 120 min, most of the FM reached the vacuole, and some Tco89p was also seen on the vacuole membrane. Thus, at least a portion of Tco89p appears to associate with endocytic compartments.
FIGURE 8.
Tco89p can be found in multiple locations. A, wild type cells expressing Tco89p-GFP were transferred to media containing fresh glucose in the presence of FM for the indicated times. Tco89p-GFP and FM were visualized with fluorescence microscopy. Cells were visualized by Nomarski optics. B, wild type cells expressing Tco89p-GFP were labeled with FM for 16 h (FM O/N) and then refed with fresh glucose in the absence of FM for the indicated times. Tco89p-GFP, FM, and cells were visualized by fluorescence microscopy. Arrows indicate the co-localization of Tco89p-GFP with FM.
We previously observed retrograde transport vesicles containing Sec28p that formed from the vacuole (25). To test whether Tco89p could also be in these vesicles, the vacuole was prelabeled with FM overnight, and cells were then shifted to glucose in the absence of FM. A portion of Tco89p (Fig. 8B, arrows) was observed in dots that were closely associated with the vacuole membrane. Thus, these results indicate that Tco89p associates with vesicles that appear to form from the vacuole membrane.
We performed the same experiments with wild type cells expressing Tor1p-GFP. When cells were labeled with FM during the glucose addition, we observed some co-localization of Tor1p with FM-containing endosomes (Fig. 9A, arrows). Likewise, when the vacuole was prelabeled with FM and then shifted to glucose, some Tor1p-GFP was localized in dots that were also labeled with FM on the vacuole membrane (Fig. 9B, arrows). Thus, these results suggest that Tor1p can be found in both endocytic vesicles and retrograde transport vesicles coming from the vacuole.
FIGURE 9.
Tor1p is distributed in multiple locations. A, wild type cells expressing Tor1p-GFP were transferred from low to high glucose in the presence of FM for the indicated times. B, the vacuole was pre-labeled with FM for 16 h (FM O/N) in the same cells expressing Tor1p-GFP. Cells were then transferred to media containing fresh glucose in the absence of FM for the indicated times. Tor1p-GFP and FM were visualized with fluorescence microscopy. Arrows indicate the co-localization of Tor1p-GFP with FM.
Rapamycin Inhibits FBPase Degradation
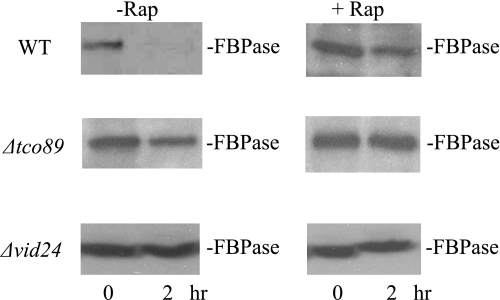
After periods of nitrogen starvation, autophagy is induced. Autophagy can also be induced by the drug rapamycin, even in the absence of nitrogen starvation (7–12). Because Tor1p is inhibitory to FBPase degradation, we anticipated that rapamycin treatment would enhance the degradation of this protein. To our surprise, the presence of rapamycin in fact blocked FBPase degradation in wild type cells that were starved for 3 days and then re-fed with glucose (Fig. 10). FBPase degradation was impaired in 3-day-starved Δtco89 and Δvid24 cells that were re-fed with glucose in the absence or presence of rapamycin.
FIGURE 10.
Rapamycin inhibits FBPase degradation. Wild type (WT), Δtco89, and Δvid24 strains were glucose starved for 3 days and transferred to media containing fresh glucose in the absence or presence of rapamycin. FBPase degradation was then examined.
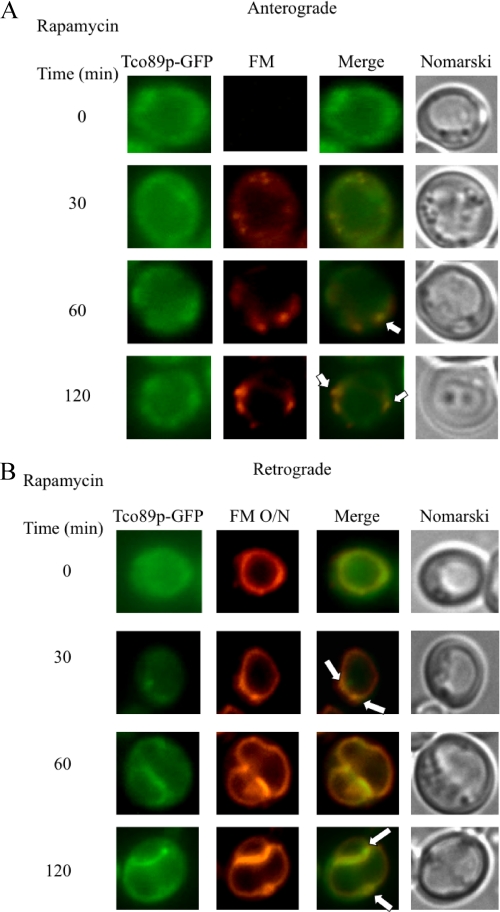
We next determined whether rapamycin might affect the distribution of Tor1p or Tco89p within cells. For these experiments, wild type cells expressing Tor1p-GFP or Tco89p-GFP were refed with glucose in the presence of rapamycin. Anterograde endocytic pathway was monitored with FM added at the same time of glucose, whereas retrograde movement from the vacuole membrane was examined by labeling of the vacuole membrane with FM for 16 h. For anterograde pathway, rapamycin appeared to cause a kinetic delay of the movement of FM to the vacuole (Fig. 11A). FM normally reached the vacuole in wild type cells in 30–60 min. In cells treated with rapamycin, a high percentage of FM was in endosomes at 60–120 min. A portion of Tco89p was also found in these FM positive endosomes at these time points (Fig. 11A, arrows).
FIGURE 11.
Tco89p distribution in response to rapamycin. A, Tco89p-GFP cells were glucose-starved for 3 days and then transferred to media containing fresh glucose and FM in the presence of rapamycin for the indicated times. Tco89p-GFP, FM, and cells were visualized by fluorescence microscopy. B, the vacuole of the Tco89p-GFP cells was pre-labeled with FM for 16 h (FM O/N). The distribution of Tco89p-GFP after the addition of glucose in the presence of rapamycin was observed by fluorescence microscopy. Co-localization of Tco89p-GFP with FM is indicated by arrows.
We next examined the effects of rapamycin in retrograde transport by labeling the vacuole with FM overnight. Cells were then treated with rapamycin and glucose at the same time. After a shift to glucose in the presence of rapamycin for 60 or 120 min, a significant amount of Tco89p was on the vacuole membrane (Fig. 11B). Interestingly, the vacuole membrane appeared to be enlarged and twisted. Note that a percentage of Tco89p could still be observed on vesicles that appeared to form from the vacuole membrane (Fig. 11B, arrows).
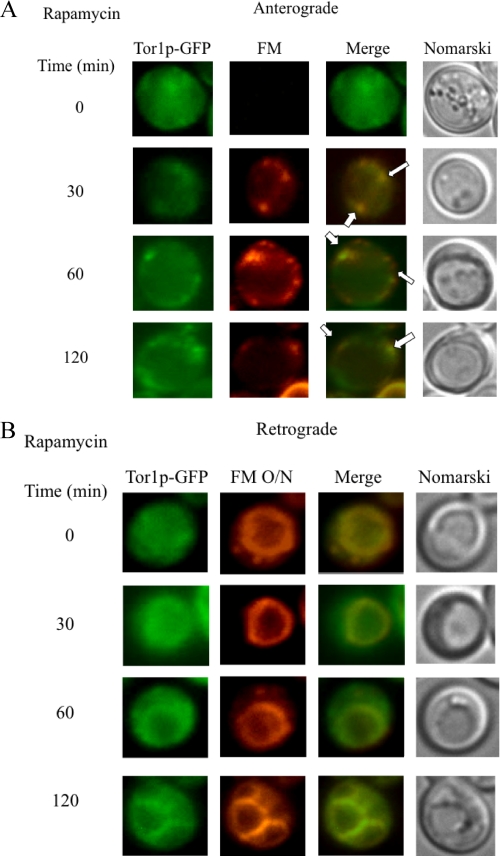
The same distribution pattern was found for Tor1p-GFP. Anterograde movement of FM appeared to be delayed in cells treated with rapamycin (Fig 12A). Most of this dye accumulated in endocytic compartments at 60–120 min upon rapamycin treatment. A percentage of Tor1p associated with these FM-positive endosomes (Fig. 12A, arrows). For the retrograde movement, Tor1p was seen on the vacuole membrane after a shift to glucose in the presence of rapamycin for 120 min (Fig. 12B).
FIGURE 12.
Tor1p distribution in response to rapamycin. A, wild type cells expressing Tor1p-GFP were glucose-starved for 3 days and transferred to media containing fresh glucose. FM and rapamycin were added to the media, and Tor1p-GFP and FM were observed using fluorescence microscopy. Co-localization of Tor1p-GFP with FM is indicated by arrows. B, the same cells were incubated with FM for 16 h (FM O/N) to pre-label vacuoles. Cells were then transferred to media containing fresh glucose in the presence of rapamycin for the indicated times. Tor1p-GFP, FM, and cells were visualized using fluorescence microscopy.
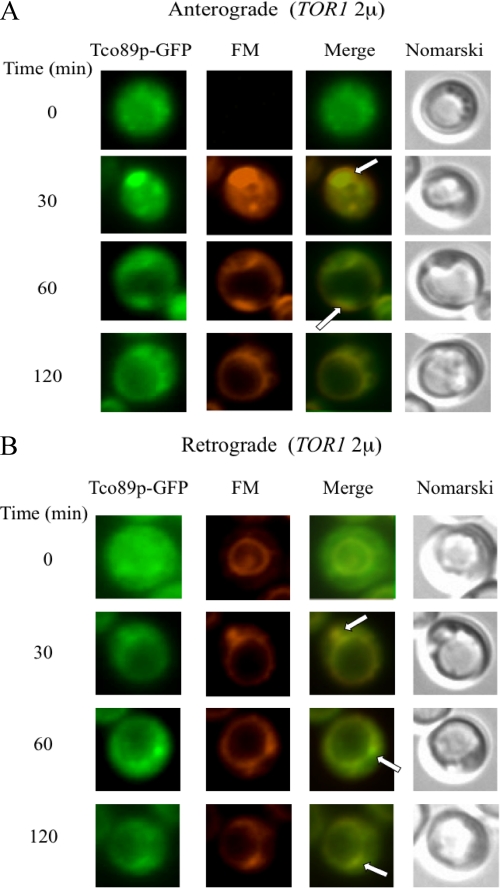
Finally, we tested whether overexpression of TOR1 affected Tco89p-GFP distribution. Wild type cells expressing Tco89p-GFP were transformed to overproduce TOR1 from a multicopy plasmid. The distribution of Tco89p-GFP was examined for both anterograde and retrograde trafficking pathways (Fig. 13, A and B). For anterograde transport, we observed that a portion of Tco89p co-localized with FM containing endosomes after the addition of glucose for 30–60 min (Fig. 13A, arrows). For retrograde transport, a percentage of Tco89p was seen in dots that appeared to form from the vacuole membrane. Thus, the distribution of Tco89p appears to be similar to that seen in wild type cells that did not overexpress TOR1(see Figs. 8, A and B).
FIGURE 13.
Tco89p-GFP distribution in cells overexpressing TOR1. A, wild type cells expressing Tco89p-GFP were transformed to overproduce TOR1 on a multicopy plasmid. Cells were starved for 3 days and transferred to media containing fresh glucose and FM for the indicated times. Co-localization of Tco89p-GFP with FM is indicated by arrows. B, FM was added to the same cells for 16 h (FM O/N) to pre-label vacuoles. Cells were then transferred to media containing fresh glucose in the absence of FM for the indicated times. Tco89p-GFP, FM, and cells were visualized using fluorescence microscopy. Arrows indicate the co-localization of Tco89p-GFP with FM.
DISCUSSION
Previously, we demonstrated that FBPase and MDH2 are targeted to the vacuole for degradation when 3-day-starved cells are shifted to glucose. Furthermore, these proteins utilize the Vid pathway for targeting to the vacuole (21). In this study we identified two additional proteins, Icl1p and Pck1p, as cargo proteins that utilize the Vid-dependent pathway for degradation in the vacuole during prolonged starvation. Both proteins were detected in the vacuoles of Δpep4 cells after a glucose shift. Furthermore, degradation of both proteins required the VID24 gene. To search for cellular factors that recognize these cargo proteins, we have identified Tco89p as an FBPase-interacting protein using affinity chromatography. In addition to FBPase, Tco89p also bound to MDH2, Icl1p, and Pck1p. Not surprisingly, other TORC1 components also bound to these proteins. We found that excessive Tor1p was inhibitory. For example, overexpression of TOR1 inhibited FBPase degradation. Kinetic studies indicated that Tor1p dissociated from FBPase upon glucose addition. We suggest that Tor1p is inhibitory, and this protein needs to be removed for FBPase degradation to occur.
When the distribution of Tco89p and Tor1p was examined, both proteins were detected in vesicles that appeared to form from the vacuole membrane. These observations support our previous findings whereby retrograde vesicles containing Sec28p could form from the vacuole membrane. In addition, these proteins can be found in endosomes coming from the plasma membrane. Therefore, we suggest that the TORC1 traffics to and from the vacuole. An increase in influx without an increase in efflux may lead to expansion of the membrane, whereas a decreased influx with an increased efflux may lead to shrinkage of the compartments. When endocytosis is induced, influx into the vacuole increases. Without an increase in efflux, the vacuole will expand. Therefore, retrograde transport provides a mechanism to maintain the size of the vacuole. In addition, retrograde transport may carry molecules back for further use. Hence, when the retrograde movement is disrupted, anterograde movement may also be affected. We observed that vacuoles became large and twisted after a rapamycin treatment. It is possible that rapamycin causes a primary defect in retrograde movement. An impaired retrograde movement then causes a secondary defect in anterograde movement due to the depletion of molecules that need to be recycled. Alternatively, rapamycin may have a direct effect on both the anterograde and retrograde trafficking pathways. Further experiments will be needed to sort out these possibilities.
Based on our results, we suggest the following model. The TORC1 complex binds to FBPase and other cargo proteins destined for the Vid pathway. After the addition of glucose, Tor1p is released from FBPase. In cells overexpressing TOR1, FBPase association with Vid vesicles was inhibited. The association of FBPase with Vid vesicles was also reduced in cells lacking TCO89, suggesting this gene plays a required role in Vid vesicle formation or Vid vesicle function. Further experiments will be needed to elucidate whether Vid vesicle formation is linked to anterograde or retrograde transport pathways and how these two trafficking pathways are coordinately regulated.
Acknowledgments
We thank Dr. McAlister-Henn (University of Texas, Houston) for the anti-MDH2 antibodies, Dr. A. Kornberg (Stanford University) for the Δpep4Δprb1Δprc1 strain, and Dr. T. Powers (University of California, Davis) for strains containing myc-tagged Tco89p and HA-tagged Tor1p, Kog1p, and Lst8p. Primers used in this study were synthesized in the Core Facility at Penn State University College of Medicine. MALDI was performed in the Core Facility at Penn State University, College of Medicine.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM 59480. This work was also supported by a grant from the Pennsylvania Tobacco Settlement Fund (to H.-L. C.).
- FBPase
- fructose-1,6-bisphosphatase
- MDH2
- malate dehydrogenase
- FM
- FM4-64
- MALDI
- matrix-assisted laser desorption ionization
- GFP
- green fluorescent protein
- HA
- hemagglutinin.
REFERENCES
- 1.Jones E. W. (1991) J. Biol. Chem. 266, 7963–7966 [PubMed] [Google Scholar]
- 2.Bryant N. J., Stevens T. H. (1998) Microbiol. Mol. Biol. Rev. 62, 230–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky D. J., Herman P. K., Emr S. D. (1990) Microbiol. Rev. 54, 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. (1992) Mol. Biol. Cell 3, 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothman J. H., Stevens T. H. (1986) Cell 47, 1041–1051 [DOI] [PubMed] [Google Scholar]
- 6.Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W. P., Klionsky D. J. (2002) Cell Struct. Funct. 27, 409–420 [DOI] [PubMed] [Google Scholar]
- 8.Klionsky D. J. (2005) J. Cell Sci. 118, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reggiori F., Klionsky D. J. (2005) Curr. Opin. Cell Biol. 17, 415–422 [DOI] [PubMed] [Google Scholar]
- 10.Wang C. W., Klionsky D. J. (2003) Mol. Med. 9, 65–76 [PMC free article] [PubMed] [Google Scholar]
- 11.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. (1998) Nature 395, 395–398 [DOI] [PubMed] [Google Scholar]
- 13.Tuttle D. L., Dunn W. A., Jr. (1995) J. Cell Sci. 108, 25–35 [DOI] [PubMed] [Google Scholar]
- 14.Dunn W. A., Jr. (1990) J. Cell Biol. 110, 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan W., Stromhaug P. E., Dunn W. A., Jr. (1999) Mol. Biol. Cell 10, 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui D. Y., Brown C. R., Chiang H. L. (2004) J. Biol. Chem. 279, 9713–9724 [DOI] [PubMed] [Google Scholar]
- 17.Chiang H. L., Schekman R. (1991) Nature 350, 313–318 [DOI] [PubMed] [Google Scholar]
- 18.Chiang H. L., Schekman R., Hamamoto S. (1996) J. Biol. Chem. 271, 9934–9941 [DOI] [PubMed] [Google Scholar]
- 19.Hoffman M., Chiang H. L. (1996) Genetics 143, 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shieh H. L., Chiang H. L. (1998) J. Biol. Chem. 273, 3381–3387 [DOI] [PubMed] [Google Scholar]
- 21.Hung G. C., Brown C. R., Wolfe A. B., Liu J., Chiang H. L. (2004) J. Biol. Chem. 279, 49138–49150 [DOI] [PubMed] [Google Scholar]
- 22.Schork S. M., Thumm M., Wolf D. H. (1995) J. Biol. Chem. 270, 26446–26450 [DOI] [PubMed] [Google Scholar]
- 23.Regelmann J., Schüle T., Josupeit F. S., Horak J., Rose M., Entian K. D., Thumm M., Wolf D. H. (2003) Mol. Biol. Cell 14, 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang P. H., Chiang H. L. (1997) J. Cell Biol. 136, 803–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown C. R., Wolfe A. B., Cui D., Chiang H. L. (2008) J. Biol. Chem. 283, 26116–26127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown C. R., McCann J. A., Chiang H. L. (2000) J. Cell Biol. 150, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown C. R., McCann J. A., Hung G. G., Elco C. P., Chiang H. L. (2002) J. Cell Sci. 115, 655–666 [DOI] [PubMed] [Google Scholar]
- 28.Brown C. R., Cui D. Y., Hung G. G., Chiang H. L. (2001) J. Biol. Chem. 276, 48017–48026 [DOI] [PubMed] [Google Scholar]
- 29.Shieh H. L., Chen Y., Brown C. R., Chiang H. L. (2001) J. Biol. Chem. 276, 10398–10406 [DOI] [PubMed] [Google Scholar]
- 30.McMahon H. T., Mills I. G. (2004) Curr. Opin. Cell Biol. 16, 379–391 [DOI] [PubMed] [Google Scholar]
- 31.Duden R. (2003) Mol. Membr. Biol. 20, 197–207 [DOI] [PubMed] [Google Scholar]
- 32.Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. (2004) Annu. Rev. Cell Dev. Biol. 20, 87–123 [DOI] [PubMed] [Google Scholar]
- 33.Wieland F., Harter C. (1999) Curr. Opin. Cell Biol. 11, 440–446 [DOI] [PubMed] [Google Scholar]
- 34.Gabriely G., Kama R., Gerst J. E. (2007) Mol. Cell. Biol. 27, 526–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daro E., Sheff D., Gomez M., Kreis T., Mellman I. (1997) J. Cell Biol. 139, 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu F., Aniento F., Parton R. G., Gruenberg J. (1997) J. Cell Biol. 139, 1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piguet V., Gu F., Foti M., Demaurex N., Gruenberg J., Carpentier J. L., Trono D. (1999) Cell 97, 63–73 [DOI] [PubMed] [Google Scholar]
- 38.Aniento F., Gu F., Parton R. G., Gruenberg J. (1996) J. Cell Biol. 133, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitney J. A., Gomez M., Sheff D., Kreis T. E., Mellman I. (1995) Cell 83, 703–713 [DOI] [PubMed] [Google Scholar]
- 40.Reinke A., Anderson S., McCaffery J. M., Yates J., 3rd, Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K. P., Powers T. (2004) J. Biol. Chem. 279, 14752–14762 [DOI] [PubMed] [Google Scholar]
- 41.Wedaman K. P., Reinke A., Anderson S., Yates J., 3rd, McCaffery J. M., Powers T. (2003) Mol. Biol. Cell 14, 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 43.Dubouloz F., Deloche O., Wanke V., Cameroni E., De Virgilio C. (2005) Mol. Cell 19, 15–26 [DOI] [PubMed] [Google Scholar]
- 44.Hämmerle M., Bauer J., Rose M., Szallies A., Thumm M., Düsterhus S., Mecke D., Entian K. D., Wolf D. H. (1998) J. Biol. Chem. 273, 25000–25005 [DOI] [PubMed] [Google Scholar]
- 45.Holzer H. (1989) Revis. Biol. Celular 21, 305–319 [PubMed] [Google Scholar]
- 46.Chiang M. C., Chiang H. L. (1998) J. Cell Biol. 140, 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aronova S., Wedaman K., Anderson S., Yates J., 3rd, Powers T. (2007) Mol. Biol. Cell 18, 2779–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berchtold D., Walther T. C. (2009) Mol. Biol. Cell 20, 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vida T. A., Emr S. D. (1995) J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]