Abstract
A multistep two-component signaling system is established as a key element of cytokinin signaling in Arabidopsis. Here, we provide evidence for a function of the two-component signaling system in cold stress response in Arabidopsis. Cold significantly induced the expression of a subset of A-type ARR genes and of GUS in ProARR7:GUS transgenic Arabidopsis. AHK2 and AHK3 were found to be primarily involved in mediating cold to express A-type ARRs despite cytokinin deficiency. Cold neither significantly induced AHK2 and AHK3 expression nor altered the cytokinin contents of wild type within the 4 h during which the A-type ARR genes exhibited peak expression in response to cold, indicating that cold might induce ARR expression via the AHK2 and AHK3 proteins without alterations in cytokinin levels. The ahk2 ahk3 and ahk3 ahk4 mutants exhibited enhanced freezing tolerance compared with wild type. These ahk double mutants acclimated as efficiently to cold as did wild type. The overexpression of the cold-inducible ARR7 in Arabidopsis resulted in a hypersensitivity response to freezing temperatures under cold-acclimated conditions. The expression of C-repeat/dehydration-responsive element target genes was not affected by ARR7 overexpression as well as in ahk double mutants. By contrast, the arr7 mutants showed increased freezing tolerance. The ahk2 ahk3 and arr7 mutants showed hypersensitive response to abscisic acid (ABA) for germination, whereas ARR7 overexpression lines exhibited insensitive response to ABA. These results suggest that AHK2 and AHK3 and the cold-inducible A-type ARRs play a negative regulatory role in cold stress signaling via inhibition of ABA response, occurring independently of the cold acclimation pathway.
Keywords: Arabidopsis, Histidine Kinases, Mutant, Plant, Signal Transduction, Arabidopsis Histidine Kinase, Arabidopsis Response Regulator, Cold, Cytokinin
Introduction
Cytokinins are plant hormones that regulate a variety of developmental and physiological processes, including cell division, cell proliferation, root and leaf differentiation, chloroplast biogenesis, and the inhibition of leaf senescence (1). Arabidopsis cytokinin signaling utilizes a multistep phospho-relay composed of a sensor kinase, a histidine phosphotransfer protein, and a response regulator similar to the TCS2 of bacterial and yeast cells (2). A hybrid-type histidine kinase referred to as CYTOKININ INDEPENDENT1 (CKI1) is essential for megagametogenesis (3). CYTOKININ RESPONSE1 (CRE1)/WOODEN LEG1 (WOL1)/ARABIDOPSIS HISTIDINE KINASE4 (AHK4) were shown to bind directly to a variety of natural and synthetic cytokinins in vitro with high specificity as well as in a yeast system and thus to be a primary receptor for cytokinins (4–8). The experiments conducted using a heterologous phospho-relay system demonstrated that AHK2 and AHK3 are also cytokinin receptors. The primary functions of these Arabidopsis histidine kinase (AHK) genes involve the triggering of cell division and the maintenance of the meristematic competence of cells to prevent subsequent differentiation (9, 10). Partially redundant functions of cytokinin receptors have also been revealed in shoot growth, root development, leaf senescence, seed size, germination, and cytokinin metabolism (11). The AHK2 and AHK3 receptors play prominent roles in the control of leaf development, whereas AHK4 functions in the roots (10). AHK3 is also involved in the cytokinin-mediated control of leaf longevity (12). AHK1 has been shown to function as an osmosensor in yeasts in a cytokinin-independent manner (13), and it has recently been demonstrated to be a positive regulator of drought and salt stress response and ABA signaling (14). AHK5 (CKI2) has histidine kinase activity independent of cytokinins and functions as a negative regulator for root elongation via an ETR1-dependent ABA and ethylene signaling pathway (15). AHK5 was found to integrate endogenous and environmental signals that generate H2O2 in guard cells (16). A recent study showed that the cytokinin-independent activity of CKI1 and cytokinin-induced AHK2 and AHK3 are important for vascular bundle formation in Arabidopsis (17).
The Arabidopsis genome harbors six genes encoding histidine phosphotransfer proteins (AHPs) that mediate the transfer of the phosphoryl group from the histidine protein kinases to the response regulators (18). Most of the Arabidopsis histidine phosphotransfer proteins act as redundant, positive regulators of cytokinin signaling, except for AHP4 and AHP6, and affect many aspects of plant development (19, 20). There are 32 genes encoding 23 putative response regulators and 9 pseudo-response regulators in Arabidopsis. The typical response regulators are classified into either type A or B (18, 21). The type B Arabidopsis response regulators (ARRs) are transcription factors that contain a receiver domain and a large C-terminal region harboring a Myb-like DNA-binding domain and a glutamine-rich domain and are not cytokinin-inducible. The middle segments of ARR1 and ARR2, the B-type ARRs, can bind to DNA in a sequence-specific manner in vitro, and their C-terminal halves function as transactivation domains in plant cells (22). ARR1, ARR2, and ARR10 have been demonstrated to function as transcriptional activators for the ARR6 promoter in Arabidopsis mesophyll cell protoplasts (23). Type B ARRs function as positive regulators of cytokinin signaling and also play a role in plant development (24, 25). By contrast, the typical type A ARRs are composed of a receiver domain and a divergent C-terminal extension. The mRNAs of A-type ARRs accumulate rapidly and transiently after cytokinin treatment. The A-type ARRs function as transcriptional repressors in Arabidopsis protoplasts (23). In transgenic Arabidopsis, ARR8 or ARR15 overexpression resulted in reduced cytokinin sensitivity, whereas ARR4 overexpression induced increased sensitivity to exogenous cytokinins (26, 27). The majority of A-type ARRs are partially redundant negative regulators of cytokinin signaling (28). The genome-wide expression profiling analysis of cytokinin response in the ARR7 overexpressor compared with the wild type demonstrated that ARR7 acts principally as a transcriptional repressor of a variety of early cytokinin-regulated genes encoding transcription factors, signal transmitters, plant development, and cellular metabolism (29). The Asp-85 mutation of ARR7 into the Asp residue blocked the phosphorylation of ARR7 by the Arabidopsis protein extracts in vitro and abolished the ability of ARR7 to suppress cytokinin-mediated responses in transgenic Arabidopsis (30), suggesting that the phosphorylation of ARR7 is essential to its function as a negative regulator of cytokinin signaling in Arabidopsis.
Recent studies have shown that AHK1 is a positive regulator of drought and salt stress responses, as well as ABA signaling (14), and is involved in water stress response during the early vegetative stages of plant growth, but it also performs a unique role in the regulation of desiccation processes during seed maturation (31). AHK2, AHK3, and CRE1 have all been implicated as negative regulators in ABA signaling and osmotic stress responses (14).
Cold temperatures exert adverse effects on plant growth and development and limit the geographic distribution of plants. Plants respond and adapt to cold via alterations in various aspects of physiological processes, including gene expression (32, 33). Abiotic stresses, including cold, induce a number of genes that encode the proteins that promote stress tolerance, mediated primarily by an ABA-dependent and/or ABA-independent pathway. Cold temperatures transcriptionally activate C-REPEAT-BINDING FACTOR/DRE-BINDING FACTORs (CBF/DREBs) that bind to the C-repeat/dehydration-responsive element in the ABA-independent pathway, inducing the expression of cold-responsive genes. Genetic and reverse genetic approaches have revealed a number of components that are involved in the cold signaling pathway (32, 34–36).
In this study, we analyzed ahk and A-type arr Arabidopsis mutants as well as ARR7-overexpressing lines under freezing temperatures and A-type ARR expression in response to cold, and we demonstrated that the TCS, which is known best as a mediator of cytokinin signal transduction, is employed as a negative regulator in the process of adaptation to cold temperatures in plants. We found that the cold-inducible expression of A-type ARR genes was severely reduced by ahk2 and ahk3 double mutations. Analyses of A-type ARR expression in ahk mutants and in cytokinin-deficient or -overproducing transgenic Arabidopsis indicate that AHK2 and AHK3 might be involved in mediating cold to express A-type ARRs independently of the influence of endogenous cytokinin levels. Moreover, the cytokinin contents were not altered significantly by cold treatment within the 4 h during which the peak expression of cold-inducible ARRs occurs. Cold did not significantly induce AHK2 and AHK3 expression, indicating that the AHK2 and AHK3 proteins may mediate cold temperatures for A-type ARR expression. The ahk2, ahk3, and cre1 (ahk4) single mutants displayed tolerance to freezing temperatures when preincubated with cytokinin prior to freezing, as compared with the wild-type plants treated with cytokinin, as well as the untreated ahk single mutants. The ahk2 ahk3 and ahk3 ahk4 double mutants exhibited enhanced freezing tolerance without cytokinin preincubation. The overexpression of the cold-inducible ARR7 in Arabidopsis caused a hypersensitivity response to freezing temperatures when cold-acclimated, whereas the arr7 mutants exhibited increased freezing tolerance prior to and after cold acclimation. The expression of CBF/DREB target genes was not affected by ARR7 overexpression as well as in ahk double mutants. In both root elongation and germination assays, ahk2 ahk3 showed hypersensitivity response to ABA. arr7 showed hypersensitive response to ABA for germination, whereas Pro35S:ARR7 lines exhibited insensitive response to ABA. These results suggest that a normal function of a part of cytokinin TCS might be to negatively regulate the cold stress adaptation response through inhibition of ABA response, independently of the CBF/DREB pathway. Our results reveal a new versatile function of the plant two-component signaling system in cold stress response.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Arabidopsis Transformation
The promoter region of ARR7 encompassing −1651 bp relative to the AUG initiation codon was isolated by PCR from the genomic DNA of Arabidopsis Col-0 and subcloned into pBI121 (Clontech) in place of the CaMV 35S promoter, and transgenic Arabidopsis containing this construct (ProARR7:GUS) was made by Agrobacterium-mediated transformation (37). T3 homozygous transformants were prepared and amplified. All constructs were verified by DNA sequencing prior to plant transformation.
ahk and arr Mutants ahk2-1, ahk3-1, ahk2-1 ahk3-1 were generously provided by Dr. Ueguchi and confirmed via genotyping prior to usage (7, 10). cre1-12, ahk2-2 ahk3-3, ahk2-2 cre1-12, and ahk3-3 cre1-12 were kindly provided by Dr. Kakimoto (9) and verified by genotyping prior to usage. arr5 (CS25267), arr6 (CS25268) (28), and arr7 were obtained from ABRC. arr7 was WiscDsLox T-DNA lines (supplemental Fig. 1). The mutants were verified by genotyping prior to usage.
Transgenic Arabidopsis Harboring 35S:AtCKX2-2 (Arabidopsis thaliana Cytokinin Oxidase/Dehydrogenase2-2), LhGR-N, or pV-ipt/LhGR-N Constructs
35S:AtCKX2-2 seeds were kindly provided by Dr. Schmülling (38) and LhGR-N(4c-S5) and pV-ipt/LhGR-N(ipt5-2) seeds were generously provided by Dr. Moore (39). The transgenic seeds were confirmed by genotyping prior to amplification and usage. The PCR conditions and primer sequences are provided in supplemental Table 1.
Cold, Hormone, and DEX Treatment
A. thaliana was grown on germination agar plates containing 0.5× Murashige-Skoog (MS) media with vitamins, 1.5% sucrose, 2.5 mm Mes, pH 5.7, and 0.8% agar at 23 °C with a 16-h photoperiod and treated essentially as described previously (40). For cold treatment, the light-grown seedlings were incubated at 1 °C with white fluorescent light. For treatment with cytokinin benzyladenine (BA), the seedlings were grown on sterile filter papers on MS agar plates and then immersed in MS medium containing 5 μm BA. For the freezing tolerance assays, the plants were germinated on MS agar plates, grown for the indicated periods, and then treated with freezing temperatures in a temperature-controlled chamber. For the DEX treatments, the plants were grown on sterile filter paper on MS plates for 10 days and then transferred to MS plates containing 10 μm DEX. The plants were then incubated for a given period.
Electrolyte Leakage Test
Freezing-induced electrolyte leakage (41) was determined from the rosette leaves of 2-week-old plants grown in soil or in MS agar plates. The excised leaflets were placed individually into 5-ml test tubes containing 100 μl of deionized water in a refrigerated circulator bath (Polysciences) at 0 °C, and the temperature of the bath was programmed to decrease to −10 °C at 1 °C decrements over 30 min. The percentage of electrolyte leakage was calculated as the percentage of the conductivity prior to autoclaving over that noted after autoclaving the leaflets. At least 8–10 assays were conducted for each sample.
Histochemical GUS Assays
Histochemical assays of GUS activity were conducted after 24 h of incubation with the treated seedlings in 5-bromo-4-chloro-3-indolyl glucuronide (Duchefa, The Netherlands) at 37 °C and removal of the chlorophyll from green tissues by incubation in 100% ethanol, as described previously (42).
Cytokinin Analysis
The procedure utilized for cytokinin purification herein was a modified version of the method described previously (43). Deuterium-labeled cytokinin internal standards (Olchemim Ltd., Czech Republic) were added, each at 1 pmol per sample, to assess the recovery during purification and to validate the determination (44). The samples were purified with a combined cation (SCX cartridge) and anion (DEAE-Sephadex C18 cartridge) exchanger and immunoaffinity chromatography based on wide range-specific monoclonal antibodies against cytokinins (45). The metabolic eluates from the immunoaffinity chromatography columns were evaporated to dryness and dissolved in 20 μl of the mobile phase utilized for quantitative analysis. The samples were analyzed by an ultra-performance liquid chromatography (Acquity UPLCTM; Waters, Milford, MA) coupled to a Quatro microTM atmospheric pressure ionization (Waters, Milford, MA) triple quadrupole mass spectrometer equipped with an electrospray interface. The purified samples were then injected into a C18 reversed-phase column (BEH C18; 1.7 μm; 2.1 × 50 mm; Waters). The column was eluted with a linear gradient (0 min, 10% B; 0–8 min, 50% B; flow-rate of 0.25 ml/min; column temperature of 40 °C) of 15 mm ammonium formate, pH 4.0 (solvent A), and methanol (solvent B). Quantification was conducted by the multiple reaction monitoring of [M + H]+ and the appropriate product ion. For the selective multiple reaction monitoring experiments, the optimal conditions, dwell time, cone voltage, and collision energy in the collision cell corresponding to exact diagnostic transition were optimized for each cytokinin (44). Quantification was conducted using Masslynx software using a standard isotope dilution method. The ratio of endogenous cytokinin to the appropriate labeled standard was determined and further used to quantify the level of endogenous compounds in the original extract, according to a known quantity of an added internal standard (45).
RNA Isolation, RT-PCR, and RNA-Gel Blot Analysis
Following treatment, the Arabidopsis plants were immediately frozen in liquid nitrogen and stored at −80 °C. Total RNA was isolated from frozen Arabidopsis samples using TRI Reagent® (Molecular Research Center, Inc.). Total RNA was separated on 1.2% agarose gel, transferred to nylon membranes, hybridized with 32P-labeled DNA probes at 68 °C for 3 h using 10 ml of QuikHyb solution (Stratagene), and then washed. The blot was subsequently exposed to x-ray film. For RT-PCR analysis, total RNA was isolated using the RNeasy plant mini kit (Qiagen) and subjected to RT-PCR analysis with an Access RT-PCR System (Promega) according to the manufacturer's instructions. RT-PCR conditions and primer sequences are provided in supplemental Table 1.
Real Time RT-PCR
Real time RT-PCR was conducted using a QuantiTect SYBR Green RT-PCR kit (Qiagen) in a Rotor-Gene 2000 real time thermal cycling system (Corbett Research). PCR conditions and primers are available upon request. To determine the copy numbers of the transcripts in the treated samples, real time PCR was conducted for each sample with a known quantity of the in vitro transcribed RNA (Promega), yielding specific threshold values (Ct). A standard curve was generated to show the linear correlation between the log (copy numbers of the RNA) and the Ct. The copy numbers of the transcripts of unknown samples subjected to real time RT-PCR were then calculated from this standard curve. Real time RT-PCR conditions and primer sequences are provided in supplemental Table 1.
RESULTS
ARR7 and ProARR7:GUS Respond to Cold Temperature with Expression Profiling Similar to Cytokinin Response
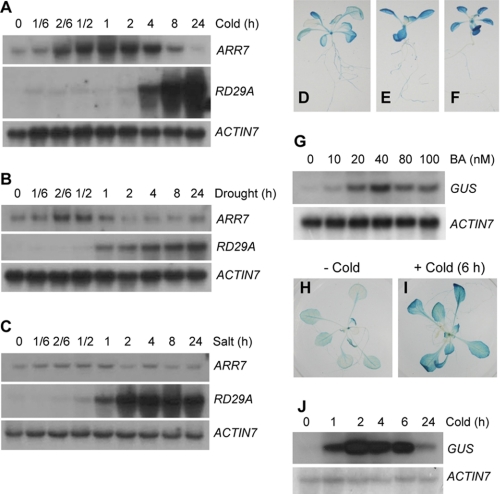
Many of the upstream components of signaling pathways are expressed quickly and primarily in response to stimuli (46, 47). We previously isolated ARR7, a cytokinin-inducible A-type ARR gene (48), as an immediate response mRNA from Arabidopsis by cold temperature at 1 °C using differential display PCR.3 We thus investigated the ARR7 gene as a starting point to understand the potential cross-talk occurring between cold response and cytokinin signaling and the role of TCS in cold response. We initially performed a time course analysis of the expression of ARR7 mRNA in response to cold via RNA-gel blot analysis (Fig. 1A). The ARR7 mRNA began to accumulate within 10 min at 1 °C, reached a peak level at 2–4 h, and then declined, displaying an early and transient expression pattern similar to that observed with cytokinin-mediated ARR7 expression. ARR7 also responds to other abiotic stresses, such as drought (Fig. 1B) or significantly high salinity (Fig. 1C and supplemental Fig. 2), but only slightly and in a transient manner, with an earlier peak. In this study, we have focused on the role of TCS in cold response. To determine whether ARR7 expression is modulated at the transcriptional level in response to cold as well as cytokinins, we fused the 1.65-kbp promoter region of the ARR7 gene to GUS and generated six transgenic Arabidopsis lines harboring the corresponding construct (ProARR7:GUS). We conducted histochemical GUS assays to assess the patterns of tissue expression. As shown in supplemental Fig. 3, A–G, we detected strong GUS expression in the meristem regions of the flowers, shoots, and roots (supplemental Fig. 3, A–C). The vascular tissues of the leaves and roots displayed strong GUS staining (supplemental Fig. 3, D, E, and G). The valves of the silique (supplemental Fig. 3F) and the pistil tip (supplemental Fig. 3A) were also strongly stained with GUS. Increasing concentrations of cytokinin benzyladenine (BA) enhanced GUS staining throughout the whole leaves (Fig. 1, D–F) and the GUS mRNA level, as demonstrated by RNA-gel blot analysis (Fig. 1G). Cold temperature significantly enhanced GUS expression (Fig. 1, H and I). The RNA-gel blot analysis also demonstrated that cold temperature transiently induced the levels of the GUS mRNA (Fig. 1J). We observed relatively stronger induction of the GUS mRNA as compared with GUS staining in response to cold temperatures. This may be attributable to the strong background of GUS staining prior to cold treatment, because of the stable GUS enzymes. These data demonstrate that exposure to cold as well as cytokinin activates ARR7 transcription. Our observations regarding the tissue-specific expression patterns of GUS and the timing of cold-responsive ARR7 expression are generally consistent with the microarray data as visualized by the Arabidopsis electronic fluorescent pictograph browser (49).
FIGURE 1.
ARR7 and ProARR7:GUS transgenics respond to abiotic stresses, including cold. A, response of ARR7 to cold. Eleven-day-old wild-type (Col-0) seedlings were incubated at 1 °C. Total RNA isolated from each treatment was subjected to RNA-gel blot analysis. RD29A was used as a marker gene (62). ACTIN7 was utilized as a loading control. B, response of ARR7 to dehydration. Three-week-old wild-type seedlings were dehydrated on filter paper in the light at 23 °C and analyzed as described in A. C, response of ARR7 to high salinity. Eleven-day-old wild-type seedlings were incubated with 300 mm NaCl in darkness and analyzed as described in A. D–G, response of ProARR7:GUS to BA. Plants grown for 16 days on MS agar plate containing 0 nm (D), 40 nm (E), and 100 nm (F) BA were subjected to histochemical GUS assays. Plants grown for 16 days on MS agar plates containing varying concentrations of BA were subjected to RNA-gel blot analysis (G). H–J, response of ProARR7:GUS to cold. ProARR7:GUS seedlings (17-day-old light-grown plants) grown on MS agar plates were treated with cold at 1 °C for 6 h and subjected to histochemical GUS assays (H and I) or treated with cold at 1 °C for the indicated time, followed by RNA-gel blot analysis (J).
Cold Temperature Induces Expression of a Subset of A-type ARR Genes
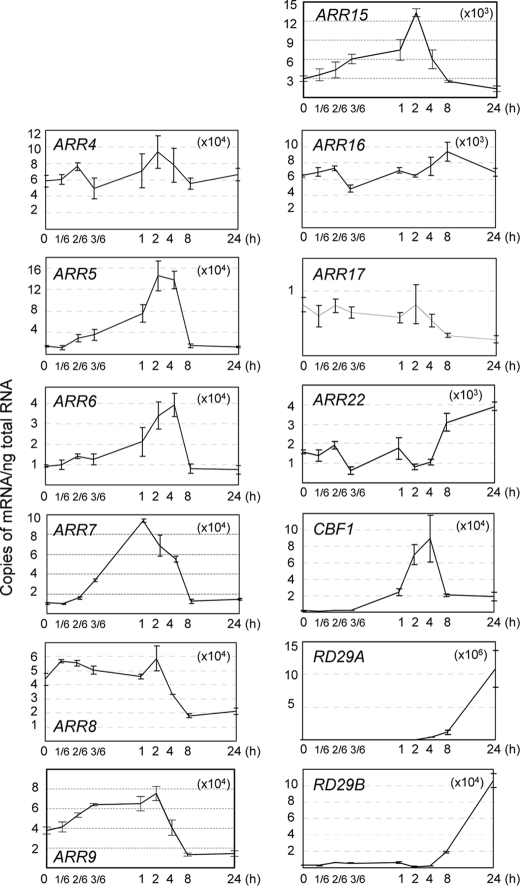
We next assessed the response of all A-type ARR genes (18, 21) to cold to determine whether or not cold-induced ARR7 expression is unique among A-type ARR genes. Both comparative RT-PCR and quantitative real time RT-PCR assays were employed and demonstrated that cold temperatures transiently induced the expression of a variety of A-type ARR genes, including ARR5, ARR6, ARR7, and ARR15 (Fig. 2). ARR9 and ARR22 showed a weak and early or late response to cold, respectively. Similar patterns were noted on comparative RT-PCR (data not shown). Although the quantification of the ARR3 mRNA could not be clearly determined due to the double bands of the PCR products, the RT-PCR data showed that ARR3 did not respond significantly to cold (data not shown). Our results are consistent with previous microarray analysis data demonstrating that ARR5 and ARR7 exhibited maximum cold response in 2–4 h with induction levels over 8- and 5-fold, respectively (50). CBF1, RD29A, and RD29B were utilized as cold marker genes to confirm the effects of our cold treatment with regard to the induction of gene expression. CBF1 was unresponsive to cytokinin BA, although BA greatly induced ARR7 expression (supplemental Fig. 4).
FIGURE 2.
Quantitative analysis of A-type ARR gene expression in response to cold. Eleven-day-old wild-type (Col-0) seedlings were incubated at 1 °C for varying periods of time (hour). Total RNA isolated from each treatment was subjected to quantitative real time RT-PCR. CBF1, RD29A, and RD29B were employed to verify stress-inducible gene expression. ACTIN7 was utilized as a loading control. Copies of the transcripts from cold-treated plants were plotted per ng of total RNA after normalization to ACTIN7 RNA. Multiplication of the number in parentheses in the graph by the number in the y axis generates copy numbers of the corresponding transcripts. The means ± S.E. from biological triplicate experiments were plotted.
Double Mutations in AHK2 and AHK3 Severely Reduced ARR5, ARR6, ARR7, and ARR15 Expression in Response to Cold but Do Not Affect the Cold-induced Expression of CBFs
The observation that ARR7 promoter-GUS and various A-type ARR genes responded to cold raised the possibility that the AHK genes encoding cytokinin receptors might be involved in mediating cold signals for the activation of A-type ARR expression. To evaluate this possibility, we analyzed the expression of a representative set of cold-inducible A-type ARR genes, ARR5, ARR6, ARR7, and ARR15 in ahk2, ahk3, and cre1 (=ahk4) mutant backgrounds in response to cold by using both quantitative real time and comparative RT-PCR methods. All ahk mutants were T-DNA insertional mutants with the Col-0 ecotype (9, 10). Plants were treated with cold for the peak expression time of 2 h. In all single ahk mutants, ahk2-1, ahk3-1, and cre1-12, ARR5, ARR6, ARR7, and ARR15 showed wild-type levels of gene expression in response to cold (Fig. 3A). In contrast, the ahk2 ahk3 double mutants, ahk2-2 ahk3-3, exhibited severely reduced transcript levels of ARR5, -6, -7, and -15 to cold, whereas the other ahk double mutants, ahk3-3 cre1-12 and ahk2-2 cre1-12, showed a normal response to cold (Fig. 3B). Similar patterns were noted on comparative RT-PCR (data not shown). The ahk2-1 ahk3-1 double mutants also exhibited similarly reduced transcript levels (data not shown). These findings indicate that AHK2 and AHK3 perform a primary role in mediating cold temperatures for the expression of A-type ARRs, whereas other AHKs not related to cytokinin reception or other mechanisms may also be involved. In these ahk mutants, CBF1, encoding the critical transcription factor that functions in the cold acclimation process, can be induced by cold treatment as efficiently as in the wild-type plants. Other CBF members, CBF2 and CBF3, also responded to cold temperatures in ahk double mutant backgrounds, similarly to the wild-type plants (data not shown). This result indicates that the CBF/DREB pathway is not coupled to cold-responsive TCS.
FIGURE 3.
Expression of ARR5, ARR6, ARR7, and ARR15 in response to cold in ahk mutants compared with the wild-type plants. A, ARR5, ARR6, ARR7, and ARR15 expression in ahk single mutant backgrounds. Eleven-day-old light-grown seedlings were treated for 2 h at 1 °C, and total RNA isolated was subjected to real time RT-PCR. Treatment and analysis of the samples were done as described in the Fig. 2 legend. Closed and open bars represent the transcript levels from the plants treated with or without cold, respectively. B, ARR5, ARR6, ARR7, and ARR15 expression in ahk double mutant backgrounds. Treatment and analysis of the samples were done as described in A. ** denotes statistically significant changes with p < 0.01 as compared with the samples without cold treatment.
Cold-responsive Expression of A-type ARRs in Transgenic Arabidopsis Deficient in Cytokinins
We next tested if cold induces the expression of the AHK genes encoding cytokinin receptors, thereby resulting in up-regulation of cold-responsive ARRs. Our quantitative time course analysis using real time RT-PCR showed that cold did not induce AHK2, AHK3, and AHK4 expression in a statistically significant manner within 24 h (supplemental Fig. 5). Previous RNA-gel blot analyses had also shown that cold did not significantly increase the expression of AHK2, AHK3, and AHK4 in general, although some expression of these AHK genes at 2 h of cold treatment had been detected, whereas dehydration strongly induced the expression of AHK2 and AHK3 (14). As we have reproducibly observed significant cold-responsive ARR expression within 30 min and strong transient gene expression (Fig. 2), both results demonstrated that cold-responsive ARR expression is not primarily due to the increase in gene expression of these AHKs.
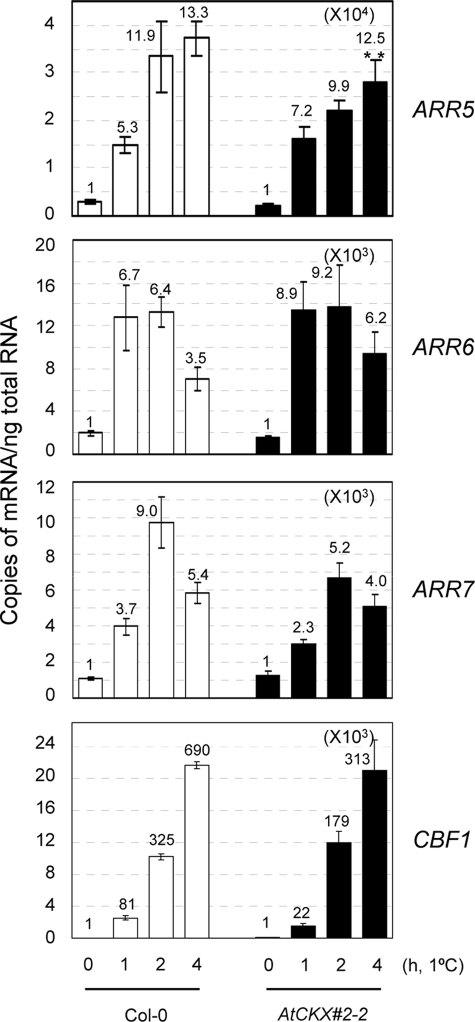
We then investigated whether cold can activate the enzymes involved in the biosynthesis of cytokinins, thereby resulting in increased cytokinin levels that subsequently activate the AHK receptors for A-type ARR expression as an early response. To test this possibility, we first attempted to determine whether decreasing levels of cytokinins can affect the cold-induced expression of A-type ARR genes via the use of cytokinin-deficient transgenic Arabidopsis plants. We utilized 35S:AtCKX2-2 plants that overexpress cytokinin oxidase and that contain less than 20% of total zeatin content and less than 40% of total cytokinin content compared with that of the wild-type plants (38). AtCKX2-2 is the most cytokinin-deficient 35S:AtCKX transgenic Arabidopsis among the 35S:AtCKX plants generated (38). We treated these cytokinin-deficient transgenic plants with cold for 0, 1, 2 or 4 h and assessed the expression of ARR5, ARR6, ARR7, and CBF1 by quantitative real time RT-PCR, as compared with the wild-type plants. Our results demonstrated that the expressions of those ARRs and CBF1 were not influenced significantly by cytokinin deficiencies, in general (Fig. 4). Although we noted a statistically significant difference in cold-responsive ARR5 expression between the wild-type and cytokinin-deficient transgenics, this difference was marginal and was observed only in cases of cold treatment applied for 4 h.
FIGURE 4.
Expression of ARR5, ARR6, and ARR7 in response to cold in 35S:AtCKX transgenic Arabidopsis deficient in cytokinins compared with the wild type. Eleven-day-old light-grown seedlings were treated at 1 °C for 1, 2, or 4 h, and the total RNA isolated was subjected to real time RT-PCR. The treatment and analysis of the samples were done as described in the Fig. 2 legend. The numbers on top of the bars indicate fold-induction relative to the control sample at 0 h. Open and closed bars represent the Col-0 wild type and 35S:AtCKX2-2 transgenic Arabidopsis, respectively. ** denotes statistically significant changes with p < 0.01 compared with the wild type.
Cold-responsive Expression of A-type ARRs in Transgenic Arabidopsis Overproducing Cytokinins in a Dexamethasone-dependent Fashion
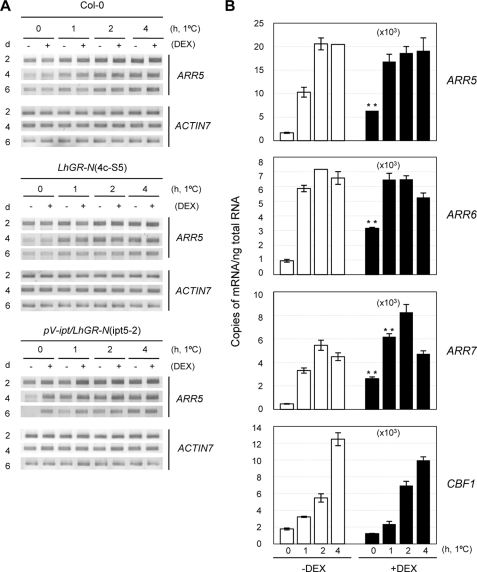
We next attempted to determine whether increased cytokinin levels might exert an effect on the cold-responsive expression of A-type ARRs. To prevent severe effects of constitutively overproduced cytokinins on plant growth and development, we utilized a transgenic Arabidopsis (pV-ipt/LhGR-N(ipt5-2)), which overexpresses isopentenyltransferase, a critical enzyme for cytokinin biosynthesis, and thus cytokinins in a DEX-inducible manner (39). The pV-ipt/LhGR-N transgenic plants contain an activator construct that expresses LhGR and a construct harboring isopentenyltransferase and GUS under the control of six copies of the ideal lac operator (39). DEX treatment results in the activation of both IPT and GUS genes (supplemental Fig. 6). Whereas DEX treatment applied for 14 days after germination induced no phenotypic change in the wild-type plants as well as the GR-overexpressing transgenic plants (LhGR-N), the same treatment induced severe growth inhibition in the pV-ipt/LhGR-N transgenic plants. However, when we applied DEX to 10-day-old pV-ipt/LhGR-N plants as well as two control plants, Col-0 and LhGR-N, for up to an additional 6 days, we could not note a significant inhibition in plant growth (supplemental Fig. 7).
Ten-day-old plants were therefore incubated with DEX for an additional 2, 4, or 6 days, followed by cold treatment at 1 °C for 0, 1, 2 or 4 h. Total RNAs extracted from the treated plants were subjected to comparative RT-PCR analysis. ARR5 was selected for expression analysis as a representative ARR because of highly inducible gene expression to both cytokinins and cold. As shown in Fig. 5A, the preincubation of the pV-ipt/LhGR-N plants with DEX enhanced ARR5 expression levels as compared with what was observed in the absence of DEX, whereas two control plants were unresponsive to DEX, thereby indicating the ability of cytokinins to activate ARR5 expression in the pV-ipt/LhGR-N plants. The application of cold treatment to all three of these plants resulted in increased ARR5 expression. We next conducted real time RT-PCR analysis to quantify the response of ARR5, ARR6, and ARR7 to cold with plants incubated for 4 days with DEX (Fig. 5B). DEX clearly induced ARR5, ARR6, and ARR7 expression (Student's t test, p < 0.001). Similar patterns were noted on comparative RT-PCR (data not shown). In the case of ARR5, DEX treatment exerted additive effects on the expression of cold-responsive genes after 1 h of cold treatment, but the expression levels reached a plateau after 2 h of treatment. ARR6 showed maximum expression levels after 1 h of cold treatment. ARR7 exhibited additive induction kinetics with both cold and DEX treatment for up to 2 h of cold treatment. These results showed that increasing levels of cytokinins exert additive effects with cold on ARR5, ARR6, and ARR7 expression at early time points of the treatment, but the maximum induction levels reached by the treatment of both cold and DEX were identical to those of cold treatment alone.
FIGURE 5.
Effects of cytokinin overproduction on cold-responsive expression of ARR5, ARR6, and ARR7. A, RT-PCR analysis of cold-responsive ARR5 expression with or without cytokinin overproduction. The pV-ipt/LhGR-N(ipt5-2) transgenic plants as well as control plants, the wild-type, and LhGR-N(4c-S5) plants were grown on sterile filter paper on MS plates for 10 days and then incubated on MS plates with or without 10 μm DEX for an additional 2, 4, or 6 days in the light. The plants were then treated with cold at 1 °C in the light for the indicated number of hours. Total RNA was isolated from treated plants and subjected to RT-PCR for ARR5. ACTIN7 was utilized as a loading control. Inverted images of the RT-PCR products are shown. LhGR-N transgenic plants contain activator construct expressing LhGR from the CaMV 35S promoter (39). pV-ipt/LhGR-N transgenic plants contain the activator construct expressing LhGR and a construct harboring IPT and GUS under six copies of an ideal lac operator (39). 4c-S5 and ipt5-2 indicate the line numbers. B, real time RT-PCR analysis of cold-responsive ARR expression with or without cytokinin overproduction. Plants were treated as described in A and analyzed by real time RT-PCR for ARR5, ARR6, and ARR7, as described in the Fig. 2 legend. CBF1 was utilized to verify cold-inducible gene expression. ** denotes statistically significant changes with p < 0.01 compared with the absence of DEX.
Cytokinin Analysis of Arabidopsis Subjected to Cold Treatment Showed Insignificant Changes in Cytokinin Levels during Peak Expression Time of Cold-responsive ARRs
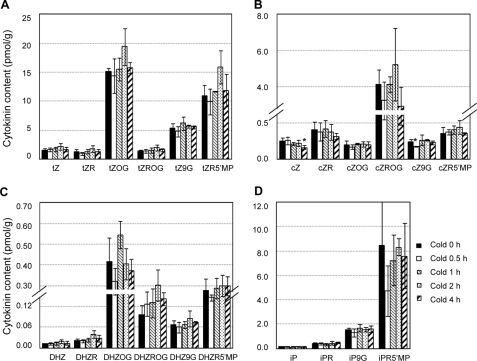
To determine whether cold could induce an increase in cytokinin levels, thereby mediating A-type ARR expression, we directly measured the amount of a variety of different forms of cytokinins in 10-day-old wild-type plants that had been subjected to 0, 0.5, 1, 2, or 4 h of cold treatment at 1 °C, at which the maximal peak expression of cold-inducible A-type ARR genes was observed, as shown in Fig. 2. We determined that none of the cytokinins evidenced a meaningful increase, which can account for the cold-inducible expression of ARR genes (Fig. 6 and supplemental Table 2). This result shows that an increase in cytokinin levels is not the cause of the expression of A-type ARR genes as an early response.
FIGURE 6.
Effects of cold on cytokinin content in the wild-type Arabidopsis plants. A, content of various forms of trans-zeatin. One gram of 10-day-old light-grown Arabidopsis Col-0 seedlings (10 days after germination) per sample was pooled, and three independent biological samples were obtained for each treatment of cold temperature at 1 °C for a given period of time. The data shown are the means (pmol/g fresh weight) ± S.D. * denotes statistically significant changes with p < 0.05 compared with the wild type. tZ, trans-zeatin; tZR, trans-zeatin riboside; tZOG, trans-zeatin O-glucoside; tZROG, trans-zeatin riboside O-glucoside; tZ9G, trans-zeatin 9-glucoside; tZR5′MP, trans-zeatin riboside 5′-monophosphate; cZ, cis-zeatin; cZR, cis-zeatin riboside; cZOG, cis-zeatin O-glucoside; cZROG, cis-zeatin riboside O-glucoside; cZ9G, cis-zeatin 9-glucoside; cZR5′MP, cis-zeatin riboside 5′-monophosphate; DHZ, dihydrozeatin; DHZR, dihydrozeatin riboside; DHZOG, dihydrozeatin O-glucoside; DHZROG, dihydrozeatin riboside O-glucoside; DHZ9G, dihydrozeatin 9-glucoside; DHZR5′MP, dihydrozeatin riboside 5′-monophosphate; iP, N6-(Δ2isopentenyl)adenine; iPR, N6-(Δ2isopentenyl) adenosine; iP9G, N6-(Δ2isopentenyl)adenine 9-glucoside; iPRMP, N6-(Δ2isopentenyl)adenosine 5′-monophosphate. B, content of various forms of cis-zeatin. The legend is the same as in A. C, content of various forms of dihydrozeatin. The legend is the same as in A. D, content of various forms of N6-(Δ2isopentenyl)adenine. The legend is the same as in A.
ARR7 Overexpression Results in Hypersensitive Response in Transgenic Arabidopsis to Freezing Temperatures after Cold Acclimation
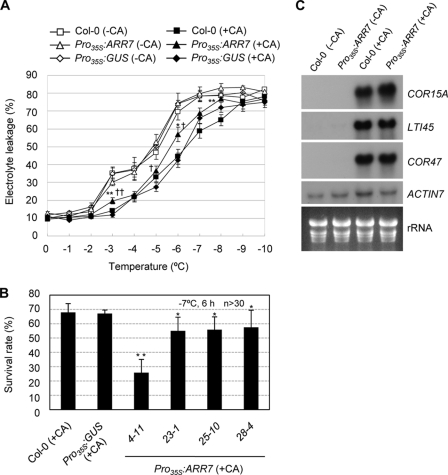
To determine whether cold-inducible A-type ARRs might perform a function in stress tolerance response, we conducted electrolyte leakage assays for transgenic Arabidopsis overexpressing ARR7 (Pro35S:ARR7) (29). Under normal conditions, Pro35S:ARR7 (line 4-11) and both control plants, Pro35S:GUS and the wild type (Col-0), showed similar ion leakage patterns (Fig. 7A). After cold acclimation, Pro35S:ARR7 exhibited hypersensitive ion leakage patterns to freezing temperatures compared with two control plants, the wild type at −3, −5, −6, and −8 °C and Pro35S:GUS at −3, −5, and −7 °C, to some extent, indicating the negative regulator activity of ARR7 against freezing stress. In planta freezing tests using additional three Pro35S:ARR7 lines yielded similar results, albeit with weaker effects than were observed in line 4-11 (Fig. 7B). The weaker effects might be attributable to the lower ARR7 activity of these transgenic lines compared with line 4-11 with regard to the repression of its target genes, as reported previously (29). Cold treatment for 2 days efficiently induced a variety of cold-regulated genes in the CBF/DREB pathway of both Pro35S:ARR7 and control plants (Fig. 7C), thereby suggesting that ARR7 may function as a negative regulator of cold signaling, independent of CBF/DREB cold signaling.
FIGURE 7.
Freezing tolerance assays and RNA-gel blot analysis of Pro35S:ARR7, Pro35S:GUS, and the wild-type plants with or without cold acclimation. A, electrolyte leakage of Pro35S:ARR7 and the wild-type plants with or without cold acclimation. Electrolyte leakage assays were conducted using the leaves from 2-week-old wild-type (Col-0) and Pro35S:ARR7 (line 4-11) (29) plants treated with or without cold acclimation (CA). For cold acclimation treatment, the plants were incubated for 3 days at 1 °C under white fluorescent light. The means ± S.E. from 10 experiments were plotted. Statistically significant changes compared with those of Col-0 plants (or Pro35S:GUS plants) are indicated by * (or †) when p < 0.05 or by ** (or ††) when p < 0.01 (Student's t test), respectively. B, freezing tolerance assays of various lines of Pro35S:ARR7 transgenic Arabidopsis, Pro35S:GUS, and the wild-type plants after cold acclimation. Ten-day-old plants were cold-acclimated at 1 °C for 3 days, treated with −7 °C for 6 h, and photographed after 2 days of incubation at 23 °C for recovery. The percentage of the plants that survived was calculated. The means ± S.E. from four independent experiments were plotted. n ≥30. # indicates line number of Pro35S:ARR7 (29). Statistically significant changes compared with the Col-0 plants as well as Pro35S:GUS plants are indicated by * when p < 0.05 or by ** when p < 0.01 (Student's t test), respectively. C, RNA-gel blot analysis of Pro35S:ARR7 and the wild-type plants. Total RNA was isolated from plants treated with or without cold acclimation and was subjected to RNA-gel blot analysis using the indicated probes. Line 4-11 of Pro35S:ARR7 (29) was utilized.
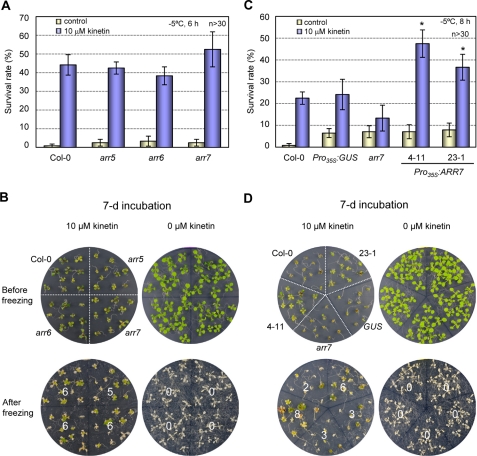
Loss-of-Function Mutations in ARR5, ARR6, or ARR7 Cause Increased Freezing Tolerance
The hypersensitive freezing tolerance of the ARR7-overexpressing lines compelled us to assess whether the loss-of-function mutations in cold-inducible ARR genes could induce enhanced freezing tolerance. Whereas arr5 and arr6 are null mutants (28), arr7 null mutants are not available. We found transposon T-DNA insertion lines of ARR7 where the mRNA levels were reduced by up to more than 20-fold compared with that of the wild type (supplemental Fig. 1). We determined the survival rates of arr5, arr6, or arr7 after 5 h of freezing at −5 °C, and we noted that arr5 and arr7 evidenced statistically significantly increased freezing tolerance, occurring to some extent even prior to cold acclimation (Fig. 8A). When these mutants were cold-acclimated, significantly increased freezing tolerance in arr5, arr6, and arr7 were noted after 7 h of freezing at −8 °C. In particular, arr6 and arr7 exhibited greatly enhanced freezing tolerance at survival rates in excess of 50% as compared with the wild-type plants at 10% (Fig. 8B). These results indicate that cold-inducible ARR genes may function as a negative regulator of cold signaling. In addition, we measured the AHK2 and AHK3 transcript levels by real time RT-PCR in arr5, arr6, arr7, and Pro35S:ARR7 as compared with the wild type, but we could not observe detectable differences in the transcript levels between these mutants and the wild-type plants (supplemental Fig. 8), suggesting that regulation of AHK2 and AHK3 gene expression is not related to alteration in cold tolerance in these mutants.
FIGURE 8.
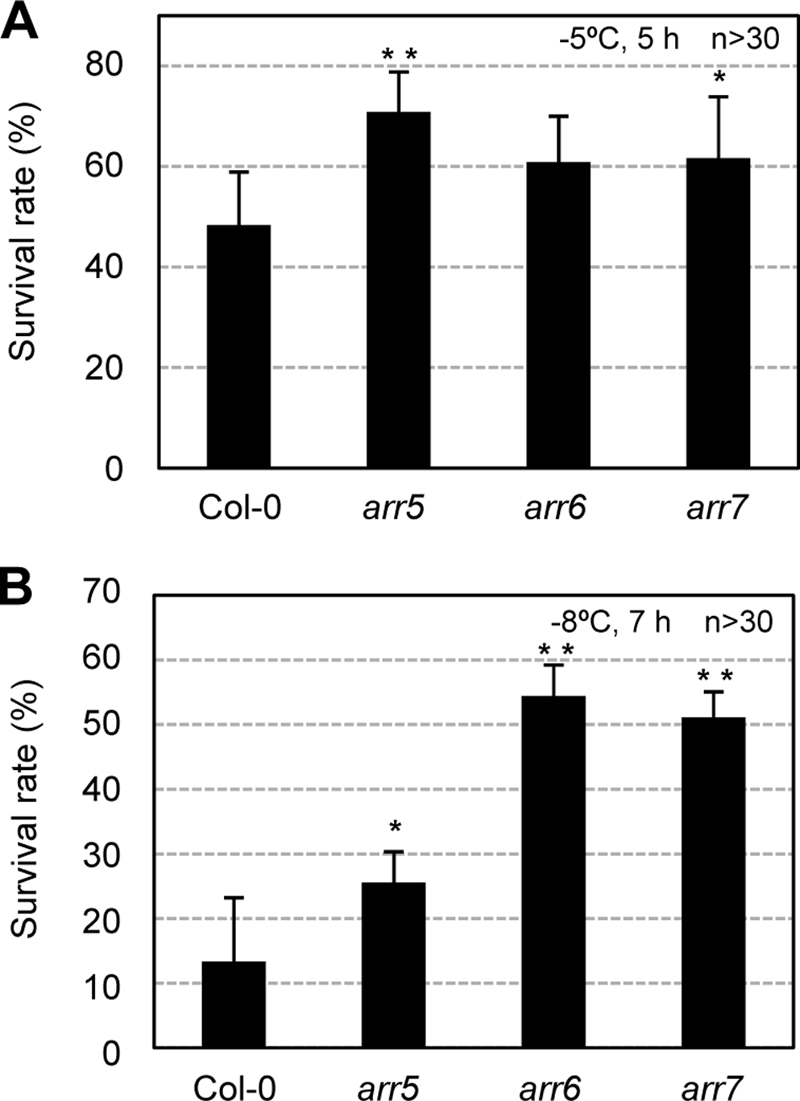
Freezing tolerance assays of A-type arr mutants compared with the wild-type plants with or without cold acclimation. A, freezing tolerance assays of the A-type arr mutants prior to cold acclimation. Freezing assays were performed on the wild-type (Col-0), arr5, arr6, and arr7 mutants, as described in the Fig. 7B legend except for 5 h of freezing treatment at −5 °C. Statistically significant changes compared with the Col-0 plants are indicated by * when p < 0.05 or by ** when p < 0.01 (Student's t test), respectively. B, freezing tolerance assays of the A-type arr mutants after cold acclimation. Freezing assays were performed on the wild-type (Col-0), arr5, arr6, and arr7 mutants after cold acclimation, as described in the Fig. 7B legend except for freezing treatment at −8 °C for 7 h. Statistically significant changes compared with the Col-0 plants are indicated by * when p < 0.05 or by ** when p < 0.01 (Student's t test), respectively.
Freezing Tolerance of ahk Mutants
We then analyzed the freezing tolerance of ahk mutants to determine whether the AHK-cytokinin receptors mediating A-type ARR expression perform a function in cold response. As the single ahk mutants did not evidence any significant phenotypes, we first evaluated the freezing tolerance of the ahk double mutants, ahk2-2 ahk3-3 and ahk3-3 cre1-12, in the Col-0 ecotype backgrounds (9). We tested a variety of freezing temperatures and incubation times. As shown in Fig. 9, A and B, ahk2-2 ahk3-3 and ahk3-3 cre1-12 exhibited significantly higher survival rates against freezing temperatures compared with the wild-type control plants, and ahk2-2 ahk3-3 displayed more profound freezing tolerance than was seen with the ahk3-3 cre1-12 line. A different allele, ahk2-1 ahk3-1 mutant, also showed similarly enhanced freezing tolerance compared with the wild-type plants (data not shown). However, the ahk2-2 cre1-12 plants showed insignificant changes in freezing tolerance compared with the wild type (data not shown). These results show that the AHK-cytokinin receptors may function partially redundantly as a negative regulator of the cold stress adaptation response.
FIGURE 9.
Freezing tolerance assays of ahk double mutants compared with the wild-type plants with or without cold acclimation. A, ahk2-2 ahk3-3 and ahk3-3 cre1-12 mutants survived after 4 h of freezing at −4 °C. Fourteen-day-old light-grown plants were treated for 4 h at −4 °C and photographed after 2 days of incubation at 23 °C for recovery. The percentage of the plants that survived was calculated. Experiments were conducted in triplicate, and the means ± S.E. were plotted. n ≥30. All mutants are derived from the Col-0 ecotype. Statistically significant changes compared with Col-0 plants were indicated by * when p < 0.05 or by ** when p < 0.01 (Student's t test), respectively. † denotes statistically significant changes with p < 0.05 among the ahk double mutants indicated. B, example plates showing plants subjected to freezing tolerance assays (A). One plate (inner diameter 150 × 20 mm) contains 10 plants per each control or mutant. C, treatment of the ahk2-2 ahk3-3 and ahk3-3 cre1-12 mutants cold-acclimated with freezing at −7 °C for 6 h. Fourteen-day-old light-grown seedlings were cold-acclimated for 3 days, treated at −7 °C for 6 h, and the plants that survived were counted after 2 days of incubation at 23 °C for recovery. The percentage of the plants that survived was calculated. Experiments were conducted in triplicate, and the means ± S.E. were plotted. n ≥30. Statistically significant changes with p < 0.01 (Student's t test) compared with the Col-0 plants are indicated by **. †† denotes statistically significant changes with p < 0.01 among ahk double mutants indicated. D, example plates showing plants subjected to freezing tolerance assays (C). One plate (inner diameter 150 × 20 mm) contains 10 plants per control or per mutant.
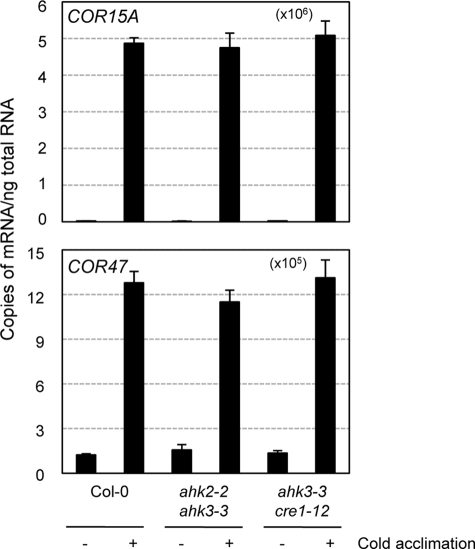
To ascertain whether the constitutive freezing tolerance of ahk mutants might be attributable to acquired cold acclimation, we investigated the ability of ahk mutants to acclimate to cold as compared with the wild-type plants. We incubated the plants for 3 days at 1 °C for cold acclimation and then treated them for 6 h with freezing temperatures of −7 °C. As shown in Fig. 9, C and D, the ahk2-2 ahk3-3 and ahk3-3 cre1-12 double mutants exhibited significantly higher survival rates against freezing temperatures compared with the wild-type control plants after cold acclimation, and the ahk2-2 ahk3-3 lines displayed higher freezing tolerance than that of ahk3-3 cre1-12. The ahk2-1 ahk3-1 double mutants also exhibited similarly enhanced freezing tolerance compared with the wild-type plants (data not shown). We also determined expression levels of COR15A and COR47, the marker genes of CBF/DREB pathway during cold acclimation, and found that the levels of these transcripts before and after cold acclimation were not changed by ahk2 ahk3 and ahk3 cre1 double mutations (Fig. 10). Similar patterns were noted on comparative RT-PCR (data not shown). These results suggest that enhanced freezing tolerance in the ahk2 ahk3 or ahk3 cre1 double mutants is not directly linked to the CBF/DREB pathway.
FIGURE 10.
Expression of COR15A and COR47 in ahk2 ahk3 mutants compared with the wild-type plants with or without cold acclimation. Eleven-day-old light-grown seedlings were treated at 1 °C for 3 days for cold acclimation, and total RNAs isolated were subjected to real time RT-PCR. Copies of the transcripts from plants treated were plotted per ng of total RNA after normalization to ACTIN7 RNA. The transcripts levels were determined as described in the Fig. 2 legend.
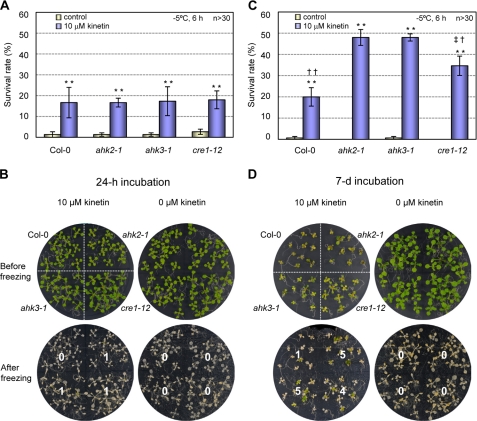
Preincubation of ahk Single Mutants with Cytokinin Caused Enhanced Freezing Tolerance Compared with Untreated Mutants as Well as the Wild Type Treated with Cytokinin
To further demonstrate that the AHK-cytokinin receptors act as a negative regulator of cold stress adaptation response, we tested the effects of cytokinin preincubation on freezing tolerance of ahk single mutants that did not show significant freezing tolerance compared with the wild type. We incubated 3-day-old ahk single mutants, ahk2-1, ahk3-1, and cre1-12, and wild-type plants in the presence of the cytokinin, kinetin, for an additional 24 h or 7 days and then conducted freezing tolerance assays. When incubated for 24 h, ahk single mutants and the wild-type plants exhibited enhanced freezing tolerance to some extent upon preincubation with cytokinin (Fig. 11, A and B). However, a longer treatment of cytokinins, for 7 days compared with 24 h, induced a profound enhancement of freezing tolerance in the ahk single mutants, particularly, ahk2 and ahk3, as compared with the wild-type plants (Fig. 11, C and D). These results bolster the notion that the AHK-cytokinin receptors may function as a negative regulator of the cold stress adaptation response and that cytokinins might be involved in adaptation response to freezing temperatures.
FIGURE 11.
Effect of cytokinin preincubation on freezing tolerance of ahk2, ahk3, and cre1 single mutants compared with the wild-type plants. A, ahk single mutants preincubated with or without cytokinin for 24 h. The ahk2-1, ahk3-1, cre1-12 mutants and the wild-type plants (Col-0) were grown for 9 days in the light, transferred to MS plate or MS plate containing 10 μm kinetin, and grown for an additional 24 h. These 10-day-old plants preincubated with or without cytokinin were subjected to freezing treatment at −5 °C for 6 h. The plants that survived after incubation at 23 °C for 2 days for recovery were counted. Experiments were conducted in triplicate, and the means ± S.E. were plotted. n ≥30. Statistically significant change with p < 0.01 (Student's t test) compared with the samples without cytokinin treatment is indicated by **. †† denotes statistically significant changes with p < 0.01 among ahk single mutants indicated. B, example plates showing the ahk2-1, ahk3-1, cre1-12 mutants, and the wild-type plants preincubated with or without cytokinin for 24 h, subjected to freezing treatment (A). C, ahk single mutants preincubated with or without cytokinin for 7 days. Plants were treated and analyzed as described in A except for 3 days growth followed by 7 days of incubation with cytokinin. D, example plates showing the ahk2-1, ahk3-1, cre1-12 mutants and the wild type plants preincubated with or without cytokinin for 7 days and then subjected to freezing treatment. Plant growth and cytokinin preincubation were conducted as described in C.
Pro35S:ARR7 Exhibited Enhanced Freezing Tolerance Compared with arr7 Mutants upon Cytokinin Preincubation
We next tested whether preincubation with cytokinin impacts the responsiveness of the arr5, arr6, and arr7 mutants and Pro35S:ARR7 lines to cold, as compared with the wild type. Preincubation of arr mutants with cytokinin for 7 days resulted in increased freezing tolerance as much as that of the wild type (Fig. 12, A and B). Interestingly, Pro35S:ARR7 plants exhibited stronger freezing tolerance compared with the arr7 mutants and two other control plants (Col-0 and Pro35S:GUS) upon cytokinin preincubation (Fig. 12, C and D). As overexpression of ARR7 in Arabidopsis reduces response to cytokinin (29), this result might indicate that proper levels of cytokinin signaling might be important for cold tolerance.
FIGURE 12.
Effect of cytokinin preincubation on freezing tolerance of arr5, arr6, arr7 single mutants and Pro35S:ARR7 lines compared with the wild-type plants. A, arr5, arr6, and arr7 mutants preincubated with or without cytokinin for 7 days. Experiments were conducted and analyzed as described in Fig. 11, C and D. B, example plates showing the arr5, arr6, arr7 mutants, and the wild-type (Col-0) plants preincubated with or without cytokinin for 7 days, subjected to freezing treatment (A). C, Pro35S:ARR7 lines compared with the arr7 mutants and Col-0 and Pro35S:GUS control plants preincubated with or without cytokinin for 7 days. Experiments were conducted and analyzed as described in A except that the plants were treated at −5 °C for 8 h for freezing. Numbers 4-11 and 23-1 indicate the line number of Pro35S:ARR7 transgenic plants. * denotes statistically significant change with p < 0.05 compared with arr7 and control plants pretreated with cytokinin. D, example plates showing Pro35S:ARR7 lines, the arr7 mutants, Col-0, and Pro35S:GUS control plants preincubated with or without cytokinin for 7 days and then subjected to freezing treatment (C).
Response of Root Growth and Germination of arr5, arr6, arr7, Pro35S:ARR7, and ahk2 ahk3 to ABA
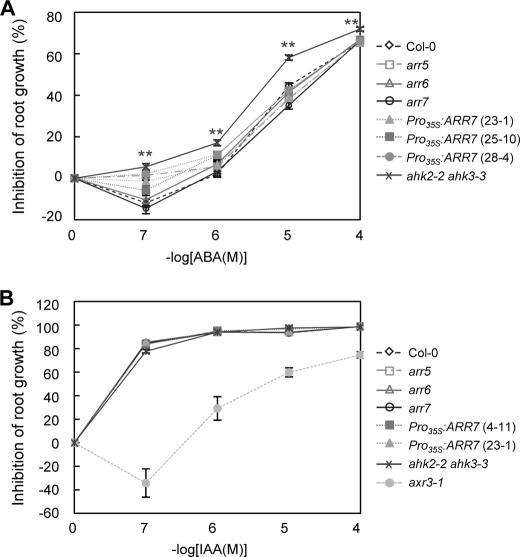
It has been reported previously that ABA-regulated genes were up-regulated in the ahk2-2 ahk3-3 mutants (14). The activation of ABA-regulated genes and observed enhancements of freezing tolerance by ahk mutations might be due to increased sensitivity in the ABA response. To test this possibility, we measured root growth in the presence of exogenous ABA compared with auxin (Fig. 13). The ahk2-2 ahk3-3 mutants were significantly sensitive to ABA compared with the wild type at the ABA concentrations tested (Student's t test, p < 0.01) (Fig. 13A), whereas this mutant showed a normal response to auxin, similar to the wild type (Fig. 13B). We also measured root growth inhibition of arr mutants and Pro35S:ARR7 lines but observed statistically insignificant difference or very marginal difference compared with the wild type (Fig. 13A).
FIGURE 13.
Root growth of arr5, arr6, arr7 mutants, Pro35S:ARR7 lines, and ahk2 ahk3 double mutants compared with the wild-type plants in the presence of exogenous ABA or auxin. A, analysis of primary root growth of arr5, arr6, arr7, Pro35S:ARR7, and ahk2 ahk3 in the presence of exogenous ABA. The seedlings, the wild type (Col-0), arr5, arr6, arr7, three different Pro35S:ARR7 transgenic lines, and ahk2-2 ahk3-3 double mutants, were grown on MS agar plates for 4 days, transferred to the medium containing increasing concentrations of ABA, and grown vertically for additional 3 days. The root lengths were measured, and the mean values were plotted. n ≥26. Bars indicate S.E. Statistically significant change with p < 0.01 (Student's t test) compared with the wild type are indicated by **. B, arr5, arr6, arr7, Pro35S:ARR7, and ahk2 ahk3 in the presence of auxin. Measurements of primary root-growth in the presence of auxin were performed as described in A. n ≥26.
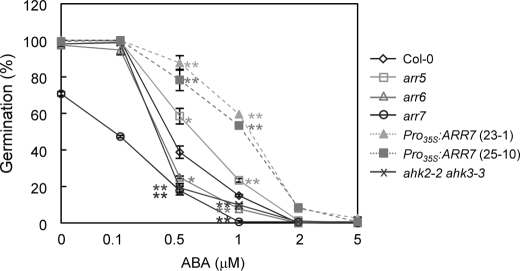
To further explore the ABA response of arr mutants, Pro35S:ARR7 lines, and ahk2 ahk3 double mutants, we measured germination of these seeds with varying concentrations of ABA (Fig. 14). Consistent with root growth inhibition results, the ahk2-2 ahk3-3 double mutants showed a sensitive response to ABA compared with the wild type. Strikingly, the arr7 mutants exhibited the highest sensitive response to ABA among the seeds tested, whereas Pro35S:ARR7 lines showed the highest insensitive response to ABA. arr6 also showed sensitive response to ABA compared with the wild type, whereas arr5 displayed insensitive response to ABA to some extent. This result further demonstrates that there is a cross-talk between cytokinin and ABA signaling. Hypersensitive response of the arr7 mutants and insensitive response of ARR7 overexpression lines to ABA suggest that ARR7 might play a role as a negative regulator in ABA signaling.
FIGURE 14.
Germination of arr5, arr6, arr7 mutants, Pro35S:ARR7 lines, ahk2 ahk3 double mutants, and the wild-type plants in the presence of exogenous ABA. Seeds of the wild type (Col-0), arr5, arr6, arr7, two different Pro35S:ARR7 transgenic lines, and ahk2-2 ahk3-3 were plated on MS medium supplemented with the indicated concentrations of ABA, chilled for 4 days at 4 °C in darkness, and incubated for 2 days at 21 °C with a 16-h light photoperiod. The number of germinated seeds was expressed as the percentage of the total number of seeds plated (63). Statistically significant change with p < 0.05 (Student's t test) or p < 0.01 compared with the wild type is indicated by * or **.
DISCUSSION
Cold-inducible A-type ARRs Function as a Negative Regulator of Cold Signaling
We noted the transient expression of a subset of A-type ARR genes, including ARR5, ARR6, ARR7, and ARR15, in response to cold temperatures. The ARR7 promoter-GUS transgenic Arabidopsis expressed GUS in response to cold, as well as to cytokinins. These results are indicative of a potential function of A-type ARRs in cold signaling. We observed the hypersensitivity response of Pro35S:ARR7 plants with decreasing freezing temperatures to some extent upon cold acclimation, as compared with the control plants (Fig. 7). Various CBF/DREB pathway genes were shown to be equally inducible by cold in both the wild-type and Pro35S:ARR7 plants (Fig. 7). These results indicated that cold-inducible ARR7 may play a role as a negative feedback regulator of cold stress signaling in the CBF/DREB-independent pathway. If a cold-inducible ARR gene does function as a negative regulator, then loss-of-function mutation in a cold-inducible ARR might induce an increase in freezing tolerance compared with the wild-type plants. Increased freezing tolerance has been noted in arr5 and arr7, although the effects were weak, prior to cold acclimation and in arr5, arr6, and arr7 after cold acclimation (Fig. 8). In particular, arr6 and arr7, after cold acclimation, showed relatively stronger freezing tolerance than was observed with the wild-type and arr5. This result further supports the proposition that the cold-inducible ARR genes might function as a negative regulator of cold signaling.
AHK-Cytokinin Receptors Function as a Negative Regulator of Cold Signaling
The ahk2 ahk3 and ahk3 cre1 mutants exhibited tolerance to freezing temperatures compared with the wild-type plants prior to or after cold acclimation (Fig. 9). This result indicates that the AHK-cytokinin receptors might function as a negative regulator of the cold stress adaptation response, which is consistent with the negative regulator function of the cold-inducible A-type ARRs. The ahk2 ahk3 mutants were shown to be more tolerant to freezing temperatures than the ahk3 cre1 mutants. However, the difference between the freezing tolerance of the ahk2 cre1 mutants and that of the wild type was not significant (data not shown). The reason that the freezing tolerance of the ahk2 ahk3 mutants was higher than that of the ahk3 cre1 or ahk2 cre1 plants remains to be elucidated, but it may be associated with the prominent functions of AHK2 and AHK3 and, more critically, AHK3 in the leaf and AHK4 in the roots (9–11). Whereas the ahk3 ahk4 or ahk2 ahk4 double mutants and ahk single mutants grew normally, the ahk2 ahk3 mutants exhibited symptoms of semi-dwarfism, including short petioles and smaller leaf blades (9, 10). The ahk4 single mutants exhibited significant resistance to cytokinin in the root elongation assays compared with the wild type, but the ahk2 or ahk3 or ahk2 ahk3 mutants did not. Consistent with this, AHK4 was expressed predominantly in the roots (9, 10). These results imply that AHK2 and AHK3 may be critical to leaf development and function, whereas AHK4 may function principally in the roots. The leaf may be the primary site used by the plants to cope with cold stress. Thus, the degree of freezing tolerance can be predicted in the following order on the basis of this rationale and the hypothesis that AHK3 might be more important than AHK2 in the cold-stress adaptation response: ahk2 ahk3 > ahk3 ahk4 > ahk2 ahk4. The inhibition of leaf development conferred by ahk2 ahk3 mutations may constitute a more stringent barrier against environmental cold stress over the ground. However, the ahk3 ahk4 double mutants grew normally (9, 10), even though this mutant exhibited enhanced freezing tolerance. The fatb mutants that exhibited significant dwarfism and small leaves (51) also showed a more sensitive phenotype to freezing stress than was observed in the wild-type plants (data not shown). Thus, the freezing tolerance exhibited by the ahk2 ahk3 double mutants appears not to be due solely to the reduction in leaf size.
To assess the connection between freezing tolerance and negative regulation of cytokinin signal transduction, we have investigated the effects of cytokinins on the freezing tolerance of ahk single mutants of the undetectable freezing tolerance phenotype, via the preincubation of the mutants with cytokinin prior to freezing. We determined that preincubation with cytokinin resulted in detectable levels of enhanced freezing tolerance in the ahk2, ahk3, or cre1 mutants as well as the wild type even at 24 h. However, longer treatment of cytokinin such as for 7 days caused significantly enhanced freezing tolerance in ahk single mutants as well as the wild type compared with untreated plants and also induced a greater increase in freezing tolerance in the ahk single mutants than in the wild-type plants (Fig. 11). This result shows that exogenous cytokinins and cytokinin signal transduction both play a role in the acquisition of freezing tolerance in ahk mutants. Whereas the relevant molecular mechanisms remain to be clearly elucidated, exogenous cytokinins applied to the ahk single mutants might strengthen the negative feedback inhibition of cytokinin signal transduction, thereby resulting in enhanced freezing tolerance. This situation may, in part, resemble higher order mutations in AHKs that induced significantly increased freezing tolerance, even in the absence of exogenous cytokinin treatment. Similar observations were also previously noted in cre1 mutants, which exhibited a salt-tolerant phenotype in the presence of exogenous cytokinins (14).
We further investigated the impact of cytokinin preincubation on the response of arr mutants to cold and observed similarly increased freezing tolerance in arr5, arr6, and arr7 as well as in the wild type (Fig. 12). However, significantly enhanced freezing tolerance in ARR7 overexpression lines was found as compared with the arr7 mutants upon cytokinin preincubation. ARR7 overexpression caused insensitive cytokinin response similar to other A-type ARR genes (29). ARR7 overexpression in Pro35S:ARR7 might have then reduced the effects of cytokinin exogenously applied by cytokinin preincubation. This inhibition effect might have been beneficial for Pro35S:ARR7 plants compared with the plants such as the arr7 mutants and the wild type exogenously added with cytokinin showing slight growth inhibition. At any rate, it is clear that certain levels of cytokinin and thus cytokinin signal transduction render the plants to be tolerant to cold.
Acquired cold acclimation might be one mechanism to explain the enhancement of freezing tolerance in ahk2 ahk3 or ahk3 ahk4 double mutants. However, our results showed that these mutants have the ability to acclimate to cold like wild-type plants and that the marker genes for cold acclimation such as COR15A and COR47 in CBF/DREB pathway are equally inducible in these mutants and the wild-type plants (Fig. 10). It is therefore unlikely that cold acclimation process is primarily involved in cold-stress response mediated by the AHK-cytokinin receptors. Another explanation is that endogenously increased levels of cytokinins in the ahk mutants might be responsible for the differing levels of freezing tolerance observed among ahk mutants. However, no linear correlation between freezing tolerance and endogenous cytokinin levels was detected in the ahk mutants (11). For example, ahk3, ahk2 ahk3, and ahk3 cre1 harbor similar cytokinin levels, although the ahk single mutants displayed wild-type like freezing tolerance and ahk2 ahk3 exhibited stronger freezing tolerance than ahk3 cre1. Thus, endogenously increased cytokinin levels due to single or double mutations in AHKs might not be responsible for enhancing freezing tolerance in these ahk mutants, as compared with exogenously added cytokinins.
Alteration of expression of biotic and abiotic stress-related and some auxin-induced genes by overexpression of ARR2D80E compared with ARR2 overexpressor or by arr2 mutation has been reported (52). Activation of ABA up-regulated genes has been observed in the ahk2 ahk3 double mutants (14). Thus enhanced ABA response might be one explanation for increased freezing tolerance in the ahk2 ahk3 mutants. This prediction is consistent with hypersensitive ABA response of the ahk2 ahk3 mutants in both root growth inhibition and germination inhibition (Fig. 13A and Fig. 14). Germination assays have previously shown that the cytokinin receptor single mutants are hypersensitive to ABA (14). It has been proposed that the increased cytokinin content might antagonistically regulate ABA synthesis, inhibit ABA action in germinating seeds through cytokinin receptors, and consequently alleviate the inhibition effect of ABA on seed germination (14). These results indicate that the AHK-cytokinin receptors might be involved in regulating interaction between cytokinin and ABA signaling. Moreover, a parallel phenotype of arr7 and Pro35S:ARR7 in germination assays (Fig. 14) has indicated that ARR7 might play a role as a negative regulator in ABA signaling. These results suggest that cross-talk might exist between cytokinin and ABA signaling in an antagonistic fashion for regulating cold stress response.
AHK-Cytokinin Receptors Mediate Cold Signals for Inducing A-type ARR Expression
Severe inhibition of cold-responsive A-type ARR expression in the ahk2 ahk3 mutants compared with the wild-type plants (Fig. 3) may be associated with the in vivo functioning of the AHK-cytokinin receptors in plants. Severely reduced expression of A-type ARRs in response to cold was noted in the ahk2 ahk3 mutants, but their normal cold response in the ahk single mutants suggests that AHK2 and AHK3 might have redundant functions and constitute the principal mediators of cold-responsive A-type ARR expression. The normal cold-responsive expression of ARR genes was noted in the ahk3 ahk4 and ahk2 ahk4 mutants (Fig. 3B). These results suggest that AHK2 may perform redundant functions with AHK3 in cold perception as well as in cytokinin reception, whereas AHK4 may perform a specific function in cytokinin reception. Thus, loss-of-function of AHK2 or AHK3 may not be replaced with AHK4 in cold perception, thereby resulting in severe inhibition of cold-responsive ARR expression than that of ahk3 ahk4 or ahk2 ahk4, which displays near wild-type levels of cold-responsive ARR expression. Although ahk3 ahk4 showed the normal cold-responsive expression of ARR genes (Fig. 3B), ahk3 ahk4 exhibited detectable freezing tolerance compared with the wild type, albeit weaker than ahk2 ahk3 (Fig. 9). These results indicate that in addition to the negative regulatory function mediated by the cold-inducible A-type ARRs, other signaling networks might be superimposed onto the cold response regulated downstream of AHKs.
The analysis of transgenic plants overproducing cytokinins in a DEX-inducible manner (Fig. 5) demonstrated that increased cytokinin levels exerted additive effects on the expression of ARR5, ARR6, and ARR7 in response to cold. The maximum induction levels achieved by treatments with both cold and DEX were identical to those treated solely with cold. These results show that cold and cytokinins might share the same AHK-cytokinin receptors for the regulation of ARR5, ARR6, and ARR7 expression. Additional AHK proteins, such as AHK1 and AHK5, may be involved in mediating cold to express A-type ARRs. Nevertheless, it is clear that AHK2 and AHK3 are the primary receptors for mediating the cold-inducible expression of A-type ARR genes.
Cold Perception of the AHK Proteins as a Primary Response Does Not Involve Changes in Cytokinin Levels for the Expression of A-type ARRs
The mechanisms by which the AHK-cytokinin receptors mediate cold temperatures for ARR expression remain to be clearly elucidated. Lack of significant cold-responsive expression of AHK2, AHK3, and AHK4 (supplemental Fig. 5) suggests that the AHK-cytokinin receptor proteins might be involved in mediating cold temperatures for the expression of cold-responsive A-type ARR genes. Cytokinins themselves may be involved in cold-inducible A-type ARR expression. However, on the basis of our observations described below, we think that increased cytokinin levels might not be involved in the cold perception process during early response of A-type ARR gene expression. ARR5, ARR6, ARR7, and ARR15 have characteristics of primary response genes in response to cold. Unfortunately, the immediate response of A-type ARR genes to cold without protein synthesis cannot be verified experimentally, because of the significant up-regulation of ARR7 resulting from treatment with cycloheximide (48). Enzymes relevant to cytokinin biosynthesis could be cold-activated and may be involved in the cold perception process. However, if cold induces A-type ARR expression via cytokinins, the same set of ARR inductions in response to cold and cytokinins would be anticipated, which was obviously not the case. Moreover, our results demonstrated that cytokinin-deficient transgenic Arabidopsis plants exhibit a cold-responsive expression of A-type ARR genes, similar to that observed with the wild-type plants (Fig. 4). Cytokinins and cold exerted additive effects on A-type ARR gene expression (Fig. 5). Finally, direct measurements of the amount of a variety of different forms of cytokinins demonstrated that cold did not cause a significant increase in cytokinin levels within the time period (∼4 h) during which a peak expression of ARR genes to cold temperatures has been observed (Figs. 2 and 6), thereby demonstrating that an increase in cytokinin levels might not be involved in the process of the cold perception of the AHK proteins for A-type ARR expression as an early response.
A potential sensor for cold perception in cyanobacteria, Synechosystis sp. PCC6803, has been proposed previously (53). Cyanobacteria modulate the composition of membrane lipids in response to shifts in temperature from 34 to 22 °C to increase the fluidity of their membranes for adaptation to cold stress by enhancing the expression of three fatty-acid-desaturase genes (des), desA, desB, and desD (54). Reductions in membrane fluidity via the catalytic hydrogenation of fatty acids in the plasma membranes of Synechosystis resulted in the induction of desA gene transcription (55), thus suggesting that the increased expression of the desaturase genes in response to cold stress might be regulated by the degree of membrane rigidity. Murata and co-workers later identified histidine kinase (Hik), Hik33 as a sensor for cold perception (56). Interestingly, Hik33 has been shown to regulate the expression of osmotic stress-inducible genes and also to bind to certain chemicals, indicating that it may function as a multifunctional sensor for a variety of stresses. Several studies have previously shown that changes in membrane fluidity appear to be a potential mechanism for the sensing of cold temperatures in higher plants as well (57–60). As proposed for bacterial Hik33 (61), it can be speculated that the plant AHK proteins might utilize a similar mechanism for the detection of cold temperatures. However, further biochemical studies will be necessary to clearly elucidate the mechanisms by which the AHK proteins contribute to the process of cold temperature recognition in higher plants.
Supplementary Material
Acknowledgments
We thank Drs. Ueguchi and Kakimoto for providing us with ahk mutant seeds, Dr. Schmülling for the 35S:AtCKX2-2 seeds, and Dr. Moore for the LhGR-N and pV-ipt/LhGR-N seeds. We also thank Dr. Myung Duk Kim for help with the Northern blot analysis shown in Fig. 7.
This work was supported by Agricultural Plant Stress Research Center Grant R11-2001-092-04001-0, by Plant Diversity Research Center of 21st Century Frontier Research Program Grant PF06302-01, and from the World Class University Project R31-2009-000-20025-0 funded by the Ministry of Education, Science and Technology of Korea (to J. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–8.
M.-O. Oh and J. Kim, unpublished observations.
- TCS
- two-component signaling system
- ABA
- abscisic acid
- CBF/DREB
- C-repeat-binding factor/dehydration-responsive element-binding factor
- DEX
- dexamethasone
- BA
- benzyladenine
- GUS
- β-glucuronidase
- Mes
- 4-morpholineethanesulfonic acid
- RT
- reverse transcription
- MS
- Murashige-Skoog
- ARR
- Arabidopsis response regulator
- AHK
- Arabidopsis histidine kinase.
REFERENCES
- 1.Davies P. J. (ed) (2004) Plant Hormones: Biosynthesis, Signal Transduction, Action! pp. 7–8, Kluwer Academic Publishers Group, Dordrecht, Netherlands [Google Scholar]
- 2.To J. P., Kieber J. J. (2008) Trends Plant Sci. 13, 85–92 [DOI] [PubMed] [Google Scholar]
- 3.Pischke M. S., Jones L. G., Otsuga D., Fernandez D. E., Drews G. N., Sussman M. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. (2001) Nature 409, 1060–1063 [DOI] [PubMed] [Google Scholar]
- 5.Suzuki T., Miwa K., Ishikawa K., Yamada H., Aiba H., Mizuno T. (2001) Plant Cell Physiol. 42, 107–113 [DOI] [PubMed] [Google Scholar]
- 6.Ueguchi C., Koizumi H., Suzuki T., Mizuno T. (2001) Plant Cell Physiol. 42, 231–235 [DOI] [PubMed] [Google Scholar]
- 7.Ueguchi C., Sato S., Kato T., Tabata S. (2001) Plant Cell Physiol. 42, 751–755 [DOI] [PubMed] [Google Scholar]
- 8.Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., Yamashino T., Mizuno T. (2001) Plant Cell Physiol. 42, 1017–1023 [DOI] [PubMed] [Google Scholar]
- 9.Higuchi M., Pischke M. S., Mähönen A. P., Miyawaki K., Hashimoto Y., Seki M., Kobayashi M., Shinozaki K., Kato T., Tabata S., Helariutta Y., Sussman M. R., Kakimoto T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. (2004) Plant Cell 16, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riefler M., Novak O., Strnad M., Schmülling T. (2006) Plant Cell 18, 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H. J., Ryu H., Hong S. H., Woo H. R., Lim P. O., Lee I. C., Sheen J., Nam H. G., Hwang I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urao T., Yakubov B., Satoh R., Yamaguchi-Shinozaki K., Seki M., Hirayama T., Shinozaki K. (1999) Plant Cell 11, 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran L. S., Urao T., Qin F., Maruyama K., Kakimoto T., Shinozaki K., Yamaguchi-Shinozaki K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwama A., Yamashino T., Tanaka Y., Sakakibara H., Kakimoto T., Sato S., Kato T., Tabata S., Nagatani A., Mizuno T. (2007) Plant Cell Physiol. 48, 375–380 [DOI] [PubMed] [Google Scholar]
- 16.Desikan R., Horák J., Chaban C., Mira-Rodado V., Witthöft J., Elgass K., Grefen C., Cheung M. K., Meixner A. J., Hooley R., Neill S. J., Hancock J. T., Harter K. (2008) PLoS One 3, e2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hejátko J., Ryu H., Kim G. T., Dobesová R., Choi S., Choi S. M., Soucek P., Horák J., Pekárová B., Palme K., Brzobohaty B., Hwang I. (2009) Plant Cell 21, 2008–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang I., Chen H. C., Sheen J. (2002) Plant Physiol. 129, 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchison C. E., Li J., Argueso C., Gonzalez M., Lee E., Lewis M. W., Maxwell B. B., Perdue T. D., Schaller G. E., Alonso J. M., Ecker J. R., Kieber J. J. (2006) Plant Cell 18, 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mähönen A. P., Bishopp A., Higuchi M., Nieminen K. M., Kinoshita K., Törmäkangas K., Ikeda Y., Oka A., Kakimoto T., Helariutta Y. (2006) Science 311, 94–98 [DOI] [PubMed] [Google Scholar]
- 21.Lohrmann J., Harter K. (2002) Plant Physiol. 128, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai H., Aoyama T., Oka A. (2000) Plant J. 24, 703–711 [DOI] [PubMed] [Google Scholar]
- 23.Hwang I., Sheen J. (2001) Nature 413, 383–389 [DOI] [PubMed] [Google Scholar]
- 24.Argyros R. D., Mathews D. E., Chiang Y. H., Palmer C. M., Thibault D. M., Etheridge N., Argyros D. A., Mason M. G., Kieber J. J., Schaller G. E. (2008) Plant Cell 20, 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason M. G., Mathews D. E., Argyros D. A., Maxwell B. B., Kieber J. J., Alonso J. M., Ecker J. R., Schaller G. E. (2005) Plant Cell 17, 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiba T., Yamada H., Sato S., Kato T., Tabata S., Yamashino T., Mizuno T. (2003) Plant Cell Physiol. 44, 868–874 [DOI] [PubMed] [Google Scholar]
- 27.Osakabe Y., Miyata S., Urao T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2002) Biochem. Biophys. Res. Commun. 293, 806–815 [DOI] [PubMed] [Google Scholar]
- 28.To J. P., Haberer G., Ferreira F. J., Deruère J., Mason M. G., Schaller G. E., Alonso J. M., Ecker J. R., Kieber J. J. (2004) Plant Cell 16, 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee D. J., Park J. Y., Ku S. J., Ha Y. M., Kim S., Kim M. D., Oh M. H., Kim J. (2007) Mol. Genet. Genomics 277, 115–137 [DOI] [PubMed] [Google Scholar]
- 30.Lee D. J., Kim S., Ha Y. M., Kim J. (2008) Planta 227, 577–587 [DOI] [PubMed] [Google Scholar]
- 31.Wohlbach D. J., Quirino B. F., Sussman M. R. (2008) Plant Cell 20, 1101–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinozaki K., Yamaguchi-Shinozaki K., Seki M. (2003) Curr. Opin. Plant Biol. 6, 410–417 [DOI] [PubMed] [Google Scholar]
- 33.Thomashow M. F. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599 [DOI] [PubMed] [Google Scholar]
- 34.Chinnusamy V., Schumaker K., Zhu J. K. (2004) J. Exp. Bot. 55, 225–236 [DOI] [PubMed] [Google Scholar]
- 35.Chinnusamy V., Zhu J., Zhu J. K. (2007) Trends Plant Sci. 12, 444–451 [DOI] [PubMed] [Google Scholar]
- 36.Kim J. (2007) J. Plant Biol. 50, 139–147 [Google Scholar]
- 37.Bechtold N., Ellis J., Pelletier G. (1993) C. R. Acad. Sci. 316, 1194–1199 [Google Scholar]
- 38.Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003) Plant Cell 15, 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craft J., Samalova M., Baroux C., Townley H., Martinez A., Jepson I., Tsiantis M., Moore I. (2005) Plant J. 41, 899–918 [DOI] [PubMed] [Google Scholar]
- 40.Kim H. J., Kim Y. K., Park J. Y., Kim J. (2002) Plant J. 29, 693–704 [DOI] [PubMed] [Google Scholar]
- 41.Ishitani M., Xiong L., Lee H., Stevenson B., Zhu J. K. (1998) Plant Cell 10, 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jefferson R. A., Wilson K. J. (1991) Plant Mol. Biol. Manual B 14, 1–33 [Google Scholar]
- 43.Faiss M., Zalubìlová J., Strnad M., Schmülling T. (1997) Plant J. 12, 401–415 [DOI] [PubMed] [Google Scholar]
- 44.Novák O., Hauserová E., Amakorová P., Dolezal K., Strnad M. (2008) Phytochemistry 69, 2214–2224 [DOI] [PubMed] [Google Scholar]
- 45.Novák O., Tarkowski P., Lenobel R., Dolezal K., Strnad M. (2003) Anal. Chim. Acta 480, 207–218 [Google Scholar]
- 46.Abel S., Theologis A. (1996) Plant Physiol. 111, 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herschman H. R. (1991) Annu. Rev. Biochem. 60, 281–319 [DOI] [PubMed] [Google Scholar]
- 48.D'Agostino I. B., Deruère J., Kieber J. J. (2000) Plant Physiol. 124, 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007) PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fowler S., Thomashow M. F. (2002) Plant Cell 14, 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonaventure G., Salas J. J., Pollard M. R., Ohlrogge J. B. (2003) Plant Cell 15, 1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hass C., Lohrmann J., Albrecht V., Sweere U., Hummel F., Yoo S. D., Hwang I., Zhu T., Schäfer E., Kudla J., Harter K. (2004) EMBO J. 23, 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikami K., Murata N. (2003) Prog. Lipid Res. 42, 527–543 [DOI] [PubMed] [Google Scholar]
- 54.Los D. A., Ray M. K., Murata N. (1997) Mol. Microbiol. 25, 1167–1175 [DOI] [PubMed] [Google Scholar]
- 55.Vigh L., Los D. A., Horváth I., Murata N. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9090–9094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki I., Los D. A., Kanesaki Y., Mikami K., Murata N. (2000) EMBO J. 19, 1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orvar B. L., Sangwan V., Omann F., Dhindsa R. S. (2000) Plant J. 23, 785–794 [DOI] [PubMed] [Google Scholar]
- 58.Sangwan V., Foulds I., Singh J., Dhindsa R. S. (2001) Plant J. 27, 1–12 [DOI] [PubMed] [Google Scholar]
- 59.Sharma P., Sharma N., Deswal R. (2005) BioEssays 27, 1048–1059 [DOI] [PubMed] [Google Scholar]
- 60.Vaultier M. N., Cantrel C., Vergnolle C., Justin A. M., Demandre C., Benhassaine-Kesri G., Ciçek D., Zachowski A., Ruelland E. (2006) FEBS Lett. 580, 4218–4223 [DOI] [PubMed] [Google Scholar]
- 61.Murata N., Los D. A. (1997) Plant Physiol. 115, 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaguchi-Shinozaki K., Shinozaki K. (1994) Plant Cell 6, 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gosti F., Beaudoin N., Serizet C., Webb A. A., Vartanian N., Giraudat J. (1999) Plant Cell 11, 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.