Abstract
The pathogen Bacillus anthracis uses the Sortase A (SrtA) enzyme to anchor proteins to its cell wall envelope during vegetative growth. To gain insight into the mechanism of protein attachment to the cell wall in B. anthracis we investigated the structure, backbone dynamics, and function of SrtA. The NMR structure of SrtA has been determined with a backbone coordinate precision of 0.40 ± 0.07 Å. SrtA possesses several novel features not previously observed in sortase enzymes including the presence of a structurally ordered amino terminus positioned within the active site and in contact with catalytically essential histidine residue (His126). We propose that this appendage, in combination with a unique flexible active site loop, mediates the recognition of lipid II, the second substrate to which proteins are attached during the anchoring reaction. pKa measurements indicate that His126 is uncharged at physiological pH compatible with the enzyme operating through a “reverse protonation” mechanism. Interestingly, NMR relaxation measurements and the results of a model building study suggest that SrtA recognizes the LPXTG sorting signal through a lock-in-key mechanism in contrast to the prototypical SrtA enzyme from Staphylococcus aureus.
Keywords: Cell Surface Protein, Cell Wall, Enzyme Mechanisms, NMR, Protein Structure, Bacillus anthracis, SrtA, Sortase, Transpeptidase
Introduction
Bacterial pathogens display proteins on their surface that enable them to evade the immune response of the host, adhere to sites of infection, acquire essential nutrients, and enter host cells (1). Gram-positive bacteria covalently attach proteins to the cell wall using sortase enzymes, a large family of membrane-associated transpeptidases (2–6). Proteins fated for cell wall attachment contain a C-terminal sorting signal that typically consists of a Leu-Pro-X-Thr-Gly motif (LPXTG, where X is any amino acid) followed by a hydrophobic segment and positively charged C-terminal amino acids. Many sortase enzymes catalyze a transpeptidation reaction that joins the threonine residue within the LPXTG motif to the free amino group within lipid II, a cell wall precursor (1). The protein is then displayed on the microbial surface when the lipid II-linked protein product is incorporated into the peptidoglycan by the transpeptidation and transglycosylation reactions of cell wall synthesis. Some members of the sortase enzyme family also assemble pili, hair-like proteinaceous structures that promote bacterial adhesion (7). These enzymes presumably function through a similar mechanism, but the transpeptidation reaction they catalyze polymerizes the protein subunits that construct the pilus. Because many clinically significant pathogens require a functioning sortase to be fully virulent, sortase enzymes are promising therapeutic targets for the development of novel antibiotics (8, 9).
Based on their primary sequences sortase enzymes can be classified into four subfamilies whose members have been shown experimentally to have distinct functions: Sortase A (SrtA)-,3 SrtB-, SrtC-, and SrtD-type enzymes (10, 11). SrtA-type enzymes are most closely related to the SrtA enzyme from Staphylococcus aureus (Sa-SrtA) and are “housekeeping” enzymes that anchor a large number of distinct proteins to the cell wall. They have attracted significant interest as potential drug targets because they are present in several clinically significant pathogens that exhibit attenuated virulence when their srtA gene is genetically eliminated (e.g. Staphylococcus aureus, Listeria monocytogenes, Streptococcus pyogenes, and Streptococcus pneumoniae among others). Other types of sortase enzymes have more specialized functions and process fewer protein substrates. SrtC-type enzymes are involved in pilin assembly, whereas the SrtB- and SrtD-type enzymes anchor proteins the cell wall involved in heme iron acquisition and sporulation, respectively (5, 8).
NMR and crystal structures of several sortase enzymes have revealed that they adopt a common eight-stranded β-barrel fold that contains conserved active site residues (12–20). The mechanism of the sortase-catalyzed transpeptidation reaction is best understood for the Sa-SrtA enzyme. All sortase enzymes contain three conserved residues that when mutated in Sa-SrtA severely reduce enzymatic activity: His120, Cys184, and Arg197 (Sa-SrtA numbering) (21–23). Transpeptidation occurs through a ping-pong mechanism that is initiated when the thiol group of Cys184 within Sa-SrtA nucleophilically attacks the carbonyl carbon of the threonine residue within the sorting signal (24, 25). This forms a transient tetrahedral intermediate, which, upon breakage of the threonine-glycine peptide bond, rearranges into a more stable thioacyl enzyme-substrate linkage. Sa-SrtA then joins the terminal amine group within the pentaglycine branch of lipid II to the carbonyl carbon of the threonine, creating a second tetrahedral intermediate that is resolved into the lipid II-linked protein product. During catalysis the Cys184 thiol covalently attaches to the carbonyl carbon of the threonine residue within the LPXTG sorting signal. Arg197 is thought to stabilize the binding of the sorting signal by hydrogen bonding to its backbone (17, 26) and may also stabilize oxyanion transition states (20, 23). The function of His120 has not been clearly established, but it may act as a general acid that protonates the amide group of the glycine residue within the sorting signal as the scissile peptide bond is broken and/or it may deprotonate the amine group of the lipid II nucleophile (17, 23, 25).
Bacillus anthracis is a spore forming Gram-positive bacterium that causes lethal anthrax disease in humans. The high mortality rate of anthrax caused by the inhalation of aerosolized bacterial spores makes it a potential bioterrorism agent and has driven the search for new therapeutics to treat and prevent infections caused by this microbe (27). B. anthracis encodes three sortase enzymes: Ba-SrtA, Ba-SrtB, and Ba-SrtC. The Ba-SrtB enzyme is involved in iron acquisition and anchors the heme-binding IsdC protein to the cell wall (28). Ba-SrtC, which is a SrtD-type enzyme, anchors two proteins required for proper spore formation (29, 30). The Ba-SrtA enzyme is a SrtA-type sortase that attaches 7 proteins to the cell wall by joining the threonine of the C-terminal LPXTG sorting signal to the amine group of meso-diaminopimelic acid (m-Dap) within lipid II (31, 32). Ba-SrtA is a potential target for new therapeutics as it is required for B. anthracis survival and replication within macrophages (33), a presumed early step in the development of inhalation anthrax.
The molecular basis of Ba-SrtA function is not well understood because it shares only limited sequence identity with previously characterized SrtA-type enzymes; the structures of the Sa-SrtA and Sp-SrtA enzymes have been determined and share 29 and 32% sequence identity with Ba-SrtA, respectively (12, 16, 17, 19). Here we report studies of the structure, dynamics, and function of Ba-SrtA. Unlike previously studied sortase enzymes, Ba-SrtA contains several unique active site features that include the presence of an N-terminal extension that contacts the catalytically essential histidine and a large structurally disordered active site loop. Moreover, in contrast to Sa-SrtA, the sorting signal binding pocket in Ba-SrtA is ordered and rigid in the apo-state and therefore presumably only needs to undergo minimal structural changes to recognize the sorting signal. The mechanistic implications of these large differences are discussed.
EXPERIMENTAL PROCEDURES
Protein Preparation
Four deletion mutants of the SrtA protein from B. anthracis were studied: Ba-SrtAΔ23 (residues Lys24–Lys210), Ba-SrtAΔ56 (residues Asp57–Lys210), Ba-SrtAΔ64 (residues Asp65–Lys210), and Ba-SrtAΔ74 (residues Asp75–Lys210). pET15b expression plasmids were used to produce the proteins in Escherichia coli BL21(DE3) cells. Subcloning made use of PCR-amplified DNA from B. anthracis Sterne genomic DNA. 13C- and 15N-labeled proteins used in the NMR studies were produced by growing cells in M9 medium supplemented with 15NH4Cl, or 15NH4Cl and [13C6]glucose, whereas standard Luria-Bertani broth was used to produce unlabeled proteins. All cell cultures were grown at 37 °C and induced for protein expression by adding isopropyl β-d-thiogalactoside to a final concentration of 1 mm when the cells reached an A600 of ∼0.6. Cells were harvested 4 h after induction by centrifugation at 6,000 × g and stored at −80 °C. Proteins were purified by resuspending the pellet in lysis buffer (50 mm NaPO4, pH 7.0, 300 mm NaCl, 2 mm phenylmethanesulfonyl fluoride, 2 mm benzamidine). The cells were then lysed by sonication and the lysate was cleared by centrifugation at 13,000 × g for 40 min at 4 °C. After filtering the supernatant with a 0.45-μm filter it was incubated with TALON His-affinity resin (Clontech). The resin was then washed with lysis buffer containing 10 mm imidazole. The histidine tag was then removed from the protein by incubating with thrombin at 37 °C for 1 h and 15 min in cleavage buffer (20 mm Tris, pH 7.9, 150 mm NaCl, 2.5 mm CaCl2). The eluate was then further purified using a Sephacryl-100 gel filtration column equilibrated with either NMR buffer (10 mm MES, 20 mm BisTris, pH 6.0) or fluorescence resonance energy transfer assay buffer (20 mm HEPES, pH 7.5). Three samples of Ba-SrtAΔ56 were studied by NMR and were dissolved in NMR buffer: 1) 4 mm [15N]Ba-SrtAΔ56 dissolved in NMR buffer containing 7% D2O; 2) 2.5 mm [15N,13C]Ba-SrtAΔ56 dissolved in NMR buffer containing 7% D2O; and 3) 2.5 mm [15N,13C]Ba-SrtAΔ56 dissolved in deuterated NMR buffer (obtained by lyophilization and redissolving in 99.999% D2O).
NMR Spectroscopy and Structure Determination
NMR spectra of Ba-SrtAΔ56 were acquired at 298 K on Bruker Avance 500-, 600-, and 800-MHz spectrometers equipped with triple resonance cryogenic probes. NMR spectra were processed using NMRPipe (34) and analyzed using the PIPP (35) and CARA (version 1.8.4) (36) software packages. Chemical shift assignments (1H, 13C, 15N) were obtained by analyzing the following experiments: HNCA, HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, HNHA, HNHB, HBHA(CO)NH, CC(CO)NH, HCCH-TOCSY, HCCH-COSY, (HB)CB(CGCDCE)HE, and (HB)CB(CGCD)HD (reviewed in Refs. 37 and 38). The majority of ϕ and ψ dihedral angle restraints were obtained using the program TALOS+ (39). Additional backbone ϕ angle restraints were obtained by analyzing HNHA spectra (40). Distance restraints were obtained from three-dimensional 15N- and 13C-edited NOESY spectra and a four-dimensional 13C,15N-edited HMQC-NOESY-HSQC spectrum. 1DNH and 1DNCO residual dipolar couplings were measured using protein samples partially aligned in PEG C12E5/hexanol, using two-dimensional 15N-coupled IPAP 1H-15N HSQC and two-dimensional carbonyl-coupled 1H-15N HSQC experiments, respectively.
NOE assignments were obtained automatically using the programs ATNOS and CANDID (41, 42). All of the NOE assignments were then verified by manually inspecting the NOESY data. During this process additional NOE restraints were identified and included in subsequent structure calculations. Restraints for hydrogen bonds were implemented using the HBDB algorithm and identified by inspecting the NOESY data for characteristic patterns combined with deuterium exchange experiments (43). In the final set of calculations a total of 200 structures were generated, of which 73 had no NOE, dihedral angle, or scalar coupling violations greater than 0.5 Å, 5°, or 2 Hz, respectively. Of these, 40 structures with the lowest overall energy were chosen to represent the structure of SrtAΔ56 and have been deposited in the protein data bank (accession code Protein Data Bank 2KW8).
Modeling of the Covalent Ba-SrtAΔ56-Sorting Signal Complex
The model was generated based on our recently determined structure of the S. aureus SrtA·LPAT* complex (17). The peptide in this structure is Cbz-LPAT*, where T* is (2R,3S)-3-amino-4-mercapto-2-butanol, and Cbz is a carbobenzyloxy protecting group (44). The Ba-SrtA·LPAT* model was calculated using artificial intermolecular distance restraints between the peptide and the Ba-SrtAΔ56 enzyme that were obtained by inspecting the intermolecular distance restraints experimentally identified for the Sa-SrtA·LPAT* complex. A total of 35 artificial intermolecular distance restraints were employed in simulated annealing calculations (supplemental Table S2). In addition, the set of restraints used to determine the structure of SrtAΔ56 in its apo-state were employed without modification. A total of 200 structures of the complex were calculated, of which 45 exhibited no NOE, dihedral angle, or scalar coupling violations greater than 0.5 Å, 5°, or 2 Hz.
Backbone Dynamics of Ba-SrtAΔ56 Determined from 15N Relaxation Data
The 15N relaxation data were collected on a Bruker Avance 600-MHz NMR spectrometer equipped with a triple resonance cryogenic probe. Data were analyzed using the program SPARKY (45) and included: 15N longitudinal relaxation rates (R1), transverse relaxation rates (R2), and {1H}-15N heteronuclear NOEs. Complete R1, R2, and {1H}-15N NOE values were available for 94 of 150 backbone amides as well as the side chain amide of Trp171. The average quantifiable R1, R2, and {1H}-15N NOE values for SrtAΔ56 are 1.18 ± 0.06, 13.07 ± 0.21, and 0.81 ± 0.01 s−1, respectively. Relaxation data were analyzed using programs kindly provided by Prof. Arthur G. Palmer III at Columbia University. The analysis procedure we used has been described previously (46, 47). Briefly, the program Pdbinertia was used to calculate the principal moments of inertia and yielded relative moments of 1.00:0.84:0.68. The program R2R1_tm was used to calculate an approximate correlation time (τm) of 10.2 ± 0.4 ns using R2/R1 ratios. Only R2/R1 ratios that met the following criteria were used in this analysis: 1) they were within one standard deviation of the average, and 2) the residue had a {1H}-15N NOE value >0.6. This data were then inputted into the program Quadric_Diffusion (48, 49) indicating the isotropic model is statistically preferred for SrtAΔ56 over the axially symmetric or anisotropic models of tumbling. The relaxation data were then interpreted using the Lipari-Szabo Model-free formalism (50) using the program FAST-Modelfree to iteratively run the program Modelfree 4.20 (51). Of the 107 amino acids that gave complete quantifiable relaxation information, data from 91 residues could be satisfactorily reproduced using the model-free approach. The data from the backbone amide nitrogen atoms of 59 residues could be fit using model 1 (S2 only), 4 residues fit model 2 (S2 and τe), 13 residues fit model 3 (S2 and Rex), 3 residues fit model 4 (S2, τe, and Rex), and 12 residues, in addition to the side chain indole Nϵ atom of Trp171 could be fit using model 5 (S2f, S2s, and τe).
Enzyme Kinetics Measurements
Substrate cleavage reaction was performed as previously described (52). The cleavage of the substrate, o-aminobenzoyl-LPETG-2,4-dinitrophenyl (abz-LPETG-DNP), was monitored by excitation at 335 nm and recording emission at 420 nm on a SpectraMax M5 spectrofluorometer (Molecular Devices). Assay conditions consisted of 20 mm HEPES, pH 7.5, and 10 μm enzyme. abz-LPETG-DNP peptide concentrations of 0, 5, 10, 20, 40, 80, 160, and 320 μm were used. Fluorescence was recorded for 10 h in 10-min increments. A standard curve was used to convert fluorescence units to rates and the steady state velocities were used to calculate Km and kcat as described previously (24).
Histidine Side Chain pKa Measurements
15N- and 13C-labeled Ba-SrtAΔ56 (0.5 mm) in buffer (10 mm MES, 20 mm BisTris in 7% D2O) was titrated using 0.2 m HCl or 0.2 m NaOH to pH 4.5–10. The chemical shift of the Hϵ1 atom from His126 and His177 were measured during the course of the titration by recording two-dimensional 1H-13C HSQC spectra. Chemical shifts recorded as a function of pH were fit to Equation 1,
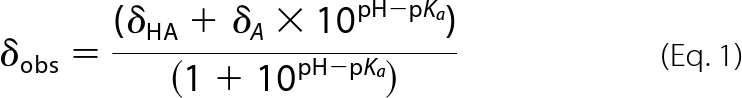 |
where δHA and δA are the chemical shifts of the fully protonated and deprotonated states of the immidizolium side chain respectively, and δobs is the observed chemical shift.
RESULTS
Structure of the Sortase A Enzyme from B. anthracis
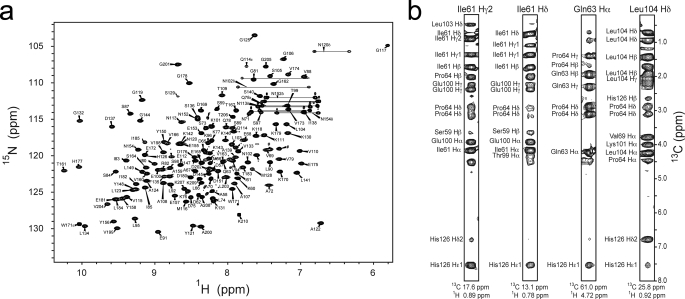
The Sortase A enzyme from B. anthracis (Ba-SrtA) is 210 amino acids in length and consists of two parts, a non-polar N terminus that presumably embeds the protein in the membrane (residues Met1–Gly23) and a C-terminal catalytic region (residues Lys24–Lys210). Previously, a deletion mutant of Ba-SrtA that removes the transmembrane region (Ba-SrtAΔ23, residues Lys24–Lys210 of Ba-SrtA) was shown to have hydrolytic activity in vitro (31). To further delineate residues that form the structured catalytic domain of the protein we used NMR spectroscopy to study a uniformly 15N-labeled sample of Ba-SrtAΔ23. Inspection of the 1H-15N HSQC spectra of [15N]Ba-SrtAΔ23 revealed that a large portion of the polypeptide was disordered as the backbone amide correlations of ∼30–40 amino acids exhibited narrow line widths and degenerate chemical shifts (data not shown). Because residues Lys24–Val56 in Ba-SrtA share only limited primary sequence homology with other sortase enzymes (supplemental Fig. S3), we purified isotopically labeled proteins missing these residues (Ba-SrtAΔ56, residues Asp57–Lys210 of Ba-SrtA). The NMR spectra of Ba-SrtAΔ56 are well resolved and have line widths that indicate that the majority of the protein is structured (Fig. 1a).
FIGURE 1.
NMR Spectra of Ba-SrtAΔ56. a, 1H-15N HSQC spectrum. The amino acid assignment for each cross-peak is indicated. Cross-peaks for residues Thr186–Tyr197, and Asn127 are broadened beyond detection. An asterisk indicates five cross-peaks that could not be assigned. # indicates cross-peaks that are folded into the spectrum that originate from side chain nitrogen atoms. b, selected panels showing proton-proton NOEs between the N-terminal extension and the side chain of His126 and other active site residues. The panels are taken from a three-dimensional 13C-edited NOESY-HSQC spectrum of Ba-SrtAΔN56 dissolved in deuterated buffer. The identity of the proton from Ba-SrtAΔN56 that gives rise to the set of NOEs and its chemical shift are shown at the top and bottom of each panel, respectively. On the left side of each cross-peak the proton within the active site that is proximal to the N-terminal extension is indicated. For clarity only protons within the active site are labeled.
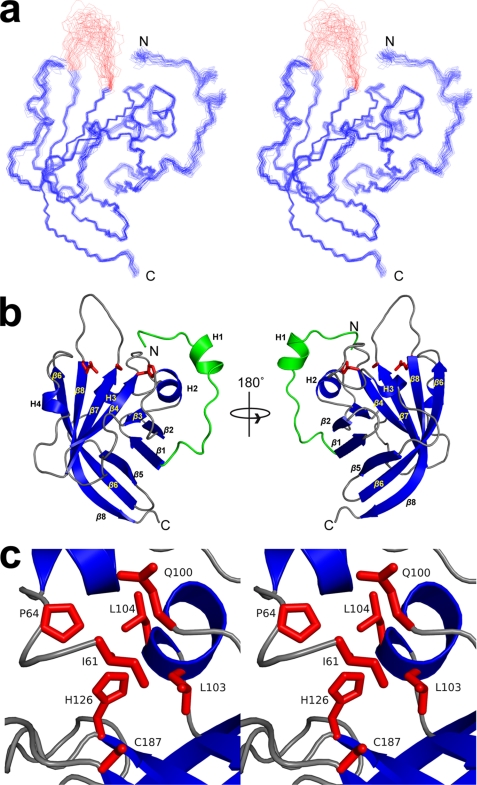
The structure of Ba-SrtAΔ56 was determined using multidimensional heteronuclear NMR and simulating annealing methods. A total of 2,812 experimental restraints were used to determine the structure, including: 2,177 inter-proton distance, 231 dihedral angle, 54 3JHNα, 264 13C secondary shifts, and 116 residual dipolar coupling restraints. An ensemble containing 40 conformers representing the structure of the protein is shown in Fig. 2a. The conformers exhibit good covalent geometry and have no NOE, dihedral angle, or scalar coupling violations greater than 0.5 Å, 5°, or 2 Hz, respectively. The enzyme is structured from residues Ile61–Thr186 and Arg197–Lys209. The backbone and heavy atom coordinates of these residues have a root mean square deviation (r.m.s. deviation) to the average structure of 0.40 ± 0.07 and 0.88 ± 0.06 Å, respectively. Complete structural and restraint statistics are presented in Table 1.
FIGURE 2.
NMR solution structure of Ba-SrtAΔ56. a, cross-eyes stereo image showing an ensemble of the 40 lowest energy structures of Ba-SrtAΔ56. Residues Ile61–Lys210 are shown. The majority of the protein is ordered and consists of residues Ile61–Thr186 and Arg196–Lys210 (colored blue). An active site loop connecting strands β7 and β8 is not defined by the NMR data and is structurally disordered (residues 187–195, colored red). The coordinates were superimposed by aligning the backbone N, Ca, and C′ atoms of Ser73–Thr186 and Arg196–Lys210. b, ribbon drawing of the structure Ba-SrtAΔ56. The structure on the left is in a similar orientation as shown in a, whereas the structure on the right has been rotated by 180°. The secondary structural elements are labeled and the conserved catalytic residues His126, Cys187, and Arg196 are shown. The N-terminal extension that is unique to Ba-SrtAΔ56 is colored green. c, cross-eyed stereo image showing an expanded view of the enzyme active site and contacts to it that are made by residues in the N-terminal extension. This interaction is unique and has not been observed in previously determined structures of other sortase enzymes.
TABLE 1.
Statistics for the NMR structure of Ba-SrtAΔ56
| 〈SA〉a | 〈SA〉a | |
|---|---|---|
| R.m.s. deviations from NOE interproton distance restraints (Å) (2204) | 0.050 ± 0.002 | 0.048 |
| R.m.s. deviations from dihedral angle restraints (degrees)b (231) | 0.489 ± 0.073 | 0.999 |
| R.m.s. deviations from 3JHNα coupling constants (Hz) (54) | 0.834 ± 0.030 | 0.814 |
| R.m.s deviations from secondary 13C shifts | ||
| 13Cα (ppm) (132) | 1.450 ± 0.064 | 1.440 |
| 13Cβ (ppm) (132) | 1.323 ± 0.066 | 1.272 |
| Residular dipolar coupling R-factors (%)c | ||
| DNH (70) | 3.2 ± 0.3 | 3.9 |
| DNCO (46) | 22.3 ± 1.1 | 22.2 |
| Devitions from idealized covalent geometry | ||
| Bonds (Å) | 0.0044 ± 0.0003 | 0.0131 |
| Angles (degrees) | 0.645 ± 0.025 | 1.258 |
| Impropers (degrees) | 0.580 ± 0.026 | 1.045 |
| PROCHECK-NMRd | ||
| Most favorable region (%) | 83.6 ± 2.0 | 84.8 |
| Additionally allowed region (%) | 15.5 ± 2.0 | 14.4 |
| Generously allowed region (%) | 0.2 ± 0.0 | 0.8 |
| Disallowed region (%) | 0.7 ± 0.0 | 0.0 |
| Coordinate precisione | ||
| Protein backbone (Å) | 0.43 ± 0.08 | |
| Protein heavy atoms (Å) | 0.90 ± 0.06 |
a The notation of the NMR structures is as follows: 〈SA〉 represent an ensemble of the 40 best structures calculated by simulated annealing. 〈SA〉 is the average energy-minimized structure. The number of terms for each restraint is given in parentheses. None of the structures exhibited distance violations greater than 0.5 Å, dihedral angle violations greater than 5°, or coupling constant violations greater than 2 Hz.
b Experimental dihedral angle restraints comprised 114 φ and 117 ψ angles.
c The dipolar coupling R-factor ranges between 0 and 100% and is defined as the ration of the r.m.s. deviation between observed and calculated values to the expected r.m.s. deviation if the vectors were randomly distributed, given by [2Da2(4 + 3η2)/5]1/2, where Da is the magnitude of the principle component of the alignment tensor, and η is the rhombicity, calculated to be −12.5 Hz and 0.44, respectively.
d PROCHECK-NMR data includes residues Ile61 to Thr186 and Arg196 to Lys209 of Ba-SrtAΔ56.
e The coordinate precision is defined as the average atomic r.m.s. deviation of the 40 individual simulated annealing structures and their mean coordinates. The reported values are for residues Ile61 to Thr186 and Arg196 to Lys209 of Ba-SrtAΔ56.
Ba-SrtAΔ56 adopts an eight-stranded β-barrel structure that is supplemented by four helices (Fig. 2b). The secondary structural elements in the protein have the following topology: H1-β1-β2-H2-β3-β4-H3-β5-β6-H4-β7-β8, where H and β refer to helices and strands, respectively. Beginning at the N terminus, an extended segment (Ala58–Gln63) is positioned in the active site cleft and is then followed by a α-helix (H1, Leu66–Asn71). The barrel structure then begins with strand β1 (Gly81–Ile85), which is connected by a short hairpin that lies anti-parallel to strand β2 (Leu90–Leu95). A 310-helix (H2, Glu100–Ser105) then joins strand β2 to β3 (Ala107–Thr109), which lay in parallel. Helix H2 also forms a wall of the active site cleft. The chain then reverses direction so as to position strand β4 (Asn120–His126) in an anti-parallel orientation next to strand β3. An extended polypeptide segment containing a 310-helix (H3, Ile138–Ser140) then wraps around the enzyme to initiate strand β5 (Lys146–Asp151) on the opposite face of the protein. This strand pairs with β1 in an anti-parallel fashion and is separated by a short hairpin from residues in strand β6 (Asn154-Glu165), whose chain also is positioned in an opposite orientation. A long loop containing a 310-helix (H4, Trp171–Val173) then connects strands β6 and β7 (Glu181–Thr186). The β7 strand runs parallel with respect to the β4 strand and is followed by a structurally disordered loop that reverses the direction of the chain thereby enabling residues in strand β8 (Typ197–Ala208) to hydrogen-bond with residues in strands β6 and β7 in an anti-parallel manner. Two adjacent β-bulges present within strands β6 (Thr109) and β8 (Val204) introduce a kink that enables extensive interactions between the chains and allows them to form opposing faces of the β-barrel structure. Residues His126, Cys187, and Arg196 are completely conserved in sortase enzymes and form the active site. They are located near the end of the sheet formed by strands β4, β7, and β8. Cys187 is situated at the C-terminal end of the β7 strand and is bracketed by side chains of His126 and Arg196 located on strands β4 and β8, respectively.
Unique Active Site Features: a Histidine Contacting N-terminal Extension and a Disordered β7/β8 Loop
All sortases contain a conserved histidine residue whose mutation in Sa-SrtA inactivates the enzyme (21). In contrast to previously studied enzymes, in Ba-SrtA the histidine residue (His126) is contacted by an N-terminal extension positioned in the groove that separates helix H2 and the β7/β8 loop (Fig. 2b). The structured extension precedes strand β1 in the primary sequence and consists of residues Ile61–Val79. At its N terminus residues Ile61–Pro64 are positioned near His126. The chain then forms a short α-helix (H1) that packs against helix H2 before changing its direction to initiate strand β1. Contacts to His126 are extensive, with the methyl groups Ile61 partially encapsulating the imidizole ring (Fig. 2c). Ile61 methyl groups also contact the methyl groups of Leu103, a residue believed to be important in recognizing the LPXTG substrate (17). Pro63 for the N-terminal extension also fits into the groove, making contacts with Leu104 in helix H2. The N-terminal tail, along with Leu103, Leu104, Met128, and Leu134 enclose catalytic His126 in a hydrophobic pocket. This is unique, as the analogous active site histidine in other sortase enzymes is exposed to the solvent (Fig. 5, a–c). As shown in Fig. 1b, active site contacts to the N-terminal extension are well supported by the NOESY spectra. For example, unambiguous NOEs are observed between the Hγ2 and Hδ1 methyl groups of Ile61 and the Hϵ and Hδ ring protons of His126. In addition, NOEs are present between Glu68 and Leu104, Val69 and Leu104, Asn79 and Leu104, Ala72 and Leu104, as well as Ala72 and Ser105.
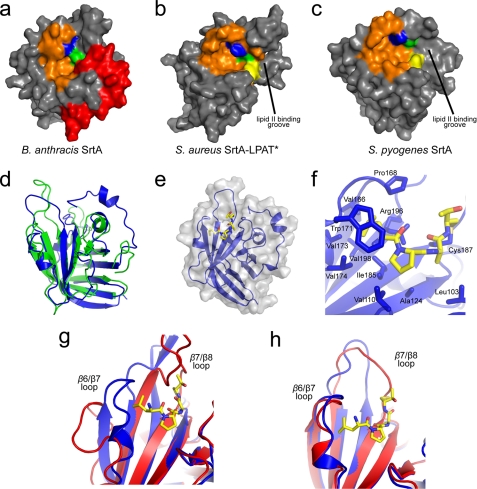
FIGURE 5.
Comparison with other SrtA-type enzymes and the model of the Ba-SrtA-sorting signal complex. The solvent accessible surfaces of SrtA-type enzymes are shown in panels a, Ba-SrtAΔ56; b, Sa-SrtA-LPAT* (with LPAT substrate removed for clarity) (PDB code 2KID) (17); and c, Sp-SrtA (PDB code 3FN7) (16). The conserved active site residues histidine (yellow), cysteine (green), and arginine (blue) are highlighted. The LPXTG binding pocket (orange) is also highlighted. The N-terminal tail in Ba-SrtAΔ56 (red) prevents His126 from being solvent exposed. The grooves leading into the active site are labeled in panels b and c. d, an overlay of the structure of apo-SrtAΔ56 (blue) and the Sa-SrtA protein in the structure of the Sa-SrtA·LPAT* complex (green) (PDB code 2KID) (17). e, a ribbon drawing of the model of the Ba-SrtA-sorting signal complex. The solvent-accessible surface of the protein is semi-transparent and colored gray. f, expanded view of protein-peptide interactions in the model of the complex. The structure of the enzyme is shown in blue with residues interacting with the substrate labeled. The sorting signal peptide is colored by atom type and is shown in a stick representation. g, apo-Sa-SrtA (red) and Sa-SrtA·LPAT* (blue) demonstrate the induced fit model of LPXTG substrate recognition. h, apo-Ba-SrtAΔ56 (red) and Ba-SrtAΔ56·LPAT* model (blue) demonstrate the lock-and-key binding model of LPXTG substrate recognition.
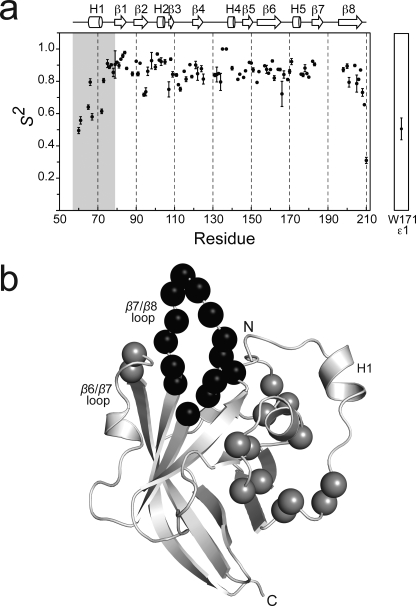
Another unique feature of the Ba-SrtA structure is the presence of a mobile active site loop that connects strands β7 to β8 (the β7/β8 loop). The loop immediately follows Cys187 in the active site and has been postulated to form a surface that recognizes the second substrate of catalysis, lipid II (17, 20). In the 1H-15N HSQC spectrum of Ba-SrtAΔ56 cross-peaks for residues Thr186–Tyr197 are absent (Fig. 3b). Their signals are presumably broadened beyond detection because they undergo conformational rearrangements that occur on the micro- to millisecond time (i.e. they are broadened by exchange processes that are intermediate on the chemical shift time scale). The notion that the β7/β8 loop is mobile is consistent with the finding that resonances for Asn127 are also absent in the NMR data. This residue is located immediately following strand β4 near the base of the β7/β8 loop and its resonances are presumably broadened as a result of fluctuations in its magnetic environment that are caused by the movement of the loop (Fig. 3b). Flexibility in the β7/β8 loop is also supported by NMR relaxation measurements for the backbone nitrogen nuclei of Ba-SrtA described in detail below.
FIGURE 3.
NMR relaxation data that defines the mobility of Ba-SrtA. a, a graph showing the general order parameter (S2) of the backbone nitrogen atoms as a function of residue number. The right panel shows data for the side chain Nϵ1 atom of Trp171. Secondary structural elements are shown above the figure, with the semi-flexible N-terminal extension shaded. Data for the β7/β8 loop is absent because many of the resonances for these residues are broadened beyond detection presumably because they are mobile. b, ribbon drawing of the structure of Ba-SrtAΔ56 showing the location of backbone amide atoms in the structure that have significant Rex values. Residues with Rex values greater than 2.0 Hz (gray spheres) include residues: Asp75, Lys76, Gln78, Val79, Leu95, Lys96, Ser98, Ser105, Ala107, Lys131, Gly132, Leu134, Val166, and Thr167. The figure also shows residues within the β7/β8 loop that presumably exhibit large amplitude motions because their backbone amide resonances could not be assigned (black spheres).
Backbone Dynamics of Ba-SrtA Revealed by 15N Relaxation Measurements
To gain insight into motions occurring within the enzyme we measured backbone {1H}-15N NOEs and 15N spin-lattice (R1) and spin-spin (R2) relaxation rates (supplemental Fig. S2). The data were then interpreted using the model-free formalism, which yields three parameters that describe the motion of the protein backbone: the general order parameter (S2), the effective correlation time for internal motions (τe), and Rex (50, 51) (supplemental Table S1). S2 describes the magnitude of fast picosecond time scale motions that the amide bond experiences and is characterized by the internal correlation time, τe. The value of S2 ranges from 0 to 1, with a value of 1 indicating that the bond is completely immobilized. Rex is the chemical (conformational) exchange contribution to R2 and reports on slower micro- to millisecond time scale motions.
The S2 data indicate that the N-terminal extension exhibits elevated mobility (Fig. 3a). S2 parameters could be measured for backbone amides within all regions of the protein, with the notable exception of residues within the β7/β8 loop whose signals as previously noted are absent from the NMR spectra. Consistent with the structure the S2 data reveals that residues forming the β-barrel beginning at strand β1 and ending at strand β8 are immobile (residues Ile61–Thr183 and Val198–Lys210 have an average S2 value of 0.85). However, the N-terminal extension that contacts the active site exhibits elevated mobility as compared with the body of the protein. Ser73 is the last highly ordered residue at the amino terminus of the protein, beginning at Ala72 immediately following helix H1 the S2 values become progressively smaller as the chain proceeds toward Ile61, the last structured residue in the tail. As numerous NOEs define the positioning of the N-terminal tail (Fig. 1b), this suggests that in solution the tail adopts two distinct conformational states, an ordered conformation observed in the NMR structure in which it contacts the active site and a less populated disordered mobile state in which it is dissociated from the body of the protein. The elevated mobility indicated by the relaxation data is presumably caused by averaging of these two states.
The Rex data suggest that the N-terminal extension transiently binds to the active site. Several residues surrounding the active site have elevated Rex values (Leu103, Ser105, Ala107, Lys131, Gly132, and Leu134, located in H2 or the β4/β3 loop) (Fig. 3b, gray spheres). In the structure they are located within the surface that contacts the N-terminal appendage and their elevated Rex values presumably report on the slow conformation changes associated with its binding and release. For some of the residues it is also possible that the Rex values are caused by changes in the tautomerization state of the His126. The notion that the N-terminal extension can open and close over His126 is further supported by the observation of significant Rex values for the backbone nitrogen atoms of Asp75, Lys76, Gln78, and Val79. These residues are located between helix H1 and strand β1 and may undergo structural rearrangements that enable them to function as a hinge about which the N-terminal extension opens. This idea is substantiated by elevated Rex values in residues Leu95, Lys96, and Ser98 within the loop that connects strand β4 to helix H2. These residues are positioned immediately adjacent to the presumed hinge, and may be indirectly broadened by fluctuations in their magnetic environment caused by opening and closing of the extension.
The relaxation data indicate that Ba-SrtA uses a rigid pocket to interact with the LPXTG sorting signal. Structural and NMR relaxation studies of Sa-SrtA have shown that it adaptively recognizes the LPXTG sorting signal by closing and immobilizing its β6/β7 loop over the substrate (12, 17, 19, 47). In contrast, residues in the analogous β6/β7 loop in Ba-SrtA adopt a rigid conformation in the absence of the sorting signal as evidenced by S2 values for its residues that are on average 0.84 ± 0.05 (residues Val166–Asp180). Based on the NMR structure of the Sa-SrtA substrate complex (17), the side chain of Trp171 located in the central 310 helix of the loop can be expected to interact with the sorting signal. Interestingly, although the backbone of Trp171 is static (S2 of 0.921 ± 0.037), its side chain indole appears to be relatively flexible. This is evidenced by data for its indole Nϵ1 atom, which are best fit using slow and fast time scale order parameters (S2 of 0.507 ± 0.068 with S2f of 0.742 ± 0.041, and S2s of 0.683 ± 0.055). Similar order parameters have been observed for other surface-exposed tryptophan residues in E. coli RNase H (51). Presumably side chain motions caused by rotations about the χ dihedral angles enable it to adjust its structure to productively contact the sorting signal.
Kinetic Measurements of Enzyme Activity
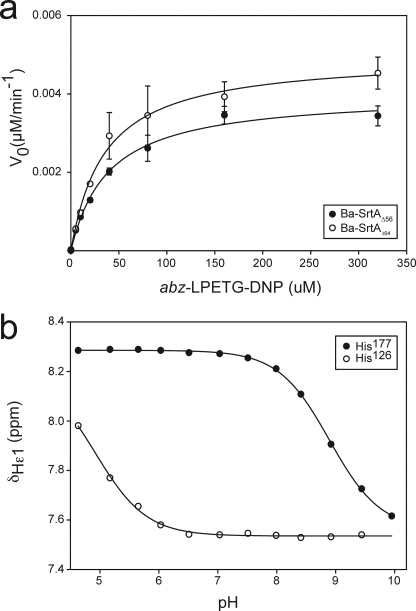
In vivo, Ba-SrtA anchors proteins to the cell wall by catalyzing a transpeptidation reaction that joins the threonine residue of the sorting signal to the free amino group of m-Dap. Previously, Schneeweind and colleagues (31) demonstrated that purified Ba-SrtAΔ23 catalyzes the hydrolysis of a fluorogenic peptide containing the amino acid sequence LPETG. In this reaction the peptide mimics the sorting signal substrate and a water molecule replaces m-Dap as the nucleophile. However, kinetic parameters for the hydrolysis reaction were not reported. We therefore adapted this assay to quantitatively measure the steady state hydrolysis kinetics of an abz-LPETG-DNP (Fig. 4a). Ba-SrtAΔ56 cleaves this peptide with kcat and Km values of 4.0 × 10−4 ± 1 × 10−5 min−1 and 38 ± 4 μm, respectively. Interestingly, our results indicate that in vitro Ba-SrtAΔ56 is unable to perform the transpeptidation reaction, because we were unable to detect transpeptidation products by mass spectrometry when Ba-SrtAΔ56 was incubated with abz-LPETG-DNP and up to a 100-fold molar excess of m-Dap (data not shown). This suggests that to be completely active, Ba-SrtA requires either the intact lipid II molecule as a substrate and/or additional cell wall components. A comparison of the activity of Ba-SrtA to other SrtA-type enzymes is provided in Table 2 and discussed later in the text.
FIGURE 4.
Enzyme kinetic and pKa measurements of Ba-SrtA. a, representative curves showing the hydrolysis kinetics of Ba-SrtAΔ56 (solid circles) and Ba-SrtAΔ64 (open circles). Each curve shows the production of product peptide after enzyme-mediated hydrolysis of the abz-LPETG-DNP sorting signal peptide substrate. The kinetic parameters extracted from this data are presented in Table 2. b, plot of the chemical shift of the 1Hϵ1 atom of His126 and His177 as a function of pH. Data were fit to Equation 1 to determine the pKa (His126 and His177 have pKa values of <5.5 and 8.9 ± 0.1, respectively). Analysis of the 1H-15N HSQC NMR spectra over the same pH range (data not shown) indicates that the protein remains folded throughout the titration study.
TABLE 2.
Summary of the enzyme kinetics of the Ba-SrtA enzyme
| kcat min−1 | Km μm | kcat/Kmμm−1 min−1 | |
|---|---|---|---|
| Ba-SrtAΔ56a | 4.0 × 10−4 ± 1 × 10−5 | 38 ± 4 | 1.1 × 10−5 ± 1 × 10−6 |
| H126ABa-SrtAΔ56 | NAb | NA | NA |
| Ba-SrtAΔ64a | 5.0 × 10−4 ± 2 × 10−5 | 35 ± 4 | 1.4 × 10−5 ± 2 × 10−6 |
| Sa-SrtAΔ56c | 7.3 × 10−3 ± 6 × 10−4 | 20 ± 4 | 3.7 × 10−4 ± 8 × 10−5 |
a kcat of Ba-SrtAΔ56 and Ba-SrtAΔ64 is significantly different with a p value of 0.0015.
b NA, not active.
c Ref. 24.
To determine whether the presence of the N-terminal extension alters enzymatic activity of Ba-SrtA we studied a truncation mutant in which amino acids preceding helix H1 are removed, Ba-SrtAΔ64 (residues Asp65–Lys210 of Ba-SrtA). Ba-SrtAΔ64 is folded based on its 1H-15N HSQC spectrum (supplemental Fig. S1). As compared with SrtAΔ54, it hydrolyzes the sorting signal with a modestly larger catalytic turnover (kcat is 20% larger, with a p value of 0.0015) and has a comparable Km for the sorting signal (Table 2). We also attempted to study SrtAΔ74 (residues Asp75–Lys210 of Ba-SrtA), a truncation mutant that completely removes the N-terminal extension. This protein failed to express in E. coli, consistent with the idea that contacts from helix H1 to the body of the protein are needed to stabilize the structure of the enzyme. The importance of the active site histidine was confirmed by showing that a H126A mutant of Ba-SrtAΔ56 has no detectable hydrolytic activity (data not shown). Combined, these data reveal that His126 is essential for the hydrolytic activity, although contacts to it from Ile61 within the N-terminal extension are dispensable. However, as the hydrolysis reaction only mimics the initial steps of transpeptidation, active site contacts from the N-terminal extension could still be important for downstream lipid II recognition events required to attach proteins to the cell surface.
pKa Measurements Reveal that His126 Is Uncharged
Knowledge of the ionization state of residues within the active site is needed to understand the mechanism of catalysis, but thus far this information has only been reported for Sa-SrtA. We therefore used NMR spectroscopy to determine the pKa values of the histidine residues within the catalytic domain of Ba-SrtAΔ56. These included His126, located in the active site, and His177 located on the β6/β7 loop. A series of 1H-13C HSQC spectra of [13C,15N]Ba-SrtAΔ56 were collected at different pH values and the chemical shifts of the side chain histidine 1Hϵ1-13Cϵ1 resonances were measured (Fig. 4b). Fitting the pH dependence of the chemical shift data reveals that His126 has a pKa less than 5.5 ± 0.1 (a more precise measurement is not possible because Ba-SrtA was not stable at lower pH values needed to complete the titration curve). This clearly indicates that the His126 side chain within the active site is uncharged at physiological pH and therefore not a participant in an imidazolium-thiolate interaction with Cys187. In contrast, the side chain of His177 located in the β6/β7 loop is fully protonated at physiological pH as it has a pKa of 8.9 ± 0.1. This finding is compatible with the structure as the Nδ1 and Nϵ2 atoms of His177 are positioned to interact with the side chain carboxyl group of Glu129 and the backbone carbonyl of Glu172, respectively. These interactions may stabilize the conformation of the β6/β7 loop and partially explain why the loop adopts an ordered structure in the absence of the sorting signal.
Model of the Sorting Signal Bound to Ba-SrtA
Recently we determined the structure of Sa-SrtA covalently bound to an analog of the LPXTG sorting signal (17). Superposition of the apo-Ba-SrtA and the Sa-SrtA-sorting signal complex reveals that the substrate-contacting β6/β7 loop in each enzyme adopts a similar conformation (Fig. 5d). This is surprising as it suggests that the Ba-SrtA enzyme may contain a preformed binding pocket for the sorting signal in contrast to Sa-SrtA, which undergoes major structural and dynamics changes upon binding the peptide (17). To investigate how the static pocket in Ba-SrtA might recognize the sorting signal we modeled the structure of the Ba-SrtA-sorting signal complex using simulated annealing and a set of 35 artificial intermolecular peptide-protein distance restraints derived from the structure of the Sa-SrtA-sorting signal complex (listed under supplemental materials). The average energy minimized model of the complex reveals that minimal structural perturbations in the enzyme are required to bind the sorting signal (Fig. 5, e and f). The peptide rests in a groove whose base is formed by residues in strands β4 and β7, and whose walls are formed by the β6/β7, β7/β8, β3/β4 and β2/H2 surface loops. The leucine methyl groups of the peptide are wedged between strand β8 and the β6/β7 loop, contacting the side chains of Val166, Pro168, Val173, and Val174 on the β6/β7 loop, and Val198 on strand β8. The indole ring of Trp171 located in the β6/β7 loop rests on top of the signal, whereas the proline in the peptide forms a kink in the substrate that is contacted by residues Ala124 and Ile185 located in strands β4 and β7, respectively, as well as Val110 on the β3/β4 loop. Contacts to the remainder of the peptide cannot be reliably predicted from the model as the positioning of residues in the β7/β8 loop of Ba-SrtA that contact this portion of the substrate are poorly defined in the NMR structure of the apo-form of the enzyme (Fig. 2a). The implications of the model with respect to the mechanism of sorting signal binding by other SrtA-type enzymes are discussed below.
DISCUSSION
Sortase enzymes are promising targets for the development of new anti-infective agents as they are required for the virulence of a range of clinically significant pathogens (8, 9). The SrtA sortase from the pathogen B. anthracis (Ba-SrtA) is required for bacterial growth within a mouse macrophage-like cell line, suggesting that it plays a critical role in the early steps of inhalation anthrax disease progression in humans, the replication of germinated spores within lung alveolar macrophages (33). Based on its primary sequence Ba-SrtA is a SrtA-type enzyme, a subfamily of the sortases that are most closely related to the SrtA enzyme from S. aureus. SrtA-type enzymes are believed to play a housekeeping role in the cell by anchoring a large number of distinct proteins that contain a LPXTG sorting signal (10). The structures of two other SrtA-type enzymes have been determined: S. pyogenes SrtA (Sp-SrtA) and S. aureus SrtA (Sa-SrtA) (12, 16, 17, 19). These proteins share only limited sequence homology with Ba-SrtA; Sa-SrtA and Sp-SrtA share 29 and 32% sequence identity with Ba-SrtA, respectively. In this study we have used a combination of NMR and enzyme kinetic measurements to investigate the structure, dynamics, and function of Ba-SrtA. Similar to previously characterized sortases, Ba-SrtA adopts a conserved β-barrel fold. However, there are substantial differences in both the structure and dynamics of its active site revealing significant mechanistic diversity.
The NMR structure of Ba-SrtA contains an N-terminal tail that forms numerous contacts to the active site histidine (His126). The tail is formed by residues that precede strand β1 and its positioning within the active site has not been previously observed in other sortase structures (12–20). If Ba-SrtA operates through a similar mechanism as the prototypical Sa-SrtA enzyme, during catalysis His126 may function as a general acid that protonates the amide group of the glycine residue within the sorting signal as the scissile peptide bond is broken and/or it may deprotonate the amine group of the lipid II nucleophile (17, 23, 25). Interestingly, in all other sortase enzymes studied to date the analogous active site histidine is exposed to the solvent and there is great structural variability in residues that precede strand β1 (Fig. 5, a–c). Notably, the Sp-SrtA enzyme also contains a structured N-terminal extension, but unlike Ba-SrtA it is positioned distal to the active site (16). This diversity is consistent with a multiple sequence alignment, which reveals that this region in the primary sequence is poorly conserved in SrtA-type enzymes (supplemental Fig. S3, green box). Interestingly, the SrtC-type sortase enzymes that catalyze pilin assembly contain an N-terminal extension that precedes the β-barrel (13, 15). However, this extension acts as “lid” that approaches the active site from a different direction and it adopts a distinct structure. Unlike Ba-SrtA, the SrtC lid does not interact with the active site histidine, but instead occludes the binding site of the sorting signal.
The N-terminal tail in Ba-SrtA may be required for later steps in the transpeptidation reaction involving lipid II. We have shown that the tail can be removed without significantly altering the ability of Ba-SrtA to hydrolyze the sorting signal, a reaction that mimics only the first half of the transpeptidation reaction by replacing the lipid II nucleophile with water (24, 25). This suggests that contacts from the tail are not required to properly position the histidine for the first steps of the transpeptidation reaction. However, it is conceivable that the tail actually inhibits the hydrolysis reaction, but it unlatches from the enzyme active site at a rate that is sufficiently fast so as not to be rate-limiting. This possibility cannot be excluded, because similar to other sortase enzymes, the isolated Ba-SrtA enzyme exhibits slow reaction kinetics in vitro (Table 2) and our 15N relaxation data show that residues within the tail exhibit elevated mobility that is compatible with a portion of the tail transiently unraveling from the enzyme (Fig. 3a).
It also possible that the N-terminal tail is involved in lipid II recognition. In the Sp-SrtA and Sa-SrtA structures two grooves lead into the active site (Fig. 5, b and c). The first groove binds to the LPXTG sorting signal and is formed by residues in strands β6, β7, and the β6/β7 loop (19, 53), whereas the second groove is located on the opposite side of the active site cysteine and is formed by residues in strands β4 and β7, helix H2, and the β7/β8 loop. The positioning of the second groove and the results of chemical shift perturbation studies has led to the suggestion that it forms the binding site for lipid II (16, 17, 20). Inspection of the Ba-SrtA structure reveals that the novel N-terminal extension masks the second groove in Ba-SrtA presumably affecting how lipid II is recognized (compare Fig. 5, a–c). In vitro the isolated Ba-SrtA enzyme was unable to catalyze the transpeptidation reaction that joins the LPETG peptide to m-DAP (a component of lipid II in B. anthracis that contains the amine to which the surface protein is attached) (31). This is in marked contrast to the Sa-SrtA and Sp-SrtA enzymes, which in vitro catalyze the transpeptidation reaction that joins the sorting signal to the appropriate peptide mimics of lipid II (Gly3 or Ala2 in S. aureus and S. pyogenes, respectively) (16, 54). Thus it appears that in vitro, Ba-SrtA requires additional protein components or larger portions of the lipid II molecule bearing m-DAP to successfully mediate transpeptidation.
The pKa of the catalytically essential His126 side chain in Ba-SrtA may explain its reduced hydrolytic activity relative to the Sa-SrtA enzyme. Ba-SrtA hydrolyzes the LPXTG sorting signal 40 times slower than the Sa-SrtA enzyme (Table 2). In Sa-SrtA the side chains of the active site cysteine and histidine are predominantly uncharged (25, 55). McCafferty and colleagues (25) have proposed that catalysis in the Sa-SrtA enzyme occurs via a reverse protonation mechanism. In this model the vast majority of Sa-SrtA is inactive containing the cysteine and histidine active site residues in their uncharged states, whereas a small percentage of the enzyme is enzymatically active (∼0.06%). The sparsely populated active form presumably contains the cysteine in its thiolate form primed to nucleophilically attack the threonine carbonyl group of the sorting signal. In addition, the histidine is in its imidazolium form poised to protonate the nitrogen of the scissile peptide bond. Our pKa measurement of the active site histidine in Ba-SrtA indicates that like Sa-SrtA it is predominantly uncharged at physiological pH. Interestingly, its pKa value is smaller than the analogous histidine in Sa-SrtA; the pKa of His126 in Ba-SrtA is less than ∼5.5, whereas the pKa of His120 in Sa-SrtA is ∼6.3–7.0 (25, 55). The lower pKa may be caused by contacts from the side chain of Ile61 on the N terminus, which partially encases the His126 side chain within a hydrophobic pocket. If the reverse protonation mechanism is operative in Ba-SrtA we would anticipate a lower percentage of the Ba-SrtA enzyme to be in its active charged state as compared with Sa-SrtA. Although there are certainly other variables to consider when comparing the activities of the enzymes, such as possible differences in the substrate binding geometry in the active site, this may help explain the lower kcat of Ba-SrtA relative to Sa-SrtA.
Another surprising finding is that the active site loop in Ba-SrtA that connects strands β7 and β8 (the β7/β8 loop) is structurally disordered. This loop is positioned immediately adjacent to the N-terminal extension and has been proposed to bind to lipid II (17, 20). It was determined to be structurally disordered in Ba-SrtA because resonances for nearly all of its residues are broadened beyond detection, presumably because the loop undergoes large amplitude motions that occur on the micro- to millisecond time scales. In principle, the broadening could be caused by protein aggregation if the loop resides at a protein-protein aggregation interface, but this seems unlikely as N15 relaxation data indicates that Ba-SrtA is monomeric based on its measured molecular correlation time of 10.2 ± 0.4 ns. In all previously determined sortase structures the analogous β7/β8 loop is structurally ordered. A notable exception is the SrtB enzyme from B. anthracis (Ba-SrtB), which is presumably flexible as many of its residues exhibit poorly defined electron density (18). Interestingly, both Ba-SrtA and Ba-SrtB presumably attach proteins to the free amino group within the m-DAP portion of lipid II, unlike other sortase enzymes of known structure that have rigid β7/β8 loops and attach proteins to structurally distinct lipid II molecules. This suggests that in B. anthracis the attachment of proteins to the m-DAP moiety of lipid II is correlated with the presence of a flexible active site loop.
The B. anthracis enzyme may recognize the sorting signal via a lock-and-key mechanism. Crystallographic and NMR studies of Sa-SrtA have shown that it binds to the LPXTG sorting signal through an induced fit mechanism in which signal binding nucleates the folding and immobilization of the β6/β7 loop (Fig. 5g) (12, 17, 19, 47). In contrast, the β6/β7 loop in apo-Ba-SrtA is immobile and appears to be in a conformation suited to interact with the sorting signal (Figs. 3a and 5h). To investigate this issue we generated a model of the Ba-SrtA-sorting complex based on the recently determined structure of the Sa-SrtA-sorting signal complex (17). This work revealed that apo-Ba-SrtA can bind to the sorting signal in a similar manner as the Sa-SrtA enzyme with only small perturbations in its structure (the backbone atoms of the protein in the model and the experimentally determined structure of Ba-SrtA can be superimposed with a r.m.s. deviation of 0.52 Å). However, to elucidate the molecular basis of sorting signal binding the structure of the Ba-SrtA-peptide complex needs to be determined. An understanding of the binding site pocket and substrate recognition mechanism by SrtA enzymes may serve to be beneficial in the rational development of sortase inhibitors. We have recently discovered several small molecules that inhibit both the Ba-SrtA and Sa-SrtA enzymes with similar potency (52). The work reported here could therefore facilitate the further development of these molecules into useful anti-infective agents to treat infections caused by S. aureus, B. anthracis, and other Gram-positive pathogens.
Supplementary Material
Acknowledgments
We thank Robert Peterson for assistance with the NMR and members of the Clubb laboratory for useful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant AI52217 and Department of Energy Grant DE-FC-03-87ER60615 (to R. T. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
The atomic coordinates and structure factors (code 2KW8) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SrtA
- Sortase A
- m-Dap
- meso-diaminopimelic acid
- HSQC
- heteronuclear single quantrum coherence
- abz-LPETG-DNP
- o-aminobenzoyl-LPETG-2,4-dinitrophenyl
- Ba-SrtA
- B. anthracis SrtA
- Sa-SrtA
- S. aureus SrtA
- Sp-SrtA
- S. pyogenes SrtA
- MES
- 4-morpholineethanesulfonic acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.
- root mean square
- Cbz
- carbobenzyloxy.
REFERENCES
- 1.Navarre W. W., Schneewind O. (1999) Microbiol. Mol. Biol. Rev. 63, 174–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazmanian S. K., Ton-That H., Schneewind O. (2001) Mol. Microbiol. 40, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 3.Ton-That H., Marraffini L. A., Schneewind O. (2004) Biochim. Biophys. Acta 1694, 269–278 [DOI] [PubMed] [Google Scholar]
- 4.Paterson G. K., Mitchell T. J. (2004) Trends Microbiol. 12, 89–95 [DOI] [PubMed] [Google Scholar]
- 5.Marraffini L. A., Dedent A. C., Schneewind O. (2006) Microbiol. Mol. Biol. Rev. 70, 192–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desvaux M., Dumas E., Chafsey I., Hébraud M. (2006) FEMS Microbiol. Lett. 256, 1–15 [DOI] [PubMed] [Google Scholar]
- 7.Mandlik A., Swierczynski A., Das A., Ton-That H. (2008) Trends Microbiol. 16, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maresso A. W., Schneewind O. (2008) Pharmacol. Rev. 60, 128–141 [DOI] [PubMed] [Google Scholar]
- 9.Suree N., Jung M. E., Clubb R. T. (2007) Mini. Rev. Med. Chem. 7, 991–1000 [DOI] [PubMed] [Google Scholar]
- 10.Comfort D., Clubb R. T. (2004) Infect. Immun. 72, 2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dramsi S., Trieu-Cuot P., Bierne H. (2005) Res. Microbiol. 156, 289–297 [DOI] [PubMed] [Google Scholar]
- 12.Ilangovan U., Ton-That H., Iwahara J., Schneewind O., Clubb R. T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6056–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manzano C., Contreras-Martel C., El Mortaji L., Izoré T., Fenel D., Vernet T., Schoehn G., Di Guilmi A. M., Dessen A. (2008) Structure 16, 1838–1848 [DOI] [PubMed] [Google Scholar]
- 14.Maresso A. W., Wu R., Kern J. W., Zhang R., Janik D., Missiakas D. M., Duban M. E., Joachimiak A., Schneewind O. (2007) J. Biol. Chem. 282, 23129–23139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neiers F., Madhurantakam C., Fälker S., Manzano C., Dessen A., Normark S., Henriques-Normark B., Achour A. (2009) J. Mol. Biol. 393, 704–716 [DOI] [PubMed] [Google Scholar]
- 16.Race P. R., Bentley M. L., Melvin J. A., Crow A., Hughes R. K., Smith W. D., Sessions R. B., Kehoe M. A., McCafferty D. G., Banfield M. J. (2009) J. Biol. Chem. 284, 6924–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suree N., Liew C. K., Villareal V. A., Thieu W., Fadeev E. A., Clemens J. J., Jung M. E., Clubb R. T. (2009) J. Biol. Chem. 284, 24465–24477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R., Wu R., Joachimiak G., Mazmanian S. K., Missiakas D. M., Gornicki P., Schneewind O., Joachimiak A. (2004) Structure 12, 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zong Y., Bice T. W., Ton-That H., Schneewind O., Narayana S. V. (2004) J. Biol. Chem. 279, 31383–31389 [DOI] [PubMed] [Google Scholar]
- 20.Zong Y., Mazmanian S. K., Schneewind O., Narayana S. V. (2004) Structure 12, 105–112 [DOI] [PubMed] [Google Scholar]
- 21.Ton-That H., Mazmanian S. K., Alksne L., Schneewind O. (2002) J. Biol. Chem. 277, 7447–7452 [DOI] [PubMed] [Google Scholar]
- 22.Marraffini L. A., Ton-That H., Zong Y., Narayana S. V., Schneewind O. (2004) J. Biol. Chem. 279, 37763–37770 [DOI] [PubMed] [Google Scholar]
- 23.Frankel B. A., Tong Y., Bentley M. L., Fitzgerald M. C., McCafferty D. G. (2007) Biochemistry 46, 7269–7278 [DOI] [PubMed] [Google Scholar]
- 24.Huang X., Aulabaugh A., Ding W., Kapoor B., Alksne L., Tabei K., Ellestad G. (2003) Biochemistry 42, 11307–11315 [DOI] [PubMed] [Google Scholar]
- 25.Frankel B. A., Kruger R. G., Robinson D. E., Kelleher N. L., McCafferty D. G. (2005) Biochemistry 44, 11188–11200 [DOI] [PubMed] [Google Scholar]
- 26.Bentley M. L., Lamb E. C., McCafferty D. G. (2008) J. Biol. Chem. 283, 14762–14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Friedlander A. M., Hauer J., McDade J., Osterholm M. T., O'Toole T., Parker G., Perl T. M., Russell P. K., Tonat K. (1999) JAMA 281, 1735–1745 [DOI] [PubMed] [Google Scholar]
- 28.Maresso A. W., Chapa T. J., Schneewind O. (2006) J. Bacteriol. 188, 8145–8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marraffini L. A., Schneewind O. (2006) Mol. Microbiol. 62, 1402–1417 [DOI] [PubMed] [Google Scholar]
- 30.Marraffini L. A., Schneewind O. (2007) J. Bacteriol. 189, 6425–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaspar A. H., Marraffini L. A., Glass E. M., Debord K. L., Ton-That H., Schneewind O. (2005) J. Bacteriol. 187, 4646–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budzik J. M., Oh S. Y., Schneewind O. (2008) J. Biol. Chem. 283, 36676–36686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zink S. D., Burns D. L. (2005) Infect. Immun. 73, 5222–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 35.Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. (1991) J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 36.Keller R. (2004) The Computer Aided Resonance Assignment Tutorial, Catina Verlag, Goldau, Switzerland [Google Scholar]
- 37.Palmer A. G., 3rd (1995) Protein NMR Spectroscopy, Academic Press, New York [Google Scholar]
- 38.Teng Q. (2005) Structural Biology: Practical NMR Applications, Springer Verlag, New York [Google Scholar]
- 39.Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vuister G. W., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 41.Herrmann T., Güntert P., Wüthrich K. (2002) J. Biomol. NMR 24, 171–189 [DOI] [PubMed] [Google Scholar]
- 42.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 43.Grishaev A., Bax A. (2004) J. Am. Chem. Soc. 126, 7281–7292 [DOI] [PubMed] [Google Scholar]
- 44.Jung M. E., Clemens J. J., Suree N., Liew C. K., Pilpa R., Campbell D. O., Clubb R. T. (2005) Bioorg. Med. Chem. Lett. 15, 5076–5079 [DOI] [PubMed] [Google Scholar]
- 45.Goddard T. D., Kneller D. G. (2006) SPARKY 3, University of California, San Francisco [Google Scholar]
- 46.Villareal V. A., Pilpa R. M., Robson S. A., Fadeev E. A., Clubb R. T. (2008) J. Biol. Chem. 283, 31591–31600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naik M. T., Suree N., Ilangovan U., Liew C. K., Thieu W., Campbell D. O., Clemens J. J., Jung M. E., Clubb R. T. (2006) J. Biol. Chem. 281, 1817–1826 [DOI] [PubMed] [Google Scholar]
- 48.Brüschweiler R., Liao X., Wright P. E. (1995) Science 268, 886–889 [DOI] [PubMed] [Google Scholar]
- 49.Lee L. K., Rance M., Chazin W. J., Palmer A. G., 3rd (1997) J. Biomol. NMR 9, 287–298 [DOI] [PubMed] [Google Scholar]
- 50.Lipari G., Szabo A. (1982) J. Am. Chem. Soc. 104, 4546–4559 [Google Scholar]
- 51.Mandel A. M., Akke M., Palmer A. G., 3rd (1995) J. Mol. Biol. 246, 144–163 [DOI] [PubMed] [Google Scholar]
- 52.Suree N., Yi S. W., Thieu W., Marohn M., Damoiseaux R., Chan A., Jung M. E., Clubb R. T. (2009) Bioorg Med. Chem. 17, 7174–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liew C. K., Smith B. T., Pilpa R., Suree N., Ilangovan U., Connolly K. M., Jung M. E., Clubb R. T. (2004) FEBS Lett. 571, 221–226 [DOI] [PubMed] [Google Scholar]
- 54.Ton-That H., Mazmanian S. K., Faull K. F., Schneewind O. (2000) J. Biol. Chem. 275, 9876–9881 [DOI] [PubMed] [Google Scholar]
- 55.Connolly K. M., Smith B. T., Pilpa R., Ilangovan U., Jung M. E., Clubb R. T. (2003) J. Biol. Chem. 278, 34061–34065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.