Abstract
Various environmental insults result in irreversible damage to proteins and protein complexes. To cope, cells have evolved dedicated protein quality control mechanisms involving molecular chaperones and proteases. Here, we provide both genetic and biochemical evidence that the Lon protease and the SecB and DnaJ/Hsp40 chaperones are involved in the quality control of presecretory proteins in Escherichia coli. We showed that mutations in the lon gene alleviate the cold-sensitive phenotype of a secB mutant. Such suppression was not observed with either clpP or clpQ protease mutants. In comparison to the respective single mutants, the double secB lon mutant strongly accumulates aggregates of SecB substrates at physiological temperatures, suggesting that the chaperone and the protease share substrates. These observations were extended in vitro by showing that the main substrates identified in secB lon aggregates, namely proOmpF and proOmpC, are highly sensitive to specific degradation by Lon. In contrast, both substrates are significantly protected from Lon degradation by SecB. Interestingly, the chaperone DnaJ by itself protects substrates better from Lon degradation than SecB or the complete DnaK/DnaJ/GrpE chaperone machinery. In agreement with this finding, a DnaJ mutant protein that does not functionally interact in vivo with DnaK efficiently suppresses the SecB cold-sensitive phenotype, highlighting the role of DnaJ in assisting presecretory proteins. Taken together, our data suggest that when the Sec secretion pathway is compromised, a pool of presecretory proteins is transiently maintained in a translocation-competent state and, thus, protected from Lon degradation by either the SecB or DnaJ chaperones.
Keywords: Protein Degradation, Protein Export, Protein Folding, Protein Secretion, Protein Targeting, DnaJ, DnaK, Lon, SecB
Introduction
Molecular chaperones comprise a large group of highly conserved proteins that aid protein folding, transit across biological membranes, and quality control as well as assembly and disassembly of protein complexes (1–3). Molecular chaperones act by repeated binding and release of aggregation-sensitive polypeptide segments normally found buried in the final folded structure of a substrate. Although predominantly found during de novo protein synthesis or after a denaturing stress leading to protein misfolding or aggregation, such aggregation-prone segments can also be transiently exposed in native multiprotein complexes. In this case molecular chaperone binding coordinates subtle conformational changes affecting biological functions (4, 5).
In the bacterium Escherichia coli, intracellular protein folding is mainly orchestrated by the chaperones Trigger Factor (TF),2 DnaK/Hsp70, and GroEL/Hsp60. The chaperone TF binds near the ribosomal polypeptide exit with a 1:1 stoichiometry, forming a protective “cradle” for emerging nascent chains. Because of its advantageous localization, TF may interact co-translationally with most nascent polypeptides (6, 7). TF cycles on and off the ribosome, and both ribosome-bound or released polypeptide chains can stay post-translationally bound to ribosome-free TF for a prolonged period (8, 9). The ATP-dependent chaperone DnaK, assisted by its dedicated cochaperones DnaJ and GrpE, provides co- and/or post-translational assistance to newly synthesized proteins released from TF (10–13). In this case the DnaJ cochaperone, which also exhibits bona fide chaperone activity, is thought to selectively bind various substrates, transfer them to DnaK, and concomitantly stimulate DnaK ATPase activity (14). In contrast, the nucleotide exchange factor GrpE stimulates ADP/ATP exchange and subsequent substrate release from DnaK (15). Like DnaK, the GroEL-GroES chaperone machine is also responsible for the post-translational folding of a substantial number of newly synthesized polypeptides inside its ATP-dependent cavity (16). Several essential proteins have been described as obligate GroEL-GroES substrates, in agreement with the observation that GroEL-GroES is the sole chaperone machine essential for E. coli viability (17, 18).
Newly synthesized presecretory proteins are generally targeted post-translationally by the SecB chaperone to the Sec translocon located at the inner membrane (19). SecB is a homotetrameric chaperone of 17-kDa monomers assembled as a dimer of dimers (20). It binds its nonnative substrates either co- or post-translationally with high affinity and without specificity for signal sequence. Productive interaction with SecB is strongly influenced by the folding rate of its substrates (21, 22). SecB transfers each bound protein substrate to the Sec translocon by interacting specifically with the peripheral translocase motor SecA. Substrate transfer to SecA and the subsequent release of SecB activates SecA for further preprotein translocation through the inner membrane (23, 24).
Functional overlaps between SecB and other cytosolic chaperones have been observed. First, SecB overexpression rescues protein aggregation and bacterial growth in the absence of both cytosolic chaperones DnaK and TF (25), and all three chaperones share potential binding sites within their polypeptide substrates (26). Second, both DnaK and GroEL assist protein export in some cases (27, 28). Third, under certain conditions, a strong antagonism has been observed between the two chaperones SecB and TF in vivo. Indeed, the presence of ribosome-bound TF generally retards protein export (29, 30). In agreement with this observation, the protein export defect, the cold-sensitive (Cs) phenotype, and the increased protein aggregation observed in the absence of SecB are fully suppressed by null mutations in the tig gene, encoding TF. In this case more SecA and ribosomes are found associated with the inner membrane, suggesting that TF directly or indirectly interferes with the process of co-translational targeting to the Sec translocon, thus driving presecretory proteins toward a post-translational mode of export that relies on the cytosolic chaperone SecB (30, 31). These observations suggest the existence of an efficient and dedicated cellular mechanism for the elimination of precursor proteins that may accumulate under various stress conditions. In this study we present genetic and biochemical evidence for the existence of such a quality control mechanism in E. coli and present a model to explain our various findings.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Phages, and Culture Conditions
Genetic experiments were carried out in E. coli strain SG20781 (32), MC4100 (33), W3110, or MG1655 (34). The deletion/replacement of the lon gene by a KanR cassette was carried out as described (35) using pKD4 as the DNA template (36). The KanR cassette was removed from the chromosome using FLP recombinase as described (36). The Δlon::KanR and Δlon alleles were confirmed by DNA sequencing and by Western blot analysis using rabbit anti-Lon antibodies (a kind gift of Tomoko Yamamoto, Chiba University). The ΔsecB::CmR (30), ΔdnaKdnaJ::KanR (37), secAcsR11 (38), secY205 (39), and secA51 (40) mutant alleles were described previously. The ΔompF::KanR, ΔclpQ::KanR, and ΔclpP::KanR alleles were from strains JWK0912, JWK3903, and JWK0427, respectively (Keio collection). DNA transformations were carried out in the strain DH5α (Invitrogen). Protein overexpression using the pET expression system was carried out in BL21 (λDE3) derivatives (Novagen). All the mutations described in this study were moved to the appropriate genetic backgrounds by bacteriophage P1-mediated transduction. Bacteria were grown in LB or M9 medium supplemented when necessary with either chloramphenicol (15 μg/ml), kanamycin (40 μg/ml), ampicillin (100 μg/ml), or tetracycline (15 μg/ml) (41). On LB agar plates, bacteria were incubated for 1 day at 30, 33, or 37 °C, for 2 days at 20 or 22 °C, and for 5 or 6 days at 16 °C.
Plasmid Constructs
Plasmids p29SEN, pSE380ΔNcoI (42), and pSE380 (Invitrogen) were described previously. To construct plasmid pSE-Lon (pSE380-Lon), the 2355-bp lon gene was PCR-amplified using primers Lon-ERI-for (5′-gcgaattctatgaatcctgagcgttctgaacg-3′) and Lon-BglII-rev (5′-cgcagatctctattttgcagtcacaacctgcat-3′) and MG1655 genomic DNA as template. The PCR fragment was digested with EcoRI-BglII and cloned into pSE380ΔNcoI, previously digested with the same enzymes. Alanine substitution of serine 679 in Lon was constructed by PCR-based site-directed mutagenesis using appropriate primers and plasmid pSE-Lon as template. The resulting plasmid was named pSE-LonSA. Plasmid pSE-LonHis6 and pSE-LonSAHis6 (Lon and Lon (S679A) with C-terminal His6 tags) were constructed by PCR using primers Lon-ERI-for and Lon-BglII-His6-rev (5′-cgagatctgttaatgatgatgatgatgatggctgctgccttttgcagtcacaacctgcatac-3′) and pSE-Lon or pSE-LonSA as the DNA template. The PCR fragments were digested with SalI and BglII enzymes then ligated into pSE-Lon previously digested with the same enzymes. Expression of the Lon proteins was confirmed by Western blot analysis using anti-Lon antibodies, and the in vivo ability of these proteins to restore ΔsecB cold-sensitivity and inhibit colanic acid synthesis on LB agar plates was confirmed. Plasmids pSE-DnaJ and pSE-DnaJ(H33Q) were constructed by subcloning the NcoI-BglII-digested dnaJ and dnaJ259(H33Q) fragments of pWKG90(dnaJ) and pWKG91(dnaJ259) (43), respectively, into pSE380 previously digested with the same enzymes.
To construct pET15b-OmpF, the entire ompF gene was PCR-amplified from MG1655 genomic DNA using primers OmpF-NdeI-for 5′-cgcacatatgatgaagcgcaatattctggcagtg-3′ and OmpF-HIII/BHI-rev (5′-caagcttggatccttagaactggtaaacgatacccacagc-3′). The PCR fragment was digested with NdeI and BamHI enzymes, then ligated into pET15b previously digested with the same enzymes. A similar cloning strategy was employed for pET15b-OmpC, pET15b-SecB, and pET15b-RpoA using primers OmpC-NdeI-for (5′-cgcacatatgaaagttaaagtactgtccctcctg-3′) and OmpC-HIII/BHI-rev (5′-caagcttggatccttagaactggtaaaccagacccagagc-3′) for pET15b-OmpC, SecB-NdeI-for (5′-cagccatatgtcagaacaaaacaacactg-3′) and SecB-BamHI-rev (5′-gcggatcctcaggcatcctgatgttcttcag-3′) for pET15b-SecB, and RpoA-NdeI-for (5′-cagccatatgcagggttctgtgacagag-3′) and RpoA-BamHI-rev (5′-gcggatccttactcgtcagcgatgcttgc-3′) for pET15b-RpoA.
The plasmid p29-OmpF containing OmpF with a C-terminal myc epitope was constructed by PCR using primers OmpF-ERI/BspHI-for (5′-cggaattctcatgatgaagcgcaatattctggcagtg-3′) and OmpF-myc-HIII/BHI-rev (5′-caagcttggatccttacagatcctcttctgagatgagtttttgttcaccaccgaactggtaaacgatacccacagc-3′) and pET15b-OmpF as template. The EcoRI-HindIII-digested fragment was then ligated into p29SEN previously digested with the same enzymes. All constructs obtained by PCR were verified by sequencing.
Protein Purification
Strain SG20781 Δlon ΔsecB::CmR ΔompF::KanR harboring pSE-LonHis6 or pSE3-LonS679AHis6 was used to purify Lon and Lon(S679A), respectively. To induce Lon(S679A) protein expression, 1 mm isopropyl-β-d-thiogalactopyranoside (IPTG) was added to mid-log phase cultures grown in LB ampicillin at 37 °C. After 2 h of incubation at 37 °C, cells were harvested by centrifugation at 7000 rpm (JLA-16.250 Beckman rotor) for 5 min at 4 °C. To obtain larger quantities of Lon wild type protein, Lon expression was also performed in continuous batch culture. In this case cells were grown until an A600 of 80, and expression was induced with 2 mm IPTG for 2 h at 37 °C. In both cases pellets were then resuspended in buffer A (20 mm Tris-HCl, pH 7.5, 400 mm NaCl, 20% (v/v) glycerol, 1 mm DTT) supplemented with 2 mg/ml lysozyme (Euromedex) and incubated on ice for 1 h. The suspension was sonicated on ice (6 pulses, 20 s each). Cell lysates were clarified by centrifugation at 13,000 rpm (JA-20 Beckman rotor) for 30 min at 4 °C and loaded onto a 2-ml Ni-NTA-agarose column (Qiagen). The column (pre-equilibrated with buffer A supplemented with 20 mm imidazole) was washed with 10 bed volumes of buffer A supplemented with 50 mm imidazole for Lon and 20 mm imidazole for Lon(S679A) following the manufacturer's protocol. Bound proteins were eluted with 300 mm imidazole in buffer A. Proteins were concentrated using Amicon Ultra centrifugal filter devices (Millipore) and stored at −80 °C in buffer containing 20 mm Tris-HCl, pH 7.5, 400 mm NaCl, 1 mm DTT, 20% (v/v) glycerol.
RpoA was purified from strain BL21 (λDE3) harboring plasmid pET15b-RpoA. At mid-log phase, RpoA expression was induced by the addition of 1 mm IPTG. After 2 h at 37 °C, cell lysis and protein purification were carried out as described for Lon(S679A). Purified RpoA used in the degradation assay was stored under denaturing conditions in buffer containing 20 mm Tris-HCl, pH 7.5, 400 mm NaCl, 1 mm DTT, 20% (v/v) glycerol, 8 m urea.
BL21 (λDE3) harboring pET15b-SecBHis6 was used to overexpress SecBHis6. Bacteria were grown at 37 °C in LB ampicillin to mid-log phase, and SecB expression was induced by the addition of 1 mm IPTG. After 2 h at 37 °C, cells were harvested at 7000 rpm (JLA-16.250 Beckman rotor) for 10 min at 4 °C and washed with 20 mm Tris, pH 7.5. Pellets were resuspended in a buffer containing 20 mm Tris, pH 7.5, 2 mm EDTA, 20 mm imidazole, protease inhibitor mixture (Roche Applied Science), 0.35 mg/ml lysozyme and kept on ice for 10 min. MgSO4 (5 mm final concentration) was then added, and samples were kept on ice for an additional 10 min. Samples were sonicated twice for 10 s to reduce viscosity. Lysates were centrifuged at 15,000 rpm for 15 min at 4 °C (JA-25.50 Beckman rotor). Cleared lysates were applied to a 1-ml Ni-NTA agarose column pre-equilibrated at 4 °C with 5 volumes of buffer containing 20 mm Tris, pH 7.5, 800 mm NaCl, 0.1 mm EDTA, 1 mm DTT, 20 mm imidazole, and 20% (v/v) glycerol. The column was washed with 15 ml of the same buffer supplemented with 50 mm imidazole. SecB was eluted with 7 ml of the same buffer containing 300 mm imidazole. Eluted protein was concentrated using a 10-kDa Amicon Ultra centrifugal filter device (Millipore) and resuspended in 20 mm Tris, pH 7.5, 2 mm EDTA, 10% (v/v) glycerol. SecB was further purified by ion exchange chromatography using a Mono Q column (GE Healthcare) and the BioLogic DuoFlow System (Bio-Rad). SecB was eluted with a linear 0–500 mm NaCl gradient in 20 mm Tris, pH 7.4. The peak fractions containing SecB were pooled and concentrated. The protein was stored in 20 mm Tris, pH 7.5, 100 mm NaCl, 1 mm DTT, 20% (v/v) glycerol.
The precursor forms of the OmpF and OmpC porins accumulate as aggregates in the cytosolic fraction of BL21 (λDE3) ΔdnaKdnaJ::KanR at 40 °C because this strain lacks the DnaK/DnaJ chaperones. To purify these precursors, BL21 (λDE3) ΔdnaKdnaJ::KanR harboring the expression vector pET15b-OmpF or pET15b-OmpC, respectively, was grown in 200 ml of LB ampicillin at 30 °C to an A600 of 0.3. The cultures were then transferred to 40 °C for 40 min in the presence of 1 mm IPTG. Cells were centrifuged at 6000 × g for 10 min at 4 °C, and aggregates were isolated as described by Tomoyasu et al. (46) except that the final pellet containing total aggregates was resuspended in 1 ml of buffer containing 100 mm NaH2PO4, 10 mm Tris, pH7.5, 8 m urea. Samples were subsequently applied to a 1-ml Ni-NTA-agarose column and shaken for 1 h at room temperature. The column was washed with 10 ml of the same buffer supplemented with 50 mm imidazole. Proteins were eluted with the same buffer containing 300 mm imidazole. Eluted proteins were precipitated and resuspended in 20 mm Tris, pH 7.5, 200 mm NaCl, 1 mm DTT, 20% (v/v) glycerol, 8 m urea, then stored at −80 °C. The proOmpFΔC-ter construct missing the last 10 amino acids (353TVAVGIVYQF362) was purified as described for proOmpF. DnaK was purified as described by Zylicz et al. (44). GrpE and DnaJ were purchased from StressGen. Protein concentrations were determined by the Bradford assay (Bio-Rad protein assay) using bovine serum albumin as the standard.
Pulse-Chase Analysis
SG20781 ΔsecB or ΔsecB Δlon mutants freshly transformed with plasmid p29-OmpF were grown overnight in M9 minimal medium supplemented with 0.2% glucose, ampicillin, and 0.2% casamino acids at 30 °C. Cultures were diluted to an A600 of ∼0.05 in the same medium and grown to approximately absorbance of 0.3 at 30 °C with aeration. Cultures were centrifuged, and the pellet was resuspended in M9 containing 0.2% glucose, ampicillin, 0.1 mm IPTG, and a methionine/cysteine-free amino acid mix. After 30 min at 30 °C, cells were pulse-labeled for 1 min at 30 °C with 10 μCi/ml of [35S]methionine/cysteine. The chase was initiated by the addition of 2 mm unlabeled methionine/cysteine and immediate transfer to 16 °C for the times indicated. After the chase, cells were precipitated with 10% (w/v) trichloroacetic acid on ice. Samples were analyzed by immunoprecipitation with anti-OmpF antibodies as described (45). Radio-labeled gels were scanned by Fuji FLA-3000 phosphorimaging.
Isolation and Separation of Protein Aggregates
Overnight cultures were diluted 1/50 in 200 ml of fresh LB medium and grown at the indicated temperature to an A600 of ∼1.0. Aggregates were isolated using the method developed by Tomoyasu et al. (46) except that all volumes were increased 10-fold (30). After separation by 12% SDS-PAGE, proteins were visualized by Coomassie Blue staining. For two-dimensional polyacrylamide gel electrophoresis analyses, aggregates were isolated from 200 ml of cultures and resuspended in 75 μl of phosphate buffer, pH 6.5, containing 1 mm EDTA. An equal volume of IDSL buffer (7 m urea, 2 m thiourea, 1% CHAPS, 0.8% ASB-14, 1.6% ampholines 5/7 (Bio-Rad 163–1152), 0.4% ampholines 3.5–10 (Bio-Rad 163-1112)) was added, and samples were incubated at 20 °C for 45 min. Isoelectric focusing was subsequently performed using the Tube Cell Model 175 (Bio-Rad) for 18 h at 500 V. Proteins were separated in the second dimension by 12.5% SDS-PAGE, stained with Coomassie Blue, excised from the gel, and identified by peptide mass fingerprinting as described (47).
In Vitro Degradation Assay
Degradation assays were performed in 50-μl reactions containing 25 mm HEPES, pH 7.5, 1 mm DTT, 10 mm MgCl2, and 50 mm KCl with or without SecB or DnaJ chaperones. Reactions were kept on ice for 10 min and transferred at 37 °C for 5 min. Protein substrates proOmpF, proOmpC, or RpoA were denatured in the presence of 8 m urea at 95 °C for 15 min and subsequently added to the reaction at a final concentration of 0.3 μm. Degradation was then initiated by addition of 10 mm ATP (GE Healthcare) and 0.16 μm Lon hexamers. Reactions were kept at 37 °C for the times indicated. At each time point, a 15-μl aliquot was transferred to Laemmli SDS loading buffer (4×) supplemented with 25 mm EDTA to stop the reaction. Proteins separated on 4–12% NUPAGE gels (Invitrogen) were transferred to nitrocellulose membrane (Invitrogen, iBlot Transfer Stack) and probed with appropriate antibodies and developed by enhanced chemiluminescence using goat anti-rabbit or anti-mouse horseradish peroxidase-conjugated IgG as secondary antibodies (Amersham Biosciences). Intensity of the bands was quantified using OptiQuant software.
RESULTS
Mutations in lon Suppress the ΔsecB Cold Sensitivity
A ΔsecB mutant is Cs for growth below 23 °C, mostly due to its defect in protein export (30, 48). Using random transposon mutagenesis, we previously isolated recessive mutations that suppress the secB Cs phenotype (30). Among these, mutations in the tig gene encoding the chaperone TF were identified, and the mechanism of suppression of the Cs phenotype was characterized (30). Through the same genetic selection, we found that 60% of total mutations isolated were either localized in the lon structural gene encoding the AAA+ stress protease Lon (49–54) or affected lon expression. To ensure that suppression of the ΔsecB Cs phenotype is indeed due to the absence of Lon, we engineered an in-frame deletion of the entire lon gene and confirmed the cold-resistant phenotype of the resulting double Δlon ΔsecB mutant (Fig. 1A). Suppression of the ΔsecB Cs phenotype by the deletion lon mutation was observed in all E. coli strain backgrounds tested, namely SG20781, MC4100, MG1655, and W3110. Furthermore, introduction of a plasmid encoding Lon fully restores the ΔsecB Cs phenotype, proving that the observed suppression is indeed Lon-dependent (Fig. 1B).
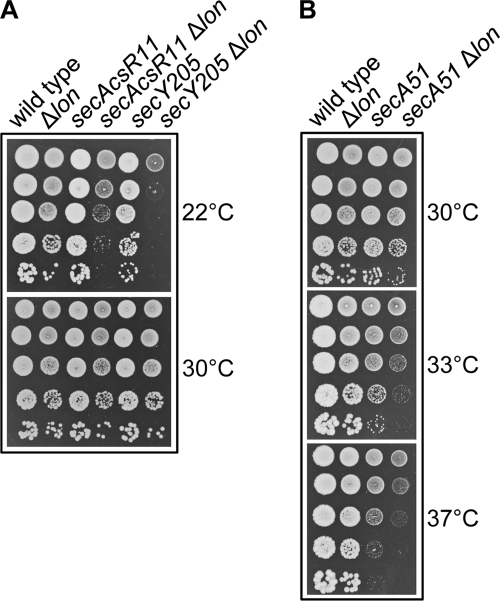
FIGURE 1.
Mutations in lon suppress the ΔsecB Cs phenotype. A, the wild type strain SG20781 and its isogenic mutant derivatives were grown to mid-log phase, serially diluted 10-fold, spotted on LB agar plates, and incubated at the indicated temperatures. Note that the Δlon and ΔsecB Δlon grow equally well at 16 °C. B, the strain SG20781 ΔsecB Δlon transformed with either the pSE380 vector, pSE-Lon, or pSE-Lon(S679A) was spotted on LB-ampicillin agar plates supplemented with 10 μm IPTG inducer and incubated at the indicated temperatures. After growth at 37 °C and a subsequent 2-h induction with 10 μm IPTG, expression of pSE-encoded Lon was measured by Western blot using anti-Lon antibodies. C, shown is growth of the strain SG20781 strains and its various isogenic single and double protease chaperone mutants as described above in A.
In the course of the complementation experiments we noticed that at 37 °C Lon overexpression is toxic in the ΔsecB mutant but not in the wild type parental strain (supplemental Fig. S1), indicating that the detrimental effect of Lon in the absence of the SecB chaperone is not solely cold-dependent. In addition to the suppression of the ΔsecB Cs phenotype by a Δlon mutation, we found that Δlon single and Δlon ΔsecB double mutants grow identically to each other but substantially slower than the wild type parental strain at low temperature (compare colony sizes in Fig. 1A). The reason for such a SecB-independent, slow-growing phenotype is unknown.
Previous work has shown that a mutation in tig also suppresses the growth defect of strains with mutations in the secA and secY genes of the Sec translocon (30). Such suppression is in agreement with the increase in co-translational translocation of SecB/SecA substrates due to the absence of ribosome-bound TF (30). We next tested whether known temperature- and cold-sensitive sec mutations (secAR11 (E276A), secA51 (L43P), and secY205 (Y429D)) that directly affect the Sec translocase at the inner membrane are also sensitive to the presence of a lon mutation (Fig. 2). Strikingly, in all cases tested, lon acts synergistically with the secA or secY genes, indicating that, as opposed to its detrimental role in the absence of cytosolic SecB, Lon function becomes critical when the downstream Sec-dependent translocation process is compromised.
FIGURE 2.
Lon is required when the Sec translocon is compromised. The Δlon::KanR mutant allele was introduced by bacteriophage P1-mediated transduction into the cold-sensitive mutant alleles secAcsR11 and secY205 (A) or into the temperature-sensitive mutant allele secA51 (B). Transductants were tested for their ability to grow on LB agar plates at the indicated temperatures. The representative set of results presented was obtained in the SG20781 strain background.
The Deleterious Effect of Lon Is Specific and Protease-dependent
To test whether the detrimental role of Lon in the absence of the SecB chaperone is due to its protease activity and not to substrate sequestration, plasmid pSE-LonSA, carrying the catalytic site mutation Ser-679 to Ala that abolishes Lon protease activity (55), was engineered and tested for its effect on growth of the ΔsecB mutant at low temperature. As shown in Fig. 1B, the plasmid-encoded Lon (S679A) is not capable of restoring the ΔsecB Cs phenotype, indicating that the deleterious effect of Lon in the ΔsecB mutant is due to its protease activity. To examine whether suppression is Lon-specific, we tested the effect of the stress proteases ClpP and ClpQ on ΔsecB growth at low temperature. The results presented in Fig. 1C clearly show that neither the ΔclpP nor ΔclpQ mutation restores the ΔsecB Cs phenotype even under less stringent temperatures of growth (i.e. 20 °C). However, we did notice that the ΔclpP mutation by itself exhibits a mild Cs phenotype, indicating that the ClpP protease may also play some role at low temperature.
The secB lon Double Mutant Accumulates Aggregated Proteins
As stated above, mutations in either the lon or tig genes were isolated as the main suppressors of the SecB-dependent Cs phenotype (Ref. 30 and this work). The fact that TF is a chaperone and Lon a protease suggests different modes of suppression. Although suppression by the tig mutation is due to a significant acceleration in protein export, the absence of Lon may increase the half-life of some essential pre-secretory substrates, thus allowing a delayed but functional SecB-independent interaction with SecA for further translocation (56). Such a mechanism predicts that SecB substrates accumulate in the absence of Lon. As shown in Fig. 3A, we found that the combined absence of Lon and SecB does in fact lead to a significant increase in protein aggregation at 37 °C. In contrast, null mutations in either clpP or clpQ exert no major effect (supplemental Fig. S2).
FIGURE 3.
The double ΔsecB Δlon mutant accumulates protein aggregates. A, protein aggregation was observed at 37 °C in strain SG20781 and its various isogenic mutant derivatives. Total cell extracts and their corresponding protein aggregates are shown. B, two-dimensional gel analysis of the aggregates isolated from the ΔsecB Δlon double mutant is shown. Exported proteins are depicted with an asterisk. UniProt references of the identified protein species are shown in supplemental Table S1.
The aggregated proteins from the double ΔsecB Δlon mutant were resolved on two-dimensional gels, and the major protein species were identified by mass spectrometry (Fig. 3B). Not surprisingly, the most abundant aggregated species were proteins destined for export (marked with asterisks). Interestingly, a significant number of identified preproteins were not detected in a previous study with a ΔsecB strain (Ref. 57; supplemental Table S1). These data taken together suggest that indeed, a significant number of SecB substrates are also bona fide Lon substrates.
Lon-dependent Degradation of SecB Substrates in Vivo and in Vitro
The results presented above suggest that Lon and SecB share some substrates and that the SecB chaperone protects them from Lon degradation at both high and low temperatures. Protection of substrates in the cytosolic compartment may be crucial when Sec-dependent protein translocation across the inner membrane is impaired, slowing down transport across the membrane. We first examined whether export and/or accumulation of proOmpF, a major SecB substrate that aggregates in the ΔsecB Δlon strain (Fig. 3B), is affected by Lon in vivo. To do so we performed pulse-chase experiments in both a ΔsecB single and a ΔsecB Δlon double mutant expressing proOmpF from a plasmid. The results presented in Fig. 4A clearly show that 35S-labeled proOmpF is more rapidly degraded in the lon+ background. Taken together, these in vivo pulse-chase experiments confirm the importance of Lon in the degradation of SecB substrates in response to export stress.
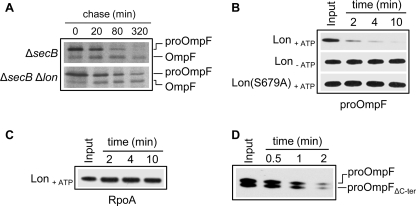
FIGURE 4.
ProOmpF is efficiently degraded by Lon both in vivo and in vitro. A, pulse-chase analysis shows the processing of 35S-labeled proOmpF expressed in SG20781 ΔsecB or ΔsecB Δlon isogenic mutants harboring plasmid p29-OmpF, grown and labeled at 30 °C, and chased for various times after incubation at 16 °C. B, in vitro degradation of urea-denatured proOmpF (0.3 μm) by Lon6 (0.16 μm) in the presence of ATP and in the absence of ATP (Lon-ATP) or by Lon6 (S679A) (0.16 μm) in the presence of ATP. C, shown are in vitro degradation assays of urea-denatured RpoA (0.3 μm) in the presence of Lon6 (0.16 μm) and ATP under the same conditions as those for proOmpF in B. D, degradation of proOmpF and proOmpFΔC-ter missing the last 10 amino acids (353TVAVGIVYQF362) by Lon6 (0.16 μm) is shown. For the assay both proOmpF (0.15 μm) and proOmpF ΔC-ter (0.15 μm) were added to the same reaction mix.
To provide direct evidence for a specific role of Lon in the quality control of presecretory proteins, we examined the affinity of Lon toward SecB substrates in vitro. The Lon protease and the SecB substrate proOmpF were purified, and Lon degradation was monitored in the presence of ATP (Fig. 4B). As a negative control, the cytosolic RpoA protein (the 40-kDa RNA polymerase α-subunit) was also purified and tested under the same conditions. Strikingly, within 2 min of incubation, more than 90% of proOmpF was degraded in the presence of Lon. The observed degradation was Lon-dependent as no degradation of proOmpF occurred in the absence of ATP or in the presence of the purified inactive Lon (S679A) mutant (Fig. 4B). In sharp contrast, the cytosolic protein RpoA did not show significant sensitivity to Lon degradation under the same conditions (Fig. 4C), indicating a Lon direct and specific role in the degradation of bona fide SecB substrates both in vivo and in vitro. The fact that Lon rapidly and efficiently degrades unfolded proOmpF suggests that in vivo Lon may recognize newly translated proOmpF before it aggregates. Note that the extreme C-terminal motif present in most porins, which serves as a specific recognition motif for either the DegS protease involved in the initiation of the cellular response to protein misfolding in the periplasm (58), or the BAM complex (the β-barrel assembly machinery) at the outer membrane (59), can be removed without affecting the sensitivity of proOmpF to Lon in vitro (Fig. 4D). This demonstrates that the motif is not responsible for degradation by Lon.
SecB Protects Its Substrates from Lon Degradation in Vitro
The results presented above suggest that the SecB chaperone protects its substrates from Lon degradation, thus favoring substrate transfer to the Sec translocon. To provide direct evidence for such a mechanism, purified proOmpF was added to a reaction containing both Lon and SecB, and degradation was monitored as before. Clearly, the presence of SecB significantly delays proOmpF degradation by Lon (Fig. 5A). The ratio of active protein species present in this assay is 3:1:0.5 of tetrameric SecB, monomeric proOmpF, and hexameric Lon, respectively. No significant protection by SecB was observed below a 1:1 ratio of SecB and proOmpF under the same conditions.3 As a control, the addition of GrpE at the same concentration does not significantly delay degradation (Fig. 5B). Analogous protection by SecB was obtained with proOmpC substrate (Fig. 5C). These results demonstrate that SecB can transiently protect its substrates from Lon degradation in a window of time that enables efficient substrate transfer to the Sec machinery.
FIGURE 5.
SecB protects proOmpF and proOmpC from degradation by Lon in vitro. A, urea-denatured proOmpF (0.3 μm) was incubated with or without SecB4 (1 μm) in the presence of Lon6 (0.16 μm) and ATP as described under “Experimental Procedures.” Lon degradation of proOmpF was monitored after 2, 4, and 10 min of incubation, and the percentage of remaining proOmpF was quantified using OptiQuant software. B, degradation of denatured proOmpF (0.3 μm) by Lon6 (0.16 μm) alone or in the presence of GrpE2 (2 μm) or SecB4 (1 μm) is shown. C, degradation of denatured proOmpC (0.3 μm) by Lon6 (0.16 μm) alone or in the presence of SecB4 (1 μm) is shown.
DnaJ Also Rescues SecB Substrates from Lon Degradation
It has been suggested by several laboratories that the DnaK/DnaJ/GrpE chaperone machine serves as a backup system for SecB function (27, 30, 60, 61). Specifically, the fact that both secB dnaK and secB dnaJ double mutations are synthetically lethal under certain conditions suggested that DnaK and DnaJ are capable of protecting SecB substrates from Lon degradation (30). To investigate this possibility, purified proOmpF was added to a reaction containing both Lon and DnaJ in the presence of ATP. We found that the DnaJ protein alone is capable of completely protecting proOmpF from Lon degradation (Fig. 6, A and B). After a 2-min incubation with DnaJ and Lon, both the DnaK chaperone and its nucleotide exchange factor GrpE were added. Surprisingly, degradation by Lon occurred rapidly after the addition of DnaK and GrpE. These results indicate that proOmpF is maintained by DnaJ in a form compatible for efficient transfer to DnaK. The fact that DnaJ alone protects proOmpF from Lon-mediated degradation suggests that under conditions of stress, SecB and DnaJ help maintain a pool of presecretory proteins in an export competent form, protected from Lon degradation, thus allowing rapid rebooting of the export machinery when permissive conditions return. Whether substrate delivery to the Sec translocon by DnaJ is direct or whether it first necessitates transfer to DnaK remains unknown.
FIGURE 6.
DnaJ efficiently protects a SecB substrate from Lon degradation. A, Lon6 (0.16 μm) degradation of proOmpF (0.3 μm) in the absence of DnaJ (−), in the presence of DnaJ2 (0.2 μm) alone, or in the presence of DnaJ2 (0.2 μm), DnaK (1 μm), and GrpE2 (0.2 μm) is shown. As indicated with an arrow, DnaK and GrpE were added after 2 min of incubation. Lon degradation of proOmpF was monitored after 2, 4, and 10 min of incubation. B, the percentage of remaining proOmpF was quantified using OptiQuant software. C, in vivo complementation of the ΔsecB Cs phenotype by plasmid-encoded wild type DnaJ or mutant DnaJ(H33Q) is shown. Mid-log phase cultures of SG20781 ΔsecB transformed with either the pSE380 vector, pSE-DnaJ, or pSE-DnaJ(H33Q) were spotted on LB-ampicillin agar plates without IPTG or with 10 μm or 50 μm IPTG inducer and incubated at the indicated temperature for 1 day at 37 °C, 2 days at 20 °C, and 5 days at 16 °C.
In separate in vivo experiments, we found that overexpression of DnaJ(H33Q), a DnaJ mutant carrying the well characterized His-33 to Gln substitution in its J-domain that abolishes all of its DnaK cochaperone functions without affecting its substrate binding capacities (62, 63), efficiently supports ΔsecB mutant growth at 16 °C (Fig. 6C). Notably, suppression by DnaJ (H33Q) is even more robust than that exhibited by SecA (30). Surprisingly, the overexpression of wild type DnaJ in this mutant results in an anomalous behavior. Specifically, wild type DnaJ overexpression is toxic at the normally permissive temperature of 37 °C and weakly supports ΔsecB growth at the less stringent temperature of 20 °C but does not support its growth at 16 °C (Fig. 6C and Ref. 12). Under these conditions, steady state expression levels of DnaJ and DnaJ (H33Q) are comparable.4 These data further suggest that DnaJ chaperone function is crucial for the protection of presecretory proteins from Lon degradation both in vivo and in vitro.
DISCUSSION
Intracellular protein quality control is performed by finely tuned networks of molecular chaperone and protease machines (2, 46, 65). Here, we have dissected the roles of the chaperone SecB and the ATP-dependent protease Lon in this process, finding that they represent a dedicated system for the quality control of presecretory proteins in E. coli. Specifically, we showed that mutations in lon, but not in the clpP or clpQ protease-encoding genes, efficiently suppress all growth defects of a secB null strain. Nevertheless, the double secB lon mutant exhibits increased levels of aggregated presecretory proteins when compared with the respective single mutants at the permissive temperature of 37 °C. In addition, the Lon protease becomes essential when the downstream Sec translocase is affected, further underscoring the significance of Lon in presecretory protein quality control. The major SecB-Lon aggregation-sensitive substrates, namely the precursor forms of the E. coli porins OmpF and OmpC, are highly sensitive to Lon degradation in vitro, in contrast to the cytosolic protein RpoA under the same conditions. The addition of SecB to these in vitro reactions transiently protects such Lon-sensitive presecretory substrates, indicating a significant overlap in substrate recognition between the chaperone and protease. In addition, we showed that DnaJ alone provides more robust protection of proOmpF from Lon degradation than either SecB or the complete DnaK/DnaJ/GrpE chaperone machine.
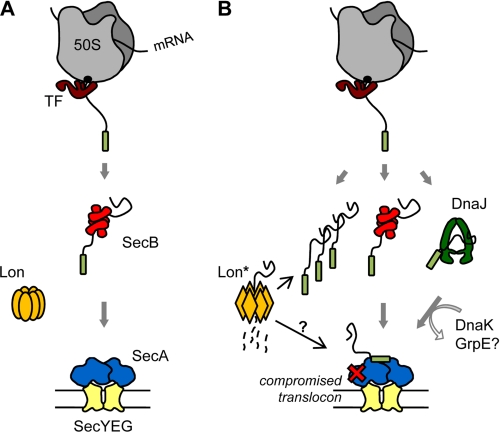
To account for these results, we propose a model for presecretory protein quality control in E. coli that includes key roles for the chaperones SecB and DnaJ and the Lon protease (Fig. 7). Under normal growth conditions (Fig. 7A), newly synthesized presecretory polypeptides can interact co- and/or post-translationally with SecB. In this case SecB binds its various substrates rapidly and with high affinity, thus preventing their premature folding (19, 21, 22). Although SecB exhibits no specificity for signal sequence, peptide scan library analysis revealed that SecB binding motifs in protein substrates are enriched in both aromatic and basic residues, with acidic residues disfavored (26). Our in vivo and in vitro data suggest that bona fide Lon “recognition tags” (67) in newly translated presecretory proteins are efficiently masked by SecB (and most likely by DnaJ), protecting the substrate from Lon degradation so substrate transfer from SecB to SecA can occur. Such a hypothesis is supported by recent work showing that the Lon protease does not recognize unfolded proteins per se but instead binds to specific amino acid clusters containing five to seven hydrophobic residues without preference for specific localization within the primary amino acid sequence (67, 68). Interestingly, the basic rules for Lon recognition (i.e. hydrophobic tags with a preference for aromatic residues, basic residues frequently present and acidic residues absent from the core region) share significant similarities with the proposed SecB substrate binding motifs (26).
FIGURE 7.
Proposed model for a presecretory protein quality control network involving the Lon protease and both chaperones SecB and DnaJ. See discussion for details.
Although severely impaired for many proteins, export can nevertheless occur without the assistance of SecB (30, 56, 66, 69). In this case the targeting of some presecretory proteins is probably assisted by other chaperones such as DnaJ, DnaK, or TF (30, 60, 61, 70), by SecA (31), or by signal recognition particle (71). Alternatively, presecretory proteins that are poor Lon substrates could be post-translationally targeted without any such chaperone assistance (56).
Upon secretion stress (Fig. 7B), as exemplified by a temperature downshift or mutation in the Sec translocon (i.e. secA or secY temperature-sensitive alleles), the fraction of newly translated unfolded precursors released in the cytosol rapidly increases, overpowering the capacity of SecB and leading to a Cs phenotype. In this case increasing concentration of exposed, chaperone-free Lon-sensitive segments in unfolded precursor proteins may cooperatively activate Lon (depicted as Lon* in Fig. 7A) as recently proposed (67), resulting in efficient degradation of unprotected substrates before they aggregate. Presumably, any amino acid substitution in a precursor protein that affects its binding to SecB would also channel it into the same degradation pathway (72), thus protecting the cell from potential harm. Meanwhile, a pool of translocation-competent presecretory proteins would be protected by SecB as well as additional chaperones, waiting for stress conditions to subside.
The role of DnaJ in protein quality control has been demonstrated in vitro by its robust protection of proOmpF from Lon degradation in a DnaK-independent manner and in vivo by the successful complementation of the ΔsecB Cs phenotype by the DnaJ (H33Q) cochaperone mutant, which has lost its ability to interact with DnaK. The fact that DnaJ (H33Q) outcompetes wild type DnaJ for complementation coupled with the fact that high expression of wild type DnaJ exhibits a toxic effect on secB mutant growth suggests that overstimulation of the DnaK machine is potentially deleterious in the absence of SecB, perhaps by accelerating substrate release and, thus, making substrate more accessible to Lon-mediated proteolysis. In contrast with DnaK (68, 73), the DnaJ and Lon substrate recognition sites may significantly overlap (74). The known synthetic lethality between secB and dnaJ mutations in the presence of TF supports such a central role for DnaJ in export quality control (27, 30).
In agreement with our proposed model, the presence of Lon is crucial when the downstream Sec translocon at the inner membrane is compromised (Fig. 2). In this case Lon activation may be required in response to the accumulation of presecretory proteins, thus preventing potential clogging of the Sec translocon. In support of the model, the previous work of Snyder and Silhavy (75) showed that protein export defects induced by LamB-LacZ hybrid protein overexpression (hybrid jamming) are relieved by the prlF1 mutation in a Lon-dependent manner. In this case the prlF1 mutation increases Lon activity and facilitates export under hybrid jamming conditions in vivo (75). How the prlF1 mutation results in Lon stimulation remains unknown. Previous work suggests that mis-localized inner membrane proteins, which are targeted co-translationally to the Sec translocon by the ribosome-bound signal recognition particle SRP (76), may follow comparable recognition/degradation pathways. Indeed, Bernstein and Hyndman (77) showed that the presence of both cytosolic proteases Lon and ClpYQ facilitates growth of SRP-depleted E. coli cells. These data support a major role for Lon in the quality control of Sec-dependent translocation for both the SecA/SecB and SRP pathways. Whether Lon-mediated protein quality control takes place directly at the inner membrane is yet to be determined. Upon return to normal growth conditions, translocation resumes, and the pool of protected SecB- or DnaJ-bound presecretory substrates can be rapidly delivered to SecA for translocation through the inner membrane. Consequently, the fraction of chaperone-free Lon substrates decreases and Lon returns to an inactive mode (67).
Intriguingly, two independent global interactome studies performed in E. coli have revealed unexpected physical interactions between Lon and SecB in vivo (64, 78). Taken together with the results presented in this study, these data suggest that a dedicated chaperone-protease network for presecretory protein quality control may exist as a complex in E. coli.
Supplementary Material
Acknowledgments
We thank Drs. Joen Luirink, Koreaki Ito, Dominique Belin, Jon Beckwith, and Tomoko Yamamoto for the kind gifts of strains and/or antibodies and Dr. Debbie Ang for critical reading of the manuscript.
This work was supported by a CNRS ATIP grant (to P. G.) and by Fond National Swiss Grant FN-3100A0-114063 (to C. G. and P. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
S. Sakr, unpublished results.
P. Genevaux, unpublished results.
- TF
- Trigger Factor
- Cs
- cold-sensitive
- IPTG
- isopropyl-β-d-thiogalactopyranoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- DTT
- dithiothreitol
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Sharma S. K., Christen P., Goloubinoff P. (2009) Curr. Protein Pept. Sci. 10, 432–446 [DOI] [PubMed] [Google Scholar]
- 2.Bukau B., Weissman J., Horwich A. (2006) Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 3.Hartl F. U., Hayer-Hartl M. (2002) Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 4.Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 5.Ellis J. (1987) Nature 328, 378–379 [DOI] [PubMed] [Google Scholar]
- 6.Lill R., Crooke E., Guthrie B., Wickner W. (1988) Cell 54, 1013–1018 [DOI] [PubMed] [Google Scholar]
- 7.Ferbitz L., Maier T., Patzelt H., Bukau B., Deuerling E., Ban N. (2004) Nature 431, 590–596 [DOI] [PubMed] [Google Scholar]
- 8.Kaiser C. M., Chang H. C., Agashe V. R., Lakshmipathy S. K., Etchells S. A., Hayer-Hartl M., Hartl F. U., Barral J. M. (2006) Nature 444, 455–460 [DOI] [PubMed] [Google Scholar]
- 9.Merz F., Boehringer D., Schaffitzel C., Preissler S., Hoffmann A., Maier T., Rutkowska A., Lozza J., Ban N., Bukau B., Deuerling E. (2008) EMBO J. 27, 1622–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teter S. A., Houry W. A., Ang D., Tradler T., Rockabrand D., Fischer G., Blum P., Georgopoulos C., Hartl F. U. (1999) Cell 97, 755–765 [DOI] [PubMed] [Google Scholar]
- 11.Kramer G., Boehringer D., Ban N., Bukau B. (2009) Nat. Struct. Mol. Biol. 16, 589–597 [DOI] [PubMed] [Google Scholar]
- 12.Genevaux P., Georgopoulos C., Kelley W. L. (2007) Mol. Microbiol. 66, 840–857 [DOI] [PubMed] [Google Scholar]
- 13.Deuerling E., Schulze-Specking A., Tomoyasu T., Mogk A., Bukau B. (1999) Nature 400, 693–696 [DOI] [PubMed] [Google Scholar]
- 14.Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegenthaler R. K., Christen P. (2006) J. Biol. Chem. 281, 34448–34456 [DOI] [PubMed] [Google Scholar]
- 16.Chapman E., Farr G. W., Usaite R., Furtak K., Fenton W. A., Chaudhuri T. K., Hondorp E. R., Matthews R. G., Wolf S. G., Yates J. R., Pypaert M., Horwich A. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. (2005) Cell 122, 209–220 [DOI] [PubMed] [Google Scholar]
- 18.Fayet O., Ziegelhoffer T., Georgopoulos C. (1989) J. Bacteriol. 171, 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randall L. L., Hardy S. J. S. (2002) Cell. Mol. Life Sci. 59, 1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekker C., de Kruijff B., Gros P. (2003) J. Struct. Biol. 144, 313–319 [DOI] [PubMed] [Google Scholar]
- 21.Diamond D. L., Randall L. L. (1997) J. Biol. Chem. 272, 28994–28998 [DOI] [PubMed] [Google Scholar]
- 22.Lilly A. A., Crane J. M., Randall L. L. (2009) Protein Sci. 18, 1860–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartl F. U., Lecker S., Schiebel E., Hendrick J. P., Wickner W. (1990) Cell 63, 269–279 [DOI] [PubMed] [Google Scholar]
- 24.Driessen A. J., Nouwen N. (2008) Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 25.Ullers R. S., Luirink J., Harms N., Schwager F., Georgopoulos C., Genevaux P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7583–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoblauch N. T., Rüdiger S., Schönfeld H. J., Driessen A. J., Schneider-Mergener J., Bukau B. (1999) J. Biol. Chem. 274, 34219–34225 [DOI] [PubMed] [Google Scholar]
- 27.Wild J., Altman E., Yura T., Gross C. A. (1992) Genes Dev. 6, 1165–1172 [DOI] [PubMed] [Google Scholar]
- 28.Bochkareva E., Seluanov A., Bibi E., Girshovich A. (1996) J. Biol. Chem. 271, 22256–22261 [DOI] [PubMed] [Google Scholar]
- 29.Lee H. C., Bernstein H. D. (2002) J. Biol. Chem. 277, 43527–43535 [DOI] [PubMed] [Google Scholar]
- 30.Ullers R. S., Ang D., Schwager F., Georgopoulos C., Genevaux P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karamyshev A. L., Johnson A. E. (2005) J. Biol. Chem. 280, 37930–37940 [DOI] [PubMed] [Google Scholar]
- 32.Painbéni E., Mouray E., Gottesman S., Rouvière-Yaniv J. (1993) J. Mol. Biol. 234, 1021–1037 [DOI] [PubMed] [Google Scholar]
- 33.Casadaban M. J. (1976) J. Mol. Biol. 104, 541–555 [DOI] [PubMed] [Google Scholar]
- 34.Bachmann B. J. (1972) Bacteriol. Rev. 36, 525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., Court D. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang P. J., Craig E. A. (1990) J. Bacteriol. 172, 2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riggs P. D., Derman A. I., Beckwith J. (1988) Genetics 118, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimohata N., Nagamori S., Akiyama Y., Kaback H. R., Ito K. (2007) J. Cell Biol. 176, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliver D. B., Beckwith J. (1981) Cell 25, 765–772 [DOI] [PubMed] [Google Scholar]
- 41.Miller J. H. (1992) A Short Course in Bacterial Genetics (Cold Spring Harbor Lab., Cold Spring Harbor, NY: ) [Google Scholar]
- 42.Genevaux P., Keppel F., Schwager F., Langendijk-Genevaux P. S., Hartl F. U., Georgopoulos C. (2004) EMBO Rep. 5, 195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley W. L., Georgopoulos C. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zylicz M., Ang D., Georgopoulos C. (1987) J. Biol. Chem. 262, 17437–17442 [PubMed] [Google Scholar]
- 45.Jong W. S., ten Hagen-Jongman C. M., Genevaux P., Brunner J., Oudega B., Luirink J. (2004) Eur. J. Biochem. 271, 4779–4787 [DOI] [PubMed] [Google Scholar]
- 46.Tomoyasu T., Mogk A., Langen H., Goloubinoff P., Bukau B. (2001) Mol. Microbiol. 40, 397–413 [DOI] [PubMed] [Google Scholar]
- 47.Scherl A., Couté Y., Déon C., Callé A., Kindbeiter K., Sanchez J. C., Greco A., Hochstrasser D., Diaz J. J. (2002) Mol. Biol. Cell 13, 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumamoto C. A., Beckwith J. (1983) J. Bacteriol. 154, 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotanova T. V., Botos I., Melnikov E. E., Rasulova F., Gustchina A., Maurizi M. R., Wlodawer A. (2006) Protein Sci. 15, 1815–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker T. A., Sauer R. T. (2006) Trends Biochem. Sci. 31, 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsilibaris V., Maenhaut-Michel G., Van Melderen L. (2006) Res. Microbiol. 157, 701–713 [DOI] [PubMed] [Google Scholar]
- 52.Gottesman S. (2003) Annu. Rev. Cell Dev. Biol. 19, 565–587 [DOI] [PubMed] [Google Scholar]
- 53.Howard-Flanders P., Simson E., Theriot I. (1964) Genetics 49, 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung C. H., Goldberg A. L. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 4931–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Melderen L., Gottesman S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6064–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gouridis G., Karamanou S., Gelis I., Kalodimos C. G., Economou A. (2009) Nature 462, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baars L., Ytterberg A. J., Drew D., Wagner S., Thilo C., van Wijk K. J., de Gier J. W. (2006) J. Biol. Chem. 281, 10024–10034 [DOI] [PubMed] [Google Scholar]
- 58.Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T. (2003) Cell 113, 61–71 [DOI] [PubMed] [Google Scholar]
- 59.Robert V., Volokhina E. B., Senf F., Bos M. P., Van Gelder P., Tommassen J. (2006) PLoS Biol. 4, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altman E., Kumamoto C. A., Emr S. D. (1991) EMBO J. 10, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi H. Y., Hyndman J. B., Bernstein H. D. (2002) J. Biol. Chem. 277, 51077–51083 [DOI] [PubMed] [Google Scholar]
- 62.Tsai J., Douglas M. G. (1996) J. Biol. Chem. 271, 9347–9354 [DOI] [PubMed] [Google Scholar]
- 63.Wall D., Zylicz M., Georgopoulos C. (1994) J. Biol. Chem. 269, 5446–5451 [PubMed] [Google Scholar]
- 64.Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 65.Gottesman S., Wickner S., Maurizi M. R. (1997) Genes Dev. 11, 815–823 [DOI] [PubMed] [Google Scholar]
- 66.van der Sluis E. O., Driessen A. J. (2006) Trends Microbiol. 14, 105–108 [DOI] [PubMed] [Google Scholar]
- 67.Gur E., Sauer R. T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18503–18508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gur E., Sauer R. T. (2008) Genes Dev. 22, 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumamoto C. A., Beckwith J. (1985) J. Bacteriol. 163, 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crooke E., Wickner W. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5216–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schierle C. F., Berkmen M., Huber D., Kumamoto C., Boyd D., Beckwith J. (2003) J. Bacteriol. 185, 5706–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei S. Q., Stader J. (1994) J. Biol. Chem. 269, 1648–1653 [PubMed] [Google Scholar]
- 73.Rüdiger S., Germeroth L., Schneider-Mergener J., Bukau B. (1997) EMBO J. 16, 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jubete Y., Maurizi M. R., Gottesman S. (1996) J. Biol. Chem. 271, 30798–30803 [DOI] [PubMed] [Google Scholar]
- 75.Snyder W. B., Silhavy T. J. (1992) J. Bacteriol. 174, 5661–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luirink J., von Heijne G., Houben E., de Gier J. W. (2005) Annu. Rev. Microbiol. 59, 329–355 [DOI] [PubMed] [Google Scholar]
- 77.Bernstein H. D., Hyndman J. B. (2001) J. Bacteriol. 183, 2187–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arifuzzaman M., Maeda M., Itoh A., Nishikata K., Takita C., Saito R., Ara T., Nakahigashi K., Huang H. C., Hirai A., Tsuzuki K., Nakamura S., Altaf-Ul-Amin M., Oshima T., Baba T., Yamamoto N., Kawamura T., Ioka-Nakamichi T., Kitagawa M., Tomita M., Kanaya S., Wada C., Mori H. (2006) Genome Res. 16, 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.