Abstract
Fibronectin, a 250-kDa eukaryotic extracellular matrix protein containing an RGD motif plays crucial roles in cell-cell communication, development, tissue homeostasis, and disease development. The highly complex fibrillar fibronectin meshwork orchestrates the functions of other extracellular matrix proteins, promoting cell adhesion, migration, and intracellular signaling. Here, we demonstrate that CagL, a 26-kDa protein of the gastric pathogen and type I carcinogen Helicobacter pylori, mimics fibronectin in various cellular functions. Like fibronectin, CagL contains a RGD motif and is located on the surface of the bacterial type IV secretion pili as previously shown. CagL binds to the integrin receptor α5β1 and mediates the injection of virulence factors into host target cells. We show that purified CagL alone can directly trigger intracellular signaling pathways upon contact with mammalian cells and can complement the spreading defect of fibronectin−/− knock-out cells in vitro. During interaction with various human and mouse cell lines, CagL mimics fibronectin in triggering cell spreading, focal adhesion formation, and activation of several tyrosine kinases in an RGD-dependent manner. Among the activated factors are the nonreceptor tyrosine kinases focal adhesion kinase and Src but also the epidermal growth factor receptor and epidermal growth factor receptor family member Her3/ErbB3. Interestingly, fibronectin activates a similar range of tyrosine kinases but not Her3/ErbB3. These findings suggest that the bacterial protein CagL not only exhibits functional mimicry with fibronectin but is also capable of activating fibronectin-independent signaling events. We thus postulate that CagL may contribute directly to H. pylori pathogenesis by promoting aberrant signaling cross-talk within host cells.
Keywords: Bacterial Signal Transduction, Cell Adhesion, Cell Motility, Fibronectin, Signal Transduction, EGF Receptor, Integrin, Pathogenesis, Pathogenicity Island, RGD Motif
Introduction
Eukaryotic cell adhesion is a fundamental process in tissue development, homeostasis, and disease and is mediated by specific interactions of cell surface receptors with extracellular matrix (ECM)2 proteins (1–5). The ECM is a meshwork of fibrillar and nonfibrillar components assembled into complex structures such as basement membranes. The latter provide a scaffold for cell adhesion, spreading, and migration. ECM regulates numerous cell functions by activating multiple signaling pathways at the adhesion sites. ECMs, composed of collagens, laminins, and other glycoproteins such as fibronectin (FN), serve as substrates for different adhesion molecules including the integrin family of transmembrane receptors. The assembly of ECM components into functional supramolecular modules is highly regulated (3–7). FN matrix assembly alone is a dynamic cell-driven process in which the soluble FN molecules assemble into insoluble fibrillar polymeric ECM structures (8).
FN and integrin receptors play crucial roles in a variety of morphogenetic processes, which are regulated by processes termed outside-in and inside-out signaling cascades (3–5). Deregulation of integrin and FN functions associates with disease development including chronic inflammation, heart failure, cancer, and metastasis (7, 9–11). The outside-in signaling triggered by ligation of integrin receptors with FN and other ECM components results in the reorganization of cytoskeletal and signaling molecules into complexes of more than 90 proteins (9–13). This occurs by synergistic processes dependent on integrin aggregation and occupancy, as well as tyrosine phosphorylation. Integrins also cooperate with growth factor receptors such as epidermal growth factor receptor (EGFR) to enhance signaling (14).
FN consists of multiple domains (classified types I–III) that show binding specificities for specific cell membrane receptors, collagen, fibrin, and heparin. FN alone is sufficient to induce highly efficient spreading of many mammalian cell types including fibroblast and epithelial cells in vitro. An important functional unit of FN is its RGD tripeptide motif, which acts in synergy with a PHSRN sequence for binding to integrins. In particular, the RGD motif is crucial for mediating eukaryotic cell adhesion and spreading (3, 7, 15, 16). Remarkably, FN is no passive adhesive molecule but actively triggers signal transduction to the F-actin cytoskeleton and focal contact formation upon binding to integrins. Binding of FN to integrin α5β1 results in the recruitment of focal adhesion kinase (FAK) and Src kinase and the subsequent activation of these kinases in the focal adhesion complexes (9, 10).
Although ECM proteins are unique to the eukaryotic kingdom, many bacterial pathogens adhere to host ECM molecules or integrins to exploit the downstream signaling pathways for entering host cells or establishing persistent infection (17–20). For example, the well characterized bacterial protein invasin (InvA) of Yersinia spp. has been shown to bind α5β1 and some other integrins in a manner similar to FN (21–24). Immobilized membrane proteins of an InvA-overproducing Yersinia strain were observed to trigger spreading of HEp-2 cells. Although this effect of InvA was attributed to its ability to bind integrin β1, the molecular mechanisms involved have not been investigated in full detail (25). Recently, we reported that the gastric pathogen and type I carcinogen Helicobacter pylori (Hp) exploit integrin receptors for the injection of virulence factors into mammalian cells (26). This is achieved by a type IV secretion system (T4SS) consisting of 11 VirB protein orthologs (encoded by virB1–11 genes) and the so-called coupling protein (VirD4, an NTPase). These proteins are encoded by a 40-kb gene cluster known as the cag (cytotoxin-associated gene) pathogenicity island, the cagPAI (27–29). We have shown that integrin α5β1 binds to a small 26-kDa protein designated CagL, which is encoded by the open reading frame HP0539 in the cagPAI (26). CagL is predicted to be a functional VirB5 ortholog and structural component of the T4SS pilus (30), as seen with other VirB5 proteins such as that of Agrobacterium tumefaciens (31). CagL has no significant sequence homology to any known eukaryotic protein. But like FN, CagL carries a RGD motif shown to be important for interaction with the α5β1 integrin (26). However, it has been recently shown in yeast two-hybrid screens that other T4SS proteins such as CagY (VirB10), CagN, and the effector protein CagA can also bind β1 integrin in vitro (32), confirming that Hp targets this integrin member as a receptor for the T4SS. However, mutation of the RGD motif in CagL had no defect in T4SS functions such as the phosphorylation of injected CagA (32). In contrast, another very recent study showed a clear role of CagL in activating ADAM17, a metalloprotease involved in catalyzing ectodomain shedding of receptor tyrosine kinase ligands (33). In nonstimulated cells, ADAM17 is normally in complex with integrin α5β1 and inactive (34). During acute Hp infection, however, it was shown that CagL dissociates ADAM17 from the integrin α5β1 and activates ADAM17 (33). This was confirmed by infection with a ΔcagL deletion mutant, which is entirely defective in the latter response, and by genetic complementation with the wild-type (wt) cagL gene or biochemical complementation by the addition of extracellular CagL restoring this function (33). These studies indicate that there is a controversy in the literature about the importance of CagL in T4SS functions and host cell signaling. Thus, the role of CagL needs to be investigated in more detail.
Investigating the contribution of each of the various cagPAI proteins for signaling during infection is certainly very difficult to perform because mutation of single genes often lead to complete abolition of T4SS functions. For this purpose, the aim of the present study was to investigate whether purified CagL alone can trigger host cell signaling. We demonstrate that purified CagL mimics a number of cellular functions of FN in the induction of eukaryotic cell spreading and focal adhesion formation, involving the activation of Src, FAK, and EGFR tyrosine kinases. Despite this functional mimicry between CagL and FN, CagL also activates the EGF receptor family tyrosine kinase Her3/ErbB3 under conditions where Her3/ErbB3 is not activated by FN. Our findings support the hypothesis that CagL may promote aberrant signaling cross-talk in the host cells by mimicking FN in the induction of cell spreading and focal adhesion formation while activating FN-independent signaling pathways. CagL may be used as a novel tool for dissecting the regulatory networks that govern FN and integrin signaling.
EXPERIMENTAL PROCEDURES
Eukaryotic Cell Culture and Bacteria
The human gastric adenocarcinoma cell line AGS, MKN45, HeLa, and several mouse fibroblast knock-out cell lines were cultured in RPMI1640 or DMEM, respectively, which were supplemented with 10% fetal calf serum (Invitrogen). Mouse knock-out cells deficient in focal adhesion kinase (FAK−/− cells) or fibroblasts derived from c-src−/−, c-yes−/−, and c-fyn−/− triple knock-out mouse embryos (SYF cells) as well as stable expression of wt FAK in FAK−/− cells or wt c-Src in SYF cells were described previously (35, 36). Generation of the floxed FN+/+ fibroblast cells and FN−/− knock-out cells has been described (37, 38). The FN−/− cells were grown in DMEM supplemented with 10% fetal calf serum or, alternatively, in serum replacement medium (Sigma-Aldrich). After reaching a confluency of ∼70%, the cells were washed two times with phosphate-buffered saline and then starved for 12 h by incubation with fresh medium without fetal calf serum before the cells were trypsinized and prepared for the spreading assays as described below. Hp strains P1 and P1ΔcagL were grown as described (26).
CagL Peptides and Purification of CagL and VirB10 Proteins
Several CagL-derived RGD peptides (peptide 1, cyclo-Arg-Gly-Asp-d-Leu-Ala-; peptide 2, cyclo-Arg-Gly-Asp-Leu-d-Ala-; and peptide 3, cyclo-Arg-Gly-Asp-Leu-d-Ala-Leu-) were synthesized as described (26). To construct the vectors for overexpression of wild-type CagL (CagLwt) in Escherichia coli, a DNA fragment corresponding to amino acid residues 21–237 of the protein (minus the predicted signal peptide) was amplified by PCR, sequenced, and ligated into pET-28a vector (Novagen). Mutagenesis of the RGD motif in the CagL sequence to CagLRGA or CagLRAD was performed using a QuikChange site-directed mutagenesis kit according to the instructions of the supplier (Stratagene). CagLwt, CagLRAD, and CagLRGA were overexpressed and purified by a standard protocol (26). Briefly, E. coli BL21(DE3) transformed with the plasmids were grown in 5 ml of LB medium at 37 °C. After overnight incubation, 500 ml of fresh LB medium were added and shaken for another 2.5–3 h up to A600 = 1. Then 1 mm isopropyl β-d-thiogalactopyranoside was added, and the bacteria were grown for 1.5 h to induce CagL expression. Bacterial pellets were collected by centrifugation and then resuspended in ice-cold buffer CW (50 mm KH2PO4-K2HPO4, pH 7.5, 200 mm NaCl) supplemented with protease inhibitor mixture (Roche Applied Science). After sonication, the overexpressed CagL present in the inclusion bodies was solubilized in buffer LW (50 mm KH2PO4-K2HPO4, pH 7.5, 200 mm NaCl, 6 m guanidine hydrochloride) and refolded in ice-cold refolding buffer (52 mm Tris-HCl, pH 8.2, 20 mm NaCl, 834 μm KCl, 1.1 mm EDTA, 2.1 mm reduced glutathione, 210 μm oxidized glutathione). After refolding, CagL was further purified by metal-chelate affinity chromatography through Talon® resin (BD Biosciences) and gel filtration in buffer CW through Sephacryl S-200 (16/60) according to the manufacturer's instructions (Amersham Biosciences). Protein concentrations of the resultant samples were determined by the BCA protein assay (Pierce) and typically yielded a total amount of ∼1.5 mg of CagL in 10 ml of buffer. Under denatured conditions, purified CagL proteins run at a size of 26 kDa. Purification of CagL was judged to be of >95% homogeneity by SDS-PAGE/Coomassie Blue staining (supplemental Fig. S2). The folded conformations of the purified CagLwt, CagLRAD, and CagLRGA were confirmed by circular dichroism (26). No indication of post-translational modifications of purified CagL such as disulfide formation or methylation was detected.
In addition to CagL, several VirB10 fragments were cloned for expression. Sequence encoding C-terminal VirB10 from Hp strain 13a (VirB10 C-term) was cloned to BamHI/EcoRI sites of pGEX-2T using forward (5′-ACGGGATCCCTAGATAAACTCATAGGCCTTGG-3′) and reverse (5′-ACGGAATTCTTAATTGCCACCTTTGGG-3′) primers. Expression of GST and GST-VirB10 C-terminal fusion proteins was induced with isopropyl β-d-thiogalactopyranoside, and the proteins were subsequently purified from clarified lysates in the presence of protease inhibitor mixture (Roche Applied Science) using glutathione-Sepharose 4 Fast Flow (GE Healthcare) according to the manufacturer's instructions. Furthermore, His-tagged VirB10 repeat proteins, VirB10 Rpt 2.1 and 2.2 from strains Q86A and 13a, respectively, were expressed and purified as described previously (39). The exact coordinates of all VirB10 fragments are given in Fig. 7A.
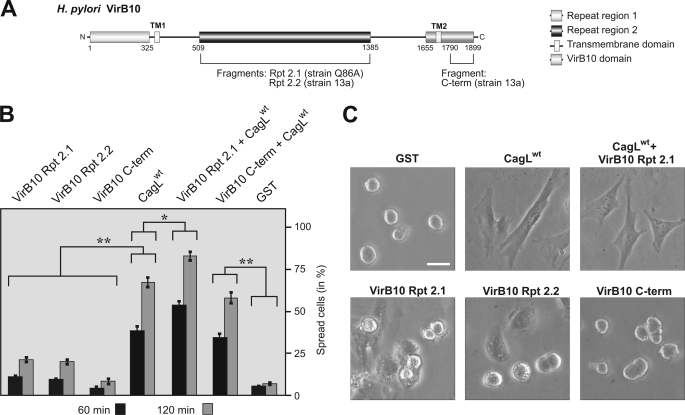
FIGURE 7.
The internal repeat region of VirB10 but not its C terminus can enhance CagL-induced cell spreading. A, schematic representation of the entire CagY (also called VirB10) protein and its domains as reported recently (39, 68). Approximate amino acid positions of each region are given. The indicated C-terminal (C-term) VirB10 domain of the CagY protein shares 31% identity with ∼55% of the A. tumefaciens VirB10 protein (39). The putative secreted T4SS pilus-associated form of VirB10 comprises the large repeat 2 region encoded between the two transmembrane domains. The following fragments were purified: VirB10 repeat region 2.1 (Rpt 2.1, Hp strain Q86A, 578 amino acids long), VirB10 repeat region 2.2 (Rpt 2.2, Hp strain 13a, 799 amino acids long), and VirB10 C-terminal region (C-term, Hp strain 13a, 109 amino acids long). To investigate whether these VirB10 proteins can also induce cell spreading, respective assays were performed with fibroblast cells. B, quantitation of spread cells at 1- and 2-h time points is shown. The results demonstrate that CagLwt can induce profound cell spreading of fibroblasts, whereas all VirB10 fragments or GST and BSA cannot. When CagL was mixed with VirB10 fragments, the repeat region 2 but not the C terminus enhanced the CagL effect significantly. C, representative phase contrast micrographs of fibroblast cells incubated with the indicated Hp proteins, respectively. Bar, 10 μm. Quantitation data of three independent experiments are shown.
Precoating of Petri Dishes and Cell Spreading Assays
Cell spreading assays and quantitation of spread cells were performed according to procedures described previously (40). Briefly, each well of the microtitre plates was coated with 100 μl of 50 μg/ml ligand (fibronectin (from human plasma; Sigma), purified CagL variants, RGD peptides, VirB10 proteins, BSA (Sigma)), or polylysine (0.01% solution; Sigma) at 4 °C overnight. In the case of heat-denatured CagL (CagLhdn), the protein was boiled for 10 min and then incubated under identical conditions. For double coatings, the plates were first incubated at 4 °C overnight each with polylysine and then with CagLwt. Nonspecific binding sites were blocked by incubation with 5% BSA in buffer CW for 2 h at room temperature. The cells were grown as described above, trypsinized, and then treated with soybean trypsin inhibitor according to the instructions of the supplier (Sigma). After washing with phosphate-buffered saline, 4 × 105 cells in RPMI or DMEM were added to the wells and incubated in a time course. One hundred cells were randomly evaluated for cell spreading. The cells were scored as spread or not spread as described (40). The percentage of spreading of each cell line from a representative experiment is shown in Fig. 2. The pharmacological inhibitors AG1478 (BIOSOURCE; 10 μm), PP2 (Calbiochem; 10 μm), and PF-573228 (Tocris; 10 μm) were added in some experiments as indicated. The experiments were repeated at least three times with similar results.
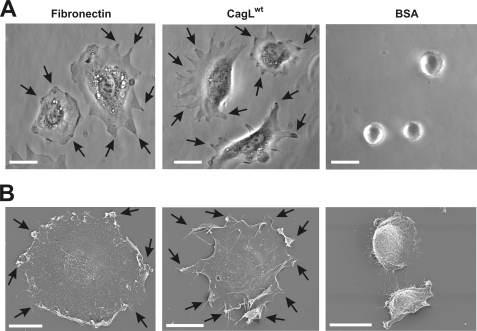
FIGURE 2.
Immobilized fibronectin and CagL induce eukaryotic cell spreading in vitro. A, phase contrast microscopy of spread cells. B, scanning electron microscopy of spread cells. Cell spreading assays were performed for 2 h on immobilized fibronectin, CagLwt, or BSA, respectively. Bars, 20 μm.
Antibodies and Western Blotting
Rabbit α-CagL antiserum was raised against the C-terminal peptide (C-RSLEQSKRQYLQER) of the protein and was prepared by Biogenes (Berlin, Germany). Western blotting experiments were performed as described (41, 42). The pan-α-phosphotyrosine antibody and the polyclonal α-phospho-Src-Y416, α-phospho-Her3/ErbB3-Y1289, and α-phospho-EGFR-Y846 antibodies were purchased from NEB. The polyclonal α-phospho-FAK-Y397 antibody was from BIOSOURCE. The α-glyceraldehyde-3-phosphate dehydrogenase antibody (Santa Cruz) served as a loading control in each Western blot. As secondary antibodies, horseradish peroxidase-conjugated α-mouse, α-rabbit, or α-goat polyvalent sheep immunoglobulin was used, and antibody detection was performed with the ECL Plus chemiluminescence kit (Amersham Biosciences). Band intensities, and the corresponding kinase activities were quantitated with the Lumi-Imager F1 (Roche Applied Science). The images were processed using Adobe Photoshop (version 6.0).
Live Cell Imaging
For live cell imaging experiments, the cells were grown in monolayers for 2 days in conventional flasks using RPMI or DMEM with 10% fetal calf serum. After reaching a confluency of ∼70%, the cells were washed two times with phosphate-buffered saline and then starved for 12 h by incubation with fresh medium without fetal calf serum. The cells were trypsinized and then treated with soybean trypsin inhibitor according to the instructions of the supplier (Sigma). Three washing steps followed using fresh RPMI or DMEM, respectively. The cells were then deposited onto precoated 35-mm Petri dishes placed in a prewarmed (37 °C), humidified, and equilibrated (5% (v/v) CO2) incubation chamber (PeCon GmbH) mounted on an inverted microscope (Leica DM IRE2, Leica Microsystems) equipped with a CCD camera (Spot RT, Diagnostic Instruments Corp.) and controlled by image acquisition software (MetaVue, Molecular Devices Corp.). A 40× phase contrast objective (Leica Microsystems) was also used to record cell spreading. The acquisition time interval was 2 min, and the transmitted light was switched on only during the exposure time of the CCD camera (1 s) controlled by the acquisition software. Alternatively, the microscopic analysis was performed using a confocal laser scanning microscope system TCS SP2 (Leica Microsystems) equipped with a DM-IRE2 inverted microscope and an incubation chamber (PeCon). Image data were obtained at time intervals of 2 min using a 63×/1.4 N.A. oil immersion objective and CLSM software (Leica Microsystems). Image data sets of both microscope systems were processed using ImageJ software.
Immunofluorescence Staining and Microscopy
Immunofluorescence staining was performed as described (43). In brief, cells fixed in 3.4% paraformaldehyde were stained with different antibodies as shown in each experiment. The samples were analyzed using a Leica TCS SP2 microscope system equipped with a DM-IRE2 microscope and different lasers (Leica Microsystems). To avoid spectral overlap and channel cross-talk, fluorescein isothiocyanate, TRITC, CY5, and Alexa-350 fluorophores were excited sequentially with argon laser (488 nm), green helium (543 nm), red helium (633 nm), and UV laser (364 nm). The images were processed using ImageJ.
Field and Immuno Field Emission Scanning Electron Microscopy (FESEM)
Procedures for FESEM of Hp and spread cells were carried out as described previously (26). For immuno-FESEM of CagL, Hp samples were incubated with purified rabbit α-CagL IgG antibodies (100 μg IgG protein/ml) followed by incubation with 15-nm protein A-gold particles as described (26). All of the samples were coated with a thin carbon film. FESEM of spread cells was performed using procedures as described previously (26). The images were processed for contrast and brightness using Adobe Photoshop.
Statistical Analysis
All of the data were evaluated using Student's t test with SigmaStat statistical software (version 2.0). Statistical significance was defined by p ≤ 0.05 (*) and p ≤ 0.005 (**). All of the error bars shown in the figures and those quoted following the ± signs represent standard deviations.
RESULTS
CagL Is a Surface Protein of Hp That Can Induce Eukaryotic Cell Spreading and Focal Adhesion Formation in Vitro
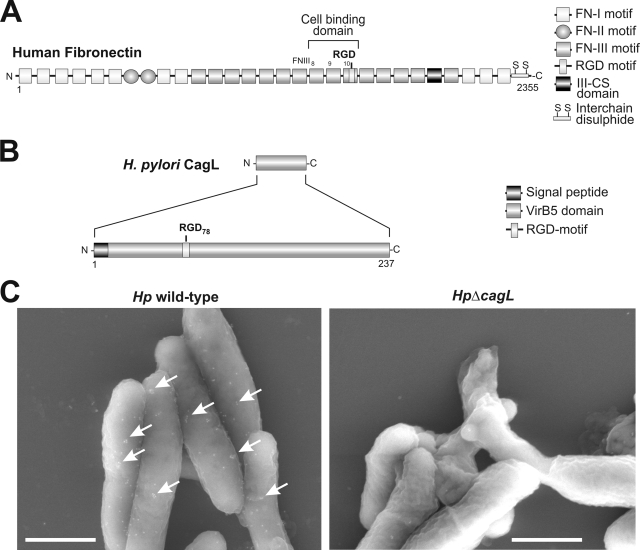
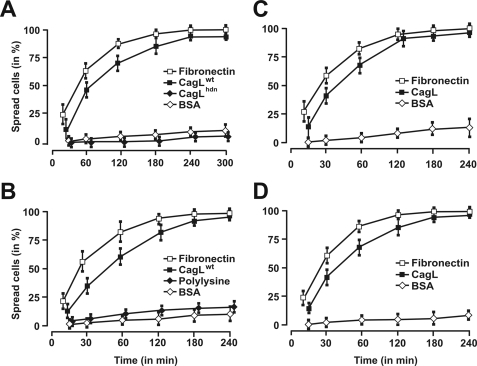
CagL has been shown to be a component of the T4SS pilus and has no significant sequence homology to any known eukaryotic protein (26, 30). However, similar to FN, CagL carries a RGD motif (Fig. 1, A and B) shown to be important for interaction with the α5β1 integrin in vivo and in vitro (26). CagL is expressed on the surface of Hp even in the absence of host cells (Fig. 1C and supplemental Fig. S1). After host cell contact, CagL decorates the T4SS pili surface (26), suggesting that the protein may act as some kind of a molecular sensor on the bacteria. Here, we further examined the ability of CagL to trigger intracellular signaling pathways. For this purpose, we overexpressed CagL in E. coli and purified the recombinant protein to homogeneity (supplemental Fig. S2). In our in vitro binding studies of AGS gastric epithelial cells, we noticed during phase contrast microscopy that eukaryotic cells not only bound to immobilized CagL on Petri dishes but that they were also triggered to spread (Fig. 2A). Cell spreading induced by CagL was surprising because such a feature is only known for some eukaryotic ECM proteins including FN. CagL-induced cell spreading was phenotypically very similar to that seen with immobilized FN (Fig. 2A) and was also verified at high resolution by scanning electron microscopy (Fig. 2B). A time course shows that AGS cell spreading is not as rapid as the cell spreading induced by FN but became very efficient after 30–60 min of interaction (Fig. 3A and supplemental Movie S1). To exclude artifacts or cell type-specific effects of the observed phenomenon, CagL-induced cell spreading was confirmed with other cultured cell lines including human HeLa, human MKN45, and mouse fibroblasts (Fig. 3, B and C). Moreover, wild-type CagL (CagLwt) also induced the formation of focal adhesions in host cells (Fig. 2, arrows) as confirmed by immunostaining using antibodies specific for the focal adhesion marker proteins vinculin and focal adhesion kinase (FAK) as well as co-staining with phalloidin (for F-actin) (Fig. 4, arrows, and data not shown). As controls, these cells do not spread on immobilized BSA, GST, polylysine, or heat-denatured CagL (CagLhdn) within 4 h (Figs. 2, 3, A and B, and 7C), suggesting that the spreading was CagL-specific and requires proper folding of CagL. Three-dimensional modeling predicts that CagL forms a three α-helix bundle with a protruding globular domain carrying the RGD motif in a surface exposed loop (30). In contrast, the crystal structure of the FN type III domain encompassing the RGD motif is composed predominantly of β-sheets (44) and is thus entirely different from the predicted structure of CagL (supplemental Fig. S3).
FIGURE 1.
Comparison of the domain structure of human fibronectin with CagL from Hp and surface localization of CagL. A, diagrammatic representation of the 250-kDa fibronectin molecule. Subdomains of the protein are highlighted to the right. Please note the short cell-binding domain of fibronectin as indicated. B, diagrammatic representation of the 26-kDa CagL-polypeptide. Both fibronectin and CagL exhibit no sequence homology to each other, except a short RGD motif. C, nonpermeabilized wild-type and ΔcagL mutant Hp were labeled with protein A-coated gold particles (15 nm) conjugated with α-CagL antibody. The signals obtained indicate surface localization of CagL in wild-type Hp (arrows) but an absence of CagL in the mutant. Bars, 1 μm.
FIGURE 3.
Immobilized CagL induces cell spreading of both human and mouse cells. To investigate whether or not CagL-induced cell spreading is a cell type-specific effect, CagL-, fibronectin-, and BSA-induced cell spreading was investigated in a time course with various cell lines including human AGS cells (A), human HeLa cells (B), human MKN45 cells (C), and mouse fibroblasts (D). The experiments in A also include CagLhdn, and B includes polylysine as controls. Quantitation data of three independent experiments for each cell line are shown.
FIGURE 4.
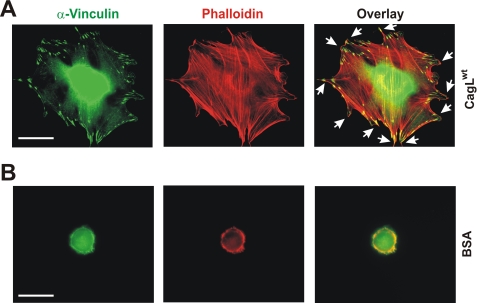
Cell spreading induced by CagL leads to focal adhesion formation and is accompanied by rearrangements of vinculin and filamentous actin. Immunofluorescence microscopy images of representative spread mouse fibroblast cells on immobilized CagLwt (A) or BSA (B). After 2 h of co-incubation, the cells were fixed and stained with α-vinculin antibody and phalloidin (for F-actin) as indicated. The arrows indicate focal adhesion structures. Bar, 10 mm. The results are representative of at least three independent experiments.
CagL Induces Focal Adhesion Dynamics and Phosphotyrosine Signaling during Cell Spreading in an RGD-dependent Manner
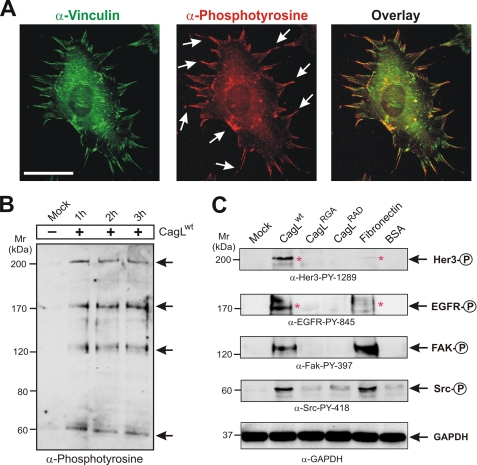
The findings above suggest that CagL may mimic FN upon interaction with host cells. To confirm this hypothesis, we next investigated whether tyrosine phosphorylation of host cell proteins is induced during CagL-mediated cell spreading. Host cells were allowed to spread on immobilized CagLwt for 1 h, fixed, and then stained using a pan-α-phosphotyrosine antibody. The results show that CagLwt induces the tyrosine phosphorylation of host cell proteins in the cytoplasm and focal adhesions, which were co-stained with an α-vinculin antibody (Fig. 5A, arrows). This suggests that upon binding to host cells, CagL induces tyrosine kinase signaling. This prompted us to investigate the pattern of phosphorylated proteins by Western blotting. The results show that immobilized CagLwt induces the tyrosine phosphorylation of four major protein species, at ∼60, 120, 170, and 200 kDa (Fig. 5B, arrows). The identity of these proteins was confirmed by Western blotting using phospho-specific antibodies against well known host signaling proteins in these size ranges. The proteins were identified as the two nonreceptor tyrosine kinases Src (60 kDa) and FAK (120 kDa) and the two growth factor receptor tyrosine kinases EGFR (epidermal growth factor receptor, 170 kDa) and Her3/ErbB3 (200 kDa), respectively (Fig. 5C, arrows). Interestingly, cell spreading on FN induced similar phosphorylated proteins as expected, but significant differences were also observed between the CagLwt- and FN-induced phospho-patterns (Fig. 5C, asterisks). Although the extents of phosphorylation of Src and FAK were similar, the activation of EGFR by FN was significantly reduced, and almost no activation of Her3/ErbB3 by FN was seen. The quantification data of tyrosine kinase activities are shown in supplemental Fig. S4.
FIGURE 5.
CagL induces focal adhesion dynamics and phosphotyrosine signaling during eukaryotic cell spreading in an RGD-dependent manner. A, immunofluorescence microscopy of fibroblast spreading on immobilized CagLwt (1 h) as visualized by immunostaining with α-vinculin and α-phosphotyrosine antibodies. The stainings show that cells spreading on immobilized CagLwt induce phosphotyrosine signaling in the cytoplasm and focal adhesion complexes (arrows). B, the global pattern of phosphorylated proteins induced by CagLwt were investigated by Western blotting using a pan-α-phosphotyrosine antibody. The results show that immobilized CagLwt induces the tyrosine phosphorylation of four major protein species, at ∼60, 120, 170, and 200 kDa (arrows). C, using commercially available phospho-specific antibodies, the proteins were identified as tyrosine kinases Src (60 kDa), FAK (120 kDa), EGFR (170 kDa), and Her3/ErbB3 (200 kDa), respectively (arrows). The α-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) blot confirms that equal amounts of protein were loaded in each lane. Parallel cell spreading experiments for 1 h were performed on immobilized fibronectin, CagLRGA, CagLRAD, and BSA as indicated. Mutagenesis of the RGD motif resulted in a profound inhibition of CagL-induced tyrosine kinase activation, suggesting that CagL uses its RGD motif to trigger similar host cell responses as human fibronectin. Quantitation data of kinase activities were done densitometrically and are shown in supplemental Fig. S4.
Next, we asked whether the RGD motif is important for CagL-induced signaling. Mutagenesis of the RGD motif to either RAD (CagLRAD) or RGA (CagLRGA) led to a profound inhibition of both CagL-induced cell spreading and tyrosine kinase activation (Figs. 5C and 6), suggesting that the RGD motif of CagL is crucial for triggering the host cell responses, which are also stimulated by human FN. We have recently shown that co-incubation of AGS cells with CagL-derived RGD peptides leads to the induction of FAK and Src phosphorylation (26). Thus, we immobilized various CagL-specific RGD peptides on Petri dishes followed by cell spreading analysis. The peptides alone did not induce cell spreading. The cells incubated on immobilized RGD peptides remained in a round shape similar to the cells incubated on heat-denatured CagL (supplemental Fig. S5). These observations strongly suggest that the RGD motif alone is not sufficient for triggering cell spreading. Taken together, our findings suggest that although the RGD motif of CagL plays a key role, other structural determinants in CagL are also required for the induction of cell spreading.
FIGURE 6.
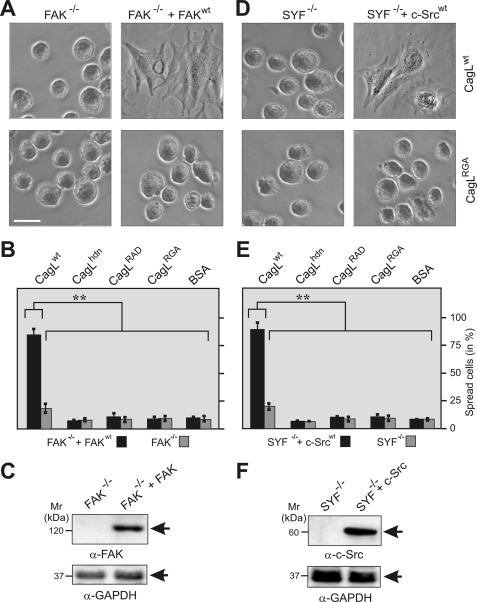
Eukaryotic cell spreading induced by immobilized CagL requires the tyrosine kinases FAK and Src. To investigate whether the kinases of the FAK and Src family are required for CagL-induced cell spreading, FAK−/− or fibroblasts derived from c-src−/−, c-yes−/−, and c-fyn−/− triple knock-out mouse embryos (SYF cells) and their respective wt control cells were used for cell spreading assays (2 h). Interestingly, neither FAK−/− cells (A and B) nor SYF cells (D and E) were able to spread efficiently on CagLwt, whereas stable expression of FAK in FAK−/− cells or c-Src in SYF cells restored cell spreading. Similar results were obtained for immobilized fibronectin as expected, whereas neither CagLRGA, CagLRAD, nor BSA were able to induce spreading of any of the cells. Bar, 10 μm. Western blotting with α-FAK (C) or α-c-Src antibodies (F) confirmed the absence or presence of the proteins in the respective knock-out cells. The α-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) blots confirm that equal amounts of protein were loaded in each lane.
Eukaryotic Cell Spreading Induced by Immobilized CagL Requires FAK, Src, and EGF Receptor Tyrosine Kinases
To characterize the cellular effects of CagL in more detail, we next tested whether CagLwt can induce spreading of mouse fibroblast cells deficient in focal adhesion kinase (FAK−/− cells) or fibroblasts derived from c-src−/−, c-yes−/−, and c-fyn−/− triple knock-out mouse embryos (SYF cells) (35, 36). Interestingly, neither FAK−/− cells (Fig. 6, A–C) nor SYF cells (Fig. 6, D–F) were able to spread efficiently on CagLwt. Stable expression of wt FAK in FAK−/− cells or wt c-Src in SYF cells restored cell spreading, suggesting that both FAK and Src kinase signaling play an important role in CagL-induced cell spreading (Fig. 6). In addition, pharmacological inhibition of Src family kinases by PP2, FAK by PF-573228, and the receptor tyrosine kinases EGFR and Her3/ErbB3 by AG1478 also suppressed cell spreading on CagL, confirming that each of these activated components are indeed necessary for CagL signaling (supplemental Fig. S6).
The Internal Repeat Region of VirB10 but Not the C terminus Can Enhance CagL-induced Cell Spreading
Next, we asked whether the observed effect of CagL is specific for this protein or whether similar observations can be made with the Hp CagY (VirB10) protein, which can also bind integrin β1 (32). For this purpose, the internal repeat region 2 and the C-terminal fragment were cloned, purified, and immobilized for cell spreading assays (Fig. 7A). The results show that neither of the two fragments was able to induce efficient cell spreading (Fig. 7, B and C). However, whereas the C-terminal VirB10 behaved like GST or BSA controls with no signs of spreading, repeat region 2 from two strains showed some weak activity. When CagLwt and either repeat region were mixed together, the cell spreading effect of CagL was enhanced (Fig. 7, B and C).
CagL Can Functionally Complement FN to Induce Cell Spreading of Fibronectin−/− Fibroblasts
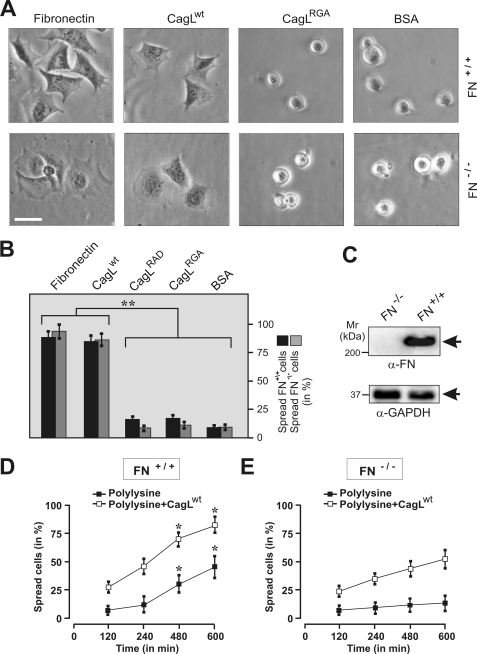
Finally, we investigated whether CagL can functionally complement FN and induce cell spreading of knock-out fibroblasts deficient in the gene encoding FN (FN−/− cells) (37, 38). Interestingly, CagLwt but not CagLRAD or CagLRGA induced spreading of FN−/− cells (Fig. 8, A–C, and supplemental Movie S2). To confirm that CagLwt can restore the function of FN, we performed cell spreading assays of trypsinized FN−/− and FN+/+ cells for 2–10 h on either polylysine, a substrate that keeps integrins in a nonactivated form (45), or polylysine mixed with CagLwt. Although neither FN−/− nor FN+/+ cells were able to spread on any of these substrates between 2 and 4 h, FN+/+ cells spread efficiently at later time points (4–10 h), which can be explained by secretion of FN at these time points. The spreading of FN+/+ cells on polylysine was significantly enhanced by the presence of CagLwt (Fig. 8D). By direct comparison, FN−/− cells were unable to spread efficiently on polylysine even at 10 h, whereas the addition of CagLwt induced cell spreading to an extent similar to that observed with the spreading of FN+/+ cells on polylysine alone (Fig. 8E). Collectively, these data strongly support the view that CagL mimics the function of FN and can even restore its function in FN−/− cells.
FIGURE 8.
CagL can functionally complement fibronectin to induce spreading of fibronectin−/− fibroblasts. To investigate whether the CagL can replace FN in the induction of cell spreading, cell spreading assays were performed with FN−/− and FN+/+ cells (2 h). A, representative phase contrast micrographs of FN−/− cells incubated with immobilized FN, CagLwt, CagLRGA, or BSA, respectively. Bar, 10 μm. B, quantitation of spread cells is shown. The results show that both FN and CagLwt can induce profound cell spreading of FN−/− and FN+/+ cells, whereas CagLRGA, CagLRAD, or BSA cannot. C, Western blotting with α-fibronectin antibody confirmed the absence and presence of FN in the FN−/− and FN+/+ cells, respectively. The α-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) blots confirm that equal amounts of protein were loaded in each lane. Cell spreading assays were performed with FN+/+ cells (D) or FN−/− cells (E) in a time course of 2–10 h. The cells were allowed to spread on immobilized polylysine or polylysine supplemented with CagLwt. Quantitation data of three independent experiments are shown.
DISCUSSION
Human FN and its RGD tripeptide motif play an important role in mediating eukaryotic cell-to-cell interactions and triggering signaling, which is important for cell adhesion, spreading, migration, and other processes (3–5, 7, 15, 16, 46). The importance of the RGD motif is underlined by the finding that mouse embryos with a mutation to an inactive RGE motif die at day 10 because of severe defects that resemble the phenotype of integrin α5β1-deficient mice (47). Other studies indicated that FN is a target of microbial pathogens because a number of bacterial proteins have been found to bind FN for adhesion to or invasion of bacteria into host cells (17–19). In the present report, we show that the Hp CagL protein mimics FN in the induction of cell spreading, focal adhesion formation, and activation of Src, FAK, EGFR, and Her3/ErbB3 in vitro. CagL therefore represents a novel example of molecular mimicry between a eukaryotic ECM protein and a virulence factor of bacterial origin.
We observed that immobilized CagLwt induced host cell spreading and focal adhesion formation that was phenotypically similar to that seen with immobilized FN. The induction of cell spreading by CagL was not as rapid as that induced by FN but became very efficient after 30–60 min of co-incubation. To exclude artifacts, we confirmed that host cells do not spread on immobilized BSA, GST, or polylysine. We also investigated whether CagL can induce cell spreading and focal adhesion formation of different human cell lines (AGS, MKN45, and HeLa) and mouse fibroblast cell lines, thus excluding cell type-specific effects of the observed phenomenon. Moreover, as another control, we could demonstrate that denatured CagLhdn was unable to induce cell spreading and focal adhesion formation, suggesting that proper folding of CagL is essential for the induction of these phenotypes. Finally, we showed that neither immobilized CagLRGA nor CagLRAD mutants were able to induce these responses. Taken together, these findings provide comprehensive evidence that Hp CagL not only interacts with the integrin member α5β1 (26) but is also capable of inducing signaling leading to cell spreading and focal adhesion formation.
In healthy tissues, maximal binding of FN to α5β1 integrin requires two internal domains, called FnIII-9 and FnIII-10 (Ref. 48 and Fig. 1A). Genetic studies indicated that residues in both domains are involved in contacting the integrin receptor, with aspartate residue 1495 in the RGD motif of FnIII-10 being the most significant contributor to binding energy (48). Several other residues located on the same face of the molecule as the RGD sequence of FN also contribute to integrin binding, including those within the so-called synergy region in FnIII-9 (48–50), as well as residues located between the synergy and RGD motif sites (51). Interestingly, CagLwt can bind to integrin α5β1 (dissociation constant Kd = 0.09) and with an affinity higher than that of CagLRGA mutant (Kd = 0.36) (26) or FN (Kd = 0.8) (52). However, except for the RGD motif, there is no sequence similarity of CagL to FN, and the three-dimensional structure model of CagL is mainly formed by a three α-helix bundle with an protruding globular domain carrying the RGD motif (30). Interestingly, the crystal structure of the FN host cell binding domain is composed predominantly of β-sheets (44) and therefore appears to be quite different from the predicted structure of CagL. The fact that cyclic CagL-derived RGD peptides alone fail to induce cell spreading confirms that additional structural determinants in CagL are required for triggering cell spreading; identification of these structural determinants by means of site-directed mutagenesis is currently underway in our labs. Determination and subsequent comparison of the three-dimensional structure of CagL with that of FN might provide novel insights into the structural basis underlying the induction of cell spreading or focal adhesion formation by external stimuli.
CagL also exhibits neither sequence nor structural homology to the well known bacterial protein invasin (InvA, Yersinia spp.), which binds α5β1 and some other integrins in a manner similar to FN but does not contain an RGD motif (21–24). It is intriguing that invasin and FN recognize similar residues on the integrin receptor, given that the solved crystal structures of their respective integrin-binding regions have very different surface contours (24, 52, 53). Three important factors have been proposed to enhance invasin-mediated uptake: (i) high affinity binding of integrin receptors by the so-called D4-D5 superdomain (54), (ii) the ability of invasin monomers to undergo homotypic interactions (23), and (iii) an increase in the concentration of integrin receptors available to bind invasin (23, 54). Mutations that lower the affinity of InvA for integrin receptors, deletion of a region of invasin necessary for homotypic interaction, and depletion of integrins from the host cell all severely depress bacterial uptake, causing extracellular adhesion of the bacteria. Most of the bacteria-host interaction studies used InvA-coupled latex beads or invA mutant bacteria (55). Nevertheless, the ability of Petri dish-immobilized purified InvA to induce cell spreading or focal adhesion formation of host cells was only marginally investigated. For example, HEp-2 cells incubated on immobilized membrane proteins of an InvA-overproducing Yersinia strain showed signs of spreading (25). In a similar study, incubation of immobilized purified InvA with T-lymphocytes led to the rapid apoptotic death of these cells (56). Despite these investigations, the specificity and mechanism by which the Yersinia protein InvA mediates cell spreading in an integrin-dependent manner remain enigmatic.
The most striking difference between Yersinia invasin and FN binding is the significantly higher affinity of invasin-receptor binding. This activity is critical both for the protein to promote uptake and as a central virulence determinant for the microorganism. Low affinity integrin ligands, coated on either particles or bacteria, allow efficient adhesion to mammalian cells but have a greatly reduced capacity to promote uptake relative to that seen with invasin (54). Given the observation that bacteria expressing active InvA blocked binding of CagL to integrin α5β1 (26), the affinity of InvA to integrin α5β1 is clearly higher (Kd = 0.005) (49) than that of CagL or FN. This might explain why CagL does not act as an invasin. Hp is essentially an extracellular pathogen with only ∼2–5% of bacteria occasionally observed intracellularly during infections in vitro (57) and in vivo (58). Thus, the majority of Hp bacteria remains extracellular and triggers intracellular host cell signaling from the outside (27–29). The latter dogma is in full agreement with these observations. Our findings support the hypothesis that CagL functions as a specialized adhesin that not only anchors the T4SS to the host surface through binding to integrin (26) but also promotes intracellular signal transduction as shown in the present study.
Finally, we demonstrated that binding of host cells to immobilized CagL triggers tyrosine phosphorylation of a number of signaling proteins that play known key roles in cell adhesion and proliferation. Western blotting using pan-phosphotyrosine antibodies indicated that immobilized CagLwt induces the tyrosine phosphorylation of four major protein species, which were identified as the two nonreceptor tyrosine kinases Src (60 kDa) and FAK (120 kDa) and the two growth factor receptor tyrosine kinases EGFR (170 kDa) and Her3/ErbB3 (200 kDa), respectively. Our finding that purified CagLwt alone can activate Src and FAK upon cell contact in vitro extends our previous observations that wt Hp but not the ΔcagL mutants induced Src and FAK activation during infection of AGS cells (26). In addition, our present study identified a long awaited bacterial factor that can activate EGFR. The fact that Hp profoundly activates EGFR has been known for some time (59–62). However, it was unclear whether EGFR can be activated by a structural T4SS component, a translocated cagPAI effector molecule, or another factor of Hp. Furthermore, the data presented here indicate that CagL alone can activate not only EGFR but also Her3/ErbB3, another member of this proto-oncogenic growth factor receptor family. The biological consequence of the activation of Her3/ErbB3 by CagL during infection is currently under investigation. In addition, we demonstrated that activation of Src, FAK, EGFR, and Her3/ErbB3 by CagL proceeds in a RGD-dependent manner that is required for cell spreading and focal adhesion formation. A putative signaling model involving CagL-triggered integrin activation, tyrosine kinase activation, and cell spreading is depicted in Fig. 9. Because CagL exhibits multiple protein signals on the bacterial cell surface (Fig. 1C) and T4SS-pili during infection in vivo (26), we propose that CagL not only binds to integrin α5β1 but may also induce integrin clustering. This conclusion is also supported by our previous observations showing that CagL-coated latex beads induced integrin α5β1 clustering when co-incubated with AGS cells (26). Clustering of integrins by extracellular substrates generates a variety of intracellular signals, including tyrosine phosphorylation of cytoskeleton-associated and other factors (9–12). We propose that binding of CagL to integrin α5β1 has similar effects. Integrins also cooperate with growth factor receptors and induce their transactivation. For example, it has been shown that EGFR transactivation can be mediated by FN (63, 64). EGFR transactivation requires metalloproteinase cleavage of proHB-EGF (65), and Hp-stimulated EGFR transactivation has the same requirement (61, 66). Interestingly, integrin α5β1 and ADAM-17 can physically interact in vitro and co-localize in HeLa cells (34) and AGS cells (33). Given that Hp promotes cell proliferation through EGFR transactivation by ADAM activation (67), our present data support the hypothesis that CagL-integrin interaction activates the metalloprotease ADAM-17 and subsequently EGFR.
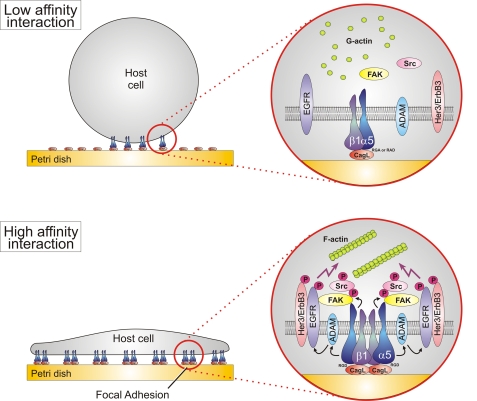
FIGURE 9.
Hypothetical model for CagL-dependent integrin targeting and activation of intracellular tyrosine kinase signaling. CagL mimics FN using its RGD motif to bind to α5β1 integrin. CagLRGA or CagLRAD mutants show low affinity binding, with cells being round and no signs of cell spreading or focal adhesion formation. We propose that CagL is able to trigger high affinity interactions, integrin clustering, and profound activation. Activated integrins can then stimulate FAK activity by phosphorylation of tyrosine 397, which is a major binding site for Src (11). CagL-stimulated integrins may also activate EGF and Her3/Neu receptors, possibly by stimulating a metalloprotease of the ADAM family (64, 66). The N-terminal domain of FAK also mediates the association with activated EGF receptor signaling complexes, and this association and FAK phosphorylation at tyrosine 397 are important for growth factor stimulated actin rearrangements, cell spreading, and cell motility (69).
Taken together, CagL is capable of mimicking a number of FN functions in vitro and can even trigger efficient spreading of FN−/− knock-out cells. The notion that convergent evolution has resulted in a molecular mimicry between CagL, a bacterial surface virulence protein, and FN, a eukaryotic extracellular matrix protein, is intriguing. Whether the persistent colonization of Hp of the human stomach epithelium and hence their co-evolution is one of the driving forces remains to be tested. Nonetheless, the FN-like properties of CagL and the stimulation of cancer-associated signaling by CagL are likely to play crucial roles in persistent Hp infection and may provide new insights into the molecular basis of Hp-induced carcinogenesis and metastasis. Based on the observation that CagL and not FN can activate Her3/ErbB3, it is tempting to propose that CagL may promote abnormal signaling cross-talk by aberrantly linking FN-dependent and FN-independent signaling pathways in the target cell. Interestingly, a yeast two-hybrid screen and GST pulldown assays revealed that T4SS-associated CagY (VirB10) and CagA proteins also bind to β1 integrin, which was proposed to facilitate the injection of CagA in an RGD-independent manner (32). However, it remained unclear whether and how binding of VirB10 and CagA to extracellular β1 integrin receptor could trigger intracellular signaling. Irrespective of the specific contribution of individual T4SS proteins (CagL, VirB10, and CagA) toward CagA injection as investigated previously (26, 32), the present data clearly demonstrate the functional importance of CagL alone in triggering transmembrane signaling to activate EGFR, Her3/ErbB3, Src, FAK, and probably other factors. Interestingly, when the purified repeat region 2 or the C terminus of VirB10 was immobilized, neither of these fragments could induce efficient cell spreading. Remarkably, however, when we mixed CagL with VirB10, the repeat region 2 (68) but not the integrin β1 interacting C terminus (32) enhanced the CagL effect (Fig. 7). This finding suggests that the internal repeat region of VirB10 and CagL may act cooperatively and that the C-terminal interaction of VirB10 with integrin β1 has a different function, further confirming that the observed cell spreading effect is specific for CagL. If other Hp factors such as extracellularly added VirB10 or CagA can also trigger similar and/or other intracellular signaling pathways and whether CagL-mediated activation of EGFR, Her3/ErbB3, Src, and Fak contributes to the injection of CagA during infection need to be investigated in future studies. Nevertheless, it seems clear that bacterial factors such as CagL, VirB10, and CagA, which interfere with host surface factors, could be used as novel tools to study integrin signaling and could be promising candidates as novel drug targets for specific intervention of the particularly severe gastric diseases caused by cagPAI-positive Hp strains.
Supplementary Material
Acknowledgments
We are very grateful to Terry Kwok (Monash University Melbourne, Australia) for help in CagL purification and critical discussion of the present data. We also thank Reinhard Faessler (Max Planck Institute Martinsried, Germany) providing the fibronectin knock-out cells, Phil Soriano (Fred Hutchinson Cancer Research Center, Seattle, WA) for the SYF cells, and David Schlaepfer (University of California, San Diego, CA) for the FAK knock-out cells. We thank Ralph Isberg for critical discussion of the data and kind advice.
This work was supported by Deutsche Forschungsgemeinschaft Grant Ba1671/8-1 and Research Grant R11408 from University College Dublin (to S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movies S1 and S2 and Figs. S1–S6.
- ECM
- extracellular matrix
- cagPAI
- cag pathogenicity island
- EGFR
- epidermal growth factor receptor
- FAK
- focal adhesion kinase
- hdn
- heat-denatured
- T4SS
- type IV secretion system
- wt
- wild-type
- FN
- fibronectin
- Hp
- H. pylori
- DMEM
- Dulbecco's modified Eagle's medium
- GST
- glutathione S-transferase
- BSA
- bovine serum albumin
- TRITC
- tetramethylrhodamine isothiocyanate
- FESEM
- field emission scanning electron microscopy.
REFERENCES
- 1.Johannson S., Svineng G., Wennerberg K., Armulik A., Lohikangas L. (1997) Front. Biosci. 2, 126–146 [DOI] [PubMed] [Google Scholar]
- 2.Magnusson M. K., Mosher D. F. (1998) Arterioscler. Thromb. Vasc. Biol. 18, 1363–1370 [DOI] [PubMed] [Google Scholar]
- 3.Humphries M. J., Travis M. A., Clark K., Mould A. P. (2004) Biochem. Soc. Trans. 32, 822–825 [DOI] [PubMed] [Google Scholar]
- 4.Larsen M., Artym V. V., Green J. A., Yamada K. M. (2006) Curr. Opin. Cell Biol. 18, 463–471 [DOI] [PubMed] [Google Scholar]
- 5.Dallas S. L., Chen Q., Sivakumar P. (2006) Curr. Top. Dev. Biol. 75, 1–24 [DOI] [PubMed] [Google Scholar]
- 6.Vakonakis I., Campbell I. D. (2007) Curr. Opin. Cell Biol. 19, 578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leiss M., Beckmann K., Girós A., Costell M., Fässler R. (2008) Curr. Opin. Cell Biol. 20, 502–507 [DOI] [PubMed] [Google Scholar]
- 8.Mao Y., Schwarzbauer J. E. (2005) Matrix Biol. 24, 389–399 [DOI] [PubMed] [Google Scholar]
- 9.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 10.Schwartz M. A., Ginsberg M. H. (2002) Nat. Cell Biol. 4, 65–68 [DOI] [PubMed] [Google Scholar]
- 11.Mitra S. K., Schlaepfer D. D. (2006) Curr. Opin. Cell Biol. 18, 516–523 [DOI] [PubMed] [Google Scholar]
- 12.Luo B. H., Carman C. V., Springer T. A. (2007) Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaidel-Bar R., Itzkovitz S., Ma'ayan A., Iyengar R., Geiger B. (2007) Nat. Cell Biol. 9, 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Streuli C. H., Akhtar N. (2009) Biochem. J. 418, 491–506 [DOI] [PubMed] [Google Scholar]
- 15.Ruoslahti E. (1996) Annu. Rev. Cell Dev. Biol. 12, 697–715 [DOI] [PubMed] [Google Scholar]
- 16.Eble J. A., Kühn K. (1997) Integrin-Ligand Interaction, Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 17.Boyle E. C., Finlay B. B. (2003) Curr. Opin. Cell Biol. 15, 633–639 [DOI] [PubMed] [Google Scholar]
- 18.Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., Briggs J. A., Gough T. S., Höök M., Campbell I. D., Potts J. R. (2003) Nature 42, 177–181 [DOI] [PubMed] [Google Scholar]
- 19.Hauck C. R., Agerer F., Muenzner P., Schmitter T. (2006) Eur. J. Cell Biol. 85, 235–242 [DOI] [PubMed] [Google Scholar]
- 20.Bierne H., Cossart P. (2007) Microbiol. Mol. Biol. Rev. 71, 377–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isberg R. R., Leong J. M. (1990) Cell 60, 861–871 [DOI] [PubMed] [Google Scholar]
- 22.Leong J. M., Fournier R. S., Isberg R. R. (1990) EMBO J. 9, 1979–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dersch P., Isberg R. R. (1999) EMBO J. 18, 1199–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamburger Z. A., Brown M. S., Isberg R. R., Bjorkman P. J. (1999) Science 286, 291–295 [DOI] [PubMed] [Google Scholar]
- 25.Isberg R. R., Leong J. M. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 6682–6686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwok T., Zabler D., Urman S., Rohde M., Hartig R., Wessler S., Misselwitz R., Berger J., Sewald N., König W., Backert S. (2007) Nature 449, 862–866 [DOI] [PubMed] [Google Scholar]
- 27.Peek R. M., Jr., Blaser M. J. (2002) Nat. Rev. Cancer 2, 28–37 [DOI] [PubMed] [Google Scholar]
- 28.Backert S., Meyer T. F. (2006) Curr. Opin. Microbiol. 9, 207–217 [DOI] [PubMed] [Google Scholar]
- 29.Amieva M. R., El-Omar E. M. (2008) Gastroenterology 134, 306–323 [DOI] [PubMed] [Google Scholar]
- 30.Backert S., Fronzes R., Waksman G. (2008) Trends Microbiol. 16, 409–413 [DOI] [PubMed] [Google Scholar]
- 31.Aly K. A., Baron C. (2007) Microbiology. 153, 3766–3775 [DOI] [PubMed] [Google Scholar]
- 32.Jiménez-Soto L. F., Kutter S., Sewald X., Ertl C., Weiss E., Kapp U., Rohde M., Pirch T., Jung K., Retta S. F., Terradot L., Fischer W., Haas R. (2009) PLoS Pathog. 5, e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha A., Backert S., Hammond C. E., Gooz M., Smolka A. J. (2010) Gastroenterology, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bax D. V., Messent A. J., Tart J., van Hoang M., Kott J., Maciewicz R. A., Humphries M. J. (2004) J. Biol. Chem. 279, 22377–22386 [DOI] [PubMed] [Google Scholar]
- 35.Sieg D. J., Hauck C. R., Schlaepfer D. D. (1999) J. Cell Sci. 112, 2677–2691 [DOI] [PubMed] [Google Scholar]
- 36.Klinghoffer R. A., Sachsenmaier C., Cooper J. A., Soriano P. (1999) EMBO J. 18, 2459–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyberg P., Sakai T., Cho K. H., Caparon M. G., Fässler R., Björck L. (2004) EMBO J. 23, 2166–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schröder A., Schröder B., Roppenser B., Linder S., Sinha B., Fässler R., Aepfelbacher M. (2006) Mol. Biol. Cell 17, 5198–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delahay R. M., Balkwill G. D., Bunting K. A., Edwards W., Atherton J. C., Searle M. S. (2008) J. Mol. Biol. 377, 956–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cary L. A., Klinghoffer R. A., Sachsenmaier C., Cooper J. A. (2002) Mol. Cell. Biol. 22, 2427–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tammer I., Brandt S., Hartig R., König W., Backert S. (2007) Gastroenterology 132, 1309–1319 [DOI] [PubMed] [Google Scholar]
- 42.Brandt S., Wessler S., Hartig R., Backert S. (2009) Cell Motil. Cytoskeleton 66, 874–892 [DOI] [PubMed] [Google Scholar]
- 43.Brandt S., Kwok T., Hartig R., König W., Backert S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9300–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dickinson C. D., Veerapandian B., Dai X. P., Hamlin R. C., Xuong N. H., Ruoslahti E., Ely K. R. (1994) J. Mol. Biol. 236, 1079–1092 [DOI] [PubMed] [Google Scholar]
- 45.Bockholt S. M., Burridge K. (1993) J. Biol. Chem. 268, 14565–14567 [PubMed] [Google Scholar]
- 46.Sakai T., Johnson K. J., Murozono M., Sakai K., Magnuson M. A., Wieloch T., Cronberg T., Isshiki A., Erickson H. P., Fässler R. (2001) Nat. Med. 7, 324–330 [DOI] [PubMed] [Google Scholar]
- 47.Takahashi S., Leiss M., Moser M., Ohashi T., Kitao T., Heckmann D., Pfeifer A., Kessler H., Takagi J., Erickson H. P., Fässler R. (2007) J. Cell Biol. 178, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aota S., Nomizu M., Yamada K. M. (1994) J. Biol. Chem. 269, 24756–24761 [PubMed] [Google Scholar]
- 49.Spitzfaden C., Grant R. P., Mardon H. J., Campbell I. D. (1997) J. Mol. Biol. 265, 565–579 [DOI] [PubMed] [Google Scholar]
- 50.Copié V., Tomita Y., Akiyama S. K., Aota S., Yamada K. M., Venable R. M., Pastor R. W., Krueger S., Torchia D. A. (1998) J. Mol. Biol. 277, 663–682 [DOI] [PubMed] [Google Scholar]
- 51.Redick S. D., Settles D. L., Briscoe G., Erickson H. P. (2000) J. Cell Biol. 149, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Nhieu G. T., Isberg R. R. (1991) J. Biol. Chem. 266, 24367–24375 [PubMed] [Google Scholar]
- 53.Leahy D. J., Aukhil I., Erickson H. P. (1996) Cell 84, 155–164 [DOI] [PubMed] [Google Scholar]
- 54.Tran Van Nhieu G., Isberg R. R. (1993) EMBO J. 12, 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isberg R. R., Tran Van Nhieu G. (1994) Trends. Microbiol. 2, 10–14 [DOI] [PubMed] [Google Scholar]
- 56.Arencibia I., Frankel G., Sundqvist K. G. (2002) Eur. J. Immunol. 32, 1129–1138 [DOI] [PubMed] [Google Scholar]
- 57.Kwok T., Backert S., Schwarz H., Berger J., Meyer T. F. (2002) Infect Immun 70, 2108–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dubois A., Borén T. (2007) Cell. Microbiol. 9, 1108–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keates S., Sougioultzis S., Keates A. C., Zhao D., Peek R. M., Jr., Shaw L. M., Kelly C. P. (2001) J. Biol. Chem. 276, 48127–48134 [DOI] [PubMed] [Google Scholar]
- 60.Keates S., Keates A. C., Nath S., Peek R. M., Jr., Kelly C. P. (2005) Gut 54, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du Y., Danjo K., Robinson P. A., Crabtree J. E. (2007) Microbes Infect. 9, 838–846 [DOI] [PubMed] [Google Scholar]
- 62.Yan F., Cao H., Chaturvedi R., Krishna U., Hobbs S. S., Dempsey P. J., Peek R. M., Jr., Cover T. L., Washington M. K., Wilson K. T., Polk D. B. (2009) Gastroenterology 136, 1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuwada S. K., Li X. (2000) Mol. Biol. Cell 11, 2485–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuo M., Sakurai H., Ueno Y., Ohtani O., Saiki I. (2006) Cancer Sci. 97, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prenzel N., Zwick E., Daub H., Leserer M., Abraham R., Wallasch C., Ullrich A. (1999) Nature 402, 884–888 [DOI] [PubMed] [Google Scholar]
- 66.Wallasch C., Crabtree J. E., Bevec D., Robinson P. A., Wagner H., Ullrich A. (2002) Biochem. Biophys. Res. Commun. 295, 695–701 [DOI] [PubMed] [Google Scholar]
- 67.Joh T., Kataoka H., Tanida S., Watanabe K., Ohshima T., Sasaki M., Nakao H., Ohhara H., Higashiyama S., Itoh M. (2005) Dig. Dis. Sci. 50, 2081–2089 [DOI] [PubMed] [Google Scholar]
- 68.Rohde M., Püls J., Buhrdorf R., Fischer W., Haas R. (2003) Mol. Microbiol. 49, 219–234 [DOI] [PubMed] [Google Scholar]
- 69.Sieg D. J., Hauck C. R., Ilic D., Klingbeil C. K., Schaefer E., Damsky C. H., Schlaepfer D. D. (2000) Nat. Cell Biol. 2, 249–256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.