Abstract
The initial encounter between a microbe and its host can dictate the success of the interaction, be it symbiosis or pathogenesis. This is the case, for example, in the symbiosis between the bacterium Vibrio fischeri and the squid Euprymna scolopes, which proceeds via a biofilm-like bacterial aggregation, followed by entry and growth. A key regulator, the sensor kinase RscS, is critical for symbiotic biofilm formation and colonization. When introduced into fish symbiont strains that naturally lack the rscS gene and cannot colonize squid, RscS permits colonization, thereby extending the host range of these bacteria. RscS controls biofilm formation by inducing transcription of the symbiosis polysaccharide (syp) gene locus. Transcription of syp also requires the σ54-dependent activator SypG, which functions downstream of RscS. In addition to these regulators, SypE, a response regulator that lacks an apparent DNA binding domain, exerts both positive and negative control over biofilm formation. The putative sensor kinase SypF and the putative response regulator VpsR, both of which contribute to control of cellulose production, also influence biofilm formation. The wealth of regulators and the correlation between biofilm formation and colonization adds to the already considerable utility of the V. fischeri-E. scolopes model system.
Introduction
20 years after the model system was established, the symbiosis between the bacterium Vibrio fischeri and its symbiotic host, the squid Euprymna scolopes, continues to provide rich and novel insights into a variety of problems in bacteria-host interactions (reviewed by (McFall-Ngai, 2008; Ruby, 2008; Stabb et al., 2000; Visick and Ruby, 2006)). Indeed, in the past year alone, researchers have uncovered numerous components of the bacteria-host interaction network, including the ability of the squid's symbiotic light organ to respond to light, colonization, and bacterially-released small molecules including autoinducers and a component of peptidoglycan, Trachael Cytotoxin (Chun et al., 2008; Tong et al., 2009; Troll et al., 2009). These studies suggest an interaction of a complexity that rivals that of traditional mammalian models. Furthermore, host defense cells called haemocytes contribute to specificity by binding to and engulfing non-native or mutant bacteria preferentially over native symbionts, thus demonstrating the utility of this system as a model for innate immunity (Nyholm et al., 2009). Understanding of the evolution and ecology of the interaction has advanced through studies of natural populations combined with experimental manipulations, which suggested that a limited number of bacteria (1–2) likely enter and populate each of the 6 internal crypts that comprise the light organ of the newly hatched and initially uncolonized squid; as a result, polyclonal but segregated populations can exist within a single squid (Wollenberg and Ruby, 2009). Finally, genomic analysis of representative symbiotic and non-symbiotic V. fischeri strains led to the recognition that a single regulatory gene, rscS, could be sufficient to alter host specificity, as it enabled a fish symbiont to colonize squid (Mandel et al., 2009). The molecular details underlying the influence of this regulator on colonization and specificity will be the main subjects of this review.
Symbiotic initiation and host specificity
The V. fischeri-E. scolopes symbiosis is highly specific. Juvenile squid hatch uncolonized but become colonized within hours following exposure to symbiosis competent bacteria. Despite the presence of numerous other bacteria in the seawater, including closely related bacteria such as Vibrio parahaemolyticus, only V. fischeri successfully colonizes the squid's symbiotic light organ (McFall-Ngai and Ruby, 1991). Furthermore, not all strains of V. fischeri are equally capable of colonizing, or are even symbiosis-competent (Nishiguchi et al., 1998; Ruby and Lee, 1998). For example, V. fischeri strains isolated from symbiosis with the fish Monocentris japonica generally fail to colonize E. scolopes (Mandel et al., 2009).
A number of factors contribute to this remarkable host specificity. Upon entering the light organ, V. fischeri must pass through mucus-filled ducts that contain outward-beating cilia (McFall-Ngai and Ruby, 1998; Visick and McFall-Ngai, 2000) and nitric oxide (Davidson et al., 2004), an anti-bacterial defense. In addition, the crypts contain halide peroxidase, a host defense protein that catalyzes the production of hypochlorous acid, which is toxic to bacteria (Small and McFall-Ngai, 1999; Tomarev et al., 1993; Weis et al., 1996). Finally, macrophage-like haemocytes circulate within the crypts (Nyholm and McFall-Ngai, 1998). V. fischeri must survive each of these host-imposed stresses that presumably decrease the chances of colonization by other microbes.
Surprisingly, host specificity appears to begin even before the bacteria enter the ducts. Newly hatched juvenile squid exhibit a short permissive period in which non-symbionts (and even particles such as beads) can enter the light organ, followed by a non-permissive period during which nothing can enter (Nyholm et al., 2002). Subsequently, mucus is secreted to the surface of the light organ (Nyholm et al., 2000; Nyholm et al., 2002). V. fischeri aggregates within this mucus, an event that is critical to colonization (Nyholm et al., 2000; Yip et al., 2006). For example, a mutant lacking the sensor kinase RscS fails to aggregate, a phenotype that correlates well with its severe defect in symbiotic initiation (Visick and Skoufos, 2001; Yip et al., 2006). Studies of additional mutants have provided further support for the connections observed between aggregation and initiation of colonization (Millikan and Ruby, 2002; Whistler et al., 2006).
Some non-symbionts, such as the closely related V. parahaemolyticus, appear to be capable of adhering to the light organ, but others, such as the Gram positive Bacillus cereus, are not, supporting the idea that specificity occurs at this stage of colonization (Nyholm et al., 2000). Furthermore, a 1:1 mixture of V. fischeri and the non-symbiont V. parahaemolyticus results in an aggregate consisting of more than 80% V. fischeri cells (Nyholm and McFall-Ngai, 2003). These latter data suggest that specificity results, in part, from a superior ability of V. fischeri to interact with the surface of the squid=s light organ.
RscS and colonization
Initiation of symbiotic colonization requires a two-component sensor kinase gene that we designated rscS, for regulator of symbiotic colonization sensor (Visick and Skoufos, 2001). A subset of juvenile squid exposed to an rscS mutant remain uncolonized, while the rest become colonized only after a significant delay (Visick and Skoufos, 2001). Initial characterization failed to reveal defects in any known or suspected colonization traits, including motility and bioluminescence; however, subsequent studies (described below) uncovered an important role for RscS in inducing biofilm formation. The symbiotic defect of the rscS mutant is likely to be due to its failure to aggregate on the surface of the light organ, an event that we hypothesize to correspond to biofilm formation (Yip et al., 2006).
In the sequenced squid symbiont strain ES114, the rscS gene is located between glpR and glpK, genes involved in glycerol regulation and metabolism, respectively (Ruby et al., 2005; Visick and Skoufos, 2001). When the genome of a second V. fischeri strain, the M. japonica isolate MJ11, became available, it was noted that rscS is absent from the glp locus (Mandel et al., 2009). Because the fish symbiont is not proficient at squid colonization, it was proposed that lack of rscS could account for the colonization deficiency of this strain. Indeed, introduction of rscS on a multi-copy plasmid permits squid colonization by MJ11 (Mandel et al., 2009). These data demonstrated that RscS alone is sufficient to extend the host range of V. fischeri from fish to squid. In further support of this notion, disruption of rscS in a variety of V. fischeri isolates from different geographic locations impairs symbiotic initiation (Mandel et al., 2009). These results demonstrate that a single regulatory gene can alter the host range of an animal-associated microorganism and point to RscS and the genes it controls as key regulators of early steps in host colonization.
RscS and the syp locus
RscS is predicted to be a member of the histidine sensor kinase class of two-component regulators (Visick and Skoufos, 2001). Sensor kinases typically sense specific environmental signals and transduce that information by initiating a phosphorelay, resulting in the phosphorylation of their cognate response regulators and thus an output such as altered transcription of target genes (Gao and Stock, 2009). The gene for rscS, however, is not linked to a gene encoding a response regulator, and the 40 response regulators recognizably encoded in the V. fischeri genome made the hunt for a partner non-trivial (Hussa et al., 2007). Furthermore, neither a partner nor a target could be readily identified based on similarity in phenotype, as the rscS mutant does not exhibit any phenotype in culture (Visick and Skoufos, 2001).
A break-through in understanding the role of RscS came with the discovery of an 18-gene symbiosis polysaccharide gene locus, syp (Yip et al., 2005; Yip et al., 2006). Mutants defective for specific syp genes exhibit phenotypes similar to those of the rscS mutant: defective for symbiotic initiation, but not for a variety of traits tested in culture (Yip et al., 2005). The syp genes encode proteins with similarity to those involved in exopolysaccharide biosynthesis, including six putative glycosyltransferase genes. Of note, the syp locus also encodes two response regulators, potential partners for RscS. One of them, SypG, is a putative a σ54-dependent enhancer binding protein, while the other, SypE, lacks any apparent DNA binding sequences.
A connection between RscS and syp was made when RscS was overproduced in a syp reporter strain. Because RscS is predicted to sense an environmental signal, potentially missing under standard laboratory conditions, a plasmid containing the rscS gene was mutagenized in an attempt to generate an allele with increased activity. This approach yielded a plasmid that substantially overproduces the RscS protein, resulting in increased transcription of the syp locus (Geszvain and Visick, 2008a; Yip et al., 2006). It also induces biofilm formation: whereas control cells produce smooth colonies, rscS-overexpressing cells produce wrinkled colonies (Fig. 1). This wrinkled colony phenotype is similar to other biofilm-forming Vibrio species such as Vibrio cholerae, which produces rugose colonies under certain conditions (Yildiz and Visick, 2009). Furthermore, rscS-overexpressing cells produce a thick pellicle at the air-liquid interface of statically grown cultures, a phenomenon not seen with wild-type V. fischeri (Yip et al., 2006); incredibly, these pellicles are so strong that the cultures can be inverted without loss of the liquid medium (Fig. 1). These biofilm phenotypes depend upon a functional syp locus as well as RscS overproduction.
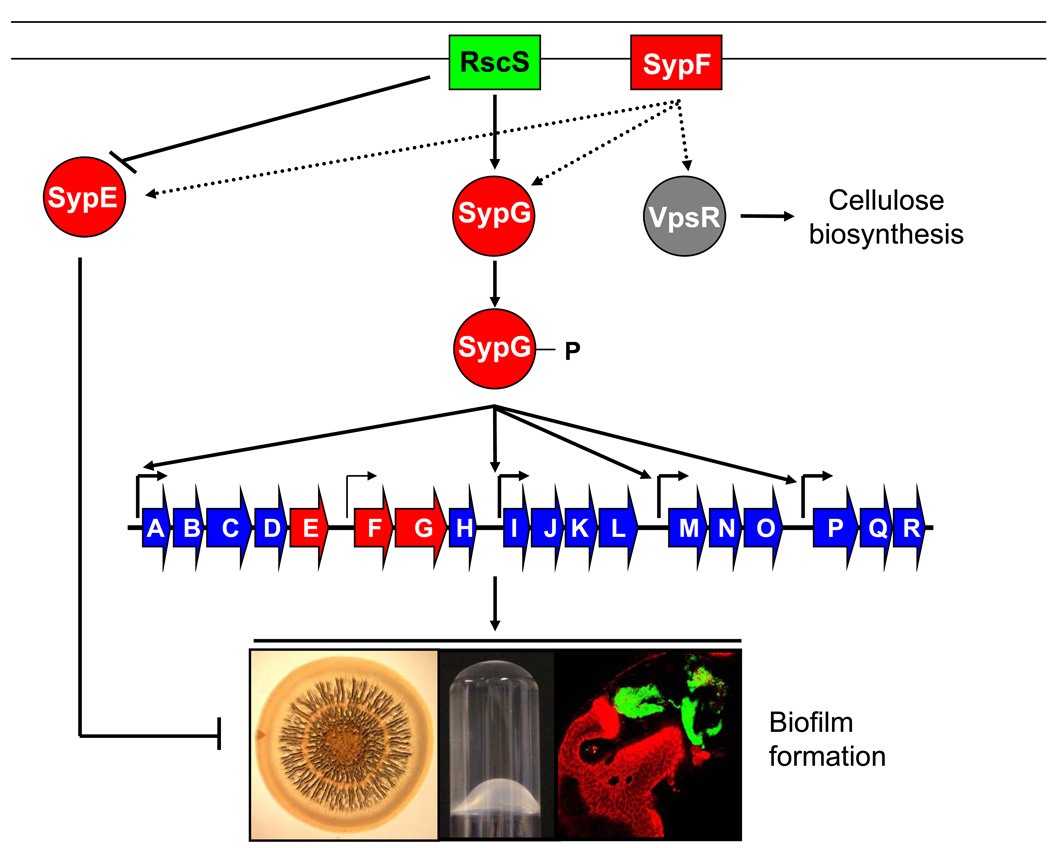
Figure 1. Model for control of biofilm formation in V. fischeri.
Depicted here is a model based on current data regarding the potential roles of the syp regulators. The sensor kinase RscS acts upstream of SypG, presumably serving as a phosphodonor in response to some as-yet unidentified environmental signal, perhaps from the squid. Once phosphorylated, SypG is predicted to directly bind to sequences upstream of each of 4 syp operons to activate syp transcription by σ54-containing RNA polymerase. The Syp proteins contribute to biofilm formation in culture, including the formation of wrinkled colonies and pellicles, as well as in situ biofilm formation and colonization. RscS overproduction also appears to inactivate SypE, which inhibits biofilm formation induced by overproduction of SypG at a level downstream from syp transcription; how RscS inactivates SypE is as yet unknown. Biofilm formation can be also induced by overexpression of a sypF allele with increased activity (SypF1) in a manner that depends in part on SypG and SypE, and in part on VpsR, which promotes cellulose biosynthesis.
The physiological importance of these biofilm phenotypes was underscored by the finding that RscS-overproducing cells form substantially enhanced aggregates on the surface of the light organ — upwards of 20X bigger than controls. As with the in vivo biofilms, disruption of the syp locus eliminates or substantially reduces the size of the symbiotic aggregates induced by RscS. Importantly, this RscS-directed activity substantially promotes symbiotic colonization, as determined by competitive colonization experiments: squid inoculated with mixtures of vector-control and RscS-overexpressing wild-type cells become colonized exclusively by those that contain the RscS expression plasmid (Yip et al., 2006)
In summary, a major function of RscS appears to be in controlling the syp locus, resulting in biofilm formation and symbiotic colonization. The ability of RscS to induce squid colonization by normally non-symbiotic strain MJ11 can likely be attributed to its ability to promote symbiotic biofilm formation, although this idea has not yet been directly tested. In support of it, however, are the findings that the syp locus is conserved in MJ11 and that RscS overexpression promotes biofilm formation by MJ11 (Mandel et al., 2009).
RscS integrates positive and negative signals
RscS is predicted to be a hybrid sensor kinase. The C-terminus contains putative histidine kinase/ATPase (HisKA/HATPase), receiver (REC), and histidine phosphotransferase (Hpt) domains (Visick and Skoufos, 2001). This domain structure is similar to such well-characterized proteins as ArcB and BvgS that undergo two internal phosphorelay events prior to donation of the phosphoryl group to the cognate response regulator (Beier and Gross, 2008; Gao and Stock, 2009). Such additional domains involved in an intramolecular phosphorelay provide opportunities for multiple levels of control. The N-terminal region of RscS contains a large periplasmic domain flanked by two transmembrane helices, and a cytoplasmic PAS domain, all of which could contribute to sensory perception. The natural ligand(s) remains unknown, however.
Mutant alleles of rscS were constructed to study the roles of the various domains (Geszvain and Visick, 2008b). Perhaps not surprisingly, alterations of key residues in the HisKA and REC domains disrupt function, as measured by induction of syp transcription and biofilm formation. These data indicate that RscS functions as a kinase. In contrast, a substitution within the Hpt domain at a histidine predicted to be critical for phosphotransfer reduces but does not eliminate function. Perhaps the specific change facilitates phosphotransfer to the downstream response regulator from the conserved histidine within the HisKA domain, or perhaps the putative Hpt domain does not function as an Hpt. Additional work will be necessary to clarify the role of the predicted Hpt domain.
Substitutions of key residues in the PAS domain also abolish RscS activity. In RscS, the predicted location of the PAS domain is on the cytoplasmic face of the inner membrane, where it might be expected to respond to an internal cue. PAS domains in other proteins detect small molecules such as FAD or FMN, light, or oxygen (Taylor and Zhulin, 1999). Alterations to residues predicted to be involved in binding FAD lead to a greater disruption in RscS activity than do substitutions in those predicted to be critical for FMN binding, suggesting that the PAS domain of RscS might bind to a FAD cofactor (Geszvain and Visick, 2008b).
Signaling by FAD, if it occurs, might be influenced by membrane localization and/or by signaling through the periplasmic loop or transmembrane regions. Indeed, membrane localization of RscS seems important, as a deletion derivative containing only the cytoplasmic portion of the protein (including PAS) exhibits reduced activity (Geszvain and Visick, 2008b). Surprisingly, however, deletions of the periplasmic loop result in increased activity, as do some substitutions within the first transmembrane helix, indicating that binding of an unidentified ligand to the periplasmic domain might inhibit, rather than activate, RscS activity. Together, these data reveal that RscS is a complex regulator, potentially integrating both inhibitory and stimulatory signals to initiate a phosphorelay that critically requires predicted phosphorylated residues in the HisKA and REC but not Hpt domains. An exciting future direction of this work will be to determine the nature of the signal(s) received by RscS.
RscS works upstream of SypG
The availability of RscS-induced phenotypes permitted a search for the response regulator(s) that functions downstream of RscS (Hussa et al., 2008). The rscS over expression plasmid was introduced into each of 35 response regulator mutants (all but five of the 40 encoded by the V. fischeri genome), and the resulting phenotypes were evaluated. Most mutants exhibit biofilm phenotypes indistinguishable from those of the wild-type overexpression control. In several cases, some measures of biofilm formation (including glass attachment and pellicles) were impacted by disruption of specific response regulators, including sypG, sypE, vpsR, flrC, arcA, and VF1401. However, wrinkled colony formation is affected only by the disruption of either sypG or sypE, and induction of syp transcription by RscS depends only on sypG. The strong requirement for SypG in all RscS-induced phenotypes suggests that RscS works upstream of SypG (Hussa et al., 2008).
Somewhat inconsistent with the above conclusion is the finding that overproduction of SypG alone does not induce the formation of wrinkled colonies or strong pellicles, although it does induce syp transcription. Furthermore, altering SypG by substituting glutamate for a conserved aspartate, a change that in many other response regulators produces a constitutively active protein (e.g., (Freeman and Bassler, 1999)), increases syp induction but does not promote biofilm formation (Hussa et al., 2008). Although it is not always the case that the phenotypes of response regulators phenocopy those of their partner sensor kinases, these data raised the possibility that additional factors might be involved. Indeed, it was subsequently determined that deletion of the response regulator gene sypE permits the SypG overproduction strain to form wrinkled colonies and pellicles (Hussa et al., 2008). This latter work thus established conditions under which overproduced SypG and overproduced RscS could induce similar phenotypes, supporting the idea that these proteins could work together. Furthermore, it appears that overproduction of RscS must somehow lead to inactivation of SypE. The role of SypE will be discussed further below.
SypG and σ54 regulate syp transcription
SypG is a putative σ54-dependent response regulator, and binding sites for σ54-containing RNA polymerase exist within the syp locus (Fig. 1). Specifically, σ54 sites exist within each of the four largest gaps between genes of the 18-gene syp locus (Yip et al., 2005). Results from primer extension experiments designed to map the start sites of three of the genes (sypA, sypI, and sypM) are consistent with the putative σ54-dependent promoters, and mutagenesis studies verify that transcription of at least sypD and sypN depends upon the gene for σ54, rpoN (Yip et al., 2005). Because σ54-containing RNA polymerase requires a transcriptional activator to provide the energy for open complex formation (Buck et al., 2000), it seems reasonable that the syp locus would require an activator such as SypG to bind and activate transcription. Indeed, bioinformatic analysis revealed the presence of a conserved 22 bp sequence upstream of each of the putative σ54 binding sites that we hypothesize to be the SypG binding site (Yip et al., 2005). Our preliminary data support the importance of the conserved sequences in syp transcription (Hussa and Visick, unpublished), but the precise role remains to be determined.
SypE, a novel response regulator
Although RscS functions upstream of SypG, the regulatory network is far from straightforward. Biofilm formation, while readily induced by overexpression of RscS, requires inactivation of sypE when SypG is overproduced. However, loss of SypE impairs biofilm formation induced by RscS. Thus, SypE appears to play both positive and negative roles in biofilm formation. Loss of sypE exerts only small effects on syp transcription, regardless of whether RscS or SypG is overproduced, suggesting that its impact is not at the level of syp transcription ((Hussa et al., 2008) and Hussa and Visick, unpublished). These data indicate that SypE controls biofilm formation at another level, such as post-transcriptional control of the synthesis of the Syp polysaccharide or via control of the synthesis of another component or regulator of the biofilm matrix.
SypE is an unusual two-component response regulator in that its REC domain is located in the center of the protein (Yip et al., 2005). In addition, instead of the typical DNA binding domain, SypE contains a putative serine kinase domain in its N-terminus and a putative serine phosphatase domain in its C-terminus. The potential opposing activities of these domains suggest a possible molecular basis for the apparent dual role of this protein in controlling biofilm formation. The phosphorylation state of the REC domain might then determine which activity is favored.
SypE thus is another complex regulator of biofilm formation in V. fischeri. Although the syp locus is conserved in many Vibrio species, sypE is generally lacking in these other syp loci (Yip et al., 2005). These data suggest that V. fischeri uses SypE to fine-tune its control of biofilm formation, perhaps to coordinate Syp production with other factors necessary for squid colonization. However, SypE itself is not a critical colonization factor, as disruption of sypE does not prevent symbiotic initiation (Hussa et al., 2007). Many intriguing questions remain to be answered about this unusual regulatory protein.
Roles for SypF and VpsR
In addition to the response regulators SypE and SypG, the syp cluster encodes a hybrid sensor kinase, SypF. Like RscS, SypF contains three domains predicted to be involved in a phosphorelay, HisKA/HATPase, REC, and Htp, as well as a putative periplasmic loop (Yip et al., 2005). Overproduction of SypF exerts no discernible effect on biofilm formation. However, an increased activity allele, sypF1, was isolated that induces wrinkled colony formation, pellicle production, and increased adherence to a glass surface (Darnell et al., 2008). SypF1 contains a serine to phenylalanine change at amino acid 247. S247 is located within the HisKA domain, three residues N-terminal to the conserved histidine predicted to be the site of phosphorylation; a change at this position could impact kinase activity. However, the involvement of these conserved residues in a phosphorelay has not yet been assessed biochemically.
Given the location of sypF between the response regulator genes sypE and sypG, it seemed likely that one or both would be necessary for SypF1-mediated induction of biofilm formation. However, the biofilm phenotypes are reduced, but not eliminated, in sypE and sypG mutants (Darnell et al., 2008). Subsequent work revealed that the residual biofilm formation induced by SypF1 overexpression in sypE and sypG mutants depends on an unlinked gene encoding VpsR, a putative response regulator. Disruption of only vpsR also fails to eliminate biofilm formation, but a vpsR sypG double mutant mimics the uninduced wild type. These data indicate that, at least under these conditions, SypF contributes to biofilm control through two distinct pathways.
VpsR exhibits sequence similarity to a protein with the same name in V. cholerae (Darnell et al., 2008; Yildiz et al., 2001). The V. cholerae protein is a major regulator of biofilm formation, through its control of the vps polysaccharide locus (Yildiz et al., 2001). Although V. fischeri contains genes similar to those within one segment of the V. cholerae vps locus (termed vps-II), this locus does not appear to be responsible for the biofilms induced by SypF1 overexpression. Instead, V. fischeri (but not V. cholerae) contains a cellulose biosynthesis locus that is necessary for biofilm formation induced by SypF1 overproduction (Darnell et al., 2008). Similarly, overproduction of VpsR also induces the formation of a cellulose-dependent biofilm. The roles of SypF, VpsR, and cellulose in biofilm formation and symbiotic colonization need to be clarified although, intriguingly, a mutant defective for vpsR exhibits a small defect in symbiotic initiation (Hussa et al., 2007).
Concluding remarks
Wild type V. fischeri does not make impressive biofilms in culture. However, through genetic analysis and an appreciation of the natural symbiotic lifestyle of the organism, remarkable progress has been made to elucidate a complex regulatory network that controls biofilm formation (Fig. 1). The integrated signaling circuitry underlying biofilm development and symbiosis includes the hybrid sensor kinase RscS (a protein that is sufficient to broaden the host range of V. fischeri to include E. scolopes) and the downstream response regulator, SypG. Together, these proteins respond to as-yet-unknown squid or environmental signals and activate transcription of the symbiosis polysaccharide (syp) locus, which leads to biofilm-dependent initiation of colonization. An additional, novel response regulator, SypE, interacts with the RscS-SypG pathway and plays both positive and negative roles in control of biofilm formation. Adding to the complexity, production of the Syp polysaccharide might be coordinated with the biosynthesis of cellulose, through the putative sensor kinase SypF and the putative response regulator VpsR. A potential model that encompasses these findings is that multiple signals are received and integrated such that V. fischeri produces one type of biofilm (e.g., Syp-produced) at some stage(s) of its symbiotic or free-living life cycle and a distinct type (e.g., cellulose) at another stage(s).
While much remains to be understood, evidence to date suggests that some aspect of the V. fischeri interaction with its host activates biofilm formationBat least during the initial encounter, if not beyond. Negative control also appears to be importantBperhaps to fine-tune synthesis of biofilm components, to produce a distinct type of biofilm under different conditions, or even to turn biofilm formation off when it is not needed. This system is one of the few in which factors involved in biofilm formation in culture have been clearly correlated with biofilm formation in a natural model of bacteria-animal associations. This ability to correlate biofilm formation in culture and during the association of V. fischeri with E. scolopes thus adds to the already considerable versatility and utility of this remarkable model system for the study of both benign and pathogenic interactions between bacteria and eukaryotes.
Acknowledgments
I thank Mark Mandel, Alan Wolfe, Jon Visick, and past and present members of my lab for many wonderful discussions and for their suggestions on this manuscript, and Satoshi Shibata for donating an image for Fig. 1. I also gratefully acknowledge funding from the National Institutes of Health (GM59690 to KLV.), which supports our work investigating RscS, the syp locus and symbiotic colonization by V. fischeri.
References
- Beier D, Gross R. The BvgS/BvgA phosphorelay system of pathogenic Bordetellae: structure, function and evolution. Adv Exp Med Biol. 2008;631:149–160. doi: 10.1007/978-0-387-78885-2_10. [DOI] [PubMed] [Google Scholar]
- Buck M, Gallegos M-T, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo Mde F, Casavant TL, Soares MB, Ruby EG, McFall-Ngai MJ. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A. 2008;105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell CL, Hussa EA, Visick KL. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J. Bacteriol. 2008;190:4941–4950. doi: 10.1128/JB.00197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means 'yes' in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell. Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological Insights from Structures of Two-Component Proteins. Annu Rev Microbiol. 2009 doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geszvain K, Visick KL. Multiple factors contribute to keeping levels of the symbiosis regulator RscS low. FEMS Microbiol lett. 2008a;285:33–39. doi: 10.1111/j.1574-6968.2008.01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geszvain K, Visick KL. The hybrid sensor kinase RscS integrates positive and negative signals to modulate biofilm formation in Vibrio fischeri. J. Bacteriol. 2008b;190:4437–4446. doi: 10.1128/JB.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa EA, O'Shea TM, Darnell CL, Ruby EG, Visick KL. Two-component response regulators of Vibrio fischeri: identification, mutagenesis and characterization. J. Bacteriol. 2007;189:5825–5838. doi: 10.1128/JB.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa EA, Darnell CL, Visick KL. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M. Host-microbe symbiosis: the squid-Vibrio association--a naturally occurring, experimental model of animal/bacterial partnerships. Adv Exp Med Biol. 2008;635:102–112. doi: 10.1007/978-0-387-09550-9_9. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Sepiolids and vibrios: when first they meet. Reciprocal interactions between host and symbiont lead to the creation of a complex light-emitting organ. BioScience. 1998;48:257–265. [Google Scholar]
- Millikan DS, Ruby EG. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 2002;68:2519–2528. doi: 10.1128/AEM.68.5.2519-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi MK, Ruby EG, McFall-Ngai MJ. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-vibrio symbioses. Appl. Environ. Microbiol. 1998;64:3209–3213. doi: 10.1128/aem.64.9.3209-3213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: Description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol. Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 2002;68:5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl. Environ. Microbiol. 2003;69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11:483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Lee K-H. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. App.. Environ. Microbiol. 1998;64:805–812. doi: 10.1128/aem.64.3.805-812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. Complete genome sequence of Vibrio fischeri: A symbiotic bacterium with pathogenic congeners. Proc Nat Acad Sci USA. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG. Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol. 2008;6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small A, McFall-Ngai M. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J. Cell. Biochem. 1999;72:445–457. [PubMed] [Google Scholar]
- Stabb EV, Visick KL, Millikan DS, Corcoran AA, Gilson L, Nyholm SV, McFall-Ngai M, Ruby EG. Recent Advances in Marine Science and Technology. Honolulu: Pacon International; 2000. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction; pp. 269–277. [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomarev SI, Zinovieva RD, Weis VM, Chepelinsky AB, Piatigorsky J, McFall-Ngai MJ. Abundant mRNAs in the squid light organ encode proteins with a high similarity to mammalian peroxidases. Gene. 1993;132:219–226. doi: 10.1016/0378-1119(93)90199-d. [DOI] [PubMed] [Google Scholar]
- Tong D, Rozas NS, Oakley TH, Mitchell J, Colley NJ, McFall-Ngai MJ. Evidence for light perception in a bioluminescent organ. Proc Natl Acad Sci U S A. 2009;106:9836–9841. doi: 10.1073/pnas.0904571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, McFall-Ngai MJ. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol. 2009;11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, McFall-Ngai MJ. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J. Bacteriol. 2000;182:1779–1787. doi: 10.1128/jb.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Skoufos LM. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 2001;183:835–842. doi: 10.1128/JB.183.3.835-842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Ruby EG. Vibrio fischeri and its host: it takes two to tango. Curr. Op. Microbiol. 2006;9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Weis VM, Small AL, McFall-Ngai MJ. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc. Natl. Acad. Sci. USA. 1996;93:13683–13688. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whistler CA, Koropatnick TA, Pollack A, McFall-Ngai MJ, Ruby EG. The GacA global regulator of Vibrio fischeri is required for normal host tissue responses that limit subsequent bacterial colonization. Cell Microbiol. 2006;9:766–778. doi: 10.1111/j.1462-5822.2006.00826.x. [DOI] [PubMed] [Google Scholar]
- Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from Two Oahu (Hawaii) populations. Appl Environ Microbiol. 2009;75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Grublesky BT, Hussa EA, Visick KL. A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 2005;57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 2006;62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]