Abstract
Casp8p41 is a protein fragment generated by cleavage of procaspase 8 by human immunodeficiency virus (HIV) protease. We measured Casp8p41 content in memory CD4 T cells and analyzed the association of Casp8p41 content with CD4 T cell count, cross-sectionally and longitudinally. Casp8p41 content was inversely correlated with CD4 T cell count, and change in Casp8p41 content was associated with absolute CD4 T cell count with change over time. Casp8p41 change was a better predictor of CD4 T cell count change than activated CD8 T cell percentage or viral load and was comparable to bacterial 16s DNA levels. This suggests that Casp8p41 is a relevant mediator of CD4 T cell death during HIV infection.
The pathophysiologic hallmark of untreated human immunodeficiency virus (HIV) infection is the gradual and progressive loss of circulating CD4+ T cells, leading to AIDS, with a resulting increase in risk for opportunistic infections and malignancies. The accelerated CD4+ T cell loss in HIV infection is due to multiple mechanisms, including increased cell death, decreased production, and redistribution. Recent evidence demonstrates that CD4+ T cell death is progressive, exists at all stages of disease, and occurs primarily within lymphoid tissues. Mechanisms of cell death include the production of proapoptotic ligands by HIV-infected accessory cells, the cytotoxic effects of HIV proteins, and the induction of anergy and apoptosis as a result of polyclonal immune activation. CD4+ T cell apoptosis is increased in HIV infection, is directionally correlated with disease progression, and is inversely associated with total CD4 T cell count and percentage [1–3]. However, apoptosis has not been consistently found to correlate with HIV levels in the peripheral blood [4, 5].
In the life cycle of the virus, HIV-1 protease cleaves the HIV Gag/Pol polyprotein into functional viral proteins and contributes to viral maturation and budding release from the infected cell. HIV-1 protease is active in the cytosol of infected cells and, in addition to cleaving Gag/Pol, also cleaves cellular procaspase 8 at amino acids 355 and 356, generating a protein fragment 41kDa in size—Casp8p41—both in vitro [6] and in vivo [7]. Casp8p41 production is specific to HIV-1 protease–induced cell death [7] and colocalizes with infected and apoptotic cells [7]. Furthermore, treatment of uninfected cells with exogenous protease does not generate Casp8p41 (unpublished observations, A.D.B.). Expression of Casp8p41 in infected CD4+ T cells is sufficient to initiate cell death, the mechanism of which is both caspase-dependent and mitochondria-dependent. Expression of procaspase 8 engineered to be resistant to HIV-1 protease cleavage significantly reduces HIV-infected cell death [8, 9]. Casp8p41 expression also enhances HIV replication by activating NF-κB–dependent HIV long terminal repeat activation [10] (Figure 1A). To date, the in vivo significance of these findings has not been well studied. Here, we questioned whether generation of Casp8p41, one mechanisms by which HIV-infected cells die, might be consistently associated with CD4 T cell absolute count and count change in HIV-infected patients.
Figure 1.
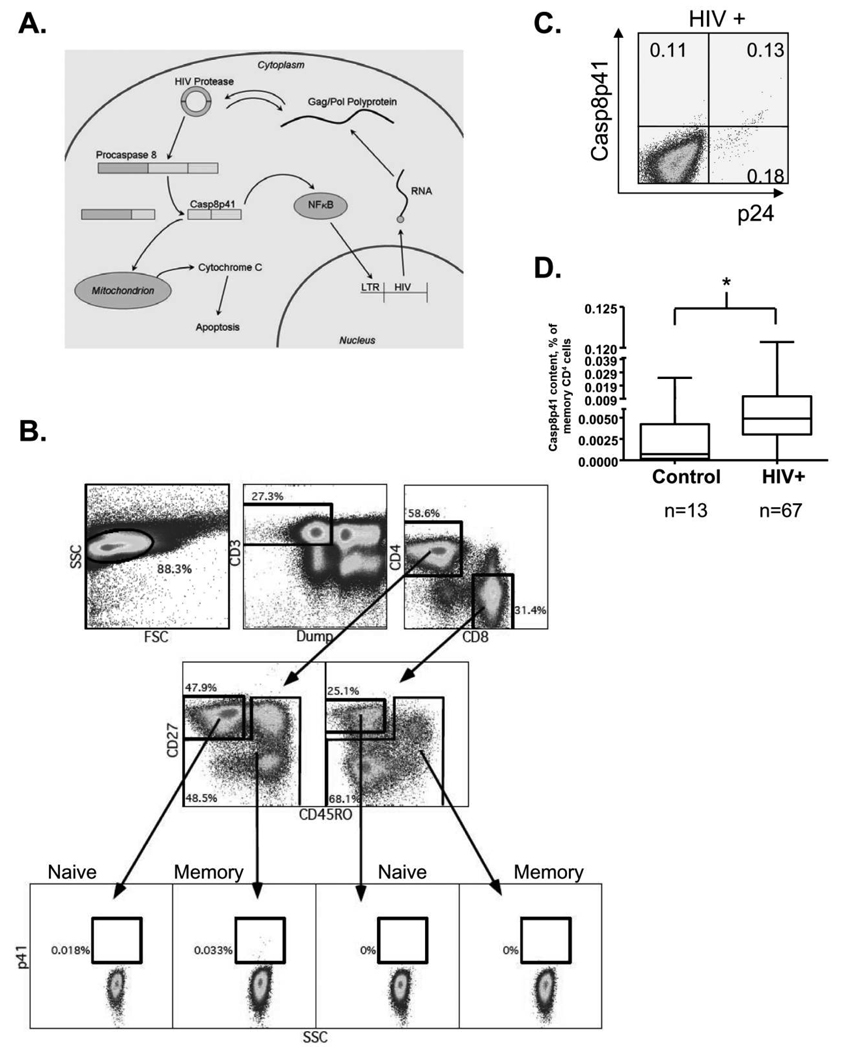
A, Diagram of the generation and effects of Casp8p41 in human immunodeficiency virus (HIV)–infected cells. B, Representative dot plots demonstrating the gating strategy for quantification of the percentage of memory CD4 T cells positive for Casp8p41 by flow cytometry. In total, 1.5 to 2 million events per patient were analyzed. C, Casp8p41 colocalizes with HIV p24 antigen in HIV-infected patients. D, Casp8p41 content was measured and compared in 13 HIV-uninfected and 67 HIV-infected individuals. *P < .002, Mann-Whitney test.
METHODS
All human studies were performed according to the guidelines of the Department of Health and Human Services and the individual participating institutional review boards.
UCSF SCOPE cohort
Fifty-three subjects from the University of California, San Francisco (UCSF) Study of the Consequences of Protease Inhibitors Era (SCOPE) cohort were included in this study. Patients who were either untreated or who were experiencing virologic failure while receiving combination antiretroviral therapy were included. Patients who had sustained virologic control in the absence of therapy and patients who had long-term nonprogression (defined as HIV infected for ≥10 years with maintenance of absolute CD4 T cell count >500 cells/µL without antiretroviral therapy) were excluded.
ACTG subjects
ACTG 5014 (ClinicalTrials.gov identifier: NCT00004855) was a randomized, open-label, pilot treatment trial involving treatment-naive HIV-infected patients who were receiving either lopinavir and ritonavir or stavudine, lamivudine, and abacavir sulfate in combination with the nonnucleoside reverse transcriptase inhibitor (NNRTI) nevirapine [11]. Details of this cohort have been described elsewhere [11]. Fourteen of the 55 patients enrolled were included in our analysis because they had available stored, frozen blood samples.
Measurements
Data on clinical indices were collected for the study subjects, including baseline demographic characteristics of age, sex, baseline CD4+ T cell count and percentage, HIV RNA levels, and baseline antiviral resistance mutations. Laboratory information included CD4+ T cell count and HIV load, measured, as described elsewhere, using the ultrasensitive Roche Amplicor assay [11]. Flow cytometric measurements of CD4 T cell counts and activated (CD38+/HLA-DR+) CD8 T cells were performed as described elsewhere [11]. Measurement of bacterial ribosomal 16S DNA was performed by quantitative polymerase chain reaction with plasma samples as described elsewhere [12].
Measurement of intracellular Casp8p41 expression in memory CD4+ T cells was performed, as described elsewhere, using a monoclonal antibody conjugated to fluorescein isothiocyanatefluorescein isothiocyanate (FITC) [7]. Briefly, peripheral blood lymphocytes were stained with the following reagents: Invitrogen Live/Dead Fixable Dead Cell Stain Kit-Aqua Blue (L34957, Molecular Probes), AntiCD3-Alexa 700 (557917, BD Pharmingen), AntiCD4-Cy5–5PE (35–0048-73, eBiosciences), AntiCD8-Pacific Blue (558207, BD Pharmingen), AntiCD27-PE (55541, BD Pharmingen), AntiCD45RO-ECD (PN18927120, Beckman Coulter). Next, cells were permeabilized, stained with Anti–Casp8p41-FITC, fixed, and analyzed on a FACSAria IIv flow cytometer (Beckton, Dickinson and Company) and data analyzed with FloJo software (TreeStar, Inc). Between 1.5 and 2 million events were collected per sample. Casp8p41 content in memory (CD45RO+ or CD27−/CD45RO−) CD4+ T cells was determined, using individual patient’s naive (CD27+/CD45RO−) CD8+ T cells as the negative gating control (Figure 1B). The investigators performing the measurements were blinded to the subjects’ group allocations, CD4 T cell counts, HIV load at the various time points, and presence or absence of treatment failure during the study periods.
Statistical methods
The primary outcome measure was Casp8p41 content, defined as the percentage of memory CD4 T cells positive for Casp8p41 at each time point. For the cross-sectional analysis, the Spearman Rank test was used to determine the correlation between Casp8p41 content or viral load and CD4 T cell count. Continuous variables were compared between patient groups using an unpaired t test or Mann-Whitney test as appropriate. For the longitudinal study, a change in Casp8p41 content and percentage of activated CD8 T cells or bacterial 16S DNA was treated as a dichotomous variable (either increase or decrease) and, in a post hoc analysis, compared with either an increase or decrease in CD4 T cell count during the coincident or subsequent time interval using standard logistic regression and generalized estimating equations. Change in HIV load was assessed similarly, except that for instances in which 2 consecutive values were <50 copies/mL, this no-change scenario was considered a decrease. A P value of <.05 was considered statistically significant. Statistical analysis was performed using GraphPad InStat 3 software (GraphPad Software Inc).
RESULTS
Validation of flow cytometric determination of Casp8p41 content
We have previously demonstrated that Casp8p41 is present only in HIV-infected cells or cells expressing HIV protease, and that our Casp8p41-specific antibody does not cross-react with either full-length Caspase-8 or Caspase-8 processing intermediates that are generated during other apoptosis signaling pathways [7]. We next sought to develop and validate a flow-based assay for Casp8p41 measurement. Cells were surface stained with monoclonal antibodies to CD3, CD4, CD8, CD27, and CD45RO, permeabilized and stained with α-Casp8p41. Because the number of infected cells is low in peripheral blood, a minimum of 1.5 million events were collected, and each patient’s own naive (CD27+/CD45R0−) CD8 T cells were used as a negative gating control. The Casp8p41-positive cells were predominantly of the memory (CD45RO+ or CD27−/CD45RO−) CD4 T cell phenotype (Figure 1B). Of note, this is the population of cells known to be preferentially infected by HIV in vivo [13]. Casp8p41-staining cells were costained for intracellular p24 antigen (Figure 1C), and HIV-infected subjects had a significantly higher Casp8p41 content than uninfected control subjects (median, 0.049% [range, 0–0.1207%] vs 0.0007% [0–0.025%]; P < .002) (Figure 1D), confirming the specificity of our staining approach.
Cross-sectional assessment of Casp8p41 in HIV-infected patients with active viral replication
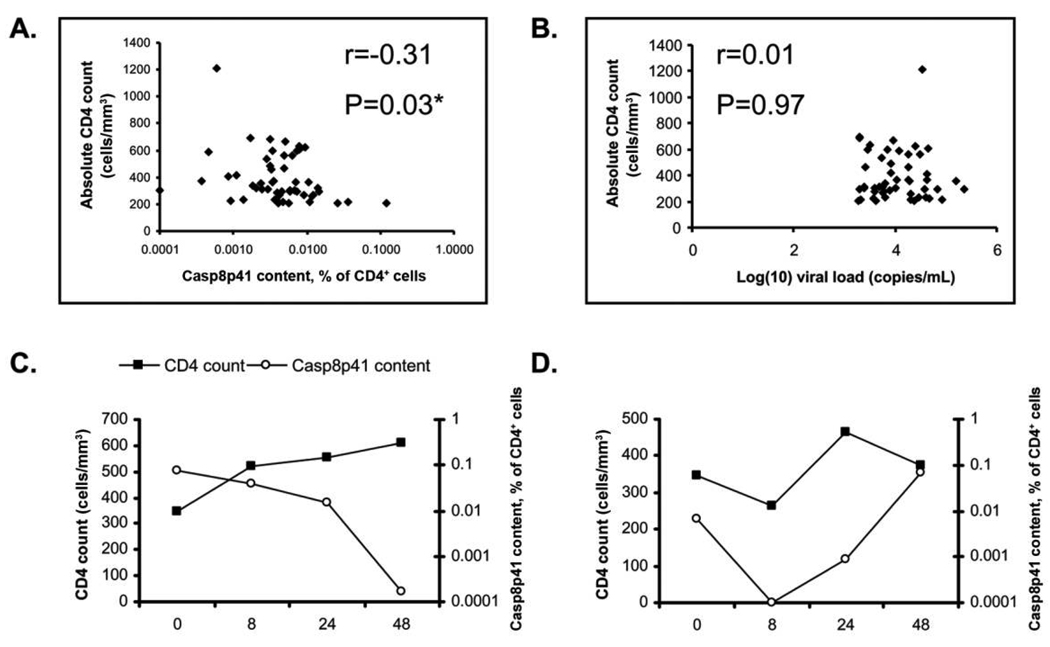
Samples from 53 patients included in the UCSF SCOPE cohort were analyzed for the percentage of memory CD4 T cells positive for Casp8p41. The mean age (± standard deviation [SD]) was 44 ± 8 years, and 74% of patients were men. Mean CD4 count (± SD) was 389 ± 187 cells/µL, and median viral load was 8423 RNA copies/mL (interquartile range [IQR], 4009–25,114 RNA copies/mL). Casp8p41 content in memory CD4 T cells was inversely correlated with absolute CD4 count (r = −0.31, P = .03) (Figure 2A). Viral load did not correlate with absolute CD4 count (r = 0.01, P = .97) (Figure 2B). Individual differences in Casp8p41 content were not associated with age, sex, race, or hepatitis C serostatus (data not shown).
Figure 2.
Cross-sectional analysis of the correlation between (A) percent memory CD4 T cells positive for Casp8p41 and absolute CD4 T cell counts (r = −0.31, P = .03) or (B) between human immunodeficiency virus level and absolute CD4 T cell counts (r = 0.006, P = .97) in 53 HIV-infected patients with active viral replication. Correlation coefficients calculated by Spearman Rank test. *P < .05 was considered statistically significant. Casp8p41 content was measured longitudinally in previously antiretroviral naive HIV-infected patients who started antiretroviral therapy and compared with the CD4 T cell count. Representative plots of the percentage memory CD4 T cells positive for Casp8p41 (circles) and CD4 T cell counts (squares) a patient with consistently decreasing Casp8p41 content (C), or a patient with increasing Casp8p41 content (D).
Longitudinal assessment of Casp8p41 in treatment-naive HIV-infected patients who started antiretroviral therapy
We next assessed in a treatment cohort whether longitudinal measurement of Casp8p41 expression change would predict CD4 T cell count change. Fifty-one individual blood samples from 14 patients obtained between weeks 0 and 48 were tested for Casp8p41 expression (median, 3.5 samples per patient [range, 2–4]); representative examples are shown in Figure 2C and 2D). Mean age (± SD) was 43 ± 12 years, and 93% of patients were male. Mean CD4 count (± SD) at baseline was 353 ± 126 cells/µL. Median baseline viral load was 18,510 copies/mL (IQR, 10,093–61,002 copies/mL).
We asked whether an increase or decrease in Casp8p41 content between 2 time points was associated with an increase or decrease in CD4 count over the subsequent time interval. A decrease in Casp8p41 was seen in 14 intervals, associated with a subsequent increase in CD4 count in 11 intervals (79%), and a subsequent decrease in CD4 count in 3 intervals (21%). Conversely, an increase in Casp8p41 was seen in 9 intervals, associated with a subsequent decrease in CD4 count in 7 intervals (78%) and a subsequent increase in CD4 count in 2 intervals (22%). Using logistic regression, the odds ratio for a subsequent increase in CD4 T cell count after Casp8p41 expression had decreased was 12.8 (95% confidence interval [CI], 1.7–97.2; P = .013), compared with the outcome if the Casp8p41 level had increased in the previous measurement interval. The positive predictive value and negative predictive value for the Casp8p41 level were 0.79 (95% CI, 0.49–0.95) and 0.78 (95% CI, 0.40–0.97), respectively, whereas for HIV load, they were 0.47 (95% CI, 0.24–0.17) and 0.00 (95% CI, 0.00–0.71). The interpretation of these predictive values, however, is limited because they do not take into consideration the within-subject correlation. In an analysis accounting for within-subject correlation across time intervals, the OR for an increase in CD4 T cell count after a decrease in Casp8p41 expression was 5.27 (95% CI, 0.76–36.7; P = .09).
Many other biomarkers have been previously correlated with HIV disease progression, including HIV load, activated (CD38+/HLA−DR+) CD8 T cell count, and bacterial 16s DNA levels in the peripheral blood. We therefore questioned whether Casp8p41 content was more closely correlated with CD4 T cell count change over time than were levels of these 3 biomarkers. The potential relative importance of Casp8p41 is underscored by the failure of changes in viral load or CD8 T cell activation to be significant predictors of CD4 increases in this small sample (Table 1). This relationship also compared favorably to bacterial ribosomal 16S DNA levels, a decrease in which was associated with a subsequent increase in CD4 T cell number (OR, 5.2; 95% CI, 1.65–16.3; P = .005 accounting for within-subject correlation).
Table 1.
Abilities of Relative Changes in Casp8p41 Content, Viral Load, Activated CD8 T Cell Percentage, and Bacterial 16s Ribosomal RNA to Predict a Subsequent Increase in the CD4 T Cell Count during Weeks 0–48 after Initiation of Antiretroviral Therapy in the ACTG5014 Cohort
| Standard logistic regression | GEE modela | |||
|---|---|---|---|---|
| Predictor | OR (95% CI) | P value | OR (95% CI) | P value |
| Decrease in Casp8p41 content | 12.8 (1.7–97.2) | .013 | 5.27 (0.76–36.7) | .09 |
| Decrease in viral loadb | 0.40 (0.01–10.8) | >.99 | NA | NA |
| Decrease in the percentage of activated CD8 T cells | 5.00 (0.18–139.0) | .40 | NA | NA |
| Decrease in bacterial 16s ribosomal DNA | 8.0 (0.75–85.3) | .09 | 5.2 (1.65–16.3) | .005 |
NOTE. P < .05 considered statistically significant. CI, confidence interval; GEE, generalized estimating equation; NA, not applicable; OR, odds ratio.
Accounting for within-subject correlation.
Or no change in viral load if 2 consecutive values were <50 copies/mL.
DISCUSSION
Although multiple mechanisms have been proposed to account for the loss of CD4 T cells in HIV-infected patients, few have been validated to occur in vivo. Identification of the HIV protease-specific Caspase 8 cleavage product, Casp8p41, in samples from HIV-infected patients is evidence of the occurrence of this cleavage effect in vivo. Moreover, demonstrating that the Casp8p41 level correlates with CD4 T cell count, and that changes in Casp8p41 content are inversely correlated with CD4 T cell count change, argue for a pathogenic role of Casp8p41 in CD4 T cell depletion during HIV disease. This model does not exclude the importance of other mechanisms of CD4 T cell loss during HIV infection. Rather, these data confirm only that Casp8p41 is present in vivo, which ultimately may be a useful predictor of CD4 T cell loss. It should be noted that our longitudinal study was restricted to patients who initiated antiretroviral therapy; it is unknown whether Casp8p41 has similar predictive use in patients who are not receiving antiretroviral therapy.
Another potential limitation of our study is the low percentage of memory CD4 T cells that stained for Casp8p41. This low frequency is, in our experience, a function of sensitivity. First, the percentage of HIV-infected cells in peripheral blood lymphocyte has been previously determined to be from 0.3% to 2%–4%. Second, Casp8p41-expressing cells undergo apoptosis within minutes to hours after expression of the protein. In our staining protocol, we excluded dead cells. Therefore, the circulating HIV-infected cells expressing Casp8p41 that were available to be measured are the small fraction that are expressing the protein but have not yet died. Finally, the clinical samples used in this study were frozen, which in our experience decreases the measured number of Casp8p410-expressing cells, compared with freshly processed samples (data not shown).
Our analysis was performed by determining the proportion of cells containing Casp8p41, as opposed to the absolute number of Casp8p41-containing cells, because the latter approach would predictably be biased by the total CD4 T cell count. By using the proportion of cells containing Casp8p41 we have de facto measured the cells that (1) have previously been infected by HIV and (2) would be predicted to die because of Casp8p41-dependent mechanisms. In fact, our results support this prediction, because patients with a higher proportion of Casp8p41-containing cells subsequently have a greater rate of CD4 T cell loss. Our study was unable to determine the effect on Casp8p41 content of a protease inhibitor–based antiviral regimen compared with a regimen not based on a protease inhibitor because of the small number of patients in both cohorts receiving a regimen not based on a protease inhibitor. Early in the era of HIV therapy, viral load assays were shown to predict CD4 cell loss; however, baseline HIV RNA levels predict an individual person’s CD4 T cell loss only minimally [14]. It will be of interest to formally compare the relative abilities of viral load and Casp8p41 level to predict within-person CD4 T cell loss in a larger cohort of patients.
It is well established that host factors affect the pace of HIV disease progression. These factors include the Δ32 mutation of CCR5, TRIM5α, CCL3LI gene duplication, PD-1, APOBEC 3G, and certain HLA types, including B*57 and B*27. Interindividual variations in Casp8p41 production, as shown by our study, might occur as a consequence of variable caspase 8 expression levels or polymorphisms within caspase 8. Alternately, altered HIV protease substrate recognition and/or cleavage, which often results from acquisition of drug resistance mutations in the protease gene, would predictably affect Casp8p41 production by HIV-infected cells. Determining whether failure of antiretroviral therapy resulting from acquired resistance to protease inhibitors is associated with relatively attenuated increases in Casp8p41 content and blunted CD4 T cell losses will be of great interest.
Acknowledgments
Financial support: Work on this paper was supported by the National Institutes of Health (NIH), the Center for AIDS Research, and the University of California, San Francisco (UCSF) (grants AI62261, AI40384, AI068636, U01 AI068634, P30 A127763, AI 69501, UL1 RR024131, P30 AI27763, and UL1 RR024131). Dr Nathan Cummins is supported by a Ruth L. Kirschstein National Research Service Award (award DK007013–32). This research was supported by the NIH (grant AI069994), Center for AIDS Research (grant P30 MH59037), and UCSF Clinical and Translational Research Institute Clinical Research Center (grant UL1 RR024131).
Footnotes
Potential conflicts of interest: A patent for Casp8p41 has been filed by Dr Andrew Badley. The remaining authors report that they do not have a commercial or other association that might pose a conflict of interest.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
- 1.Meyaard L, Otto SA, Keet IP, Roos MT, Miedema F. Programmed death of T cells in human immunodeficiency virus infection: no correlation with progression to disease. J Clin Invest. 1994;93:982–988. doi: 10.1172/JCI117105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandolfi F, Pierdominici M, Oliva A, et al. Apoptosis-related mortality in vitro of mononuclear cells from patients with HIV infection correlates with disease severity and progression. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;9:450–458. [PubMed] [Google Scholar]
- 3.Gougeon ML, Lecoeur H, Dulioust A, et al. Programmed cell death in peripheral lymphocytes from HIV-infected persons: increased susceptibility to apoptosis of CD4 and CD8 T cells correlates with lymphocyte activation and with disease progression. J Immunol. 1996;156:3509–3520. [PubMed] [Google Scholar]
- 4.Samuelsson A, Brostrom C, van Dijk N, Sonnerborg A, Chiodi F. Apoptosis of CD4+ and CD19+ cells during human immunodeficiency virus type 1 infection–correlation with clinical progression, viral load, and loss of humoral immunity. Virology. 1997;238:180–188. doi: 10.1006/viro.1997.8790. [DOI] [PubMed] [Google Scholar]
- 5.Rothen M, Gratzl S, Hirsch HH, Moroni C. Apoptosis in HIV-infected individuals is an early marker occurring independently of high viremia. AIDS Res Hum Retroviruses. 1997;13:771–779. doi: 10.1089/aid.1997.13.771. [DOI] [PubMed] [Google Scholar]
- 6.Nie Z, Phenix BN, Lum JJ, et al. HIV-1 protease processes procaspase 8 to cause mitochondrial release of cytochrome c, caspase cleavage and nuclear fragmentation. Cell Death Differ. 2002;9:1172–1184. doi: 10.1038/sj.cdd.4401094. [DOI] [PubMed] [Google Scholar]
- 7.Nie Z, Bren GD, Vlahakis SR, et al. Human immunodeficiency virus type 1 protease cleaves procaspase 8 in vivo. J Virol. 2007;81:6947–6956. doi: 10.1128/JVI.02798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Algeciras-Schimnich A, Belzacq-Casagrande AS, Bren GD, et al. Analysis of HIV protease killing through Caspase 8 reveals a novel interaction between Caspase 8 and mitochondria. Open Virol J. 2007;1:39–46. doi: 10.2174/1874357900701010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie Z, Bren GD, Rizza SA, Badley AD. HIV protease cleavage of procaspase 8 is necessary for death of HIV-infected cells. Open Virol J. 2008;2:1–7. doi: 10.2174/1874357900802010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bren GD, Whitman J, Cummins N, et al. Infected cell killing by HIV-1 protease promotes NF-kappaB dependent HIV-1 replication. PLoS ONE. 2008;3:e2112. doi: 10.1371/journal.pone.0002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landay AL, Spritzler J, Kessler H, et al. Immune reconstitution is comparable in antiretroviral-naive subjects after 1 year of successful therapy with a nucleoside reverse-transcriptase inhibitor– or protease inhibitor–containing antiretroviral regimen. J Infect Dis. 2003;188:1444–1454. doi: 10.1086/379041. [DOI] [PubMed] [Google Scholar]
- 12.Jiang W, Lederman MM, Hunt P, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199(8):1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenchley JM, Hill BJ, Ambrozak DR, et al. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol. 2004;78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez B, Sethi AK, Cheruvu VK, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. JAMA. 2006;296:1498–1506. doi: 10.1001/jama.296.12.1498. [DOI] [PubMed] [Google Scholar]