Abstract
Small RNAs modulate gene expression by forming a ribonucleoprotein complex with Argonaute proteins and directing them to specific complementary sites in target nucleic acids. However, the interactions required for the recruitment of the target nucleic acid to the ribonucleoprotein complex are poorly understood. In the present manuscript we have investigated this question by using let-7a, Argonaute2 and a fully complementary mRNA target. Importantly, we have found that recombinant Argonaute2 is sufficient to direct let-7a guided cleavage of mRNA. Thus this model system has allowed us to investigate the mechanistic basis of silencing in vitro and in vivo. Current models suggest that Argonaute proteins bind to both the 5′ and 3′ termini of the guide RNA. We have found that the termini of the let-7a microRNA are indeed critical, since circular let-7a does not support mRNA cleavage. However, the 5′ end is the key determinant, since its deletion abrogates activity. Surprisingly, we have found that alteration of the 5′ terminal uracil compromises mRNA cleavage. Importantly, we have found that substitution of this base has little effect upon the formation of the binary let-7a–Argonaute2 complex, but inhibits the formation of the ternary let-7a–Argonaute2–mRNA complex. Thus we conclude that the interaction of the 5′ uracil base with Argonaute2 plays a critical and novel role in the recruitment of mRNA.
Keywords: Argonaute, microRNA, let-7, mRNA cleavage
INTRODUCTION
It is now well appreciated that small RNAs can modulate gene expression through the formation of a ribonucleoprotein complex that interacts with complementary elements in nucleic acid targets [1–8]. The interaction of these small RNAs to their target nucleic acids results in a plethora of silencing events, including DNA methylation, mRNA cleavage, mRNA deadenylation and repression of translation [9–15]. The primary protein component of these ribonucleoprotein complexes is typically a member of the Argonaute family [16–23]. This family of proteins was first discovered in the identification of Arabidopsis mutants that developed an aberrant leaf structure that resembles squid tentacles [24,25]. Subsequently, other mutant Argonaute alleles were found in a screen to identify genes involved in plant post-transcriptional gene silencing [26]. A direct role in RNA-directed silencing was later provided by the observation that an Argonaute homologue was necessary and sufficient for the siRNA (small interfering RNA)-mediated cleavage of mRNA [20,27].
At present, Argonaute proteins are understood to contain three functional domains, theMIDdomain that binds to the 5′ phosphate of the small RNA, the PIWI domain that in some cases catalyses cleavage of the mRNA, and the PAZ domain, which is thought to bind to the 3′ end of the guide RNA [27–35]. However, most of our current understanding arises from systems that employ a model siRNA of somewhat arbitrary sequence. Importantly, cellular small RNAs are extraordinarily conserved in sequence from worm to man [36–38]. Moreover, there is a large family of closely related, but functionally distinct, Argonaute proteins in most organisms [39–47]. Therefore we anticipated that there may be sequence-specific interactions between small RNAs and their Argonaute cofactors. Thus, we elected to study the human let7amicroRNA, its Argonaute effector and a fully complimentary target mRNA. We anticipated that this model system would allow us to uncover any sequence-specific interactions in vitro and in vivo. We have found that the 5′ terminal nucleotide of let-7a is involved in a sequence-specific interaction with Argonuate2 which is critical for silencing activity.
EXPERIMENTAL
Synthetic RNAs were obtained from Dharmacon Research or the University of Calgary UCDNA services (Calgary, Alberta, Canada). All wild-type and mutant microRNAs were synthesized with a 5′ phosphate terminus. Synthetic siRNAs and antagomirs were obtained from Dharmacon Research. The GST (glutathione transferase)–Argonaute2 and GST–Argonaute2 active site mutants were a gift from Professor Leemor Joshua-Tor (HHMI/W.M. Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, U.S.A.). Anti-Argonaute2 antibody was obtained from Upstate Biochemicals and monoclonal antibodies against GAPD (glyceraldehyde-3-phosphate dehydrogenase) and vimentin were obtained from Abcam.
Luciferase reporter assay of let-7a activity
Target elements were subcloned into the SacI and BsteII sites of the pSENSOR dual luciferase reporter plasmid by the ligation of the appropriate DNA duplexes. pSENSOR is a derivative of psiCHECK-2 (Promega) in which 5′-TCGAGGAGCTCTATACGCGTCTCAAGCTTACTGGTTACCGTTCTAGAGTCGGGCCCGGGAATTCGTTTCAGCCTAGGC-3′ was inserted into the Xho1/Not1 sites within the multiple cloning site of psiCHECK-2, creating the SacI and BsteII sites used for cloning. The reporter plasmids, siRNA duplexes, antagomirs and microRNA duplexes were transfected into HeLa human cervical carcinoma cells using Lipofectamine™ 2000 (Invitrogen), according to the manufacturer’s instructions. After 36 h, dual luciferase activities were determined by assaying the cell lysates according to the manufacturer’s protocol (Promega). Renilla luciferase activity was determined by quantitative titration and normalized for transfection efficiency to firefly luciferase activity.
Purification of GST–Argonaute2 protein
An overnight culture of Escherichia coli XL1Blue, transformed with full-length human Argonaute2 cDNA tagged with GST, was diluted 1:50 in LB (Luria–Bertani) medium and grown at 37°C. At a D600 of 0.4, the culture was induced with isopropyl β-d-thiogalactoside (1 mM). After 16 h of further growth at 25°C, cells were spun down and resuspended in 5 ml of Buffer A (50 mM Tris, pH 8.0, and 1 mM EDTA). The cells were lysed by adding lysozyme and Triton X-100 to a final concentration of 0.2 mg/ml and 1% respectively [48]. The lysate was centrifuged at 12000 g for 30 min. The resultant supernatant was incubated with glutathione–agarose (20 mg of protein/ml of resin) for 2.5 h at 4°C prior to addition to the column. After washing the column with Buffer B (50 mM Tris, pH 8.0, 200 mM NaCl, 1 mM EDTA and 0.1% Triton X-100), GST–Argonaute2 was eluted with 50 mM Tris, pH 8.0, and 5 mM glutathione. Active protein was determined by let-7-directed mRNA cleavage activity, pooled and stored at −80°C.
Preparation of labelled RNA
RNAs were labelled using T4 polynucleotide kinase and [γ - 32P]ATP (Amersham Bioscience) to a typical specific activity of 106 c.p.m./pmol. After phenol/chloroform extraction, the labelled RNA was gel purified followed by chloroform extraction and ethanol precipitation [49].
mRNA cleavage
Reaction mixtures (20 µl) contained 50 mMTris, pH 7.5, 50 mM KCl, 1 mM MgCl2, let-7a microRNA and protein as indicated. Mixtures were preincubated at 37°C for 30 min. Following preincubation, 32P-end-labelled target RNA (106 c.p.m./pmol) was added to a final concentration of 1 nM. Mixtures were then incubated at 37°C for 15 min. Following incubation, 80 µl of a dye mixture (98% formamide, 10 mM EDTA, 1 mg/ml Bromophenol Blue, 1 mg/ml Xylene Cyanole) was added. Samples were incubated at 60°C for 2 min and 4%of the reaction mixture was analysed on a 12%(20:1) denaturing polyacylamide gel in TBE buffer. The gel was fixed in 10%acetic acid, dried on DE81 chromatography paper (Whatman) with a backing of gel drying paper and exposed to BioMax MS film.
Identification of the let-7a–Argonaute2 and let-7a–Argonaute2–mRNA complexes by native gel electrophoresis
Reaction mixtures (20 µl) contained 50 mMTris, pH 7.5, 50 mM KCl, 1 mMMgCl2, 0.005%Nonidet P40, 0.2 µg of tRNA, GST–Argonaute2 D597A (active site mutant deficient for cleavage activity) as indicated and 0.1 nM 5′ 32P-end-labelled let-7a RNA (106 c.p.m./pmol). Mixtures were preincubated at 37°C for 30 min. Following preincubation, target RNA was added to the reactions. Mixtures were then incubated at 37°C for 15 min. Then, 4 µl of native loading buffer (50% glycerol, 0.1 M Tris, pH 8.0, 0.1%Bromophenol Blue and 0.1%Xylene Cyanole) was added. Next, 50% of the reaction was analysed by 1% agarose gel in TAE buffer. The gel was dried on DE81 chromatography paper (Whatman) with a backing of gel drying paper and exposed to BioMax MS film.
RESULTS
Human Argonaute2 can utilize let-7a to silence gene expression
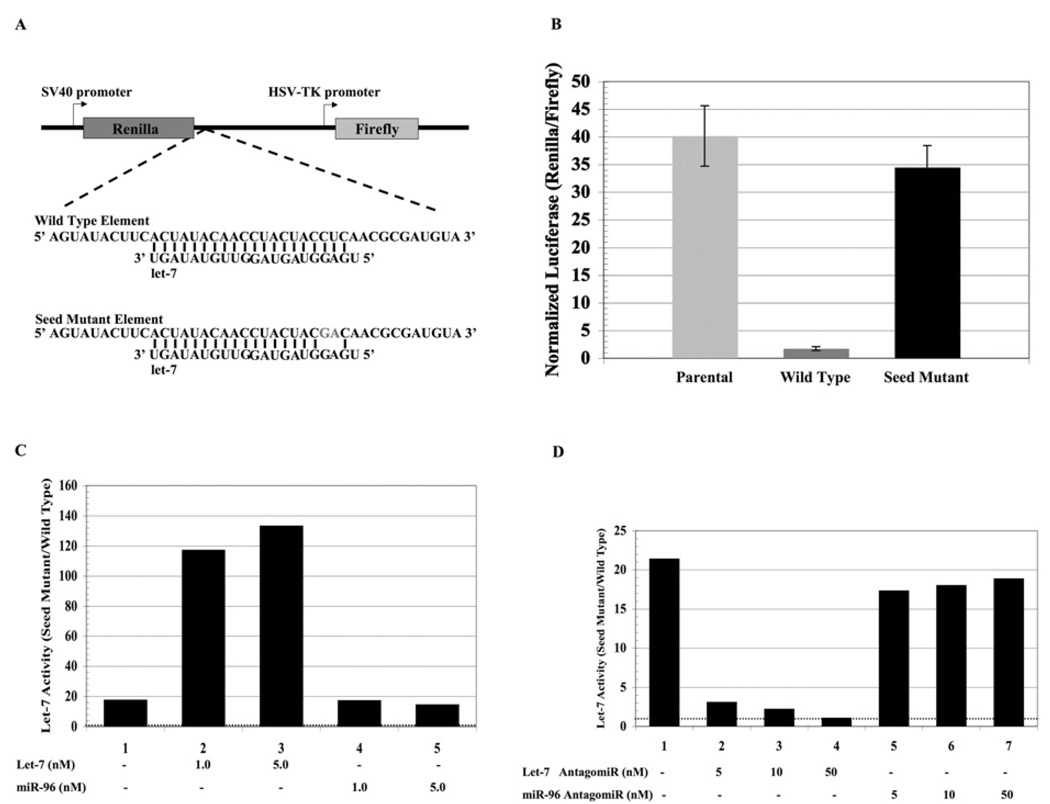
We first designed a model element to measure the suppressive activity of Argonaute in human cells. The residues of the 42 nucleotide element from Lin-41 mRNA [50] were made fully complementary to let-7a (Figure 1A). To test whether this element can silence gene expression in human cells, we subcloned it into the 3′-UTR (3′-untranslated region) of Renilla luciferase mRNA using a dual luciferase reporter (pSENSOR) in which transfection efficiency can be normalized by the simultaneous measurement of firefly luciferase. This resulted in a significant (15-fold) decrease of Renilla luciferase expression compared to the parental Renilla luciferase mRNA (Figure 1B). Such silencing might have been exerted by protein factors, so we generated a mutant element in which the putative interaction with the seed sequence of let-7a would be disrupted. The insertion of this seed mutant element into Renilla luciferase mRNA provoked remarkably little silencing (Figure 1B) and was comparable with the parental vector. In the subsequent experiments, we have measured let-7a activity as the fold repression between the seed mutant and wild-type sensor reporters.
Figure 1. Let-7a robustly silences mRNA containing a fully complementary target site derived from lin-41 mRNA.
(A) The sequence of the let-7a target elements inserted into the 3′-UTR of the Renilla luciferase gene of a dual luciferase reporter. Altered residues in the seed mutant are shown in grey. (B) HeLa cells were transfected with luciferase plasmids (50 ng) containing either the parental let-7a wild-type or let-7a seed mutant target elements. Dual luciferase activities were measured and Renilla luciferase was normalized to firefly luciferase. These results are the average of three independent experiments. (C) HeLa cells were co-transfected with luciferase reporters containing either the let-7a wild-type or the let-7a seed mutant element along with microRNA let-7a or miR-96 (as indicated). Dual luciferase activities were measured and Renilla luciferase was normalized to firefly luciferase. Let-7a activity was determined by its ability to suppress the expression of the reporter harbouring the wild-type element compared with the reporter harbouring the seed mutant element as a reference point. The dotted line at a value of 1 denotes no let-7a activity. Transfection of an irrelevant microRNA (miR-96) serves to measure endogenous let-7a activity. (D) HeLa cells were co-transfected with luciferase reporters containing either the let-7a wild-type or the let-7a seed mutant element along with antagomirs directed against let-7a or miR-96 (as indicated). Dual luciferase activities were measured and Renilla luciferase was normalized to firefly luciferase. Let-7a activity was determined as described above. The dotted line at a value of 1 denotes no let-7a activity.
Evidence that the silencing was mediated by let-7a microRNA was provided by the observation that the addition of exogenous let-7a microRNA further stimulated endogenous let-7a silencing activity (Figure 1C). Similarly, the addition of antagomirs against let-7a relieved the silencing effect, whilst an antagomir against an irrelevant microRNA (miR-96) had no effect (Figure 1D). Thus we concluded that this model element significantly silenced expression, and that its effects can be attributed to let-7a. Next, we down-regulated Argonaute2 and ascertained its effect upon the silencing activity of let-7a. siRNA-mediated down-regulation of Argonaute2 significantly attenuated let-7a silencing activity (Figure 2A). On the other hand, the down-regulation of GAPD, as shown by Western blot analysis (Figure 2B), had little effect upon let-7a activity. Thus, we concluded that let-7a can use Argonaute2 to silence gene expression in HeLa cells.
Figure 2. Argonaute2 is necessary for let-7a activity.
(A) HeLa cells treated with siRNA directed against either Argonaute2 or GAPD (concentration indicated) were transfected with luciferase reporters containing either the let-7a wild-type or let-7a seed mutant element. The cells were analysed for dual luciferase activity 36 h post transfection. Renilla luciferase was normalized to firefly luciferase. Let-7a activity was determined as described above. The dotted line at a value of one denotes no let-7a activity. The results are the average of two independent experiments. (B) Western blot of the cells confirms down-regulation of Argonaute2 and GAPD. Vimentin was used as a loading control.
Recombinant Argonaute2 is sufficient to support the let-7a directed cleavage of mRNA
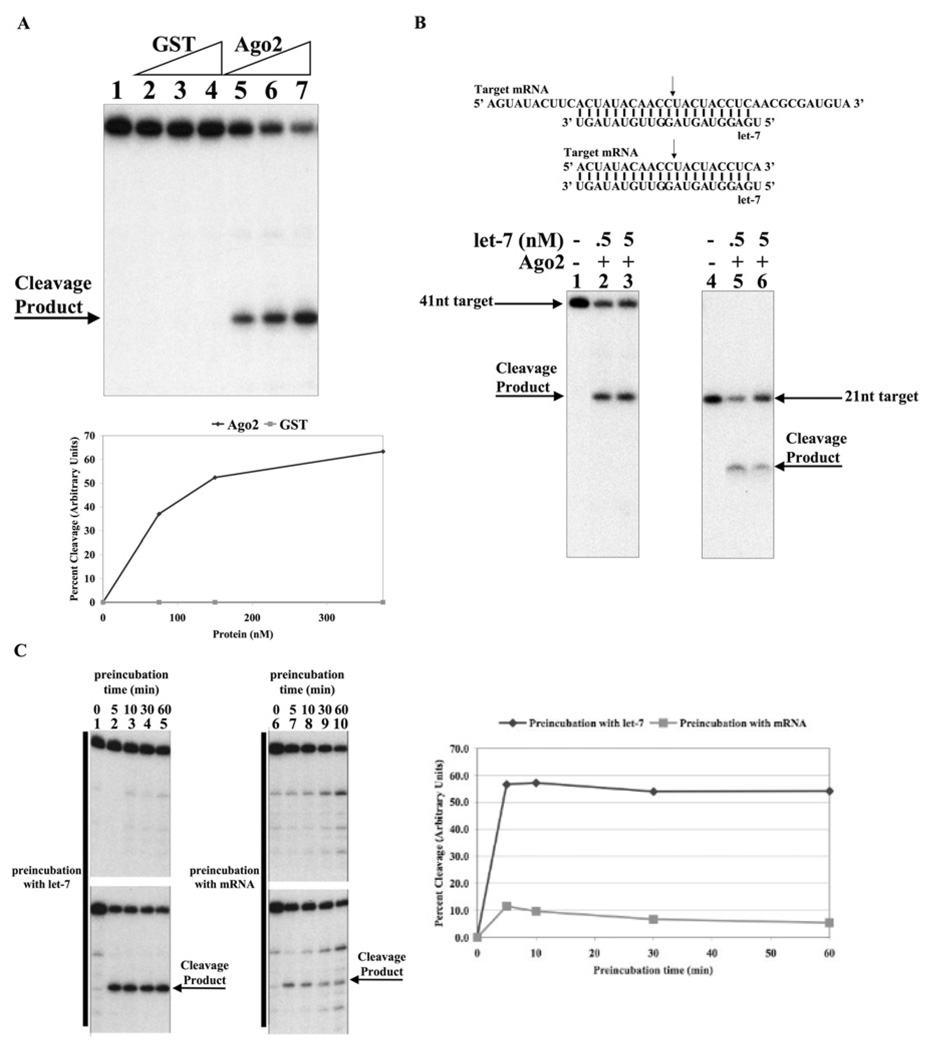
Given that let-7a can use Argonaute2 to silence gene expression, the next key question was whether purified recombinant human Argonaute2 was sufficient to recapitulate silencing activity in vitro. MicroRNAs are thought to silence gene expression by either translational repression, deadenylation of mRNA or mRNA cleavage [3,10,11,14,15,51]. Currently, the accurate cleavage of mRNA is the most robust and unambiguous in vitro measure of microRNA/Argonaute ribonucleoprotein activity. Thus, we have used mRNA cleavage as the principal assay for the formation of an active let-7a–Argonaute2 ribonucleoprotein complex. Accordingly, we affinity purified a human GST–Argonaute2 fusion protein from E. coli [27]. This preparation was preincubated with let-7a microRNA and then incubated with an end radiolabelled mRNA corresponding to the target element (Figure 3A). Impressively, even after a short incubation (15 min), the mRNA was efficiently cleaved at a position consistent with the scissile phosphate opposite the tenth and eleventh nucleotides from the 5′ end of the let-7a microRNA(Figure 3A). Preincubation of an irrelevant protein (GST) with let-7a did not result in mRNA cleavage. Importantly, cleavage of a smaller target RNA (21 nucleotides in length) resulted in the formation of a smaller cleavage product also corresponding to a position between the nucleotides complementary to the tenth and eleventh nucleotides from the 5′ end of the let-7a microRNA (Figure 3B). The current belief is that the guide RNA–Argonaute complex forms first and then recruits the target mRNA. To test this directly, we performed an order of addition experiment. Figure 3(C) shows that the incubation of Argonaute2 with let-7a followed by the addition of target mRNA leads to a much greater reaction than in the scenario where the mRNA is added first, followed by the addition of let-7a microRNA. Thus, we conclude that the formation of a let-7a–Argonaute2 complex is indeed the obligate first step in the silencing reaction.
Figure 3. Recombinant Argonaute2 is sufficient to support the let-7a-directed cleavage of mRNA.
(A) Let-7a guides recombinant Argonaute2 to cleave mRNA. Reactions (20 µl) containing 50 mM Tris, pH 7.5, 50 mM KCl, 1 mM MgCl2 and 2 nM let-7a RNA were preincubated for 30 min with 75, 150 or 375 nM GST–Argonaute2 or GST (as indicated). 32P-5′-end-labelled target mRNA (1 nM) was added to each reaction followed by a 15 min incubation. Formamide loading buffer was added and reactions were analysed by 12% polyacrylamide gel run under denaturing conditions. Quantification of the cleavage activity is shown in the bottom panel. (B) Mapping of the cleavage site. Sequence of the 41 nucleotide and 21 nucleotide fully complementary target mRNA and let-7a RNA used in the following experiments. The arrow indicates the expected cleavage site. Reactions containing 200 nM GST–Argonaute2 and let-7a (as indicated) were carried out as described above. (C) Formation of the let-7a–Argonaute2 complex is the obligate first step for mRNA cleavage. Reactions (20 µl) containing 50 mM Tris, pH 7.5, 50 mM KCl, 1 mM MgCl2 and 200 nM GST–Argonaute2 were preincubated with either 2 nM microRNA (let-7a 5′ mutant or let-7a) or 1 nM labelled target mRNA for 30 min. The requisite RNA (microRNA or labelled target) was added to each reaction followed by a 15 min incubation. In each set, the top panel was conducted using let-7a 5′ mutant microRNA, whereas the bottom panel contains let-7a microRNA. Quantification of the cleavage activity is shown in the right-hand panel.
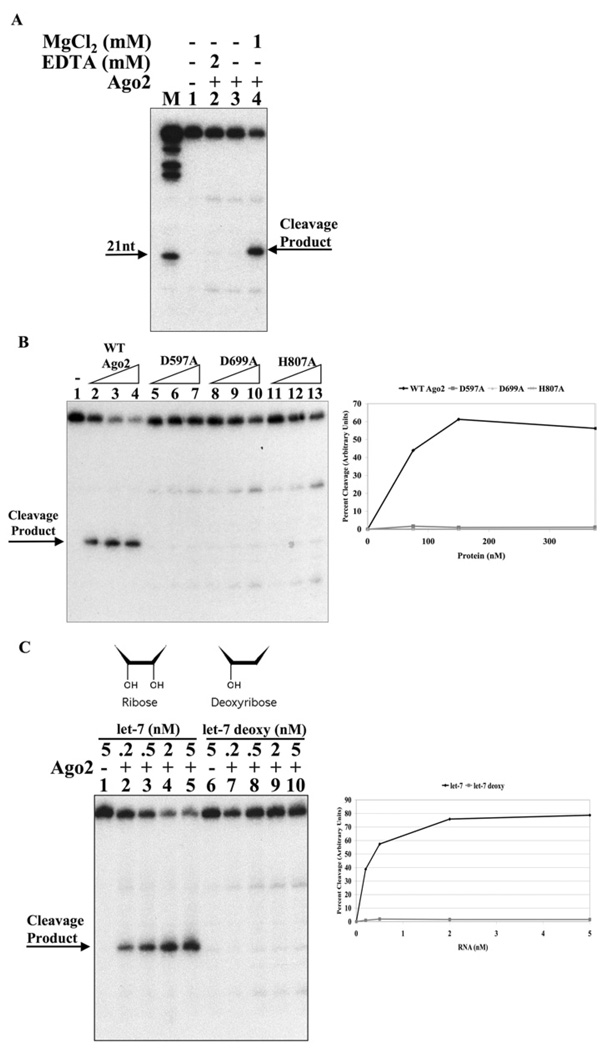
Cleavage required a divalent cation, since no activity was evident in the absence of magnesium or in the presence of EDTA (Figure 4A). Studies on the cleavage reaction directed by the guide strand of an siRNA have indicated that it is probably catalysed by the Argonaute2 DDH (aspartate-aspartate-histidine) catalytic triad in the PIWI domain [27]. Thus we examined whether these residues were also critical for let-7a microRNA-directed cleavage of mRNA. Indeed, we observed that the alteration of any one of these residues to alanine completely abrogated cleavage activity (Figure 4B). Previous studies have drawn attention to the structural similarities between Argonaute2 and ribonuclease H, a DNA-directed RNA endonuclease [27,30]. Similarly, many of the existing structural models for the Argonaute protein employ proteins from archea bacteria that are DNA-directed endonucleases [30–32,35]. To test whether recombinant Argonaute2 is an RNA-directed endonuclease, we provided Argonaute2 with DNA corresponding to the let-7a sequence (Figure 4C). We observed that DNA is unable to support cleavage of the target mRNA. Thus, we concluded that Argonaute2 is indeed an RNA-dependent endonuclease. It is important to note that in this experiment and indeed in most of our assays, there is a very minor band that migrates close to, but is distinguishable, from the cleaved mRNA. This minor band probably arises from a contaminant activity, as it is present on incubation with mutant let-7a or catalytically inactive Argonaute2.
Figure 4. Argonaute2 is a RNA-dependent endonuclease that requires magnesium and the integrity of the DDH catalytic domain.
(A) Let-7a-directed cleavage of mRNA by Argonaute2 requires magnesium. Reactions (20 µl) containing 50 mM Tris, pH 7.5, 50 mM KCl, 2 nM let-7a RNA and 200 nM GST–Argonaute2 were preincubated for 30 min in the presence of either EDTA or MgCl2 (as indicated). 5′-end-labelled target mRNA (1 nM) was added to each reaction followed by a 15 min incubation. Formamide loading buffer was added and reactions were analysed by 12% polyacrylamide run under denaturing conditions. M, marker nucleotide ladder. (B) The DDH domain of Argonaute2 is essential. Reactions containing 2 nM let-7a and 75, 150 or 375 nM GST–Argonaute2 or active site mutants D597A, D699A and H807A (as indicated) were carried out as described above. (C) Argonaute2 is a RNA-dependent endonuclease. Reactions containing 200 nM GST–Argonaute2 and let-7a ribose or let-7a deoxyribose (as indicated) were carried out as described above.
The 5′ end of let-7a is critical but the 3′ end is dispensable
The current models of the interaction between the guide strand and Argonaute proteins suggest binding pockets for both the 5′ and 3′ termini [27–29,31–33]. To test the requirement for the ends of the microRNA in the formation of an active ribonucleoprotein complex, we generated circular let-7a microRNA using RNA ligase [52]. Linear let-7a was treated with RNA ligase and the resultant circles were gel purified. Linear let-7a, not treated with RNA ligase, was carried through the same regimen as a comparison control. Importantly, circular let-7a RNA did not direct mRNA cleavage (Figure 5C), even though both linear and circular let-7a were fully capable of annealing to the target mRNA (Figure 5B). Thus we conclude that indeed the termini of the let-7a microRNA are critical for silencing activity.
Figure 5. Circular let-7a does not support Argonaute2-catalysed cleavage of mRNA.
(A) Generation of circular let-7a. Let-7a RNA was circularized using T4 RNA ligase and gel purified. Linear let-7a RNA was prepared in an identical fashion and gel purified from a reaction lacking T4 RNA ligase. Linear and circularized 32P-end-labelled let-7a prior to gel purification. Samples were analysed by 10%polyacrylamide gel under denaturing conditions. (B) Both linear and circular let-7aRNAcan anneal to the mRNA target. Reactions (20 µl) containing 50 mM Tris, pH 7.5, 200 mM NaCl, 1 mM MgCl2, radiolabelled target mRNA (1 nM) and circular or linear let-7a (0.5, 2.5 or 5 nM) were incubated for 30 min. Samples were analysed by 10% polyacrylamide gel under native conditions. (C) Circular let-7a RNA does not support mRNA cleavage. Reactions (20 µl) containing 50 mM Tris, pH 7.5, 50 mM KCl, 1 mM MgCl2 and 200 nM GST–Argonaute2 were preincubated for 30 min with linear or circularized let-7a RNA (as indicated). 5′-end-labelled target mRNA (1 nM) was added to each reaction followed by a 15 min incubation. Formamide loading buffer was added and reactions were analysed by 12%polyacrylamide gel run under denaturing conditions. Quantification of the cleavage activity is shown in the right-hand.
To elucidate whether the 5′ and 3′ ends of let-7a are both important, we created let-7a microRNAs harbouring a four base mismatch at either the 5′ or 3′ end. Since the seed sequence of a microRNA is important for target recognition, it was not surprising that a four base mismatch in the seed sequence of let-7a abrogated its ability to direct mRNA cleavage (Figure 6A). Importantly, a similar alteration at the 3′ end had no visible effect (Figure 6A). Using our reporter assay system, we introduced the corresponding alterations into the target element and measured its effect on let-7a activity in vivo. Similar to the results in vitro, we find that complementarity at the 5′ end of let-7a is critical for silencing activity, whereas mutation of the 3′ had no discrete effect (Figure 6B). These observations suggest that the let-7a–Argonaute2 ribonucleoprotein complex can silence a partially complementary mRNA target in vitro and in vivo.
Figure 6. The 5′ end of let-7a is critical but the 3′ end is dispensable for Argonaute2-catalysed cleavage.
(A) Sequence of the wild-type, 5′ and 3′ mutant let-7a microRNAs annealed to the target mRNA. Reactions containing 50 mM Tris, pH 7.5, 50 mM KCl, 1 mM MgCl2 and 200 nM GST–Argonaute2 were preincubated for 30 min with wild-type, 5′ mutant or 3′ mutant let-7a RNA (as indicated). 5′-end-labelled target mRNA (1 nM) was added to each reaction followed by a 15 min incubation. Formamide loading buffer was added and reactions were analysed by 12% polyacrylamide gel run under denaturing conditions. Quantification of the cleavage activity is shown in the right-hand panel. (B) HeLa cells were transfected with a luciferase reporter containing a wild-type, seed mutant, 5′ mutant or 3′ mutant element to let-7a. Dual luciferase activities were measured. Renilla luciferase was normalized to firefly luciferase. Let-7a activity was determined by its ability to suppress the expression of the reporter harbouring the experimental (wild-type, 5′ mutant or 3′ mutant) element compared to the reporter harbouring the seed mutant element as a reference point. The dotted line, at a value of 1, denotes no let-7a activity. The results shown are the average of three independent experiments. (C) Right, a schematic representation of the let-7a RNAs used in the assays below. The dotted line indicates the cleavage site. It is important to note that all wild-type and mutant microRNAs are phosphorylated at the 5′ terminus. Left, reactions containing 200 nM GST–Argonaute2 and let-7a RNA or let-7a deletion mutants (5′ deletion, 3′ deletion or 3′ major deletion) as indicated were carried out as described above. Quantification of the cleavage activity is shown in the adjacent graph.
To further study the interactions of the 5′ and 3′ end of let-7a, we generated let-7a mutants containing deletions at either the 5′ or 3′ end. A five nucleotide deletion at the 3′ end had little effect whereas a more extensive deletion significantly attenuated silencing activity (Figure 6C). Thus this extends our previous observation and asserts that the let-7a–Argonaute complex can silence a partially complementary target mRNA. Strikingly, deletion of the 5′ end of let-7a abrogated its ability to direct mRNA cleavage. Previous studies have shown that deletion of the 5′ end of a guide siRNA did not preclude mRNA cleavage, but resulted in the formation of a new cleavage site [27]. This has been attributed to the ability of the de novo terminus of the siRNA to ‘slide’ into the MID domain phosphate-binding pocket and thereby direct a new cleavage site. Since deletion of the 5′ end of let-7a microRNA abrogated cleavage and no new cleavage site was created, we speculate that let-7a is unable to ‘slide’ in the binding groove of Argonaute2.
The 5′ uracil residue of let-7a is critical to direct mRNA cleavage in vitro and in vivo
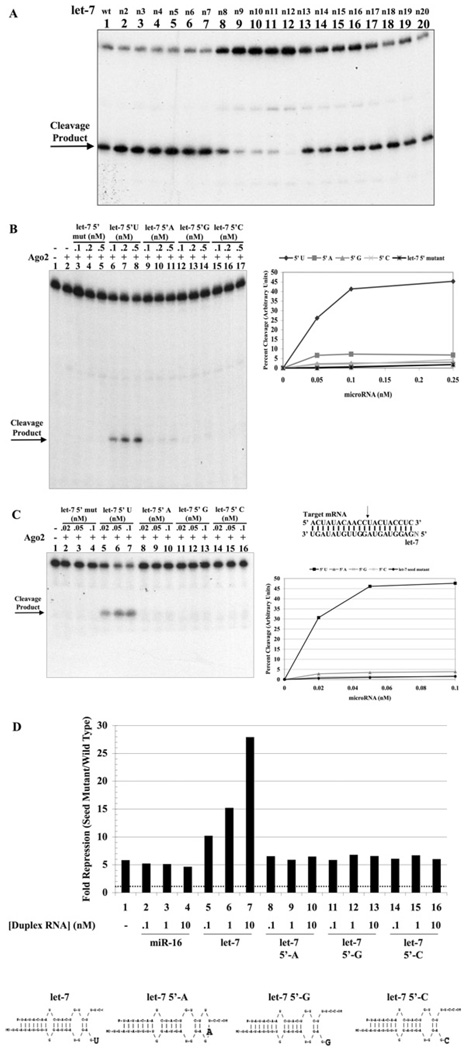
Finally, we examined each residue of the let-7a microRNA. Sequential mutation of residues 2–20 had little effect (Figure 7A) on mRNA cleavage. Only the residues which surround the cleavage site (9–12), had any significant effect upon silencing activity. However, to our surprise, the 5′ terminal nucleotide (residue 1) was critical. We substituted the 5′ terminal uracil of let-7a with adenine, guanine or cytosine and found that Argonaute2-directed cleavage required a uracil terminus (Figure 7B). Although a very small amount of activity was seen with an adenine terminus, no activity was apparent with let-7a microRNA containing cytosine or guanine at the terminus. Thus, we concluded that we had probably disrupted a critical interaction between the terminal base of let-7a and amino acids surrounding the phosphate-binding pocket. The 5′ terminal nucleotide is not thought to interact with the target mRNA [34,53]; however, our target mRNA contains an adenine residue that could potentially base pair to the terminal uracil. It was possible that the disruption of this interaction was responsible for the loss of mRNA cleavage activity. To test this, we utilized a shorter mRNA target that lacks this residue. This truncated mRNA was also robustly cleaved by the let-7a/Argonaute2 complex and displayed the same requirement for uracil at the 5′ terminus of let-7a (Figure 7C). Thus we conclude that the uracil base is important for an interaction with Argonaute2, rather than an interaction with mRNA.
Figure 7. The 5′ uracil residue of let-7a is critical to direct mRNA cleavage in vitro and in vivo.
(A) Reaction mixtures (20 µl) containing 50 mM Tris, pH 7.5, 50 mM KCl, 1 mM MgCl2, wild-type let-7a or let-7a RNA containing single-nucleotide alterations (as indicated) and 200 nM GST–Argonaute2 were carried out as described above. (B) Reactions containing 200 nM GST–Argonaute2 and let-7a RNAs containing a 5′ terminal U, A, G or C (as indicated) were carried out as described above. Quantification of the cleavage activity is shown in the adjacent panel. (C) Reactions containing let-7a RNA with a 5′ terminal U, A, G or C (as indicated) and 200 nM GST–Argonaute2 were carried out as described above. 5′-end-labelled 20-nucleotide target mRNA lacking the nucleotide, which would base-pair to the 5′ terminal nucleotide of let-7a, as illustrated above, was added to each reaction (1 nM). Quantification of the cleavage activity is shown in the adjacent panel. (D) Let-7a activity in HeLa cells (column 1) was determined by its ability to suppress the expression of a luciferase reporter harbouring the model element compared with a reporter harbouring a mutant element as a reference point. The dotted line, at a value of 1, denotes no let-7a activity. The activity of the exogenously provided duplexes (columns 5–16) was determined by co-transfection with the same reporters and determination of let-7a activity is described above. The transfection of an irrelevant microRNA duplex (miR-16) (columns 2–4) serves to measure endogenous let-7a activity. Lower panels, a schematic representation of the let-7a duplex RNAs used in the assay.
To confirm these observations at the cellular level, we generated let-7a microRNA duplexes in which the 5′ terminus of the microRNA had been similarly altered. These microRNA duplexes were transfected into HeLa cells and their silencing activity was measured by their ability to stimulate endogenous let-7a activity as measured by the reporter assay. As observed in vitro, only the let-7a duplexes with a uracil at the 5′ end of the microRNA were capable of efficiently silencing expression in HeLa cells (Figure 7D). It is important to note these cellular experiments could not be conducted with single-stranded microRNA. Thus, there is a possibility that the 5′ terminal alterations may affect the loading of let-7a into Argonaute. However, in the case of the uracil to cytosine alteration the 5′ end of the guide strand remains in an open configuration. Thus, we attribute the effects of the mutants to the reduced cleavage activity of the let-7a–Argonaute2 complex.
The 5′ uracil residue of let-7a is critical for the recruitment of mRNA to the Argonaute2 silencing complex
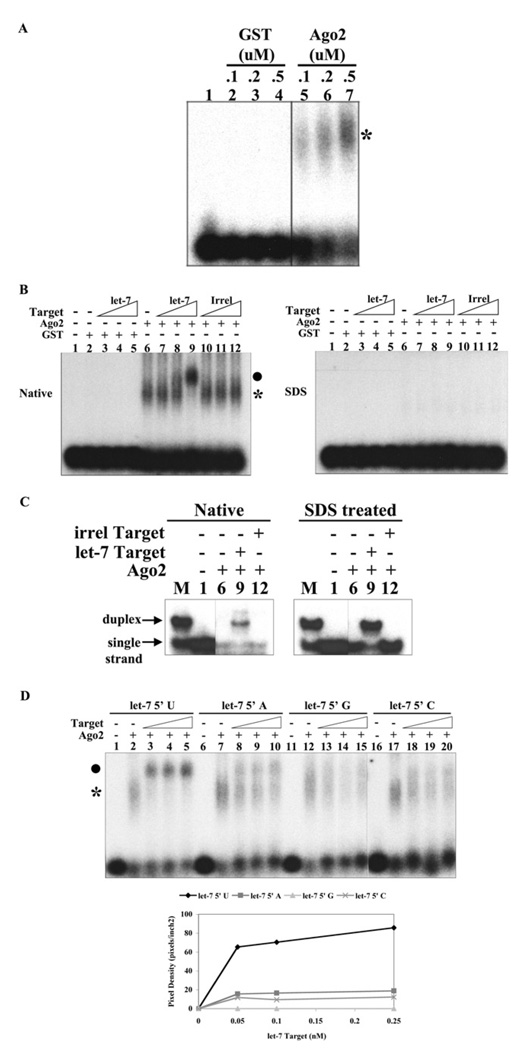
Next we investigated whether the 5′ terminal uracil was critical for the formation of the let7a–Argonaute complex itself or for the subsequent step of the recruitment of mRNA. Although small RNA–Argonaute complexes have previously been identified by crosslinking analysis [27,54], it has not yet been possible to resolve the postulated binary microRNA–Argonaute complex from the ternary microRNA–Argonaute–mRNA complex. Thus, we investigated whether the binary and ternary complexes could be resolved by native gel electrophoresis. To more readily visualize these reaction intermediates, we used the Argonaute2 catalytic mutant (D597A), which we anticipated may trap the let-7a–Argonaute2–mRNA complex. Incubation of radiolabelled let-7a with recombinant human Argonaute2 led to the appearance of a slow migrating species indicative of the let-7a–Argonaute2 complex (Figure 8A). Such a complex was not apparent on incubation with an irrelevant protein (GST). Importantly, this putative let7a–Argonaute2 complex could be shifted to a slower migrating species with the addition of the target mRNA in a concentration-dependent fashion (Figure 8B). This complex was not identified on incubation with GST or upon addition of an irrelevant target mRNA. To investigate whether the mRNA was annealed to let-7a in the complex, we treated the reaction with SDS and looked to see whether the let-7a–mRNA duplex was released. As predicted, SDS dissolved complex formation (Figure 8B) and indeed led to an increased amount of a let-7a–mRNA duplex on analysis by polyacrylamide gel electrophoresis (Figure 8C). The relatively small amount of let-7a–mRNA duplex observed in the absence of SDS may arise from the turnover of Argonaute2 complexes or the direct annealing of let-7a to mRNA. Thus we conclude that this approach can resolve the let-7a–Argonaute2 and let-7a–Argonaute2–mRNA complexes and may be utilized to study the interactions required for the recruitment of mRNA to the Argonaute silencing complex.
Figure 8. The 5′ uracil residue of let-7a is critical for the recruitment of mRNA to the Argonaute2 silencing complex.
(A) Identification of the let-7a–Argonaute2 ribonucleoprotein complex. Reaction mixtures (20 µl) containing 50 mM Tris, pH 7.5, 50 mM KCl, 1 mM MgCl2, 0.005% Nonidet P40, 0.2 µg of tRNA and 5′ end-labelled let-7a microRNA (0.1 nM) were incubated for 30 min with 0.1, 0.2 or 0.5 µM GST or catalytically deficient GST–Argonaute2 (D597A) (as indicated). Native loading buffer was added and the reactions were analysed by 1% agarose gel. The position of the let-7a–Argonaute2 complex is indicated by an asterisk. (B) Identification of the let-7a–Argonaute2–mRNA complex. Reaction mixtures (20 µl) containing 50 mM Tris, pH 7.5, 50 mM KCl, 1 mM MgCl2, 0.005% Nonidet P40, 0.2 µg of tRNA and 5′ end-labelled Let-7a microRNA (0.1 nM) were incubated for 30 min with 0.2 µM GST or catalytically deficient GST–Argonaute2 (D597A) (as indicated). Let-7a target RNA or irrelevant (Irrel) target RNA (1, 10 or 100 pM) was added to each reaction, followed by a 15 min incubation. Reactions were split and analysed by 1% agarose gel, using both native loading buffer (50% glycerol, 0.1 M Tris, pH 8.0, 0.1% Bromophenol Blue and 0.1% Xylene Cyanaole) or SDS loading buffer (50% glycerol, 0.1 M Tris, pH 8.0, 0.5% SDS, 20 mM EDTA, 0.1% Bromophenol Blue and 0.1% Xylene Cyanole). The position of the let-7a–Argonaute2 and let-7a–Argonaute2–mRNA complexes are indicated by * and ● respectively. (C) The let-7a–Argonaute2–mRNA complex contains RNA duplex. Samples from (B) were analysed via 12% PAGE under native conditions. M, let-7a–mRNA duplex formed in 200 mM NaCl. (D) The 5′ terminal uracil of let-7a is critical for mRNA recruitment. Reactions (20 µl) containing 50 mM Tris, pH 7.5, 50 mM KCl, 1 mM MgCl2, 0.005% Nonidet P40, 0.2 µg tRNA and 0.2 µM catalytically deficient GST–Argonaute2 were preincubated for 30 min with 5′-end-labelled let-7a or let-7a containing 5′ nucleotide alterations (as indicated). Let-7a target RNA (50, 100 or 250 pM) was added to each reaction, followed by a 15 min incubation. Native loading buffer was added and the reactions were analysed by 1% agarose gel. The position of the let-7a–Argonaute2 and let-7a–Argonaute2–mRNA complexes are indicated by * and ● respectively.
Next we sought to establish whether the 5′ terminal uracil was important for the formation of the let-7a–Argonaute2 complex itself, or for a subsequent step in the silencing reaction. Alteration of the uracil base did not affect the formation of the let-7a–Argonaute2 complex (Figure 8D). However, only the microRNA that contained uracil at the terminus was capable of efficiently forming the let-7a–Argonaute2–mRNA complex. This observation suggested that an appropriate occupancy of the 5′ binding pocket by let-7a is necessary to recruit mRNA and direct mRNA cleavage.
DISCUSSION
It is well established that small-RNA-directed silencing plays a critical role in the regulation of gene expression [3–6]. However, the fundamental steps in this pathway, the formation of the microRNA ribonucleoprotein complex and its recruitment of mRNA, have remained poorly understood. In the present study, we have chosen to study the mechanism of action of let-7a and Argonaute2 using a fully complementary model target mRNA. Importantly, we show that recombinant human Argonaute2 is sufficient to direct mRNA silencing. Similar to studies with siRNA [20,27,55], let-7a-directed cleavage of mRNA requires the divalent cation, magnesium, and the integrity of the Argonaute2 DDH catalytic domain.
Little attention has been paid to the possibility that human microRNAs may silence via mRNA cleavage. Largely, this is because it has been thought that complete complementarity is required, and contemporary sequence analysis indicates that there are very few human mRNAs that contain elements that are fully complementary to microRNAs [56–61]. However, we show here that complete complementarity is not critical and therefore it is quite likely that let-7amay silence some cellular mRNAs. It is also thought that the ability to cleave mRNA is unique to Argonaute2. This has also contributed to the notion that microRNA-directed silencing via mRNA cleavage would be a rare event. However, the experiments to test the cleavage activity of Argonautes 1, 3 and 4 were conducted with a guide RNA that contained a uracil at its 5′ terminus [23,62]. Thus, it is quite possible that these Argonautes have a different specificity and it might be interesting to re-examine their catalytic potential by providing them with guide RNAs that have other nucleotides at their 5′ terminus.
Previous studies have also illuminated the critical role of the 5′ ends of microRNA and the guide strand of siRNA [3,59,61,63–66]. However, in those studies, the deletion of the 5′ end of the guide strand of siRNA did not preclude mRNA cleavage, but resulted in a new cleavage site [27]. The most reasonable interpretation of those studies is that the new 5′ terminus of the siRNA can enter the phosphate-binding pocket and thus ‘move’ the cleavage site accordingly. This is also consistent with the observation that the 3′ end of the guide RNA can be deleted without compromising cleavage activity. However, in our experiments, deletion of the 5′ end of let-7a microRNA abrogated cleavage, and no new cleavage site was created. Importantly, the deletion recreated a uracil base at the 5′ end. Despite this, it would appear that the new terminus cannot occupy the 5′ phosphate-binding site and redirect cleavage. This suggests that there are likely to be specific interactions between the other residues of let-7a and Argonaute2, and that these are stronger than those involved in the 5′ pocket binding. Thus the development of dynamic analytical techniques that can distinguish interactions at the 5′ end from those at the body of the let-7a microRNA is a critical future endeavor. In summary, our observations suggest that sequence-specific interactions between the let-7a microRNA and Argonaute2 might be more important than have been suspected.
The major observation here is that the 5′ terminus of the guide RNA plays a unique sequence-specific role in the recruitment of mRNA. Structural studies on Archaeoglobus fulgidus and Thermus thermophilus PIWI complexes have shown that the 5′ phosphate of the guide molecule is complexed to amino acid residues and Mg2+ [27,32,35]. Interestingly, these studies show that the terminal base (in this case thymine) can interact with the side chain of an arginine residue. It is likely that substitution of the terminal base will weaken this interaction and perhaps preclude the engagement of the 5′ end with the binding pocket. Alternatively, this substitution may result in an inappropriate engagement that alters the accessibility of the guide RNA to mRNA. At present, we cannot distinguish these possibilities. It is important to point out that there are many functional microRNAs and siRNAs that do not have a uracil residue at the 5′ terminus. From our studies in the present manuscript, it would appear unlikely that these RNAs efficiently utilize the Argonaute2 family member. Indeed, one suspects that, as is the case in plant cells, each Argonaute family member will associate with particular classes of small RNAs directed by the nature of the 5′ terminus [67,68].
Finally, our observations suggest an additional consideration to the models that seek to explain the selective loading of the let-7a guide strand of the precursor microRNA duplex into Argonaute2 [69]. Given that the two strands of microRNA precursor duplexes usually have different bases at the 5′ termini, it is now plausible that Argonaute2 itself exerts some specificity in strand uptake. Indeed, we have recently shown that let-7*, which contains a cytosine residue at its 5′ terminus, is remarkably inefficient in supporting mRNA cleavage (D. W. Salzman, K. M. Felice and H. M. Furneaux, unpublished work).
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [grant numbers R03 DA022226, P01HL70694].
Abbreviations used
- DDH catalytic triad
aspartate-aspartate-histidine catalytic triad
- GAPD
glyceraldehyde-3-phosphate dehydrogenase
- GST
glutathione transferase
- siRNA
small interfering RNA
- 3′-UTR
3′-untranslated region
Footnotes
AUTHOR CONTRIBUTION
Kristin Felice performed research, designed experiments, analysed data and wrote the manuscript. David Salzman performed research, designed experiments and analysed data. Jonathan Shubert-Coleman performed research. Kevin Jensen performed research. Henry Furneax wrote the manuscript, designed experiments and analysed data.
REFERENCES
- 1.Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Slack FJ. microRNAs: small molecules with big roles – C. elegans to human cancer. Biol. Cell. 2008;100:71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Ekwall K. The RITS complex-A direct link between small RNA and heterochromatin. Mol. Cell. 2004;13:304–305. doi: 10.1016/s1097-2765(04)00057-7. [DOI] [PubMed] [Google Scholar]
- 8.Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 9.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 11.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 12.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 13.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 14.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 16.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 19.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 22.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 24.Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- 26.Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2 and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 28.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 31.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 32.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 37.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 38.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 39.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 40.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 42.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 43.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 45.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 47.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Z, Chang FC, Furneaux HM. The identification of an endonuclease that cleaves within an HuR binding site in mRNA. Nucleic Acids Res. 2000;28:2695–2701. doi: 10.1093/nar/28.14.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silber R, Malathi VG, Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl. Acad. Sci. U.S.A. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 55.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 56.Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev. Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 57.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem. Biol. 2004;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 58.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 60.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 62.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 63.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 64.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 65.Lai EC. Predicting and validating microRNA targets. Genome Biol. 2004;5:115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 67.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 69.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]