Abstract
This study evaluated whether the gap junction protein connexin (Cx) 43 could serve as a negative cell surface marker for human corneal epithelial stem cells. Cx43 expression was evaluated in corneo-limbal tissue and primary limbal epithelial cultures. Immunofluorescent staining and laser scanning confocal microscopy showed that Cx43 was strongly expressed in the corneal and limbal suprabasal epithelial cells, but the basal cells of the limbal epithelium were negative. Cx43 antibody stained mainly large cells but not small cells in primary limbal epithelial cultures. As determined by semiquantitative reverse transcription polymerase chain reaction (PCR) and real-time PCR, Cx43 mRNA was more abundant in the corneal than limbal epithelia, and it was expressed in higher levels in mature limbal epithelial cultures. Using GAP11, a rabbit polyclonal antibody against the Cx32 extracellular loop 2 (151–187), a sequence that is highly homologous in Cx43, the Cx43dim and Cx43bright cells were selected from primary limbal epithelial cultures by fluorescence-activated cell sorting and were evaluated for stem cell properties. These Cx43dim and Cx43bright cells were confirmed by their expression levels of Cx43 protein and mRNA. The Cx43dim cells were found to contain higher percentages of slow-cycling bromodeoxyuridine (BrdU)-label retaining cells and the cells that were positive for stem cell-associated markers p63, ABCG2, and integrin β1 and negative for differentiation markers K3 and involucrin. The Cx43dim cells possessed a greater proliferative potential than Cx43bright cells and nonfractionated cells as evaluated by BrdU incorporation, colony-forming efficiency, and growth capacity. Our findings suggest that human limbal basal cells do not express connexin 43, which could serve as a negative cell surface marker for the stem cell-containing population of human limbal epithelial cells.
Keywords: Connexin 43, Cornea, Limbus, Epithelia, Stem cells
Introduction
It has been known for more than a decade that corneal epithelium homeostasis is governed by a small subpopulation of corneal epithelial stem cells, which are located in the basal layer of the limbus (reviewed in refs. [1–3]). Limbal basal epithelial cells are not homogeneous but consist of diverse cell populations of stem cells, transit amplifying cells (TACs) and terminally differentiated cells (TDCs) [4–7]. Limbal epithelial stem cells exhibit unique characteristics that satisfy the widely accepted criteria for defining adult stem cells, which include 1) slow-cycling or long cell-cycle time during homeostasis in vivo; 2) small size and a poor state of differentiation with primitive cytoplasm; 3) high proliferative potential after wounding or placement in culture; and 4) the ability for self-renewal and functional tissue regeneration. The limbal microenvironment is believed to be important in maintaining the “stemness” of these stem cells.
There is no single specific molecular marker that can identify limbal stem cells to date, although several stem cell-associated markers have been proposed. Our previous study showed that the basal cells at limbal epithelium are small primitive cells expressing three patterns of molecular markers: 1) certain basal cells are exclusively positive for p63, ABCG2, and integrin α9; 2) most basal cells show relatively higher expression of integrin β1, epidermal growth factor receptor, K19, and α-enolase than suprabasal cells; and 3) lack of expression of nestin, E-cadherin, connexin 43, involucrin, K3, and K12 [8].
Connexin (Cx) gap junction proteins are specialized cell surface membrane structures that directly connect the cytoplasms of adjacent cells [9]. Gap junction plaques consist of clusters of intercellular aqueous channels that permit bidirectional passage of ions, metabolites, and molecules of size up to 1 kDa between two neighboring cells and have been implicated to be important in the control of cell proliferation, differentiation, and regeneration [10]. These proteins have highly conserved transmembrane and extracellular regions [11]. Currently, there are more than 20 related connexin isoforms identified by humans and mice [9, 12]. Each connexin is characterized by its molecular mass, and three subgroups (α, β, and γ) have been identified based on similarities at the nucleotide and amino acid levels [13]. Irrespective of their molecular mass, all connexins share a common architecture. Each has four hydrophobic amino acid regions that correspond to transmembrane domains, two extracellular loops that share high homology among all connexins, and the carboxy terminus, which projects into the cytoplasm and controls the specificity and interacts with a number of accessory proteins [14, 15].
Most cell types express several different connexin isoforms in a temporal-, spatial-, and differentiation-specific manner. At least 10 connexins have been found to be expressed in the human eye, at the protein level and/or the mRNA level. Cx43, Cx46, and Cx50 were found in the lens, and Cx36, Cx37, Cx43, and Cx45 were found in the retina. There are several connexins, such as Cx26, Cx31, Cx32, and Cx40, which have been found in the retina at mRNA level only [16]. Using electron microscopy, immunohistochemistry, and flow cytometry, previous studies have revealed that Cx26 [17], Cx43, and Cx50 [18] were expressed in the corneal epithelial cells. Shurman et al. surveyed 13 connexins and found that Cx26, Cx30, Cx31.1, and Cx43 were expressed in the human corneal epithelium [19].
Cx43 has been found in a number of epithelial tissues, including human and mouse follicular and interfollicular epidermis [18] and corneal epithelium [8, 18, 19], and in these tissues it is expressed in the suprabasal epithelia and is absent in the basal epithelial cells of the limbus [18] and epidermis [20]. Based on this property of the limbal basal epithelial cells, it has been proposed that the apparent lack of connexin expression may be a marker of the stemness of these cells [18]. The purpose of this study was to evaluate the expression of Cx43 by human limbal epithelial cells, to investigate whether Cx43 negative cells possess stem cell properties, and thus to determine whether Cx43 could serve as a negative marker for the limbal basal epithelial cells that contain the stem cells of the corneal epithelium.
Materials and Methods
Material and Reagents
Cell culture dishes, plates, centrifuge tubes and other plastic ware were purchased from Becton, Dickinson and Company (Bedford, MA, http://www.bdbiosciences.com). Nunc Lab-Tek II eight-chamber slides were from VWR International (West Chester, PA, http://www.vwr.com). Dulbecco's modified Eagle's medium, Ham's F-12 medium, amphotericin B, gentamicin, and 0.25% trypsin-0.03% ethylenediaminetetraacetic acid solution were from Gibco (Grand Island, NY, http://www.invitrogen.com). Fetal bovine serum (FBS) was from Hyclone (Logan, UT, http://www.hyclone.com). Mouse monoclonal antibodies (mAbs) against integrin β1, p63, and involucrin were purchased from Lab Vision (Fremont, CA, http://www.labvision.com). Human ABCG2 mAb and AE5 mAb for keratin 3 (K3) were from Calbiochem (San Diego, http://www.emdbiosciences.com) and ICN Pharmaceuticals (Costa Mesa, CA), respectively. Connexin 32 and 43 mAbs were from Zymed (San Francisco, http://www.invitrogen.com). Rabbit anti-bromodeoxyuridine (BrdU) polyclonal antibody was from Megabase Research Products (Lincoln, NE, http://www.pcrjet.com). Fluorescein Alexa Fluor 488 conjugated goat anti-mouse or rabbit IgG were from Molecular Probes (Eugene, OR, http://probes.invitrogen.com). Antifade Gel/Mount was from Fisher Scientific International (Hampton, NH, http://www.fisherscientific.com). GeneAmp RNA-polymerase chain reaction (PCR) kit and Tagman Universal PCR master mix were from Applied BioSystems (Foster City, CA, http://www.appliedbiosystems.com). Ready-To-Go You-Prime First-Strand Beads was from Amersham Biosciences (Piscataway, NJ, http://www.amersham.com). Hoechst 33342, propidium iodide, rhodamine B, DNA size marker, and other reagents were from Sigma-Aldrich (St. Louis, http://www.sigmaaldrich.com).
Human Corneal and Limbal Tissue and Limbal Epithelial Cell Culture
Fresh human corneoscleral tissues (less than 72 hours post mortem) that were not suitable for clinical use, from donors aged 19–67 years, were obtained from the Lions Eye Bank of Texas (Houston, TX) and from the National Disease Research Interchange (Philadelphia). They were cut through the horizontal meridian, frozen and sectioned for immunostaining, and whole mounted for laser scanning confocal microscopy (LSCM). Primary corneal epithelial cells were cultured from limbal explants using a previously described method [21]. Limbal epithelial cells used for flow cytometry analysis were isolated from fresh limbal tissue and from the confluent limbal explant culture using a previously described method [22].
Immunofluorescent Staining and LSCM
Immunofluorescent staining was performed with primary mAbs against Cx43 (clone CX-1B1, 1:50), Cx32 (clone CX-2C2, 1:50), integrin β1 (clone 4B7R, 1:200), p63 (clone 4A4, 1:1000), ABCG2 (clone BXP-21, 1:25), K3 (AE5, 1:50), or involucrin (SY5, 1:40) or with polyclonal rabbit antibody GAP11 (1:10) using a previously reported method [8, 22, 23] on whole mounted corneal tissues, frozen sections, cultured limbal epithelial cells on the confluent day and fluorescence-activated cell sorting (FACS) selected cells in eight-chamber slides after seeding at a density of 1 × 104 cells per chamber in a 37°C incubator for overnight. Sections were examined and photographed with an epifluorescent microscope (Eclipse 400; Nikon, Tokyo, http://www.nikon.com) with a digital camera (model DMX 1200; Nikon). Cells were observed under a Nikon TE200 inverted microscope (Nikon). Whole mounted corneal tissues were observed with a laser scanning confocal microscope (LSM 510; Carl Zeiss, Thornwood, NY, http://www.zeiss.com) with excitation at 488 and 543 nm and emission filters LP505 and LP560, respectively [22]. Images were acquired by using ×40 oil-immersion objectives with ×10 eyepiece and were processed using Zeiss LSM-PC software and Adobe Photoshop 6.0.
Flow Cytometry Analysis
Limbal epithelial cells harvested from fresh human limbal tissue and primary limbal epithelial cultures were used for flow cytometry. These cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% saponin for 5 minutes at room temperature. Following three washes in phosphate-buffered saline (PBS), cells at 0.5 × 106 cells per 100 μl were incubated with IgG1 anti-Cx43 mAb for 20 minutes on ice. After three washes in PBS-1% FBS and incubation with Alexa Fluor 488-conjugated goat anti-mouse IgG1 antibody for 20 minutes, all cells were washed with PBS-1% FBS and then resuspended in supplemental hormonal epithelial medium (SHEM). The cells were kept on ice until flow cytometry was performed at the Baylor College of Medicine Core Facility using a Beckman Coulter EPICS XL-MCL flow cytometer (Hialeah, FL, http://www.beckmancoulter.com). The data were analyzed by Windowsbased Expo32 software (Beckman Coulter).
GAP11 Antibody Preparation and FACS
Since there is no commercially available antibody generated to the external loop region of Cx43 and useful for separation of living cells [24], we used a unique functional rabbit polyclonal antibody, GAP11. GAP11 was generated originally against an external loop peptide of Cx32 protein, which has high homology to the Cx43 extracellular loop 2 and a further antibody to these cell surface exposed amino acid sequence identified the protein by Western blot and FACS analysis [24]. GAP11 was raised in rabbits to a protein-conjugated peptide corresponding in sequence to amino acid residues 151–187 of rat Cx32 [25]. The peptide was synthesized by 9-fluorenyl methyloxy carbonyl-polyamide solid-phase chemistry. It was cleaved from the resin and deported by treatment with 95% aqueous trifluoroacetic acid. Purity was routinely assessed by high performance liquid chromatography and amino acid analysis. Sandylop rabbits were immunized intramuscularly at 10-day intervals with two equal amounts of 150 μg of peptide coupled to keyhole limpet hemocyanin in a conjugate-Freund's complete emulsion (1:1). After 6 weeks, rabbits were boosted with a similar amount of antigen administered subcutaneously at multiple sites. Animals were bled 10 days later at 4-day intervals. Antiserum was affinity purified by immunoabsorption. This antibody was identified to bind specially to the extracellular loop of Cx32 and Cx43 in dissociated living cells by FACS analysis and immnoprecipitation [25]. A very similar extracellular loop rabbit polyclonal antibody that included the peptide sequence used to generate GAP11 was used to sort T and B lymphocytes [24].
Human limbal epithelial cells from primary explant cultures were used for FACS with GAP11 antibody immunolabeling without fixation as previously described [22]. Two subpopulations differing in their expression of Cx43 were sorted: Cx43 positive (Cx43bright) and negative (Cx43dim) cells. FACS was performed at the Baylor College of Medicine Core Facility using a triple-laser Beckman Coulter Altra high-pressure, high-speed cell sorter. A 488-nm Argon laser was used to excite fluorescein isothiocyanate, and a band-pass filter of 525/20 was used to measure emitted light. All FACS data were analyzed with Expo 32 software.
The cells from unfractionated and sorted Cx43bright and Cx43dim populations were seeded at approximately 1 × 104 cells per chamber in eight-chamber culture slides, incubated at 37°C overnight, and then fixed for immunofluorescent staining with antibodies to stem cell-associated markers. The positive cells were counted using the same way as BrdU labeling index. The selected cell populations were also lysed in 4 M guanidium thiocyanate solution for total RNA extraction to evaluate gene expressions.
Semiquantitative Reverse Transcription-PCR and Relatively Quantitative Real-Time PCR
Total RNA was isolated by acid guanidium thiocyanate-phenolchloroform extraction using our previously described method [23]. The RNA was quantified by its absorption at 260 nm and stored at −80°C before use. With a housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as an internal control, the mRNA expression of different molecular markers was analyzed by semiquantitative reverse transcription (RT)-PCR as previously described [8, 23]. The specific primer pairs for each gene were designed from published human gene sequences (Table 1).
Table 1.
Human primer sequences used for semiquantitative RT-PCR
| Gene | Accession | Sense primer | Antisense primer | PCR product |
|---|---|---|---|---|
| Cx43 | M_65188 | CCTTCTTGCTGATCCAGTGGTAC | ACCAAGGACACCACCAGCAT | 154 bp |
| ΔNp63 | XM_036421 | CAGACTCAATTTAGTGAG | AGCTCATGGTTGGGGCAC | 440 bp |
| ABCG2 | AY_017168 | ACCATTGCATCTTGGCTGTC | CGATGCCCTGCTTTACCAAA | 181 bp |
| Integrin β1 | X_07979 | AGTGAATGGGAACAACGAGGTC | CAATTCCAGCAACCACACCA | 104 bp |
| Involucrin | NM_005547 | GGACTGCCTGAGCAAGAATGTG | TAAGCTGCTGCTCTGGGTTT | 121 bp |
| K3 | NM_057808 | GGCAGAGATCGAGGGTGTC | GTCATCCTTCGCCTGCTGTAG | 145 bp |
| GAPDH | M_33197 | GCCAAGGTCATCCATGACAAC | GTCCACCACCCTGTTGCTGTA | 498 bp |
Abbreviations, bp, base pair(s); GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RT-PCR, reverse transcription polymerase chain reaction.
Relatively quantitative real-time PCR was performed by a previously described method [26] with modification. In brief, the first-strand cDNA was synthesized from 1 μg of total RNA with random hexamer and M-MuLV reverse transcriptase using Ready-To-Go You-Prime First-Strand Beads. Real-time PCR was performed in a Smart Cycler (Cepheid, Sunnyvale, CA) with a 25-μl reaction volume containing cDNA, TaqMan primers, and reporter probe (M_65188, sequence 918–942: TGGT-GCGCTGAGCCCTGCCAAAGAC) for Cx43 (TaqMan Gene Expression Assays; Applied BioSystems), and TaqMan Universal PCR Master Mix Assays were performed in duplicate. A nontemplate control was included in all the experiments to evaluate DNA contamination of the reagent used. The results of relatively quantitative real-time PCR were analyzed by the comparative threshold cycle method and normalized by GAPDH as an internal control.
BrdU Retention Assay
To identify the label-retaining (slow-cycling) stem cells in cultures for the selected populations, BrdU retention assay was performed using a previously reported method [21, 27] before FACS. In brief, early limbal explant cultures in 35-mm dishes at day 3–5, when cells showed outgrowth to 2–3 mm in diameter, were incubated with fresh SHEM medium containing 10 μM BrdU. After continuous labeling with BrdU for 72 hours, the cultures were switched to a BrdU-free medium and chased for 18 days. The cells were then labeled with GAP11 antibody and sorted by FACS. The unfractionated cells and the sorted Cx43bright and Cx43dim cells were plated at 5 × 104 cells per well on eight-chamber slides to perform BrdU immunofluorescent staining. The BrdU labeling index was assessed by point counting through a Nikon TE200 inverted microscope. A total of 500–900 nuclei were counted in six to eight representative fields because this number was considered a minimum requirement to obtain a representative sample [21]. The labeling index was expressed as the number of positively labeled nuclei ÷ the total number of nuclei × 100%.
BrdU Incorporation, Colony-Forming Efficiency, and Growth Capacity
BrdU can be incorporated into DNA in place of thymidine to measure DNA synthesis and cell proliferation. The unfractionated cells, the sorted Cx43bright cells, and the sorted Cx43dim cells (n = 5 each) were incubated with fresh medium containing 10 μM BrdU for 30 minutes, and then immunofluorescent staining for BrdU and label index counting were performed as described above.
To evaluate growth capacity of the Cx43 selected cell populations, a mitomycin C-treated 3T3 fibroblast feeder layer was used as previously described [27, 28]. Each cell population was seeded, at least in triplicate, at 1 × 103 cells per cm2 into six-well culture plates. The colony-forming efficiency (CFE) was calculated as the percentage of the number of colonies generated at day 6 divided by the number of epithelial cells plated in a well. The growth capacity was evaluated on day 12 when cultured cells were stained with 1% Rhodamine.
Results
Cx43 Expression in Human Corneal-Limbal Epithelia and Primary Limbal Epithelial Cultures
Immunofluorescent stainings of corneal and limbal sections and whole mount of corneal and limbal tissue by LSCM showed that Cx43 was strongly expressed by the corneal and limbal suprabasal epithelia (Fig. 1A, 1C, 1D), and the majority of basal cells of limbal epithelium were negative (Fig. 1B, 1E). Flow cytometry analysis showed that Cx43-positive cells accounted for 91.5% ± 1.7% of limbal epithelial cells isolated from fresh limbal tissue (Fig. 1J). Semiquantitative RT-PCR showed that the Cx43 mRNA was expressed at much lower levels by limbal epithelia than the corneal epithelia, whereas the levels of GAPDH mRNA were similar in these samples (Fig. 1L). Real-time PCR confirmed and relatively quantified this difference, showing that the levels of Cx43 transcripts expressed by corneal epithelia were 2.3-fold greater than those of the limbal epithelia (Fig. 1M; n = 3; p < .05).
Figure 1.
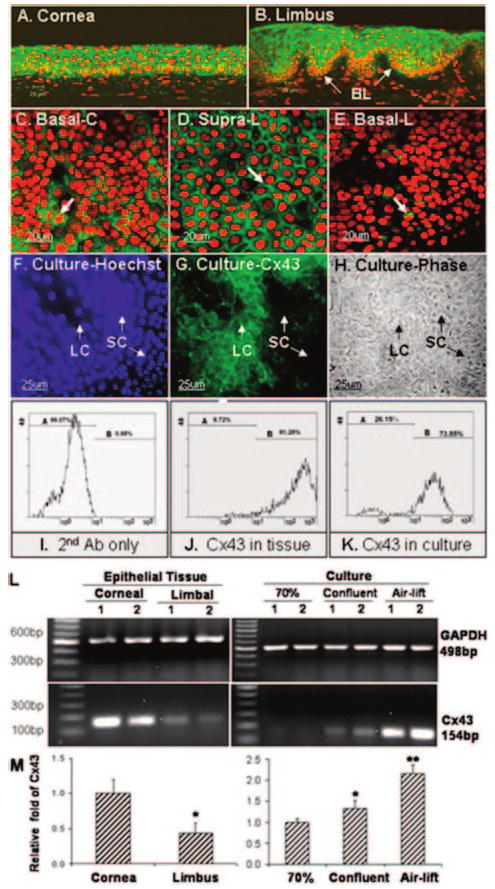
Expression of Cx43 protein and mRNA. (A–H): Immunofluorescent staining (A–G) and laser scanning confocal microscopy (C–E) for Cx43 protein (green) localization in frozen sections of human cornea (A) and limbus (B), in corneal basal (C), limbal suprabasal (D), and limbal basal (E) layers of whole mount cornea with propidium iodide counterstaining (red) and in primary human limbal epithelial cultures (G) with Hoechst 33342 counterstaining (F) (Blue) and phase contrast (H). (I–K): Flow cytometry for Cx43 on human limbal epithelial cells from tissue (J) and cultures (K) with second antibody only as control (I). A, negative; B, positive. (L, M): Semiquantitative reverse transcription polymerase chain reaction (PCR) (L) and relatively quantitative real-time PCR (M) profiles showing expression of Cx43 mRNA in corneal and limbal epithelial tissues and primary limbal epithelial cultures at 70% confluent, confluent, and airlift stages. *, p < .05; **, p < .01 (n = 3; compared with the cornea or 70% confluent culture). Abbreviations: Ab, antibody; Basal-C, corneal basal; Basal-L, limbal basal; bp, base pair(s); BL, basal layer; Cx, connexin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LC, large cells (>20 μm); SC, small cells (<15 μm); Supra-L, limbal suprabasal.
In the primary limbal epithelial explant cultures, the Cx43 antibody stained the membranes of large cells, whereas only a few small-sized cells were stained by this antibody (Fig. 1F–1H). The Cx43 positive cells accounted for 60.8% ± 11.3% (n = 3) by immunofluorescent staining and 73.8% ± 5.2% (n = 3) by flow cytometry analysis of cells in primary limbal epithelial cultures (Fig. 1K). Cx43 expression was analyzed in primary limbal epithelial cell cultures at different growth stages, ranging from 70%–100% confluent and after 7 days of airlift after reaching confluence by RT-PCR (Fig. 1L) and real-time PCR (Fig. 1M). Cx43 mRNA was barely detectable in 70% confluent cultures, and it increased 1.34-fold in confluent cultures and 2.16-fold in the airlifted stratified limbal epithelial cultures (Fig. 1M; n = 3; p < .05 and p < .01, respectively).
Selection of Cx43-Positive and Cx43-Negative Populations by FACS with GAP11 Ab
To test our hypothesis that the Cx43 could serve as a negative marker for the putative corneal epithelial stem cells, we selected Cx43-positive and Cx43-negative cells from primary cultured limbal epithelia by FACS using the GAP11 antibody [25]. Taking advantage that Cx32 protein is not present in the epithelial cells on ocular surface, we used GAP11 antibody to label the Cx43 protein in these cells. Figure 2A shows that GAP11 stained the corneo-limbal epithelial tissue in the same pattern as a commercial Cx43 antibody (clone 1B1) from Zymed (Fig. 1A), whereas a commercial Cx32 mAb (clone CX-2C2, Zymed) did not stain the human corneal and limbal epithelia (Fig. 2A). In primary limbal epithelial cultures, GAP11 positively labeled 61.5% ± 2.4% (n = 3) of cells by flow cytometry. The percentage of GAP11 positive cells was slightly lower than that labeled by the commercial Cx43 mAb (Zymed), which recognizes cytoplasmic C terminal peptide of Cx43 protein. This may be due to the difference in labeling living cells with GAP11 antibody and fixed dead cells with Cx43 mAb, or it may be due to the limited homology between two extra loop sequences of Cx43 and Cx32. These experiments were repeated several times, and the results were averaged. One of the representative experiments using primary cultured limbal epithelial cells is shown in Figure 2B. Based on the levels of functional labeling with the GAP11 antibody, we selected two populations from primary cultured limbal epithelial cells: strongly positive (Cx43bright) with a fluorescein isothiocyanate (FITC) density more than 100 and negative (Cx43dim) with a FITC density less than 0.2, which accounted for 11.9% ± 2.4% and 9.3% ± 1.7% of the population, respectively (Fig. 2B, panels 2, 3).
Figure 2.
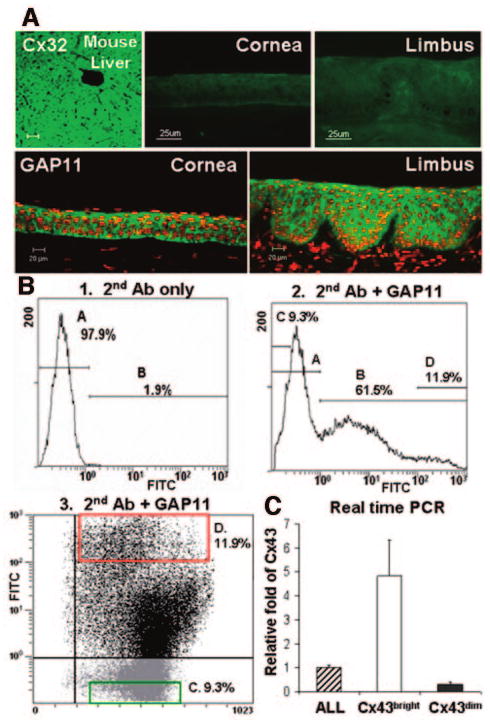
Immunostainings and fluorescence-activated cell sorting by GAP11. (A): Immunofluorescent staining with Cx32 and GAP11 antibodies (green) on frozen sections of human cornea and limbus with mouse liver as Cx32 positive control and PI counterstaining (red). (B): Flow cytometry and fluorescence-activated cell sorting of primary cultured human limbal epithelial cells by GAP11 (panels 2 and 3), with second Ab only as negative control (panel 1). A, negative cells; B, positive cells; C, selected negative cells (Cx43dim) with FITC intensity less than 0.2; D, selected positive cells (Cx43bright) with FITC intensity higher than 100. (C): Real-time PCR for Cx43 expression by ALL and selected Cx43bright and Cx43dim populations. Abbreviations: Ab, antibody; ALL, unfractioned; Cx, connexin; FITC, fluorescein isothiocyanate; PCR, polymerase chain reaction.
To confirm that Cx43-positive and Cx43-negative populations selected by GAP11 are real, the sorted cells were evaluated for their expression of Cx43 by immunofluorescent staining using Cx43 antibody from Zymed and by RT-PCR and real-time PCR for Cx43. The immunofluorescent staining results showed that 80.1% ± 7.8% of cells stained positively for Cx43 mAb in the Cx43bright group, 5.1% ± 3.0% cells stained positively in the Cx43dim group, and 59.3% ± 15.2% of cells were positive in the unfractionated whole population (ALL) (Fig. 3; n = 5; p < .01). Semiquantitative RT-PCR (Fig. 4) and real-time PCR showed that the Cx43bright cells expressed Cx43 mRNA at levels that were fivefold higher than ALL cells and 15-fold higher than Cx43dim cells, which expressed only one-third the level of Cx43 mRNA of ALL cells (Fig. 2C).
Figure 3.
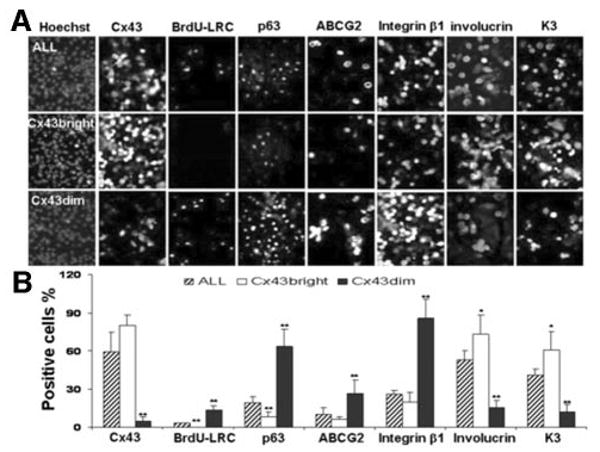
Immunofluorescent staining images (A) and percentage of positive cells (B) for Cx43, BrdU label-retaining cells, p63, ABCG2, integrin β1, involucrin, and K3 in ALL (top row) and selected Cx43bright (second row) and Cx43dim (third row) populations from primary human limbal epithelial cultures with Hoechst 33342 as counterstaining. *, p < .05; **, p < .01 (n = 3; compared with ALL cells). Abbreviations: ALL, unfractioned; BrdU, bromodeoxyuridine; Cx, connexin; LRC, label-retaining slow-cycling cell.
Figure 4.
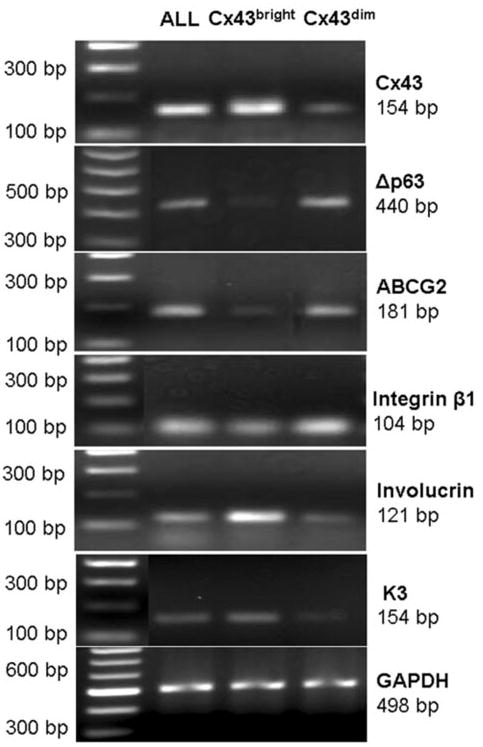
Semiquantiative reverse transcription polymerase chain reaction profiles showing mRNA expression of Cx43 (154 bp), Δp63 (440 bp), ABCG2 (181 bp), integrin β1 (104 bp), involucrin (121 bp), and K3 (154 bp), with GAPDH (498 bp) as internal control, by ALL and by selected Cx43bright and Cx43dim populations from primary human limbal epithelial cultures. A 100-bp DNA ladder is shown in the left lane. Abbreviations: ALL, unfractioned; bp, base pair(s); Cx, connexin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Cx43dim Population Enriched for BrdU Label-Retaining Cells
It is widely accepted that slow-cycling is a characteristic of epithelial stem cells [4, 29]. BrdU label retention assay was used to evaluate the label-retaining slow-cycling cells (LRCs). Our results showed that the labeling index was 3.37% ± 0.01% in ALL cells after 72 hours of labeling and after chasing for 18 days. The BrdU labeling index in the Cx43dim population was 13.49% ± 3.55% (Fig. 3), approximately fourfold greater than unfractionated ALL cells (p < .01; n = 5). In contrast, the Cx43bright population contained no BrdU label-retaining cells.
Cx43dim Population Expressed Higher Levels of p63, ABCG2, and Integrin β1 and Lower Levels of K3 and Involucrin
The cell phenotypes of the Cx43bright and Cx43dim cells were evaluated based on their expressions of proposed limbal epithelial stem cell-associated markers, such as nuclear protein p63, membrane protein ABCG2, and integrin β1, and differentiation markers including K3 and involucrin. As shown in Fig. 3, with Hoechst 33342 counterstaining and the ALL cells used as a control, immunofluorescent staining showed that the Cx43dim population contained 63.3% ± 13.7% p63 protein positive cells, whereas the Cx43bright population had only 8.3% ± 3.5% p63 positive cells (p < .01; n = 5). ABCG2-positive cells accounted for 26.7% ± 10.5% of the Cx43dim population and only 6.5% ± 2.1% of the CX43bright population (p < .05; n = 5). Integrin β1-positive cells were 85.4% ± 15.2% in Cx43dim group and 20.1% ± 7.7% in Cx43bright group (p < .01; n = 5). In contrast, expression of differentiation markers was much lower in the Cx43dim population (15.2% ± 5.9% positive for involucrin and 12.5% ± 5.3% positive for K3) than in the Cx43bright population, which contained 73.3% ± 14.8% (p < .01; n = 5) and 60.7% ± 14.3% (p < .05; n = 5) positive cells for these markers, respectively (Fig. 3).
Semiquantitative RT-PCR further confirmed this expression pattern of above stem cell-associated markers at the transcriptional level. Using GAPDH as an internal control, the Cx43dim cells expressed higher levels of ABCG2, p63, and integrin β1 and lower levels of involucrin and K3 than the Cx43bright cells (Fig. 4).
The Cx43dim Population Possessed Greater Proliferative Potential
The proliferation potential of the Cx43dim and Cx43bright populations and unfractionated ALL cells was assessed by BrdU incorporation, CFE, and growth capacity. BrdU incorporation rate after 30 minutes of labeling was 40.8% ± 6.7% in the Cx43dim group, 24.6% ± 3.5% in the ALL cells, and 8.5% ± 1.9% in the Cx43bright cells (Fig. 5A; p < .01; n = 3).
Figure 5.
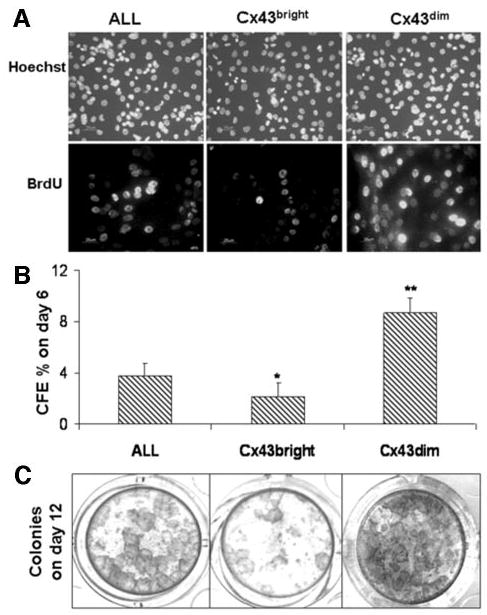
Growth potential of ALL and selected Cx43bright and Cx43dim populations from primary human limbal epithelial cultures. (A): Evaluated by BrdU incorporation with Hoechst 33342 as counter-staining on the day after fluorescence-activated cell sorting. (B): The CFE on day 6. (C): Growth capacity evaluated by 1% Rhodamine staining at day 12. *, p < .05; **, p < .01 (compared with ALL cells). Abbreviations: ALL, unfractioned; BrdU, bromodeoxyuridine; CFE, colony-forming efficiency; Cx, connexin.
To evaluate their growth capacity, the cells in each of the selected populations were seeded in triplicate at a density of 1 × 103 cells per cm2 into six-well culture plates containing a 3T3 fibroblast feeder layer. Figure 5B summarizes the CFE on day 6 from five separate experiments. Cx43dim cells generated the greatest CFE (8.7% ± 1.15%), approximately fourfold higher than the Cx43bright cells (2.15% ± 1.05%; p < .01) and 2.3-fold higher than ALL cells (3.78% ± 0.98%; p < .01). In addition, the colonies generated by Cx43dim cells were much larger than those by Cx43bright cells. The colonies from the CX43dim group grew to confluence by day 12, whereas they became confluent between day 18 and day 24 in the ALL cells population. The Cx43bright cell generated fewer and smaller colonies that failed to grow. Figure 5C shows the representative 1% Rhodamine B-stained cultures from these three populations at day 12.
Discussion
Connexins are the building blocks of gap junctions. In forming a gap junction, six connexins oligomerize to form a hexameric torus called a connexon. Gap junctions provide coupled cells with a direct pathway for sharing ions, nutrients, and small metabolites, thus helping to maintain homeostasis in various tissues. However, it should be noted that unopposed connexin hemichannels also exist on cell surface and are involved in paracine intercellular signaling, implicating the release by cells of signaling molecules such as ATP [30]. These single channels constructed of connexins and biogenetic precursors of gap junctions are diffusely distributed around cells, where their channels, when open, complement communication occurring directly across gap junctions.
Connexins play an important role in the control of cell proliferation, differentiation, and regeneration. Abnormal function and expression of specific connexin genes has been linked to several diseases, including genetic deafness, skin disease, peripheral neuropathies, and cataracts (reviewed in refs. [10, 11, 14, 16, 31]). Cx43, which is an α1 connexin, has been found to be expressed abundantly among several connexins including Cx26, Cx30, Cx31.1, and Cx50 expressed in corneal epithelium [8, 17–19]. Matic et al. [18, 20] have demonstrated that stem cells of the corneal epithelium and epidermis lack connexins and metabolite transfer capacity and that the label-retaining cells (presumptive stem cells) of mice vibrissae do not express gap junction protein connexin 43 [32]. Wolosin et al. [33] found that the isolated Cx43-negative cells represent the precursors of the basal and putative stem cells of the limbal epithelium at a very early stage in ocular development. These previous reports suggest that the apparent incongruity of connexin expression in the cornea and limbus may reflect the limbal basal epithelial cells to retain its “stemness” in a microenvironmental niche.
The Basal Cells of Human Limbal Epithelium Lack Connexin 43 Expression
In this study, the Cx43 expression pattern in human corneal and limbal tissues and cultures was observed from multiple donors using immunofluorescent staining, LSCM, and flow cytometry (Fig. 1). Cx43 antibody strongly stained the corneal epithelia and limbal suprabasal epithelia, but not the basal cells of limbal epithelia, where the corneal stem cells are located. We found that more than 91% of human limbal epithelia were Cx43 positive by flow cytometry. In primary human limbal epithelial cultures, most large-sized cells, representing the differentiated cells, stained strongly for Cx43, whereas the small-sized cells, representing less differentiated proliferating cells, did not express Cx43. Almost 74% of cultured limbal epithelial cells were noted to express Cx43. The expression of Cx43 mRNA (Fig. 1L–1M) was also noted to be greater in the corneal epithelia than the limbal epithelia. In primary cultured limbal epithelial cells, the level of Cx43 mRNA was correlated to the growth potential and state of differentiation. Cx43 mRNA was barely detected in the early cultures at 70% confluence where the cells were still in the proliferative stage. The levels of Cx43 mRNA increased in completely confluent cultures, and the highest levels were observed in the air-lifted stratified cultures that were terminally differentiated. This suggests that as the cells differentiate, they form more gap junction to communicate with each other.
Cx43dim Population Enriched for Certain Properties of Putative Limbal Epithelial Stem Cells
Matic et al. [20] have used Cx43 as a negative marker to isolate epidermal stem cells in neonatal foreskin by FACS and obtained 10% Cx43 negative cells. The antibody they used recognized the cytoplasmic C-terminal peptide (252–270) of Cx43 and could only be used for fixed cells. To the best of our knowledge, sorting living limbal epithelial cells based on Cx43 expression has not been reported. This may have been due to lack of an antibody that recognizes the Cx43 extracellular loop peptide.
The GAP11 antibody used in this study was generated to a linear amino acid sequence located in the second extracellular loop of Cx32 (residues 151–187), and it has been well characterized [24, 25]. For example, it was shown to cross-react by Western blotting to Cx43 in the heart and with Cx32 but not Cx26 in liver tissue. It is a contentious issue whether polyclonal anti-peptide antibodies recognize linear amino acid sequences (as in Western blotting), the spatial contours, or both of extracellular epitopes of connexins expressed on cells. Therefore, the possibility cannot be excluded that the Gap 11 antibody may also recognize other connexins previously detected in limbal epithelial cells, where Cx26, 30, and 30.1 are expressed in addition to Cx43 [19]. It is considered unlikely that Gap11 will recognize Cx30 and 30.1 since they are more closely related to Cx26 [12]. In addition, Cx32 and Cx50 proteins were found to be undetectable on human corneal epithelia [19]. Thus, Cx43, abundantly expressed by human corneal epithelium, is a major form of connexins recognized by GAP11 antibody.
Based on the facts that Cx43 is not expressed by human limbal basal epithelia that contain stem cells and that the corneo-limbal epithelia express abundant Cx43 but not Cx32, we performed FACS using this unique GAP11 antibody, which binds to the extracellular loops of Cx32 and Cx43 [24, 25] and displays the same staining pattern (Fig. 3) as the Cx43 mAb from Zymed, to select Cx43bright and Cx43dim populations from live cultured limbal epithelial cells. The validity of the Cx43bright and Cx43dim populations was confirmed by immunofluorescent staining, RT-PCR, and real-time PCR for Cx43 (Figs. 3, 4).
It is believed that limbal epithelial cultures contain stem cells, TACs, and TDCs, although some properties of the stem cells in vivo may be changed or lost in culture due to the different microenvironment. We evaluated the selected populations for properties that are used to define stem cells. As summarized in Table 2, our studies showed that the Cx43dim population contained a fourfold greater number of BrdU LRCs than the unfractionated whole population. BrdU label retention is a characteristic feature of stem cells, indicating that these cells are slow-cycling. We previously reported that limbal basal epithelial cells express higher levels of stem cell-associated markers, such as p63, ABCG2, and integrin β1, and low or absent levels of differentiation markers, including K3 and involucrin [8]. In our current study, these markers were analyzed in the unfractionated, Cx43dim, and Cx43bright populations. We found that Cx43dim cells expressed higher levels of p63, ABCG2, and integrin β1, and lower or barely detectable levels of involucrin and K3 than the unfractionated whole populations and Cx43bright cells. A high proliferative potential is another important characteristic of stem cells. Our results indicate that Cx43dim cells showed the highest proliferative rate among the three groups by BrdU incorporation and CFE assay. The Cx43dim cells were capable of generating large, actively expanding colonies that grew to confluence in a short time (12–14 days).
Table 2.
Properties of ALL cells and selected populations (Cx43bright and Cx43dim) of human limbal epithelial cells by FACS with GAP11 antibodies
| Properties | All | Cx43bright | Cx43dim |
|---|---|---|---|
| Population %a | 100 | 11.9 ± 2.4 | 9.3 ± 1.7 |
| Label-retaining cells % | 3.37 ± 0.01 | 0 | 13.49 ± 3.55 |
| Positive (+) cells % | |||
| Cx43+b | 59.3 ± 15.2 | 80.1 ± 7.8 | 5.1 ± 3.0 |
| p63+ | 19.6 ± 4.5 | 8.3 ± 3.5 | 63.3 ± 13.7 |
| ABCG2+ | 10.2 ± 5.5 | 6.5 ± 2.1 | 26.7 ± 10.5 |
| Integrin β1+ | 25.9 ± 3.0 | 20.1 ± 7.7 | 85.4 ± 15.2 |
| Involucrin+ | 53.3 ± 6.8 | 73.3 ± 14.8 | 15.2 ± 5.9 |
| K3+ | 40.9 ± 5.0 | 60.7 ± 14.3 | 12.5 ± 5.3 |
| mRNA (RT-PCR) | |||
| Cx43 | +++ | ++++ | + |
| p63 | ++ | – | +++ |
| ABCG2 | ++ | – | +++ |
| Integrin β1 | ++ | + | +++ |
| Involucrin | ++ | ++++ | + |
| K3 | + | ++ | – |
| GAPDH | ++++ | ++++ | ++++ |
| Proliferative capacity | |||
| BrdU incorporation % | 24.6 ± 3.5 | 8.3 ± 1.98 | 40.8 ± 6.7 |
| CFE % | 3.78 ± 0.98 | 2.15 ± 1.05 | 8.7 ± 1.15 |
| Confluent days | 18–24 | Not confluent | 12–14 |
FACS sorted by GAP11.
Immunofluorescent staining with Cx43 monoclonal antibody.
Abbreviations: ALL, unfractionated whole population; BrdU, bromodeoxyuridine; CFE, colony-forming efficiency; FACS, fluorescence-activated cell sorting; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RT-PCR, reverse transcription polymerase chain reaction.
Interestingly, adult stem cells have been identified to possess two properties: they are slow-cycling or have a long cell cycle time during homeostasis in vivo ([3H]-thymidine- or BrdU-label retaining cells) [2–4], and they possess great proliferative potential after wounding or placement in culture in vitro [34]. These properties may be due to the ability of stem cells to divide asymmetrically to produce one daughter cell and one progenitor cell. The daughter cell retains the slow-cycling stem cell property for self-renewal. Once the slow-cycling cells have been labeled, they retain this label for a much longer period and they are identified as label-retaining cells. The progenitor cells are mitotically active cells and proliferate continually to become TACs. Thus, stem cells are able to self-renew and regenerate tissue. In addition, the Cx43dim cells are a partially enriched stem cell-containing population of limbal epithelial cells, which may also contain TACs because most limbal basal cells are negative to Cx43 and stem cells are only a small portion of the basal cells. Future studies are needed to see whether the Cx43dim cells give rise to the Cx43bright cells and display a change in all appropriate markers as they mature, and to investigate the relationship between Cx43 expression and the switch to a differentiation pathway in limbal epithelial stem cells. It is also our future plan to develop a more specific monoclonal antibody against a sequence derived directly from the human connexin 43 extracellular loop for living cell studies.
In conclusion, our results demonstrated that the gap junction protein Cx43 is not expressed by the basal cells of human limbal epithelia and by small cells in limbal epithelial cultures, and that the Cx43dim cells selected by FACS were enriched for certain stem cell-associated properties. Thus, Cx43 appears to serve as a novel negative cell surface marker for human limbal epithelial cells that contain the putative stem cells. The putative human limbal stem cells could be partially selected by Cx43 expression with FACS. Compared with positive FACS selection based on the expression of stem cell-associated markers such as ABCG2 [22] and integrin β1 [35], the negative selection provides an alternative approach for stem cell studies. Although they were not a pure stem cell population, the Cx43dim cells may prove to be useful in the future clinical applications to reconstruct the corneal surface after chemical, thermal, or inflammatory insults.
Acknowledgments
We thank the Lions Eye Bank of Texas for their great support in providing human corneosceral tissues. This study was supported by grants from NIH National Eye Institute, Bethesda, MD EY014553 (D.-Q.L.) and EY11915 (S.C.P.), Lion Eye Bank Foundation, an unrestricted grant from Research to Prevent Blindness, the Oshman Foundation, and the William Stams Farish Fund. Presented in part as abstract at the annual meeting of the Association for Research in Vision and Ophthalmology, April 25–29, 2004, Fort Lauderdale, FL.
Footnotes
Disclosures: The authors indicate no potential conflicts of interest.
For reprints contact Reprints@AlphaMedPress.com
References
- 1.Dua HS, Azuara-Blanco A. Limbal stem cells of the corneal epithelium. Surv Ophthalmol. 2000;44:415–425. doi: 10.1016/s0039-6257(00)00109-0. [DOI] [PubMed] [Google Scholar]
- 2.Tseng SC. Concept and application of limbal stem cells. Eye. 1989;3:141–157. doi: 10.1038/eye.1989.22. [DOI] [PubMed] [Google Scholar]
- 3.Lavker RM, Sun TT. Epidermal stem cells: Properties, markers, and location. Proc Natl Acad Sci U S A. 2000;97:13473–13475. doi: 10.1073/pnas.250380097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotsarelis G, Cheng SZ, Dong G, et al. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 5.Schermer A, Galvin S, Sun TT. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62. doi: 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehrer MS, Sun TT, Lavker RM. Strategies of epithelial repair: Modulation of stem cell and transit amplifying cell proliferation. J Cell Sci. 1998;111:2867–2875. doi: 10.1242/jcs.111.19.2867. [DOI] [PubMed] [Google Scholar]
- 7.Tseng SCG. Regulation and clinical implications of corneal epithelial stem cells. Mol Biol Rep. 1996;23:47–58. doi: 10.1007/BF00357072. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, de Paiva CS, Luo L, et al. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004;22:355–366. doi: 10.1634/stemcells.22-3-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon AM, Goodenough DA. Diverse functions of vertebrate gap junctions. Trends Cell Biol. 1998;8:477–483. doi: 10.1016/s0962-8924(98)01372-5. [DOI] [PubMed] [Google Scholar]
- 10.Donaldson P, Eckert R, Green C, et al. Gap junction channels: New roles in disease. Histol Histopathol. 1997;12:219–231. [PubMed] [Google Scholar]
- 11.Bennett MV, Verselis VK. Biophysics of gap junctions. Semin Cell Biol. 1992;3:29–47. doi: 10.1016/s1043-4682(10)80006-6. [DOI] [PubMed] [Google Scholar]
- 12.Willecke K, Eiberger J, Degen J, et al. Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem. 2002;383:725–737. doi: 10.1515/BC.2002.076. [DOI] [PubMed] [Google Scholar]
- 13.Evans WH, Martin PE. Gap junctions: Structure and function (Review) Mol Membr Biol. 2002;19:121–136. doi: 10.1080/09687680210139839. [DOI] [PubMed] [Google Scholar]
- 14.Chang EH, Van Camp G, Smith RJ. The role of connexins in human disease. Ear Hear. 2003;24:314–323. doi: 10.1097/01.AUD.0000079801.55588.13. [DOI] [PubMed] [Google Scholar]
- 15.Herve JC. The connexins. Biochim Biophys Acta. 2004;1662:1–2. doi: 10.1016/j.bbamem.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Gerido DA, White TW. Connexin disorders of the ear, skin, and lens. Biochim Biophys Acta. 2004;1662:159–170. doi: 10.1016/j.bbamem.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Ratkay-Traub I, Hopp B, Bor Z, et al. Regeneration of rabbit cornea following excimer laser photorefractive keratectomy: A study on gap junctions, epithelial junctions and epidermal growth factor receptor expression in correlation with cell proliferation. Exp Eye Res. 2001;73:291–302. doi: 10.1006/exer.2001.1040. [DOI] [PubMed] [Google Scholar]
- 18.Matic M, Petrov IN, Chen S, et al. Stem cells of the corneal epithelium lack connexins and metabolite transfer capacity. Differentiation. 1997;61:251–260. doi: 10.1046/j.1432-0436.1997.6140251.x. [DOI] [PubMed] [Google Scholar]
- 19.Shurman DL, Glazewski L, Gumpert A, et al. In vivo and in vitro expression of connexins in the human corneal epithelium. Invest Ophthalmol Vis Sci. 2005;46:1957–1965. doi: 10.1167/iovs.04-1364. [DOI] [PubMed] [Google Scholar]
- 20.Matic M, Evans WH, Brink PR, et al. Epidermal stem cells do not communicate through gap junctions. J Invest Dermatol. 2002;118:110–116. doi: 10.1046/j.0022-202x.2001.01623.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim HS, Jun SX, de Paiva CS, et al. Phenotypic characterization of human corneal epithelial cells expanded ex vivo from limbal explant and single cell cultures. Exp Eye Res. 2004;79:41–49. doi: 10.1016/j.exer.2004.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Paiva CS, Chen Z, Corrales RM, et al. ABCG2 transporter identifies a population of clonogenic human limbal epithelial cells. Stem Cells. 2005;23:63–73. doi: 10.1634/stemcells.2004-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li DQ, Tseng SC. Three patterns of cytokine expression potentially involved in epithelial-fibroblast interactions of human ocular surface. J Cell Physiol. 1995;163:61–79. doi: 10.1002/jcp.1041630108. [DOI] [PubMed] [Google Scholar]
- 24.Oviedo-Orta E, Hoy T, Evans WH. Intercellular communication in the immune system: Differential expression of connexin40 and 43, and perturbation of gap junction channel functions in peripheral blood and tonsil human lymphocyte subpopulations. Immunology. 2000;99:578–590. doi: 10.1046/j.1365-2567.2000.00991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman S, Evans WH. Topography of connexin32 in rat liver gap junctions. Evidence for an intramolecular disulphide linkage connecting the two extracellular peptide loops. J Cell Sci. 1991;100:567–578. doi: 10.1242/jcs.100.3.567. [DOI] [PubMed] [Google Scholar]
- 26.Luo L, Li DQ, Doshi A, et al. Experimental dry eye stimulates production of inflammatory cytokines and MMP-9 and activates MAPK signaling pathways on the ocular surface. Invest Ophthalmol Vis Sci. 2004;45:4293–4301. doi: 10.1167/iovs.03-1145. [DOI] [PubMed] [Google Scholar]
- 27.Li DQ, Chen Z, Song XJ, et al. Partial enrichment of a population of human limbal epithelial cells with putative stem cell properties based on collagen type IV adhesiveness. Exp Eye Res. 2005;80:581–590. doi: 10.1016/j.exer.2004.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tseng SC, Kruse FE, Merritt J, et al. Comparison between serum-free and fibroblast-cocultured single-cell clonal culture systems: Evidence showing that epithelial anti-apoptotic activity is present in 3T3 fibroblast-conditioned media. Curr Eye Res. 1996;15:973–984. doi: 10.3109/02713689609017643. [DOI] [PubMed] [Google Scholar]
- 29.Sun TT, Lavker RM. Corneal epithelial stem cells: Past, present, and future. J Investig Dermatol Symp Proc. 2004;9:202–207. doi: 10.1111/j.1087-0024.2004.09311.x. [DOI] [PubMed] [Google Scholar]
- 30.Goodenough DA, Paul DL. Beyond the gap: Functions of unpaired connexon channels. Nat Rev Mol Cell Biol. 2003;4:285–294. doi: 10.1038/nrm1072. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson BJ. Gap junctions: From cell to molecule. J Cell Sci. 2003;116:4479–4481. doi: 10.1242/jcs.00821. [DOI] [PubMed] [Google Scholar]
- 32.Matic M, Simon M. Label-retaining cells (presumptive stem cells) of mice vibrissae do not express gap junction protein connexin 43. J Investig Dermatol Symp Proc. 2003;8:91–95. doi: 10.1046/j.1523-1747.2003.12179.x. [DOI] [PubMed] [Google Scholar]
- 33.Wolosin JM, Schutte M, Zieske JD, et al. Changes in connexin43 in early ocular surface development. Curr Eye Res. 2002;24:430–438. doi: 10.1076/ceyr.24.6.430.8599. [DOI] [PubMed] [Google Scholar]
- 34.Barrandon Y, Green H. Three clonal types of keratinocytes with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]


