Abstract
Purpose
To evaluate the role of cyclin-dependent kinase inhibitors p57 and p15 in transforming growth factor (TGF)-β1 or TGF-β2 inhibited proliferation of primary cultured human limbal epithelial cells using short interfering RNA (siRNA).
Methods
Primary cultured human limbal epithelial cells were treated with TGF-β1 or TGF-β2 for 6 and 24 h, and total RNA extracted for RT-PCR and real-time PCR using primers for p21, p27, and p57 (CipP/Kip family) and p15 and p19 (INK4 family). Proteins were extracted for western blot analysis of p57 and p15. For RNA interference, primary cultured human limbal epithelial cells were transfected with annealed double-stranded siRNA (67 nM) specific for p57, p15, or siRNA-Fluorescein (siRNA-F; as a negative control) followed by treatment with TGF-β1 or TGF-β2 at 1 ng/ml. P57 and p15 were quantitatively detected by real-time PCR and western blot; and immunolocalized by immunofluorescent staining. The effects of TGF-β1 or TGF-β2 on cell proliferation were evaluated by BrdU incorporation and MTT assay.
Results
TGF-β1 or TGF-β2 significantly inhibited primary cultured human limbal epithelial cell proliferation measured by BrdU incorporation and MTT assay. TGF-β1 or TGF-β2 upregulated the expression of p57 and p15 mRNA and protein, but did not effect the expression of p19, p21, or p27. The siRNA transfection efficiency of these cells was 75% and no cellular toxicity was observed by 24 h. The TGF-β1 or TGF-β2 stimulated expression of p57 and p15 mRNA were markedly blocked by siRNA-p57 or siRNA-p15, respectively, but not by siRNA-F. The TGF-β1 or TGF-β2 suppression of epithelial proliferation measured by BrdU incorporation and MTT generation was increased to near normal levels by siRNA-p57 or siRNA-p15. Western blot and immunofluorescent staining showed that levels of p57 and p15 proteins were equally reduced in the cytoplasm and nucleus.
Conclusions
These findings demonstrate that TGF-β1 and/or TGF-β2 inhibit proliferation of primary cultured human limbal epithelial cells and that p57 and p15 play roles in this process.
The transforming growth factor (TGF)-β family comprises a large number of structurally related growth factors, each capable of regulating an array of cellular processes including proliferation, lineage determination, differentiation, motility, and cell death [1]. TGF-β is one of a few known negative regulators of epithelial cell growth, yet the mechanisms by which it affects cell cycle arrest in epithelial cells are poorly understood [2].
Previous reports have shown that TGF-β induces cell migration after wounding and inhibits proliferation of corneal epithelial cells either in vivo or in vitro [3–5]. It has been demonstrated that TGF-β antagonizes the ability of epidermal growth factor to stimulate corneal epithelial proliferation [5,6]. TGF-β comprises three closely related isoforms in mammals known as TGF-β1, TGF-β2, and TGF-β3 which have been detected in corneal epithelium and stroma [7]. Among them, TGF-β1 and TGF-β2 are the predominant forms in the ocular surface and they play important roles as negative modulators of corneal cell proliferation [8]. In addition, they induce G1-phase arrest in limbal basal cells through an autocrine or a paracrine mechanism [9]. Hayashida-Hibino et al. [10] have demonstrated that TGF-β1 control the differentiation and proliferation of corneal epithelial cells through downregulation of various targets, including plasminogen activator inhibitor type 2, transferrin, integrin α3, and cyclin D1. However, the effects of TGF-β1 and TGF-β2 on cell cycle regulation in primary cultured human limbal epithelial cells have not been fully clarified.
Cell division consists of two consecutive processes; mitosis (M) and interphase (including G1, S, and G2 phases). M, G1, S, and G2 form the cell cycle. In the process of cell cycle regulation, cyclin-dependent kinase (CDK) inhibitors (CDKI) play an important role. Two major classes of CDKI have been identified: Cip/Kip and INK4. The Cip/Kip family contains the more general CDKI consisting of p21, p27, and p57, which specifically inactivate G1 cyclinE/CDK2 and cyclinD/CDK4/6. Member of the INK4 family, including p15, p16, p18, and p19, possess 4 ankyrin repeats and inhibit the G1 cyclinD/CDK4/6 complexes [11]. The expression of p15, p21, and p27 increases in response to TGF-β in many cell types [12]. Increased levels of these CDKIs result in a major inhibition of CDK activities associated with the early G1 phase progression, thus locking the cell cycle prior to the G1 restriction point [11,13]. It has been reported that p15, p16, p21, p27, and p57 are expressed by mammary epithelial cells [14–18]. However, CDKI expression in the human limbal epithelial cells has not been completely investigated.
RNA interference (RNAi) is a phenomenon where double-stranded RNA (dsRNA) induces the sequence-dependent degradation of a cognate mRNA in cells [19]. It has been found that mRNA produced by the RNAi-targeted gene is absent from the cytoplasm and reduced in the nucleus. So RNAi exerts its effect during or following RNA processing, but before protein translation [20]. Several techniques have been developed to improve the effects of RNAi including short interfering RNAs (siRNAs), hairpinRNAs (hpRNAs), tiny non-coding RNAs (tncRNAs), and small modulatory RNA (smRNA) [21], among which siRNA is the most popular method. SiRNA uses dsRNAs of 21–22 nucleotides in length and silences genes by promoting the cleavage of mRNAs with near complementary sequences. SiRNA possesses enough sequence complexity to silence specific genes, but does not achieve cell death [22]. It has become a powerful tool for silencing gene expression in mammalian cells and it has wide clinical potential. The efficiency of siRNA in inhibiting gene expression in primary cultured human limbal epithelial cells has not been fully explored.
The goal of the current study was to evaluate CDKI expression; TGF-β1 or TGF-β2 regulation of cell cycle in primary cultured human limbal epithelial cell through CDKI, including CipP/Kip family members p21, p27, and p57 and INK4 family members p15 and p19. We optimized the use of siRNA on primary cultured human limbal epithelial cell to produce minimum toxicity and maximal transfection efficiency, and used siRNA to inhibit the stimulated expression of p57 and p15 by TGF-β. We also investigated whether this technique can reverse the TGF-β inhibition of cell proliferation in primary cultured human limbal epithelial cells.
METHODS
Materials and reagents
Keratinocyte serum-free medium (KSFM) was purchased from Invitrogen-GIBCO BRL (Grand Island, NY). Rabbit anti-BrdU, p15, and p57 polyclonal antibodies were from Megabase Research Products (Lincoln, NE) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Fluorescein Alexa Fluor 488 conjugated goat anti anti-rabbit IgG was from Molecular Probes (Eugene, OR). Hoechst 33342, propidium iodide (PI), DNA size marker, and other reagents were from Sigma (St. Louis, MO). RNeasy Mini Kit, RNAi kit and siRNA-Fluorescein (siRNA-F) were from Qiagen (Valencia, CA). SiCONTROL TOX was from Dharmacon (Lafayette, CO). GeneAmp RNA-PCR kit and Taqman® Universal PCR master mix were from Applied Biosystems (Foster City, CA). Ready-To-Go-Primer First-Strand Beads were from American Pharmacia Biotech Inc. (Piscataway, NJ). NEPER Nuclear and Cytoplasmic Extraction Reagents and the BCA protein assay kit were from Pierce (Rockford, IL). Ready Gel for protein electrophoresis (4–15% Tris-HCl) was from Bio-Rad (Hercules, CA). Immobilon-P polyvinylidendifluoride (PVDF) membrane was from Millipore (Billerica, MA). Luminol Reagent was from Santa Cruz Biotechnology (Santa Cruz, CA). TGF-β1, TGF-β2, and MTT cell proliferation assay kits were from R&D Systems, Inc. (Minneapolis, MN).
Human limbal epithelial cell culture and treatment with TGF-β
Fresh human corneoscleral tissues (less than 72 h post mortem) that were not suitable for clinical use, from donors aged 19–67 years, were obtained from the Lions Eye Bank of Texas (LEBT, Houston, TX). They were prepared for explant culture using a modification of a previously described method [23,24].
The confluent primary cultured human limbal epithelial cells were treated with TGF-β1 or TGF-β2 at 0.1, 1, and 10 ng/ml for 6 and 24 h, respectively. mRNA was extracted following 6 and 24 h treatment for RT-PCR and real-time PCR. Nuclear and cytoplasmic proteins were extracted separately from 24 h 1 ng/ml TGF-β1 or TGF-β2 treated groups for western blot analysis. Primary cultured human limbal epithelial cells treated with 1 ng/ml TGF-β1 or TGF-β2 were fixed for immunofluorescent staining, BrdU incorporation, and MTT assay.
siRNA transfection conditions
To maximize the transfection efficiency and minimize toxicity, primary cultured human limbal epithelial cells were treated with siRNA at different concentrations (nM) at different ratios of siRNA to HiperFect (µg/µl) by conventional or Fast-Forward techniques. Primary cultured human limbal epithelial cells were passaged at 4 × 104 cells/cm2 into 12 well plates in KSFM, and they were transfected by annealed siRNA-F at different concentrations (33, 67, 133, or 267 nM) at a 1:6 or 1:8 ratios of siRNA to HiperFect reagent 24 h after seeding as the conventional method, or they were transfected immediately after cell seeding by the Fast-Forward technique. SiRNA transfection efficiencies were analyzed by flow cytometry. In addition, the transfection efficiencies by siRNA transfection at 67 nM were determined at 3, 6, 12, 16, and 24 h of treatment.
To better determine the transfection efficiency of different concentrations of siRNA in primary cultured human limbal epithelial cells, siCONTROL TOX transfection control from Dharmacon [25] was performed according to the manufacturer’s protocol. Briefly, primary cultured human limbal epithelial cells were treated with 267 nM, 133 nM, 100 nM, 67 nM or 33 nM siCONTROL TOX with HiFect transfection reagent (Qiagen) at a ratio of 1:8 or 1:6 for 24 h by the conventional or Fast-Forward methods. MTT was detected at 24, 48, and 72 h after transfection.
To determine if the transfection reagent and siRNA-F could influence the mRNA level of target genes of this experiment, real-time PCR was performed to detect GAPDH, p57, and p15 following treatment with different concentrations of siRNA (33, 67, 100, 133, or 267 nM) at a 1:6 or 1:8 ratio of siRNA to HiperFect by the conventional or Fast-Forward method.
Transfection of siRNA-p57 and siRNA-p15
To design target-specific siRNA duplexes, a sequence of the type AA (N9) dTdT was selected from the open reading frame and the CTGF mRNA in order to obtain a 21 nucleotide antisense strand with symmetrical-2 nt 3' identical overhanging sequences. The selected siRNA sequence was also submitted to a BLAST search against the human genome sequence to ensure that only one gene was targeted. The dsRNA sequence targeted mRNA for CDKN1C/p57 (GenBank NM_000076) corresponding to position exon 2 or 3, and dsRNA sequence targeted CDKN2B/p15 mRNA (GenBank NM_004936) in exon 2. Two different sets of dsRNA were designed for each gene. DsRNAs were synthesized by Ambion (Austin, TX; Table 1).
TABLE 1.
Human siRNA sequences used in the experiments.
| Gene | Accession | Site | Sequence | target position |
|---|---|---|---|---|
| CDKN1C (p57) | NM_000076 | 1 | sense 5' GGUACACUGGUCCCAAAGUtt 3' antisense 5' ACUUUGGGACCAGUGUACCtt 3' |
exon 2 and 3 |
| 2 | sense 5' GCCAAUUUAGAGCCCAAAGtt 3' antisense 5' CUUUGGGCUCUAAAUUGGCtc 3' |
exon 2 and 3 | ||
| CDKN2B (p15) | NM_004936 | 1 | sense 5' GGGAUAUUUAGGAGUGUGUtt 3' antisense 5' ACACACUCCUAAAUAUCCCtg 3' |
exon 2 |
| 2 | sense 5' GGUCAGACUAAGAAAUAUUtt 3' antisense 5' AAUAUUUCUUAGUCUGACCtc 3' |
exon 2 |
Primary cultured human limbal epithelial cells were transfected by annealed dsRNA at a final concentration at 67 nM with the Fast-Forward technique following the Qiagen protocol. The cells were also treated with transfection reagent alone or with the nonsilencing siRNA-F as negative controls for 24 h, followed by treatment with 1 ng/ml TGF-β1 or TGF-β2 for 24 h, and then mRNA and protein were extracted. Primary cultured human limbal epithelial cells were also fixed for immunofluorescent staining, BrdU incorporation and MTT assay.
RNA extraction, RT-PCR and real-time PCR
Total RNA was isolated from different groups of primary cultured human limbal epithelial cells by a selective silica-gel-based membrane extraction (Qiagen, Valencia, CA).
With a housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as internal control, the mRNA expression of different CDKIs by primary cultured human limbal epithelial cells were analyzed by semi-quantitative RT-PCR with primers listed in Table 2 using the previous protocol [26].
TABLE 2.
Human primer sequences used for semi-quantitative RT-PCR.
| Gene | Accession | Sense primer | Antisense primer | PCR Product |
|---|---|---|---|---|
| P15CDK4B | NM_004936 | CGTTAAGTT TACGGCCAACG | GGTGAGAGTGGCAGGGTCT | 302bp |
| P16INK4 | U26727 | TCGTGCTGATGCTACTGAGG | TTCTTTCAATCGGGGATGTC | 386 bp |
| P19INK4D | U40343 | CTGCAGGTCATG ATG TTTGG | CAGCAGTGTGACCCTCTT GA | 229 bp |
| P21CIP1 | BC000312.2 | GGA AGACCATGTGGACCTGT | AAGATGTAGAGCGGGCCTTT | 238 bp |
| P27KIP1 | BC001971 | AATAAGGAAGCGACCTGCAA | CCTCCCTTCCCCAAAGTTTA | 451 bp |
| p57 | U22398 | AGA TCAGCGCCTGAGAAGTC | GGGACCAGTGTACCTTCTCG | 329bp |
| GAPDH | M_33197 | GCCAAGGTCATCCATGACAAC | GTCCACCACCCTGTTGCTGTA | 493 bp |
Relatively quantitative real-time PCR was performed as previously described [27]. In brief, first-strand cDNA was synthesized from 1 µg of total RNA with random hexamers using M-MuLV reverse transcriptase using Ready-To-Go First-Strand Beads. Real-time PCR was performed in a Smart Cycler (Cepheid, Sunnyvale, CA) with a 25 µl reaction volume containing 2 µl of cDNA, 200 mM of each primer, and 400 nM of TaqMan® Universal PCR Master Mix. TaqMan® primers and reporter probe for p57 (GenBank NM_000076, sequence 1080–1105: TCT GAT CTC CGA TTT CTT CGC CAAG) or p15 (GenBank NM_004936, sequence 512–539: GGC GCG CGA TCC AGG TCA TGA TGA T; TaqMan® Gene Expression Assays; Applied Biosystems). The thermocycler parameters were 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The TaqMan probe, an oligonucleotide complimentary to the amplified segment, was labeled with FAM fluorescent dye at the 3' end of labeled forward primer sequences. Assays were performed in duplicate. A non-template control was included in all the experiments to evaluate DNA contamination of the reagent used. The results of the relative-quantitative real-time PCR were analyzed by the comparative threshold cycle (CT) method [28] and normalized by GAPDH as an internal control.
Western blot analysis
Nuclear and cytoplasm proteins were extracted by NE-PER Nuclear and Cytoplasmic Extraction Reagents kit from Pierce, and protein concentration was determined using the Pierce BCA method calibrating against standards of known bovine serum albumin concentration. Western blot was performed using a previously described method [29] with modifications. The extracted protein samples (50 µg/lane) were mixed with 6X SDS reducing sample buffer and boiled for 5 min before loading. Proteins were separated by SDS polyacrylamide gel electrophoresis, and transferred electronically to PVDF membranes. The membranes were blocked with 5% non-fat milk in tris buffered saline with 0.1% Tween20 (TTBS) for 1 h at room temperature (RT), and then incubated 2 h at RT with rabbit antibodies against p57 and p15 (1:100). The membranes were washed with TTBS and then incubated for 1 h at RT with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:1000 dilution). After washing the membranes, the signals were detected with an ECL advanced chemiluminescence reagent (Santa Cruz Biotechnology, Santa Cruz, CA) using a Kodak image station 2000R (Eastman Kodak, New Haven, CT).
Immunofluorescent staining, laser scanning confocal microscopy (LSCM), and deconvolution microscopy
Immunofluorescent staining was performed with primary polyclonal rabbit p57 and p15 antibodies (1:150 and 1:300) as reported by Stenoien [30]. Briefly, passaged cells from primary cultured human limbal epithelial cells were seeded on poly-D-lysine coated coverslips and treated with or without siRNA for 24 h followed by TGF-β1 or -β2 for 24 h. Before immunofluorescent staining, cells were fixed in 4% formaldehyde and quenched with ammonium chloride. The primary antibodies were applied at RT for 2 h, washed with blocking solution and then anti-rabbit Alexa Fluor 488 conjugated goat anti-rabbit IgG secondary antibody was applied for 1 h at RT. After washing with TTBS, post-fixing in 4% formaldehyde and countainstaining with DAPI, coverslips were observed under the LSCM (LSM 510, Zeiss, Thornwood, NY) and using the precision DeltaVision deconvolution microscope system located at the Integrated Microscopy Core at Baylor College of Medicine. A 488 nm excitation and 505 nm (LP505) emission filter were used for LSCM [31]. LSCM images were acquired using 40× oil immersion objectives with a 10× eye piece and were processed using Zeiss LSM-PC software and Adobe Photoshop. Deconvolution microscopy was performed on a Carl Zeiss AxioVert S100 TV microscope and a DeltaVision Restoration Microscopy System (Applied Precision, Inc., Issaquah, WA). A Z-series of focal planes were digitally imaged and deconvolved with the DeltaVision constrained iterative algorithm [30,32] to generate high resolution images. All image files were digitally processed for presentation using Adobe Photoshop.
BrdU incorporation and MTT assay
A BrdU incorporation assay was performed to measure DNA synthesis and cell proliferation. All groups (n=3 in every group) with or without siRNA and TGF-β1 or TGF-β2 were incubated with fresh medium containing 10 µM BrdU for 30 min, and then BrdU immunofluorescent staining and label index counting were performed. The BrdU labeling index was assessed by point counting through a Nikon TE200 inverted microscope using a 40× objective lens. A total of 500 to 951 nuclei were counted in 6–8 representative fields. This number was considered as a minimum requirement to obtain representative figures [33]. The labeling index was expressed as the number of positively labeled nuclei/total number of nuclei times 100%.
Cellular toxicity and proliferation was further measured by the MTT assay. The MTT assay measures the activity of the mitochondrial enzyme succinyl dehydrogenase, which is expressed in living cells and the signal generated is dependent on the degree of activation of the cells. Therefore, this method can detect cell cytotoxicity or proliferation [34]. It was performed per the manufacturer’s instructions. Briefly, for each experimental condition, cells were seeded at 2 × 105 cells/ml in a volume of 100 µl per well on a 96 well plate (in triplicate). All groups (n=3 in every group) with or without siRNA and TGF-β1 or TGF-β2 were incubated with MTT (3-[4,5-dimethylthiazol-2-yl]2,5-diphenyltetrazolium bromide) reagent at 37 °C for 2 h, then detergent reagent to develop in the dark at RT for 2 h. Well absorbance was measured at 570 nm with a reference wavelength of 650 nm using VERSAmax spectrophotometer (Molecular Devices, Sunnyvale, CA). Media in wells without cells was used as a negative control.
Statistical analysis
Statistical analyses of the real-time PCR data, western blot density, BrdU incorporation, and MTT data for control and treated cells were performed with Student t-test using Excel 7.0 software (Microsoft, WA), with a considered significant at p<0.05.
RESULTS
CDKI expression and regulation by TGF-β in primary cultured human limbal epithelial cells
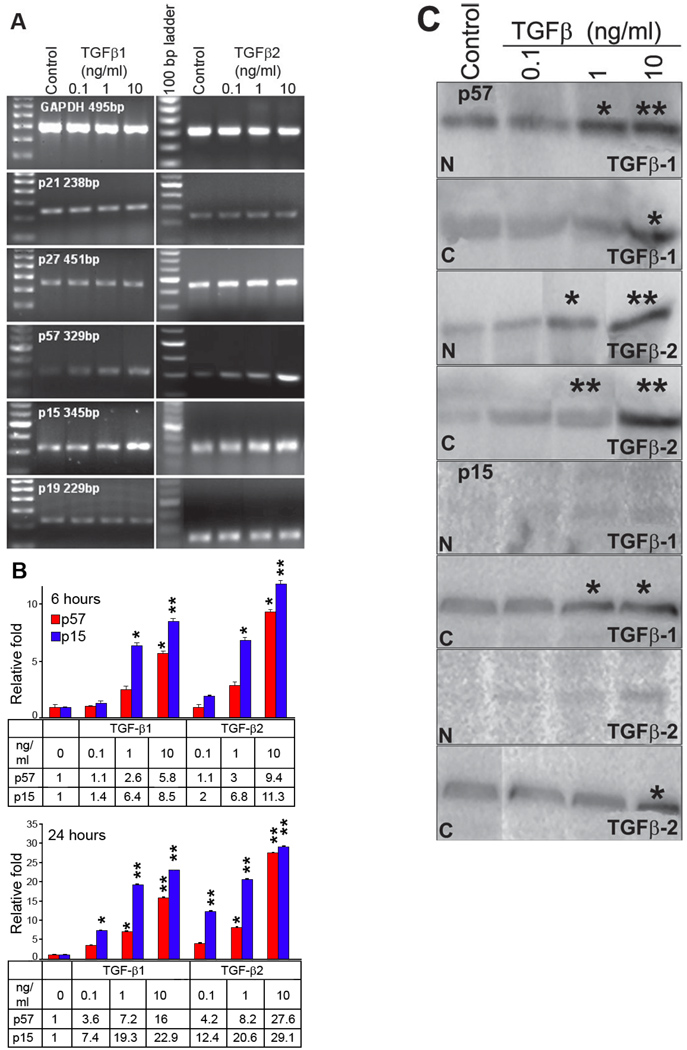
A total of 5 CDKI mRNAs, p15, p19, p21, p27, and p57, were detected by semi-quantitative RT-PCR in primary cultured human limbal epithelial cells. To asses whether TGF-βs can affect CDKIs in the primary cultured human limbal epithelial cells, total RNA and protein were extracted and quantified. As shown in Figure 1, TGF-β1 or TGF-β2 stimulated mRNA of CDKI p15 and p57 in a dose-dependent manner after 6 h of treatment, but they did not change mRNA of other CDKIs, p19, p21, and p27. The stimulated effect of TGF-β2 on mRNA of p15 and p57 was stronger than TGF-β1 treatment (Figure 1A).
Figure 1.
TGF-β1 and TGF-β2 regulation of cyclin-dependent kinase inhibitors (CDKI) in primary cultured human limbal epithelial cells. A: Semi-quantitative RT-PCR of different doses of TGF-β1 and TGF-β2 treatment for 6 h. B: Relative quantitative real-time PCR for p57 and p15 in primary cultured human limbal epithelial cells before and following TGF-β1 and TGF-β2 treatment for 6 and 24 h. The asterisk indicates a p<0.05 and the double asterisk indicates a p<0.01. C: Western blot analysis showed the location of p57 and p15 before and following TGF-β1 and TGF-β2 treatment of nuclear (N) and cytoplasmic (C) fractions for 24 h. The asterisk indicates a p<0.05 and the double asterisk indicates a p<0.01.
Relatively quantitative real-time PCR confirmed that following TGF-β1 treatment of primary cultured human limbal epithelial cells for 6 h, both p57 and p15 mRNA increased, while p15 mRNA increased more significantly than p57 mRNA, especially in the 1 ng/ml and 10 ng/ml groups (n=3, Figure 1B). The changes in p57 and p15 mRNA following TGF-β2 treatment for 6 h showed the same pattern as TGF-β1 treatment, but p57 and p15 mRNA increased more significantly than TGF-β1 treatment (n=3, Figure 1B). At 24 h, there was a similar increase in levels of p57 and p15 mRNA by TGF-β1 or TGF-β2 treatment. The stimulated increase in p57 and p15 were stronger at 24 h than 6 h post-treatment, with a greater increase in p15 than p57 mRNA (n=3, Figure 1B).
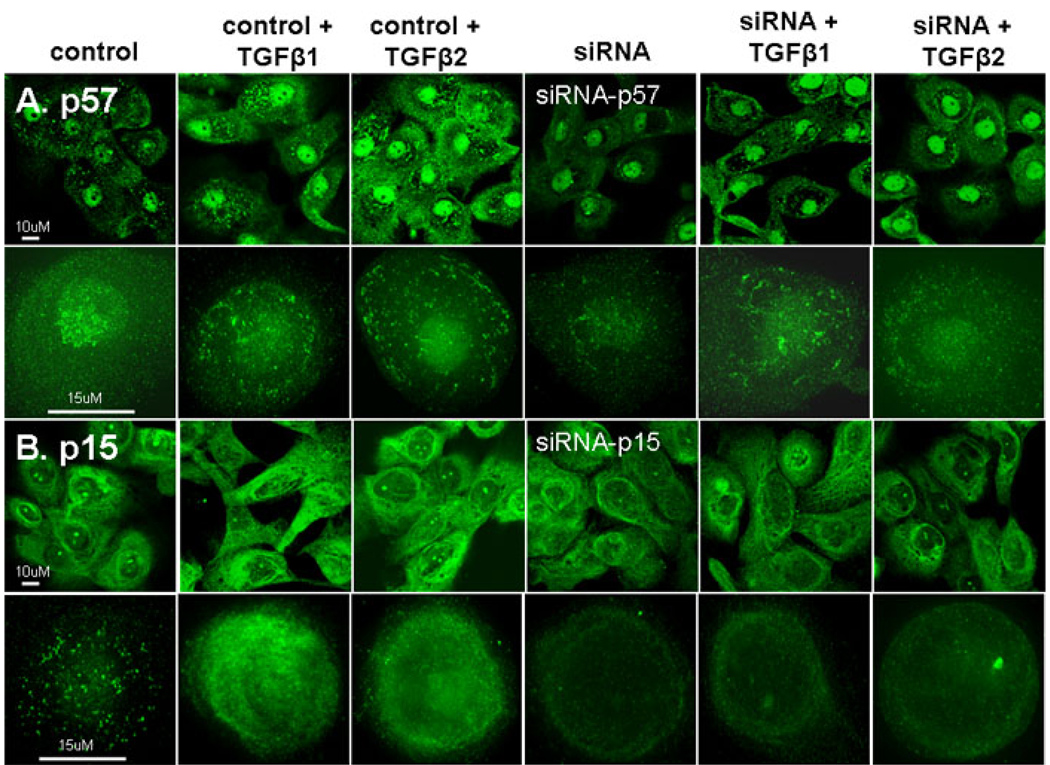
Immunofluorescent staining showed more p57 protein localized in the nucleus than in the cytoplasm. TGF-β1 or TGF-β2 increased p57 protein equally in the nucleus and cytoplasm, and this effect was greater for TGF-β2 than TGF-β1 (Figure 2A). P15 protein primarily localized to the cytoplasm and TGF-β2 caused a greater increase in p15 then than TGF-β1 (Figure 2B).
Figure 2.
Immunofluorescent staining, LSCM (first row) and deconvolution microscopy (second row) for A: p57 in control, siRNA-p57 treated before and following TGF-β1 or TGF-β2 treated (1 ng/ml) in primary cultured human limbal epithelial cells. B: p15 in control, siRNA-p57 treated before and following TGF-β1 or TGF-β2 treated in primary cultured human limbal epithelial cells.
Western blot showed that TGF-β1 or TGF-β2 increased levels of p57 protein in the nucleus and cytoplasm in a dose-dependent manner (Figure 1C). In contrast, p15 protein was found only in the cytoplasm of primary cultured human limbal epithelial cells. TGF-β1 or TGF-β2 induced p15 protein production in a dose-dependent manner (Figure 1C).
Efficiency of siRNA transfection in primary cultured human limbal epithelial cells
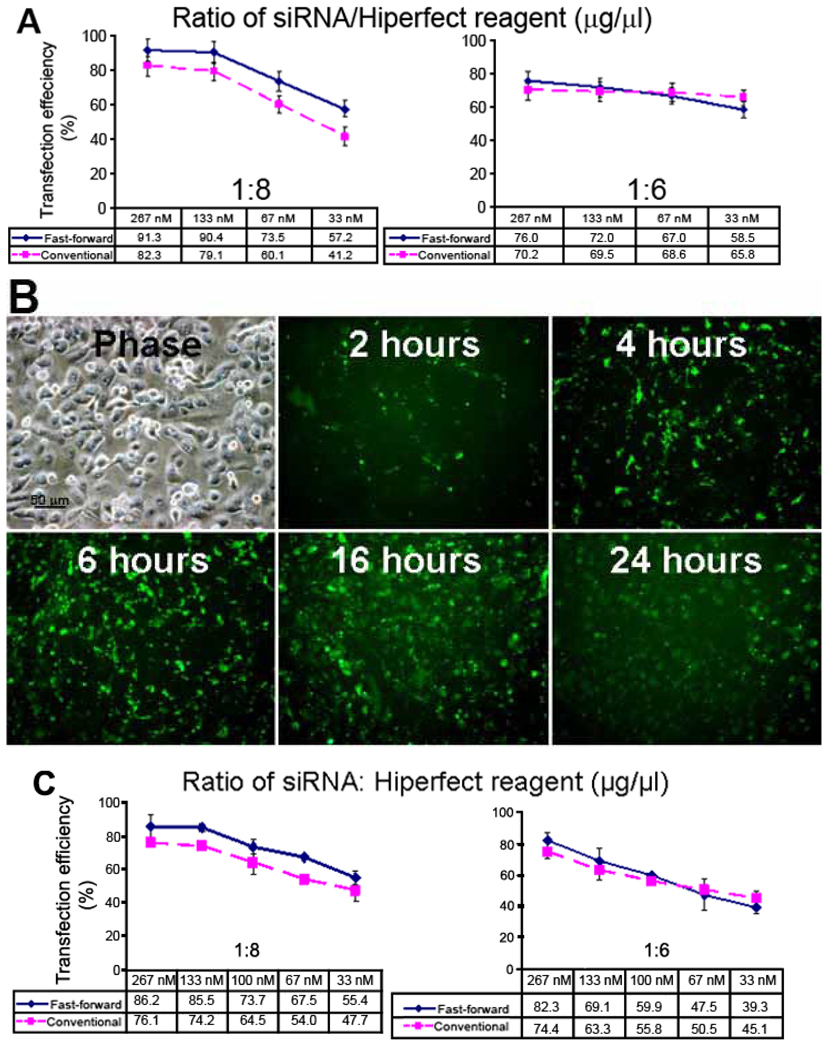
Flow cytometry analysis showed that after siRNA transfection for 24 h, transfection efficiencies were 91.3±2.5%, 90.4±2.4%, 73.5±2.1%, and 57.2±2.3% with a 1:8 ratio of siRNA to Hiperfect transfection reagent (µg/µl), and 76.0±2.7%, 72.0±2.4%, 67.0±1.8%, and 58.5±1.9% with a 1:6 ratio of siRNA to transfection reagent with different concentrations of siRNA-F (267, 133, 67, and 33 nM) by the Fast-Forward technique where the siRNA mixture was added at the same time as cells were seeded. By the conventional transfection method where transfection was performed 24 h after cell seeding, the siRNA transfection efficiencies were 82.3±3.2%, 79.1±3.5%, 60.1±2.9%, and 41.2±1.7% with a 1:8 ratio of siRNA to transfection reagent, and 70.2±2.8%, 69.5±2.6%, 68.6±1.7%, and 65.8±1.9% with a 1:6 ratio of siRNA to transfection reagent corresponding to 267, 133, 67, and 33 nM siRNA-F (Figure 3A). By the Fast-Forward method at a concentration of 67 nM with a 1:8 ratio of siRNA to reagent, transfection efficiencies were 21±2.5%, 41.5±2.7%, and 63±3.4% at 2 h, 4 h, and 6 h, respectively (Figure 3B).
Figure 3.
Transfection efficiency of different ratios of siRNA/Hiperfect reagent and at different times. A: Comparison of transfection efficiency of siRNA-F in primary cultured human limbal epithelial cells using Fast-Forward and conventional methods with different concentrations of siRNA-F and different ratios of siRNA to Hiperfect reagent by flow cytometry. B: Fluorescent microscopy showing fluorescent (siRNA-F) transfection efficiency at different times following the Fast-Forward technique using 67 nM with a 1:8 ratio of siRNA to transfection reagent (µg/µl). C: Comparison of transfection efficiency of siCONTROL TOX in primary cultured human limbal epithelial cells using Fast-Forward and conventional methods with different concentrations of siCONTROL TOX and different ratios of siCONTROL TOX to Hiperfect reagent.
The siCONTROL TOX transfection control is a cytotoxic, RNA-based reagent that can be used to optimize nucleic acid transfection efficiencies in cells. After entering the cell, the reagent triggers a strong, adverse response, leading to apoptosis and cell death within 24–72 h, the degree of cell viability detected by MTT correlates with the transfection efficiency. Our results showed that after siCONTROL TOX transfection with different concentrations (267, 133, 100, 67, and 33 nM) by the Fast-Forward technique for 48 h, transfection efficiencies reached levels as high as 86.2±7%, 85.5±2%, 73.7±4.4%, 67.5±1.5%, and 55.4±4% with a 1:8 ratio of siCONTROL TOX to Hiperfect transfection reagent (µg/µl), and 76.1±2.9%, 74.2±0.76%, 64.5±7.3%, 54±2%, and 47.7±6.3% with a 1:6 ratio of siCONTROL TOX to transfection reagent. By the conventional transfection method, the transfection efficiencies were 82.3±4.6%, 69.1±7.8%, 59.9±1.2%, 47.5±10.1%, and 39.3±3.4% with a 1:8 ratio of siCONTROL TOX to transfection reagent, and 74.4±3.5%, 63.3±6.8%, 55.8±1.95%, 50.8±3.5%, and 45.1±5.2% with a 1:6 ratio of siCONTROL TOX to transfection reagent corresponding to 267, 133, 100, 67, and 33 nM siCONTROL TOX (Figure 3C). Transfection efficiencies were similar between the siRNA-F measured by flow cytometry and siCONTROL TOX.
Relatively quantitative real-time PCR data showed that different concentrations of siRNA-F with a 1:8 or 1:6 ratio of siRNA to Hiperfect transfection reagent (µg/µl) used to treat primary cultured human limbal epithelial cells did not change the mRNA level of GAPDH, p57, and p15 compared to non siRNA-F treated cells (data no shown).
siRNA-p57 and siRNA-p15 knock-down the TGF-βinduced expression and production of p57 and p15 in primary cultured human limbal epithelial cells
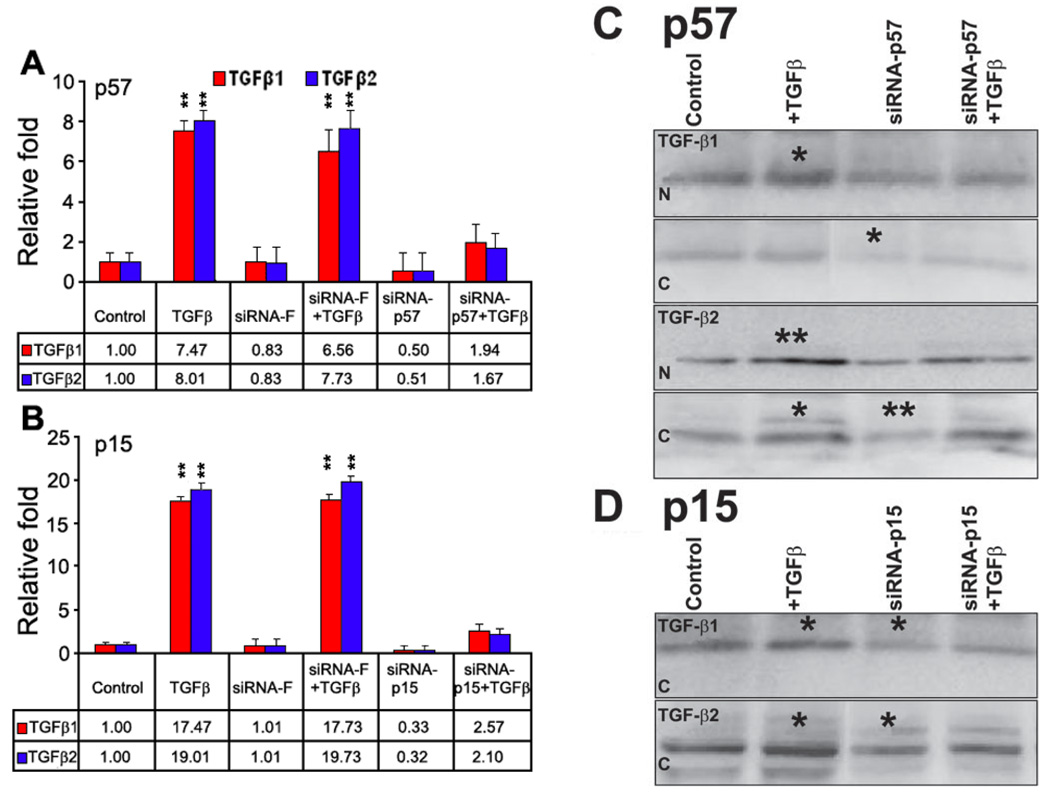
Real-time PCR showed that siRNA-p57 could specifically reduced the level of p57 mRNA and markedly blunt the stimulated increase of p57 in the primary cultured human limbal epithelial cells following TGF-βs treatment (Figure 4A). No effects on control or TGF-βs stimulated levels of p57 were observed with the control siRNA-F (Figure 4A). SiRNA-p57 decreased basal expression of p57 mRNA by 50% (Figure 4A) and siRNA-p15 decreased basal p15 mRNA expression at almost 70% (Figure 4B). Both siRNA-p57 and siRNA-p15 caused a marked inhibition of the stimulatory effects of both TGF-β1 and TGF-β2 on levels of p57 and p15, respectively (Figure 4A,B).
Figure 4.
mRNA and protein analysis of primary cultured human limbal epithelial cells followed by TGF-β treatment after siRNA-p15 and siRNA-p57. A: Relative quantitative real-time PCR analysis showing the effects of siRNA-F and siRNA-p57 on p57 mRNA expression in control and TGF-β (1 ng/ml) stimulated primary cultured human limbal epithelial cells. B: Relative quantitative real-time PCR analysis showing the effects of siRNA-F and siRNA-p15 on p15 mRNA expression in control and TGF-β stimulated (1 ng/ml) primary cultured human limbal epithelial cells. C: Western blot analysis of p57 protein in nuclear (N) and cytoplasmic (C) preparations following TGF-β1 and TGF-β2 treatment (1 ng/ml) for 24 h in primary cultured human limbal epithelial cells with or without siRNA-p57. D: Western blot analysis of p15 protein in cytoplasmic preparation following TGF-β1 and TGF-β2 treatment (1 ng/ml) for 24 h in primary cultured human limbal epithelial cells with or without siRNA-p15. No p15 protein was detected in the nucleus, as shown in Figure 1C. In both C and D, the asterisk indicates a p<0.05 and the double asterisk indicates a p<0.01.
Both western blot and immunofluorescent staining showed that TGF-β1 and TGF-β2 increased p57 protein in the nucleus and cytoplasm after 24 h of treatment (Figure 2A, Figure 4C; <0.05 and <0.01 in the nucleus in TGF-β1 and TGF-β2 treatment, respectively; <0.05 in the cytoplasm in TGF-β2 treatment). siRNA-p57 decreased p57 protein in the nucleus and particularly the cytoplasm (Figure 2A, Figure 4C; <0.05 in TGF-β1 treatment and <0.01 in TGF-β2 treatment). siRNA-p57 treated cells, also showed less of an increase in p57 in the nuclear and cytoplasm following TGF-β1 and TGF-β2 treatments (Figure 2A, Figure 4C; no significant changes were observed after TGF treatments in siRNA groups). Similarly, siRNA-p15 caused a decreased in basal levels of p15 protein and inhibited some of the stimulatory effects of TGF-β1 and TGF-β2 (Figure 2B, Figure 4D; <0.05 in TGF-β1 and TGF-β2 treatment). A greater effect was observed on TGF-β2 treated cells (Figure 2, Figure 4C,D).
siRNA-p57 and siRNA-p15 blocked the inhibitory effect of TGF-βs on proliferation of limbal epithelial cells
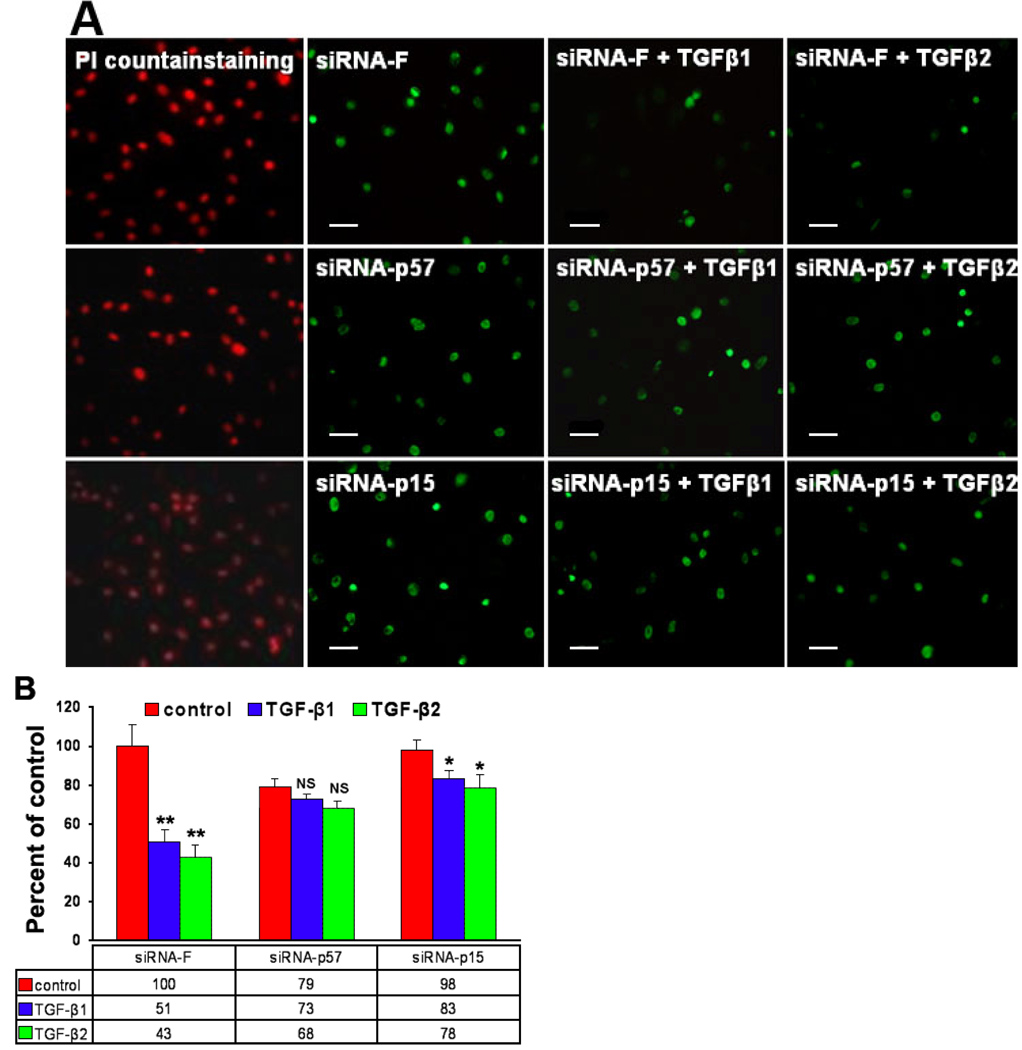
TGF-β1 or TGF-β2 significantly reduced the BrdU incorporation index from 41.95±2.75% in the siRNA-F control group to 10.5±0.71% or 8.5±0.71%, respectively. In the siRNA-p57 groups, the BrdU index recovered to 37.05±4.31% and 33.5±2.12% following TGF-β1 or TGF-β2 treatment, respectively, compared to a 40.35±3.32% in siRNA-p57 treated only. In siRNA-p15 treated groups, the BrdU index recovered to 35.3±6.22% and 31.05±2.76% following TGF-β1 or TGF-β2 treatments, respectively, compared to 41.74±5.03% in siRNA-p15 only (Figure 5A, Table 3).
Figure 5.
Cell proliferation ability assay. A: Representative BrdU incorporation in primary cultured human limbal epithelial cells treated with siRNA-F, siRNA-p57, and siRNA-p15 with or without TGF-β1 or TGF-β2 treatment (1 ng/ml). B: MTT assay showing effects of siRNA-F, siRNA-p57, and siRNA-on cell proliferation of TGF-β1 or TGF-β2 treatment (1 ng/ml) in primary cultured human limbal epithelial cells. In the image, NS indicates not significant, the asterisk indicates a p<0.05, and the double asterisk indicates a p<0.01.
TABLE 3.
BrdU incorporation index and MTT assay results.
| BrdU incorporation index (%) | MTT assay (%) | |||||
|---|---|---|---|---|---|---|
| siRNA-F | siRNA-p57 | siRNA-p15 | siRNA-F | siRNA-p57 | siRNA-p15 | |
| NONE | 41.95 ± 2.75 | 40.35 ± 3.32 | 41.7 ± 5.03 | 100 ± 11 | 79 ± 6 | 98 ± 6 |
| TGF-β1 | 10.5 ± 0.71 | 37.05 ± 4.31 | 35.3 ± 6.22 | 52 ± 4 | 73 ± 2 | 83 ± 4 |
| P value | <0.01 | NS | NS | <0.01 | NS | <0.05 |
| TGF-β2 | 8.5 ± 0.71 | 33.5 ± 2.12 | 31.05 ± 2.76 | 53 ± 5 | 68 ± 4 | 78 ± 7 |
| P value | <0.01 | NS | NS | <0.01 | NS | <0.05 |
MTT assay showed that TGF-β1 or TGF-β2 treatment reduced MTT absorbance to 52±4% or 53±5% of the control group. In the siRNA-p57 groups, MTT recovered to 73±2% or 68±4% of control, respectively, with TGF-β1 or TGF-β2 treatment (Figure 5B). In the si-RNA-p15 groups the MTT recovered to 83±4% or 78±4%, respectively, with TGF-β1 or TGF-β2 treatment (Figure 5B, Table 3).
DISCUSSION
In this study we found that TGF-β1 or TGF-β2 treatments of primary cultured human limbal epithelial cells stimulated production of the CDKIs p57 and p15 and inhibited proliferation of these cells. These findings suggest that TGF-βs play a role in cell cycle regulation in vivo, especially TGF-β2. p57 and p15 play different role in response to TGF-βstimulation.
TGF-β is a 25 kDa protein that is secreted by nucleated cells. Three isoforms of TGF-β (TGF-β1, TGF-β2, and TGF-β3) have been identified in mammalian cells and they produce similar biological effects. Among them, TGF-β2 has been found to be strongly expressed in corneal cells, including epithelia, keratocytes, and endothelia, while latent TGF-β1 is the predominant isoform detected in human tears. In contrast, TGF-β3 is detected only faintly in corneal cells [7,35,36]. Both TGF-β1 and TGF-β2 inhibited corneal epithelial cell proliferation promoted by KGF and HGF in a dose-dependent fashion [3]. The inhibitory effect of TGF-β2 is greater than TGF-β1 [3]. Therefore, in this study we have only evaluated the effect of TGF-β1 or TGF-β2 on primary cultured human limbal epithelial cells.
Previous studies have shown that TGF-β can upregulate p15, p21 and p27 CDKIs in epithelial cells [12,37], inhibit the activities of both cyclin D-CDK4/6 and cyclin E/CDK2 complexes resulting in hypophosphorylation of pRbs and decreased transcriptional activity by E2Fs in lung epithelial cells [38,39] and human corneal epithelial cells [15–18]. It has been shown that active transcriptional repression by the Rb-E2F complex mediates G1 arrest by TGF-β [40,41]. However, there is not much evidence on weather TGF-βs regulate other CDKI besides p15 in epithelial cells [12]. Due to its pleiotropic nature, the mechanisms of TGFβ-mediated growth inhibition varies between cell types and they have not been fully studied in corneal epithelial cells. To the best of our knowledge, there has been no thorough investigation of CDKI expression and location in human corneal epithelial cells, nor have the effects of TGF-βs on regulation of CDKIs besides p15 in these cells been evaluated.
In this study, we found that mRNA of 3 members of the Cip/Kip family, p21, p27, and p57, and 2 members of INK4 family, p15 and p19, were detected by semi-quantitative RT-PCR in primary cultured human limbal epithelial cells. Among them, only mRNA from p57 and p15 increased following TGF-β1 and TGF-β2 treatments for 6 and 24 h. Semi-quantitative and relative quantitative real-time RT-PCR, western blot, and immunofluorescent staining showed that TGF-β1 or TGF-β2 stimulated the expression of CDKI p57 and p15 at the mRNA and protein levels in a dose-dependent manner in primary cultured human limbal epithelial cells, and inhibited cell proliferation measured by BrdU incorporation and an MTT assay. The strong TGF-β mediated upregulation of p57 observed in primary cultured human limbal epithelial cells has not been reported in other epithelial cells types [42].
Generally, INK4 proteins function as proliferation suppressor by phosphorylation of Rb protein which is found in the cytoplasm. Cip/Kip proteins are mainly related to DNA and programmed cell death such as apoptosis which happens in the nucleus, they also participate in Rb phosphorylation in the cytoplasm [43]. In this work, we found that INK protein p15 primarily locates in the cytoplasm, and Cip/Kip protein p57 locates in the nucleus, and slightly expresses in the cytoplasm of human cultured limbal epithelial cells. These finding suggest that although p57 and p15 are both CDKIs, their roles in response to TGF-βstimulation in the human limbal epithelial cells are different. p15 is a major CDKI for Rb phospholylation and p57 is related to cell DNA damage.
RNA silences gene expression through specific degradation of mRNAs. Using siRNA duplexes <30 bp long circumvents both the induction of dsRNA induced cell death and nonspecific gene silencing initially observed with longer sequences [22]. Lee and colleagues [44] have reported a study employing siRNA transfection in primary cultured human limbal epithelial cells using Ambion transfection reagent, but the transfection efficiency was not reported in that study. siRNA experiments in mammalian cells have used siRNAs at varying concentrations, typically ranging from 20 nM [45] to 200 nM [46]. Most published RNA interference studies report using siRNAs at 100 nM or less, which is a concentration where nonspecific effects may occur [47]. In our study, we used this technology to specifically knock down expression of endogenous p57 and p15 in these cells with 67 nM. Using siRNA-F as a reporter we show that siRNA can be successfully transfected into approximately 75% of primary cultured human limbal epithelial cells, and we have optimized the Qiagen protocol on primary cultured human limbal epithelial cells to minimize toxicity and maximize transfection efficiency, confirmed by siCONTROL TOX.
In our study, silencing of p57 and p15 is associated with marked phenotypic changes in the nucleus and cytoplasm in response to TGF-βs, indicating that p57 and p15 are critical downstream mediators of TGF-βs in primary cultured human limbal epithelial cells. Transfection of siRNA-p57 and siRNA-p15 resulted in reduced levels of p57 and p15 mRNA and protein 24 h post-transfection. LSCM and deconvolution microscopy data show that siRNA-57 and siRNA-p15 decrease the p57 and p15 protein levels equally in the nucleus and cytoplasm.
These data presented in this paper support the contention that 5 CDKI members are produced by human corneal epithelial cells, and besides p15, p57 is a key CDKI in TGF-β1 and TGF-β2 growth inhibition proliferation of these cells. TGF-β1 or TGF-β2 induced cell cycle arrest in human corneal epithelial cells depend upon upregulation of the CDKIs p57 and p15. Our study confirms that a TGF-β induced inhibition of human corneal epithelial cell proliferation can be reversed by knock down of p57 and p15. It is likely that other cell signaling pathways affected by TGF-βs such as Smad, MAPK, RhoA, PP2A, and the TAK/MEKK pathways also play a role in this process. To dissect the biological response to TGF-βs at the molecular level, future investigations are needed. Our results also underscore the potential utility of siRNA in regulating of human corneal epithelial cell proliferation in ocular surface diseases and corneal wound healing.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the contribution of Dr. Paul A. Overbeek to our work and the Lions Eye Bank of Texas for their great support in providing human corneoscleral tissues. This study was supported by a grant from Lion Eye Bank Foundation, NIH Grants, EY014553 (DQL) and EY11915 (SCP), National Eye Institute, Bethesda, MD, an unrestricted grant from Research to Prevent Blindness, the Oshman Foundation, and the William Stams Farish Fund. This data was presented in part as poster abstract at the annual meeting of the Association for Research in Vision and Ophthalmology, May 1-May 5, 2005, Fort Lauderdale, FL.
REFERENCES
- 1.Massague J. The transforming growth factor-beta family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 2.Scandura JM, Boccuni P, Massague J, Nimer SD. Transforming growth factor beta-induced cell cycle arrest of human hematopoietic cells requires p57KIP2 upregulation. Proc Natl Acad Sci U S A. 2004;101:15231–15236. doi: 10.1073/pnas.0406771101. Erratum in: Proc Natl Acad Sci U S A 2004; 101:16707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honma Y, Nishida K, Sotozono C, Kinoshita S. Effect of transforming growth factor-beta1 and -beta2 on in vitro rabbit corneal epithelial cell proliferation promoted by epidermal growth factor, keratinocyte growth factor, or hepatocyte growth factor. Exp Eye Res. 1997;65:391–396. doi: 10.1006/exer.1997.0338. [DOI] [PubMed] [Google Scholar]
- 4.Wilson SE, He YG, Weng J, Zieske JD, Jester JV, Schultz GS. Effect of epidermal growth factor, hepatocyte growth factor, and keratinocyte growth factor, on proliferation, motility and differentiation of human corneal epithelial cells. Exp Eye Res. 1994;59:665–678. doi: 10.1006/exer.1994.1152. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura T, Toda S, Mitsumoto T, Oono S, Sugihara H. Effects of hepatocyte growth factor, transforming growth factor-beta1 and epidermal growth factor on bovine corneal epithelial cells under epithelial-keratocyte interaction in reconstruction culture. Exp Eye Res. 1998;66:105–116. doi: 10.1006/exer.1997.0419. [DOI] [PubMed] [Google Scholar]
- 6.Mishima H, Nakamura M, Murakami J, Nishida T, Otori T. Transforming growth factor-beta modulates effects of epidermal growth factor on corneal epithelial cells. Curr Eye Res. 1992;11:691–696. doi: 10.3109/02713689209000742. [DOI] [PubMed] [Google Scholar]
- 7.Li DQ, Lee SB, Tseng SC. Differential expression and regulation of TGF-beta1, TGF-beta2, TGF-beta3, TGF-betaRI, TGF-betaRII and TGF-betaRIII in cultured human corneal, limbal, and conjunctival fibroblasts. Curr Eye Res. 1999;19:154–161. doi: 10.1076/ceyr.19.2.154.5321. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita S, Adachi W, Sotozono C, Nishida K, Yokoi N, Quantock AJ, Okubo K. Characteristics of the human ocular surface epithelium. Prog Retin Eye Res. 2001;20:639–673. doi: 10.1016/s1350-9462(01)00007-6. [DOI] [PubMed] [Google Scholar]
- 9.Joyce NC, Zieske JD. Transforming growth factor-beta receptor expression in human cornea. Invest Ophthalmol Vis Sci. 1997;38:1922–1928. [PubMed] [Google Scholar]
- 10.Hayashida-Hibino S, Watanabe H, Nishida K, Tsujikawa M, Tanaka T, Hori Y, Saishin Y, Tano Y. The effect of TGF-beta1 on differential gene expression profiles in human corneal epithelium studied by cDNA expression array. Invest Ophthalmol Vis Sci. 2001;42:1691–1697. [PubMed] [Google Scholar]
- 11.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue J, Mulder KM. Transforming growth factor-beta signal transduction in epithelial cells. Pharmacol Ther. 2001;91:1–34. doi: 10.1016/s0163-7258(01)00143-7. [DOI] [PubMed] [Google Scholar]
- 13.Moustakas A, Pardali K, Gaal A, Heldin CH. Mechanisms of TGF-beta signaling in regulation of cell growth and differentiation. Immunol Lett. 2002;82:85–91. doi: 10.1016/s0165-2478(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 14.Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 15.Zieske JD. Expression of cyclin-dependent kinase inhibitors during corneal wound repair. Prog Retin Eye Res. 2000;19:257–270. doi: 10.1016/s1350-9462(99)00018-x. [DOI] [PubMed] [Google Scholar]
- 16.Zieske JD, Hutcheon AE, Guo X, Chung EH, Joyce NC. TGF-beta receptor types I and II are differentially expressed during corneal epithelial wound repair. Invest Ophthalmol Vis Sci. 2001;42:1465–1471. [PubMed] [Google Scholar]
- 17.Guo X, Hutcheon AE, Zieske JD. TAT-mediated protein transduction into human corneal epithelial cells: p15(INK4b) inhibits cell proliferation and stimulates cell migration. Invest Ophthalmol Vis Sci. 2004;45:1804–1811. doi: 10.1167/iovs.03-1164. [DOI] [PubMed] [Google Scholar]
- 18.Zieske JD, Francesconi CM, Guo X. Cell cycle regulators at the ocular surface. Exp Eye Res. 2004;78:447–456. doi: 10.1016/s0014-4835(03)00205-7. [DOI] [PubMed] [Google Scholar]
- 19.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 20.Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 22.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 23.Kim HS, Jun Song X, de Paiva CS, Chen Z, Pflugfelder SC, Li DQ. Phenotypic characterization of human corneal epithelial cells expanded ex vivo from limbal explant and single cell cultures. Exp Eye Res. 2004;79:41–49. doi: 10.1016/j.exer.2004.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng SC, Kruse FE, Merritt J, Li DQ. Comparison between serum-free and fibroblast-cocultured single-cell clonal culture systems: evidence showing that epithelial anti-apoptotic activity is present in 3T3 fibroblast-conditioned media. Curr Eye Res. 1996;15:973–984. doi: 10.3109/02713689609017643. [DOI] [PubMed] [Google Scholar]
- 25.Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, Karpilow J, Marshall WS, Khvorova A. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12:988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li DQ, Tseng SC. Three patterns of cytokine expression potentially involved in epithelial-fibroblast interactions of human ocular surface. J Cell Physiol. 1995;163:61–79. doi: 10.1002/jcp.1041630108. [DOI] [PubMed] [Google Scholar]
- 27.Corrales RM, Galarreta DJ, Herreras JM, Calonge M, Chaves FJ. [Normal human conjunctival epithelium expresses MUC13, MUC15, MUC16 and MUC17 mucin genes] Arch Soc Esp Oftalmol. 2003;78:375–381. [PubMed] [Google Scholar]
- 28.Lowe B, Avila HA, Bloom FR, Gleeson M, Kusser W. Quantitation of gene expression in neural precursors by reverse-transcription polymerase chain reaction using self-quenched, fluorogenic primers. Anal Biochem. 2003;315:95–105. doi: 10.1016/s0003-2697(02)00695-4. Erratum in: Anal Biochem 2003; 319:177. [DOI] [PubMed] [Google Scholar]
- 29.Li DQ, Lee SB, Gunja-Smith Z, Liu Y, Solomon A, Meller D, Tseng SC. Overexpression of collagenase (MMP-1) and stromelysin (MMP-3) by pterygium head fibroblasts. Arch Ophthalmol. 2001;119:71–80. [PubMed] [Google Scholar]
- 30.Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA. Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Mol Endocrinol. 2000;14:518–534. doi: 10.1210/mend.14.4.0436. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z, Evans WH, Pflugfelder SC, Li DQ. Gap junction protein connexin 43 serves as a negative marker for a stem cell-containing population of human limbal epithelial cells. Stem Cells. 2006;24:1265–1273. doi: 10.1634/stemcells.2005-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agard DA. Optical sectioning microscopy: cellular architecture in three dimensions. Annu Rev Biophys Bioeng. 1984;13:191–219. doi: 10.1146/annurev.bb.13.060184.001203. [DOI] [PubMed] [Google Scholar]
- 33.Selvamurugan N, Kwok S, Alliston T, Reiss M, Partridge NC. Transforming growth factor-beta 1 regulation of collagenase-3 expression in osteoblastic cells by cross-talk between the Smad and MAPK signaling pathways and their components, Smad2 and Runx2. J Biol Chem. 2004;279:19327–19334. doi: 10.1074/jbc.M314048200. [DOI] [PubMed] [Google Scholar]
- 34.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 35.Pasquale LR, Dorman-Pease ME, Lutty GA, Quigley HA, Jampel HD. Immunolocalization of TGF-beta 1, TGF-beta 2, and TGF-beta 3 in the anterior segment of the human eye. Invest Ophthalmol Vis Sci. 1993;34:23–30. [PubMed] [Google Scholar]
- 36.Kokawa N, Sotozono C, Nishida K, Kinoshita S. High total TGF-beta 2 levels in normal human tears. Curr Eye Res. 1996;15:341–343. doi: 10.3109/02713689609007630. [DOI] [PubMed] [Google Scholar]
- 37.Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 38.Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 39.Hocevar BA, Howe PH. Mechanisms of TGF-beta-induced cell cycle arrest. Miner Electrolyte Metab. 1998;24:131–135. doi: 10.1159/000057360. [DOI] [PubMed] [Google Scholar]
- 40.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 41.Zhang HS, Postigo AA, Dean DC. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- 42.Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 43.Lodish HF, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Molecular Cell Biology. 4th ed. New York: W.H. Freeman; 2000. [Google Scholar]
- 44.Lee JH, Ryu IH, Kim EK, Lee JE, Hong S, Lee HK. Induced expression of insulin-like growth factor-1 by amniotic membrane-conditioned medium in cultured human corneal epithelial cells. Invest Ophthalmol Vis Sci. 2006;47:864–872. doi: 10.1167/iovs.05-0596. [DOI] [PubMed] [Google Scholar]
- 45.Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci U S A. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, Hait WN, Yang JM. Small interfering RNA-induced suppression of MDR1 (P-glycoprotein) restores sensitivity to multidrug-resistant cancer cells. Cancer Res. 2003;63:1515–1519. [PubMed] [Google Scholar]
- 47.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]