Abstract
This study investigated whether cell size correlates with phenotype and proliferative capacity in human corneal epithelial cells. Primary cultured human corneal epithelial cells were sorted by flow cytometry based on forward scatter profile in comparison with the profile of beads of known size. Four fractions (A, B, C, and D) of cells ranging in size from 10 to 16, 17 to 23, 24 to 30, and ≥31 μm in diameter, respectively, were collected to evaluate their 5-bromo-2-deoxyuridine (BrdU) label retention properties, cell phenotype, and clonal growth capacity on a 3T3 fibroblast feeder layer. Among these four populations, cell size was shown to positively correlate with the expression of the differentiation markers keratin (K) 3, K12, and involucrin and inversely with the levels of stem cell–associated markers ΔNp63 and ABCG2 and with colony-forming efficiency (CFE) and growth capacity. Population A with the smallest size, accounting for 11.0% ± 4.5% of the entire population, contained the greatest number of BrdU label-retaining slow-cycling cells, displayed the highest percentage of cells immunopositive to p63 and ABCG2 and negative to K3 and involucrin, expressed the highest levels of ΔNp63 and ABCG2 mRNA and the lowest levels of K3, K12, and involucrin, and possessed the highest CFE and growth capacity. These findings suggest that cell size correlates with cell differentiation phenotypes and proliferative capacity in human corneal epithelial cells. The smallest cells in population A seem to be enriched for putative stem cells, and small cell size may represent one of the important properties of adult corneal epithelial stem cells.
Keywords: Cornea, Limbus, Epithelium, Stem cells, Cell size, BrdU
Introduction
Although the role and mechanism of cell size in stem cell regulation have not been completely defined, it has been reported that cell size is related to cell cycle [1], cell proliferation [2–4], and differentiation [5–7]. The cycle time between successive cell divisions in higher eukaryotes has been shown to depend on cell size, which under normal conditions is divided into two phases, corresponding to a sizer and timer [8]. A size-sensing mechanism in animal cells has been recently proposed, suggesting that an active size threshold mechanism exists in the G1 phase, which induces adjustment of cell-cycle length in the next cycle, thus ensuring maintenance of a proper balance between growth and proliferation rates in vertebrates [8, 9]. Recently, spore-like cells were reported as adult stem cells in virtually every tissue of the body. These small cells, measuring 3 to 7 μm, seem to lie dormant until activated by injury or disease. Being dormant, they survive in extremely low oxygen environments and have an exceptional ability to survive in hostile conditions, known to be detrimental to mammalian cells, including extremes of temperature. They are undifferentiated cells consisting predominantly of the nucleus within a small amount of cytoplasm containing a few mitochondria, and they have the potential to generate numerous adult stem cells [10].
Human corneal epithelial stem cells are believed to be located at the limbus. A recent study determined that the basal epithelial layer of the human limbus harbors cells of the smallest size (10.1 ± 0.8 μm) and with the fewest cytoplasmic granules, measured by a combination of in vivo confocal microscopy and flow cytometry [11]. Transmission electron microscopy has shown that the basal limbal epithelial cells are small and have a larger nucleus/cytoplasm ratio [12]. Cultured human corneal epithelial cells display a heterogeneity of cell sizes. The smaller cells are preferentially immunostained with antibodies for stem cell-associated markers, whereas the larger cells are strongly stained with antibodies against differentiation markers [13]. This study was conducted to evaluate the relationship between cell size and cell phenotype and growth potential in human corneal epithelial cells.
Materials and Methods
Materials and Reagents
Cell culture dishes, plates, centrifuge tubes, and other plastic ware were purchased from Becton, Dickinson and Company (Franklin Lakes, NJ, http://www.bd.com). Nunc Lab-Tek II eight-chamber slides were from Nalge Nunc International Corp (Naperville, IL, http://www.nalgenunc.com). Dulbecco modified Eagle's medium (DMEM), Ham F-12, HEPES, amphotericin B, gentamicin, and 0.25% trypsin/0.03% EDTA solution were from Invitrogen-GIBCO BRL (Grand Island, NY, http://www.invitrogen.com). Fetal bovine serum (FBS) was from Hyclone (Logan, UT, http://www.hyclone.com). Mouse NIH 3T3 fibroblasts (ATCC CCL 92) were from American Type Culture Collection (Rockville, MD, http://www.atcc.org). Dispase II, 5-bromo-2-deoxyuridine (BrdU), and anti-BrdU monoclonal antibody (mAb) (clone BMC 9318) were from Roche Molecular Biochemicals (Indianapolis, http://www.roche-applied-science.com). mAb against ABCG2 (clone BXP-21) was from Calbiochem (San Diego, CA, http://www.emdbiosciences.com). p63 (clone 4A4) and involucrin (clone SY5) mAbs came from Lab Vision (Fremont, CA, http://www.labvision.com), and K3 mAb (clone AE5) was from ICN Pharmaceuticals (Costa Mesa, CA, http://www.mpbio.com). Fluorescein Alexa Fluor 488-conjugated goat anti-mouse secondary antibody was from Molecular Probes (Eugene, OR, http://probes.invitrogen.com). Ready-To-Go You-Prime First-Strand Beads were purchased from Amersham Pharmacia Biotech (Piscataway, NJ, http://www.amersham.com). GeneAmp RNA-PCR and Taqman Universal PCR Master Mix AmpErase UNG kits were from Applied Biosystems (Foster City, CA, http://www.appliedbiosystems.com). Mitomycin C, bovine insulin, human transferrin, sodium selenite, hydrocortisone, human epidermal growth factor (EGF), cholera toxin A subunit, propidium iodide (PI), DNA size marker, and all other reagents came from Sigma-Aldrich (St. Louis, http://www.sigmaaldrich.com).
Corneal Limbal Tissues and Human Corneal Epithelial Cultures
Fresh human corneoscleral tissues (less than 72 hours postmortem) that were not suitable for clinical use, from donors aged 29 to 65 years, were obtained from the Lions Eye Bank of Texas (Houston, http://www.lebct.org). Corneal epithelial cultures were established from a single-cell suspension of limbal epithelia by a previously described method [13, 14] with modification. In brief, corneoscleral tissues were rinsed with Hanks' balanced solution containing 50 μg/ml gentamicin and 1.25 μg/ml amphotericin B. After careful removal of the central cornea, excess sclera, iris, corneal endothelium, conjunctiva, and Tenon's capsule, the remaining limbal rims were incubated with dispase II (5 mg/ml) at 37°C for 1 hour. The limbal epithelial sheets were collected and treated with 0.25% trypsin/0.03% EDTA at 37°C for 5 to 10 minutes to isolate single cells. Mouse NIH 3T3 fibroblasts, grown in DMEM containing 10% FBS at confluence, were treated with mitomycin C (5 μg/ml) for 2 hours and then trypsinized to prepare a feeder layer at 3 × 104 cells/cm2. The limbal epithelial cells were seeded at 1 × 103 cells/cm2 in supplemented hormonal epidermal medium (SHEM), which was a 1:1 mixture of DMEM and Ham's F12 medium containing 5 ng/ml EGF, 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml sodium selenite, 0.5 μg/ml hydrocortisone, 30 ng/ml cholera toxin A, 0.5% DMSO, 50 μg/ml gentamicin, 1.25 μg/ml amphotericin B, and 5% FBS, on 3T3 cells at 37°C under 95% humidity and 5% CO2. The cells were allowed to grow for 17 to 20 days to confluence before size-sorting for immunostaining, gene expression, and growth assays, whereas the cells cultured for 24 to 26 days were sorted for the BrdU retention assay.
Cell Size Sorting by Flow Cytometry
Cultured corneal epithelial cells were sorted based on the size, comparing the forward scatter (FS) profile to beads of known size, using a triple-laser Beckman-Coulter Altra (Hialeah, FL, http://www.beckmancoulter.com). Briefly, the confluent cultures were treated with 0.25% trypsin/0.03% EDTA at 37°C for 5 to 10 minutes to isolate single cells. The cell suspension was centrifuged and resuspended in cold SHEM with HEPES containing PI (2 μg/ml) on ice until flow cytometry was performed.
Flow cytometry sorting was performed at the Baylor College of Medicine Core Facility. A 488-nm argon laser was used as the probing beam. Sorting was based on FS, which is an indirect measurement of size, versus light scatter (LS), which is a measurement of the cell's granularity. Four gates (A, B, C, and D) were chosen comparing the FS/LS diagrams to diagrams of beads of known size. Only the cells within gate limits were sorted. The percentage of cells within each gate was calculated in relation to all cells, including excluded cells. Debris were excluded by gates in the right angle scatter versus FS diagrams and by PI fluorescence, which was read using a 610/20 bandpass filter. At least 100,000 events were collected before analysis. All flow cytometric data were analyzed with Expo 32 software (Beckman-Coulter). Four fractions of cells ranging in size from 10 to 16, 17 to 23, 24 to 30, and ≥31 μm in diameter (A, B, C, and D, respectively) were sorted and collected. The size-sorting experiments were repeated a total of eight times for BrdU retention (n = 3) and phenotype and growth assays (n = 5).
BrdU Label-Retention Assay
To evaluate the correlation between size and cell cycle, BrdU label retention assay was performed using a previously reported method [13] with modification. In brief, single-cell cultures in 35-mm dishes, at days 4 and 5, were incubated with fresh SHEM medium containing 10 μM BrdU. After labeling with BrdU for 72 hours continuously, the cultures were switched to BrdU-free medium, chased for 18 days, and then used for sorting experiments as described above. All selected populations were plated in duplicate at 5 × 104 cells per chamber on eight-chamber slides overnight, fixed in cold methanol at 4°C for 10 minutes. After rehydration in phosphate-buffered saline (PBS) for 5 minutes, samples were incubated with 2N HCl at 37°C for 60 minutes to denature DNA and neutralized in boric acid (pH 8.5) for 20 minutes. Incorporated BrdU was detected by immunofluorescent staining with a mAb as described below. The percentages of positively stained cells were assessed by point counting in digital images. A total of 500 to 951 nuclei were counted in six to eight representative fields. This number (500 counted nuclei) was considered a minimum requirement to obtain representative data [15]. The BrdU labeling index was expressed as the number of positively labeled nuclei/the total number of nuclei × 100%.
Immunofluorescent Staining
Immunofluorescent staining was performed as previously described [12, 16]. In brief, the selected populations of corneal epithelial cells sorted by size were seeded at 5 × 104 cells per chamber in eight-chamber slides overnight. They were fixed with cold methanol (for p63, K3, involucrin, and BrdU staining) or freshly prepared 2% paraformaldehyde (for ABCG2 staining) at 4°C for 10 minutes. After blocking with 5% normal goat serum in PBS for 60 minutes, primary mAbs against nuclear p63 (1:1,000, 1 μg/ml), ABCG2 (1:50, 5 μg/ml), involucrin (1:100, 2 μg/ml), K3 (1:100, 10 μg/ml), or BrdU (1:40, 25 μg/ml) were applied and incubated for 1 hour at room temperature. A secondary antibody, Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G (1:300), was then applied and incubated in a dark chamber for 1 hour, followed by counterstaining with Hoechst 33342 (1 μg/ml in PBS) or PI (2 μg/ml in PBS) DNA-binding dye for 5 minutes. After washing with PBS, Antifade Gel/Mount (Fisher, Atlanta, http://www.fisherscientific.com) and a cover slip were applied. Sections were examined and photographed with an epifluorescent microscope (Eclipse 400, Nikon, Tokyo, http://www.nikon.co.jp) with a digital camera (model DMX 1200, Nikon).
Semiquantitative Reverse Transcription–Polymerase Chain Reaction and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from the selected populations of corneal epithelial cells immediately after sorting by cell size, using acid guanidium thiocyanate-phenol-chloroform extraction [17]. The RNA was quantified by its absorption at 260 nm and stored at −80°C before use. Using a housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as an internal control, the mRNA expression of different molecular markers was analyzed by semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) as previously described [17, 18]. Briefly, first-strand cDNAs were synthesized from 0.5 μg of total RNA with murine leukemia virus (MuLV) RT. PCR amplification was performed with specific primer pairs designed from published human gene sequences for different markers (Table 1) using a GeneAmp PCR System 9700 (Applied Biosystems). Semiquantitative RT-PCR was established by terminating reactions at intervals of 20, 24, 28, 32, 36, and 40 cycles for each primer pair to ensure that the PCR products formed were within the linear portion of the amplification curve. The fidelity of the RT-PCR products was verified by comparing their size to the expected cDNA bands and by sequencing the PCR products.
Table 1.
Human primer sequences used for semiquantitative reverse transcription–polymerase chain reaction (PCR)
| Gene | Accession no. | Sense primer | Antisense primer | PCR product |
|---|---|---|---|---|
| ΔNp63 | NM_003722 | CAGACTCAATTTAGTGAG | AGCTCATGGTTGGGGCAC | 440 bp |
| ABCG2 | AY017168 | ACCATTGCATCTTGGCTGTC | CGATGCCCTGCTTTACCAAA | 187 bp |
| Involucrin | NM_005547 | GGACTGCCTGAGCAAGAATGTG | TAAGCTGCTGCTCTGGGTTT | 121 bp |
| K12 | D78367 | ACATGAAGAAGAACCACGAGGATG | TCTGCTCAGCGATGGTTTCA | 150 bp |
| K3 | NM_057808 | GGCAGAGATCGAGGGTGTC | GTCATCCTTCGCCTGCTGTAG | 145 bp |
| GAPDH | M33197 | GCCAAGGTCATCCATGACAAC | GTCCACCACCCTGTTGCTGTA | 498 bp |
Relatively quantitative real-time PCR was performed as a previously described method [19] with modification. In brief, the first-strand cDNA was synthesized from 1 μg of total RNA with random hexamer and M-MuLV RT using Ready-To-Go You-Prime First-Strand Beads. The quantitative real-time PCR was performed in a Smart Cycler (Cepheid, Sunnyvale, CA, http://www.cepheid.com) with 25 μl reaction volume containing cDNA, TaqMan primers, and probes for ABCG2, ΔNp63, K3, K12, involucrin, or GAPDH and Universal PCR Master Mix (TaqMan Gene Expression Assays, Applied Biosystems). Assays were performed in duplicate for each of three repeated experiments. A nontemplate control was included in all of the experiments to evaluate DNA contamination of the reagent used. The GAPDH gene was used as an endogenous reference for each reaction to correct differences in the amount of total RNA and cDNA added. The results of relatively quantitative real-time PCR were analyzed by the comparative threshold cycle (CT) method using population A as a reference.
Colony-Forming Efficiency
To evaluate proliferative capacity of the size-selected cell populations, the colony-forming efficiency (CFE) was assessed using a previous method [13, 14, 20]. Each selected population was seeded in triplicate at 1 × 103 cells/cm2 into six-well culture plates (each well received approximately 1 × 104 cells) containing a mitomycin C-treated 3T3 fibroblast feeder layer. A colony was scored when it had at least eight cells. Colonies were counted at days 4, 6, and 8, and the number of colonies per well was divided by 10,000 (number of epithelial cells seeded in a well) and then multiplied by 100 to calculate the CFE percentage. The growth capacity was evaluated on day 14 when cultured cells were stained with 1% rhodamine.
Statistical Analysis
The Student's t-test or analysis of variance (ANOVA) with Tukey's post-hoc testing was used for statistical comparisons. p ≤ .05 was considered statistically significant. All of these tests were performed using the GraphPad Prism 3.0 software (GraphPad Prism, Inc., San Diego, CA, http://www.graphpad.com).
Results
The primary corneal epithelial cultures showed a mosaic of stratified epithelial cells, with heterogeneous morphologies and different sizes, including very small cells and large cells (Fig. 1A). Measured by flow cytometry based on FS/LS, comparing the FS/LS diagrams to those obtained with beads with known size, the cell size of most primary corneal epithelial cultures confluent in 3 weeks ranged from 10 to 36 μm in diameter. Larger cells (>36 μm) were observed in the cultures that had grown longer than 3 weeks. Four populations (A, B, C, and D) were sorted by FS profile, with cell sizes ranging from 10 to 16 μm, 17 to 23 μm, 24 to 30 μm, and ≥31 μm, respectively (Fig. 1B). Summarized from eight sorting experiments, population A with the smallest cell size accounted for 11.0% ± 4.5% (mean ± standard deviation) of the entire population, and the other three populations, B, C, and D, with increasing cell size, accounted for 23.4% ± 10.6%, 19.6% ± 7.9%, and 16.8% ± 6.8%, respectively. The total sorted cells including the four populations accounted for 70.9% ± 11.1% of the entire population. We were unable to sort the pure viable cells smaller than 10 μm in diameter because this population was mixed with dead cells and cell debris, which were identified by PI counterstaining in our preliminary flow cytometry analysis.
Figure 1.
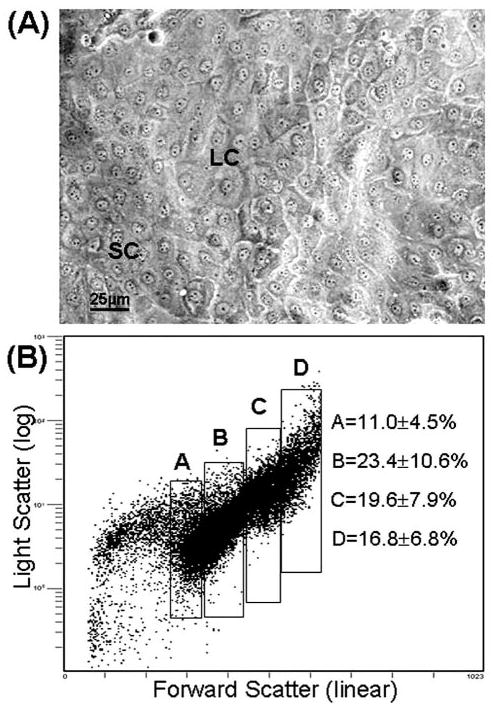
Human corneal epithelial cells in confluent culture. (A): Phase microscopy image. (B): Cell size plot by flow cytometry analysis showing representative fields of the four populations selected by cell size sorting. (A): 10 to 16 μm; (B): 17 to 23 μm; (C): 24 to 30 μm; (D): ≥ 31μm. Abbreviations: LC, large cells; SC, small cells.
Cell Size Correlates with Number of BrdU Label-Retaining Cells
The label-retaining cells (LRCs) are slow cycling cells. This has been widely accepted as a characteristic of epithelial stem cells. In the present study, population A with smallest cell size contained the highest number of LRCs (11.6% ± 1.5%, n = 3), significantly higher than the B population (7.48% ± 1.12%, p < .05, n = 3). The larger-sized populations (C and D) contained no LRCs (Fig. 2).
Figure 2.
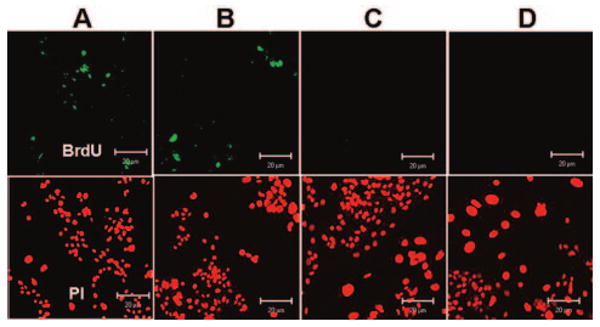
Representative immunofluorescent staining for 5-bromo-2-deoxyuridine (BrdU) label-retaining cells (green) with propidium-iodide (PI) counterstaining (red) in the size-selected populations (A-D) of primary cultured human corneal epithelial cells sorted from the smallest (A) to the largest (D) cells after the cultures were labeled with 10 μM BrdU for 72 hours and chased for 18 days. Scale bar = 20 μm.
Cell Size Correlates Inversely with Percentage of p63- and ABCG2-Positive Cells and Involucrin- and K3-Negative Cells
To evaluate the phenotype of the size-selected cell fractions, each cell population obtained from cell sorting from primary cultures was seeded into wells of eight-chamber culture slides for immunofluorescent staining with antibodies against stem cell–associated markers, nuclear transcription factor p63 [21], and membrane transporter protein ABCG2 [16] and the differentiation markers involucrin and K3. As shown in Figure 3, with Hoechst 33342 counterstaining, population A with the smallest cells showed the highest percentage of p63-positive cells (37.26% ± 5.22%, n = 3), significantly higher than the other populations (B, 12.46% ± 0.71%, p < .01; C, 7.92% ± 4.4%, p < .05; D, 2.54% ± 0.79%, p < .001; all n = 3). The percentage of ABCG2-positive cells was also significantly higher in population A (38.12% ± 1.63%, n = 3), with decreasing positive staining in the other populations containing larger cells (B, 21.25% ± 1.25%, p < .05; C, 5.92% ± 0.98%, p < .01; D, 3.19% ± 0.31%, p < .01; all n = 3).
Figure 3.
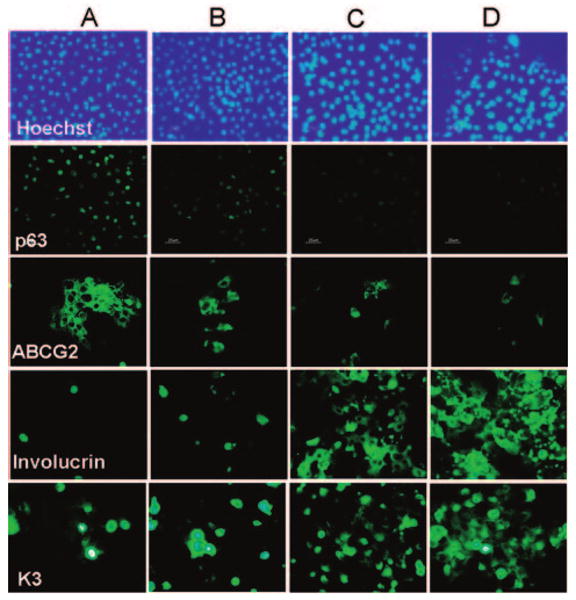
Representative immunofluorescent staining profiles for p63, ABCG2, involucrin, and K3 with Hoechst 33342 counterstaining in the size-selected populations (A-D) of primary cultured human corneal epithelial cells sorted from the smallest (A) to the largest (D) cells. Scale bar = 25 μm.
In contrast, the immunostaining for involucrin, a differentiation marker, was very low (1.86% ± 0.2% positive cells; n = 3) in population A with the smallest cell size, significantly lower than 8.78% ± 0.31% positive cells (p < .01; n = 3) in population B, 37.41% ± 4.59% (p < .01; n = 3) in C, and 72.84% ± 3.83% (p < .01; n = 3) in D. The expression of corneal differentiation marker K3 was observed in a similar manner, with the smallest cells in population A showing the lowest number of K3-positive cells (5.21% ± 0.50%; n = 3) and increasing in frequency in the cell populations with the larger size, 8.3% ± 0.37% in population B (p < .05; n = 3), 35.24% ± 2.74% in C (p < .01; n = 3), and 46.07% ± 3.93% in D (p < .01; n = 3).
Cell Size Correlates Inversely with Expression of ΔNp63 and ABCG2 mRNA and Positively with Levels of Involucrin, K12, and K3 mRNA
Semiquantitative RT-PCR disclosed a differential expression pattern of stem cell and differentiation-associated markers in these size-selected populations of corneal epithelial cells (Fig. 4A). ABCG2 mRNA was expressed at the highest level by the smallest cells in population A, dropping sharply in populations B and C with the larger cell size and to undetectable levels in population D with the largest cell size. A similar expression pattern of nuclear p63 was observed in these populations. Population A with the smallest cells expressed the highest levels of ΔNp63 mRNA (a truncated dominant-negative isoform of p63), whereas the B and C cell populations expressed much lower levels of ΔNp63 mRNA, and ΔNp63 expression was barely detectable in population D that contained the largest cells. In contrast, the mRNAs encoding the differentiation markers involucrin, K12, and K3 were detected at very low levels in population A. They were expressed at higher levels by the intermediate-sized population (B and C) and at the highest levels by the largest cells in D. No difference in the mRNA levels of GAPDH, a housekeeping gene used as an internal control, was observed among the four fractions.
Figure 4.
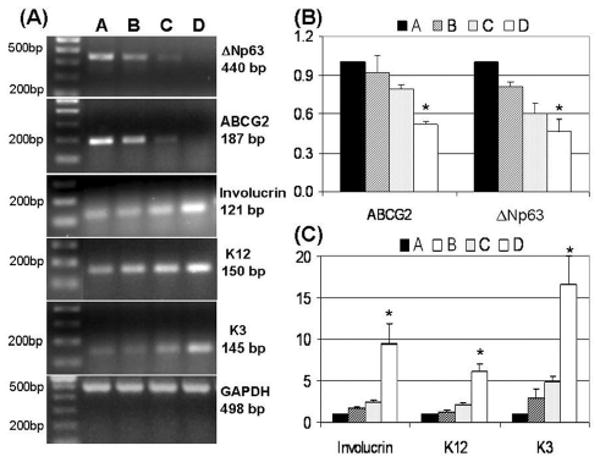
(A): Representative semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) profiles showing mRNA expression of ΔNp63, ABCG2, involucrin, K12, and K3, with GAPDH as an internal control in the size-selected populations (A-D) of primary cultured human corneal epithelial cells sorted from the smallest (A) to the largest (D) cells. (B, C): Real-time PCR analysis showing the relatively quantitative expression of ABCG2, ΔNp63, K3, K12, and Involucrin mRNA in the size-selected populations (A-D) of primary cultured human corneal epithelial cells sorted from the smallest (A) to the largest (D) cells. The comparative CT method was used to determine the fold of the targeted gene expression normalized by internal control GAPDH. Data are the mean ± standard deviation of results with mRNA levels in population A as a reference (equal to 1) in three experiments. *p < .05, n = 3, analyzed by analysis of variance with Tukey's post-hoc testing.
The levels of ABCG2, ΔNp63, K3, K12, and involucrin mRNA were quantified by real-time PCR with GAPDH as an internal control. The results of real-time PCR (Figs. 4B, 4C) confirmed the findings of conventional RT-PCR. The expression of stem cell–associated markers, ABCG2 and ΔNp63 mRNA (Fig. 4B), were highest in population A that contained the smallest sized cells (used as reference, therefore equal to 1) with decreasing levels in populations B (0.92 ± 0.14 and 0.81 ± 0.03, respectively; p > .05, n = 3) and C (0.79 ± 0.03 and 0.60 ± 0.09, respectively; p > .05, n = 3). The lowest mRNA levels were found in population D that contained the largest sized cells (0.53 ± 0.02 and 0.46 ± 0.10, respectively; p < .05, n = 3). On the other hand, mRNA for the differentiation markers involucrin, K12, and K3 (Fig. 4C) were detected at the lowest levels in population A (as reference, equal to 1), with increasing expression in the intermediate-sized population B (1.73 ± 0.08, 1.30 ± 0.16, 2.85 ± 1.25, respectively; all p > .05, n = 3) and C (2.41 ± 0.24, 2.14 ± 0.17, 4.92 ± 0.64, respectively; all p > .05, n = 3) and at the highest levels by the largest cells in D (9.44 ± 2.48, 6.20 ± 0.83, 16.59 ± 3.46; all p < .05, n = 3).
Cell Size Correlates with the CFE and Growth Capacity
To evaluate their growth capacity, the cells of each size-selected population were seeded in triplicate at a density of 1 × 103 cells/cm2 into six-well culture plates containing a 3T3 fibroblast feeder layer. A summary of the CFE at days 4, 6, and 8 from three separate size-sorting experiments is shown in Figure 5A. At day 4, the CFE was highest (2.67% ± 0.96%) in population A with the smallest cell size, significantly higher than in populations B (1.2% ± 0.22%; n = 3, p < .05) and C (0.43% ± 0.06%; n = 3, p < .01). The lowest CFE was seen in population D (0.19% ± 0.09%; n = 3, p < .01; ANOVA difference between groups, p < .005). At day 6, the CFE was highest in population A (5.43% ± 0.98%), significantly higher than populations B and C (2.18% ± 0.27% and 0.93% ± 0.26%, respectively; both n = 3, p < .001). The lowest CFE was seen in population D (0.40% ± 0.14%; n = 3, p < .001). At day 8, population A showed the highest CFE (7.26% ± 1.27%), and this was significantly higher than populations B, C, and D (2.57% ± 0.22%, 1.07% ± 0.27%, and 0.52% ± 0.14%, respectively; all p < .001, n = 3). Furthermore, the smallest cells of population A grew to confluence in 12 to 14 days, showing a small, mosaic pattern of the cells, whereas the B group grew to confluence at 18 to 24 days and showed larger differentiated cells. The cells in C and D generated colonies that failed to reach confluence (Fig. 5B).
Figure 5.
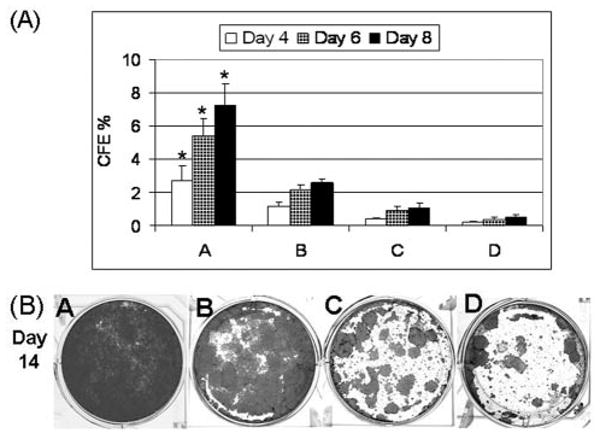
(A): Colony-forming efficiency (CFE) on a 3T3 fibroblast feeder layer at days 4 through 8 generated by the size-selected populations (A-D) of primary cultured human corneal epithelial cells sorted from the smallest (A) to the largest (D) cells. The percentages of CFE were shown as mean ± standard deviation averaged from three separate experiments (p < .005 at day 4 and p < .0001 at days 6 and 8 by analysis of variance [ANOVA]). Asterisks are shown where the differences were significant among population A and the other selected populations by ANOVA. (B): Representative cultures stained with 1% rhodamine showing growth capacity of the four size-selected populations (A-D) of human corneal epithelial cells at day 14.
Discussion
Previous studies have demonstrated that cell size correlates with some features of cell differentiation and proliferation in different cell types, including human keratinocytes and fibroblasts. The differentiation marker involucrin has been reported to correlate with increasing cell size and terminal differentiation in human epidermal cultures [7]. Human epidermal keratinocytes with the smallest size, sorted by centrifugal elutriation, expressed the highest levels of basal cell markers (p63 and basonuclin) and possessed the greatest clonogenicity in culture [2, 4, 6]. The proliferative potential of human fibroblasts and keratinocytes was shown to be inversely dependent on cell size [2, 3]. Based on these important findings, the present study was conducted to evaluate the correlations between cell size, cell phenotype, and proliferative capacity with the intention of linking cell size of human corneal epithelial cells to some properties of adult stem cells.
Adult stem cells have been observed to have several characteristic features in morphology, phenotype, and growth potential. These include slow cycling or long cell cycle time during homeostasis in vivo; small size with poor differentiation and primitive cytoplasm; high proliferative potential after wounding or placement in culture; and the ability for self-renewal and functional tissue regeneration (see review articles [22–26]). Human corneal epithelial stem cells are believed to be located at the limbus. According to these criteria, we have determined that basal epithelial cells at the limbus are small primitive cells expressing three patterns of molecular markers: (a) certain basal cells are exclusively positive for p63, ABCG2, and integrin α9; (b) most basal cells show relatively higher expression of integrin β1, EGFR, K19, and α-enolase than suprabasal cells, and (c) all lack of expression of nestin, E-cadherin, connexin 43, involucrin, K3, and K12 [12]. In human corneal epithelial cultures, we have observed that a greater number of the small cells express stem cell–associated markers such as p63, EGFR, K19, and integrin β1, whereas larger cells stained positively for differentiation markers such as K3, involucrin, and connexin 43. Slow-cycling BrdU label-retaining cells that are characteristic of stem cells were identified in 2.3%–3.7% of cells in these cultures [13].
Consistent with previous reports, the present study provides further evidence that size is an important marker related to cell behavior and function. Among the populations selected from corneal epithelial cultures by size, the smallest corneal epithelial cells (10 to 16 μm) in population A, which accounted for approximately 11% of the entire population, contained the highest number of BrdU LRCs (11.6% ± 1.5%) and displayed the highest percentage of cells positive for p63 (37.26% ± 5.22%) and ABCG2 (38.12% ± 1.63%) protein and the lowest number of cells stained with involucrin (1.86% ± 0.2%) and K3 (5.21% ± 0.50%). They also expressed the highest levels of ΔNp63 and ABCG2 mRNAs and the lowest levels of involucrin, K12, and K3 mRNAs and possessed the greatest CFE and growth capacity on a 3T3 feeder layer. It has been identified that adult stem cells are slow cycling or have a long cell cycle time during homeostasis in vivo ([3H]-thymidine or BrdU label-retaining cells) [24, 27, 28], and they possess great proliferative potential after wounding or placement in culture in vitro [29]. These properties may be due to the ability of stem cells to divide asymmetrically to produce one daughter cell and one progenitor cell. The daughter cell retains the slow-cycling stem cell property. Once the slow cycling cells have been labeled, they retain this label for a much longer period and are identified as label-retaining cells. The progenitor cell is a mitotically active cell and proliferates continually. Thus, stem cells are able to self-renew and regenerate tissue.
On the other hand, the largest cells in population D, which comprised approximately 17% of the entire population, contained no BrdU LRCs and displayed the highest percentage of cells positive for involucrin (72.84% ± 3.83%) and K3 (46.07% ± 3.93%) and the lowest number of cells positive for p63 (2.54% ± 0.79%) and ABCG2 (3.19% ± 0.31%). They also expressed the highest levels of involucrin, K12, and K3 mRNAs and lacked proliferative potential. The intermediate-sized cells in populations B and C showed a cell phenotype and growth capacity between the patterns of A and D.
These findings demonstrate that cell size correlates with cell phenotype, state of differentiation, and proliferative capacity. Small size seems to represent one of the important features of adult stem cells. The smallest cells in population A show properties that satisfy some of the criteria of adult stem cells (Table 2), suggesting that they are enriched with putative stem cells. The largest cells in population D showed the most properties of differentiation, indicating that they are terminally differentiated cells. The properties of cells with intermediate sizes in populations B and C indicate that they contain some transient amplifying cells that are capable of limited proliferation. Further studies are necessary to determine how the size regulates or determines the cell phenotypes and growth characteristics and to determine the environmental factors that maintain the small cell size of the stem cells in the basal limbal region of the cornea.
Table 2.
Properties of cell size-selected populations (A-D) of human corneal epithelial cells
| A | B | C | D | |
|---|---|---|---|---|
| Cell size (μm) | 10–16 | 17–23 | 24–30 | ≥ 31 |
| Population (%) | 11.0 ± 4.5 | 23.4 ± 10.6 | 19.6 ± 7.9 | 16.8 ± 6.8 |
| BrdU-LRC (%) | 11.6 ± 1.5 | 7.48 ± 1.12 | 0 | 0 |
| Positive (+) cells (%) | ||||
| p63+ | 37.26 ± 5.22 | 12.46 ± 0.71 | 7.92 ± 4.4 | 2.54 ± 0.79 |
| ABCG2+ | 38.12 ± 1.63 | 21.25 ± 1.25 | 5.92 ± 0.98 | 3.19 ± 0.31 |
| Involucrin+ | 1.86 ± 0.2 | 8.78 ± 0.31 | 37.41 ± 4.59 | 72.84 ± 3.83 |
| K3+ | 5.21 ± 0.50 | 8.3 ± 0.37 | 35.24 ± 2.74 | 46.07 ± 3.93 |
| Fold change of mRNA | ||||
| ΔNp63 | 1 | 0.81 ± 0.03 | 0.60 ± 0.09 | 0.46 ± 0.10 |
| ABCG2 | 1 | 0.92 ± 0.14 | 0.79 ± 0.03 | 0.53 ± 0.02 |
| Involucrin | 1 | 1.73 ± 0.08 | 2.41 ± 0.24 | 9.44 ± 2.48 |
| K3 | 1 | 2.85 ± 1.25 | 4.92 ± 0.64 | 16.59 ± 3.46 |
| K12 | 1 | 1.30 ± 0.16 | 2.14 ±0.17 | 6.2 ± 0.83 |
| Growth capacity | ||||
| CFE on day 6 (%) | 5.43 ± 0.98 | 2.18 ± 0.27 | 0.93 ± 0.26 | 0.40 ± 0.14 |
| Confluency (days) | 12–14 | 18 | no | no |
Data are shown as mean ± standard deviation.
Abbreviations: BrdU, 5-bromo-2-deoxyuridine; CFE, colony-forming efficiency; LRC, label-retaining cell.
Acknowledgments
We thank the Lions Eye Bank of Texas for their great support in providing human corneoscleral tissues and Jeffrey M. Scott for his technical help in performing cell sorting. This study was supported by NIH grants EY014553 (to D.Q.L.) and EY11915 (to S.C.P.), the National Eye Institute, Bethesda, MD, a grant from Lions Eye Bank of Texas, a post-doctoral research fellowship from Fight for Sight (to C.S.P.), an unrestricted grant from Research to Prevent Blindness, the Oshman Foundation, and the William Stamps Farish Fund. This article was presented in part as an abstract at the annual meeting of the Association for Research in Vision and Ophthalmology, April 25–29, 2004, Fort Lauderdale, Florida.
Footnotes
Disclosures: The authors indicate no potential conflicts of interest.
References
- 1.Gao FB, Raff M. Cell size control and a cell-intrinsic maturation program in proliferating oligodendrocyte precursor cells. J Cell Biol. 1997;138:1367–1377. doi: 10.1083/jcb.138.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrandon Y, Green H. Cell size as a determinant of the clone-forming ability of human keratinocytes. Proc Natl Acad Sci U S A. 1985;82:5390–5394. doi: 10.1073/pnas.82.16.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angello JC, Pendergrass WR, Norwood TH, et al. Proliferative potential of human fibroblasts: An inverse dependence on cell size. J Cell Physiol. 1987;132:125–130. doi: 10.1002/jcp.1041320117. [DOI] [PubMed] [Google Scholar]
- 4.Parsa R, Yang A, McKeon F, et al. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J Invest Dermatol. 1999;113:1099–1105. doi: 10.1046/j.1523-1747.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 5.Dazard JE, Piette J, Basset-Seguin N, et al. Switch from p53 to MDM2 as differentiating human keratinocytes lose their proliferative potential and increase in cellular size. Oncogene. 2000;19:3693–3705. doi: 10.1038/sj.onc.1203695. [DOI] [PubMed] [Google Scholar]
- 6.Tseng H, Green H. Association of basonuclin with ability of keratinocytes to multiply and with absence of terminal differentiation. J Cell Biol. 1994;126:495–506. doi: 10.1083/jcb.126.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watt FM, Green H. Involucrin synthesis is correlated with cell size in human epidermal cultures. J Cell Biol. 1981;90:738–742. doi: 10.1083/jcb.90.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu Z, MacLellan WR, Weiss JN. Dynamics of the cell cycle: Checkpoints, sizers, and timers. Biophys J. 2003;85:3600–3611. doi: 10.1016/S0006-3495(03)74778-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolznig H, Grebien F, Sauer T, et al. Evidence for a size-sensing mechanism in animal cells. Nat Cell Biol. 2004;6:899–905. doi: 10.1038/ncb1166. [DOI] [PubMed] [Google Scholar]
- 10.Vacanti MP, Roy A, Cortiella J, et al. Identification and initial characterization of spore-like cells in adult mammals. J Cell Biochem. 2001;80:455–460. [PubMed] [Google Scholar]
- 11.Romano AC, Espana EM, Yoo SH, et al. Different cell sizes in human limbal and central corneal basal epithelia measured by confocal microscopy and flow cytometry. Invest Ophthalmol Vis Sci. 2003;44:5125–5129. doi: 10.1167/iovs.03-0628. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, de Paiva CS, Luo L, et al. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004;22:355–366. doi: 10.1634/stemcells.22-3-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HS, Jun SX, de Paiva CS, et al. Phenotypic characterization of human corneal epithelial cells expanded ex vivo from limbal explant and single cell cultures. Exp Eye Res. 2004;79:41–49. doi: 10.1016/j.exer.2004.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng SC, Kruse FE, Merritt J, et al. Comparison between serum-free and fibroblast-cocultured single-cell clonal culture systems: Evidence showing that epithelial anti-apoptotic activity is present in 3T3 fibroblast-conditioned media. Curr Eye Res. 1996;15:973–984. doi: 10.3109/02713689609017643. [DOI] [PubMed] [Google Scholar]
- 15.Goodson WH, III, Moore DH, Ljung BM, et al. The functional relationship between in vivo bromodeoxyuridine labeling index and Ki-67 proliferation index in human breast cancer. Breast Cancer Res Treat. 1998;49:155–164. doi: 10.1023/a:1005926228093. [DOI] [PubMed] [Google Scholar]
- 16.de Paiva CS, Chen Z, Corrales RM, et al. ABCG2 transporter identifies a population of clonogenic human limbal epithelial cells. Stem Cells. 2005;23:63–73. doi: 10.1634/stemcells.2004-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li DQ, Tseng SC. Three patterns of cytokine expression potentially involved in epithelial-fibroblast interactions of human ocular surface. J Cell Physiol. 1995;163:61–79. doi: 10.1002/jcp.1041630108. [DOI] [PubMed] [Google Scholar]
- 18.Li DQ, Lokeshwar BL, Solomon A, et al. Regulation of MMP-9 production by human corneal epithelial cells. Exp Eye Res. 2001;73:449–459. doi: 10.1006/exer.2001.1054. [DOI] [PubMed] [Google Scholar]
- 19.Luo L, Li DQ, Doshi A, et al. Experimental dry eye stimulates production of inflammatory cytokines and MMP-9 and activates MAPK signaling pathways on the ocular surface. Invest Ophthalmol Vis Sci. 2004;45:4293–4301. doi: 10.1167/iovs.03-1145. [DOI] [PubMed] [Google Scholar]
- 20.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: The formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 21.Pellegrini G, Dellambra E, Golisano O, et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci U S A. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blau HM, Brazelton TR, Weimann JM. The evolving concept of a stem cell: Entity or function? Cell. 2001;105:829–841. doi: 10.1016/s0092-8674(01)00409-3. [DOI] [PubMed] [Google Scholar]
- 23.Watt FM, Hogan BL. Out of Eden: Stem cells and their niches. Science. 2000;287:1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 24.Lavker RM, Sun TT. Epidermal stem cells: Properties, markers, and location. Proc Natl Acad Sci U S A. 2000;97:13473–13475. doi: 10.1073/pnas.250380097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watt FM. Epidermal stem cells as targets for gene transfer. Hum Gene Ther. 2000;11:2261–2266. doi: 10.1089/104303400750035799. [DOI] [PubMed] [Google Scholar]
- 26.Cotsarelis G, Kaur P, Dhouailly D, et al. Epithelial stem cells in the skin: Definition, markers, localization and functions. Exp Dermatol. 1999;8:80–88. doi: 10.1111/j.1600-0625.1999.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 27.Cotsarelis G, Cheng SZ, Dong G, et al. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell. 1989;157:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 28.Tseng SC. Concept and application of limbal stem cells. Eye. 1989;3:141–157. doi: 10.1038/eye.1989.22. [DOI] [PubMed] [Google Scholar]
- 29.Barrandon Y, Green H. Three clonal types of keratinocytes with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]


