Abstract
A previously described Taenia saginata HDP2 DNA sequence, a 4-kb polymorphic fragment, was previously used as the basis for developing PCR diagnostic protocols for the species-specific discrimination of T. saginata from T. solium and for the differentiation of T. saginata from T. asiatica. The latter was shown subsequently to lack the required specificity, so we undertook genetic studies of the HDP2 sequence from T. saginata and T. asiatica to determine why, and to develop a novel HDP2-PCR protocol for the simultaneous unambiguous identification of human taeniids. Sequencing and further analysis of the HDP2 DNA fragments of 19 Asiatic isolates of T. saginata and T. asiatica indicated that the HDP2 sequences of both species exhibited clear genomic variability, due to polymorphic variable fragments, that could correspond to the non-transcribed region of ribosomal DNA. This newly observed polymorphism allowed us to develop a novel, reproducible and reliable HDP2-PCR protocol which permitted the simultaneous discrimination of all T. saginata and T. asiatica isolates examined. This species-specific identification was based on, and facilitated by, the clear size difference in amplicon profiles generated: fragments of 1300 bp, 600 bp and 300 bp were produced for T. asiatica, amplicons of 1300 bp and 300 bp being obtained for T. saginata. Control T. solium samples produced one amplicon of 600 bp with the HDP2-PCR protocol. The assay has the potential to prove useful as a diagnostic tool in areas such as South East Asia where T. saginata, T. asiatica and T. solium coexist.
Findings
Human taeniasis results from intestinal infection with the adult tapeworms Taenia saginata, Taenia asiatica or Taenia solium. Ingestion of eggs of T. saginata and T. solium/ T. asiatica causes cysticercosis in cattle and pigs, respectively. Importantly, T. solium eggs can also infect man, giving rise to human cysticercosis, with the frequent localization of the metacestodes in the brain, causing potentially fatal neurocysticercosis (NCC) [1-3]. Infection with T. solium is therefore a serious public health problem, notably in endemic areas (Latin America, Africa, Asia), but also in non-endemic areas due to imported cases [4]. Hence, and as an essential part of transmission control programs, there is an immediate and practical need for reliable and species-specific diagnosis of human taeniasis caused by T. saginata, T. asiatica and T. solium in order to identify tapeworm carriers, particularly those with T. solium, thus helping to avoid transmission of cysticercosis/NCC.
Until recently, species-specific diagnosis of taeniasis was unsatisfactory. Conventional clinical and morphological identification has low specificity and sensitivity [5]. Moreover, adult worms of T. saginata and T. asiatica are frequently confused due to their morphological similarities [6]. Copro-antigen diagnosis by ELISA, that detects parasite antigens in patient feces, is based on the use of polyclonal antibody reagents although it does not distinguish among the human intestinal taeniids [7,8]. More recently, serological diagnosis for T. solium taeniasis has been reported with recombinant ES antigens [9,10], but this assay not necessarily indicates active infection.
Various PCR approaches have been developed for species-specific identification of DNA from T. saginata, T. asiatica and T. solium [6,11-26]. Thus, the previously described T. saginata HDP2 DNA sequence [12,27], a 4-kb polymorphic fragment, was the basis for various PCR-based diagnostic protocols for the species-specific detection of T. saginata and T. solium taeniasis [12,17,25,26], as well as for the specific differentiation of T. saginata and T. asiatica samples [22]; however, the developed HDP2-PCR protocol was later shown to lack the required specificity to discriminate T. saginata from T. asiatica.
Although the genomic structure of the HDP2 fragment had been partially described for T. saginata and T. solium [12], its homologue in the taxonomically closely related T. asiatica had not [19,28-32]. Therefore, the aims of this study were: (i) to undertake genetic analysis of the HDP2 DNA sequence [12,27] from T. saginata and T. asiatica isolates, and (ii) to develop a novel HDP2-PCR protocol for the simultaneous species-specific identification of T. saginata and T. asiatica, using a total of 19 taeniid cestode proglottid isolates from taeniasis patients of Asian origin (Table 1). Morphological identification of the proglottid samples was based on the number of uterine branches, and confirmed by genetic characterization, according to published protocols [5,19]. Genomic DNA (gDNA) was extracted from the ethanol-preserved proglottids samples using the method described by Sambrook et al. [33]. Two purified gDNA samples from T. solium (Venezuelan and Mexican isolates) and one from T. saginata (Spanish isolate) were used as positive controls.
Table 1.
Geographical origin of the 6 Taenia saginata and 13 Taenia asiatica samples analyzeda.
| Sample origin | Sample code number and species* | |
|---|---|---|
| Taenia saginata | Taenia asiatica | |
| Indonesia | #12 | |
| Korea | #16, #19 | |
| Philippines | #7, #11 | |
| Taiwan | #4, #8, #10, #14 | #1, #3, #6, #9, #13, #15, #17, #18 |
| Thailand | #2 | |
| Asia (country unknown) | #5 | |
Previously described multiplex PCRs were used to amplify the gDNA obtained from the 19 Asian taeniid proglottids [6,22] and the positive controls. In addition, the gDNAs from the 19 Asian samples were examined by three new HDP2-based PCRs, using HDP2-derived primers (Fig. 1, table 2). HDP2 PCRs I and II were used to analyze the genomic characteristics of the HDP2 DNA sequence within the Asiatic taeniid isolates, while the novel HDP2 PCR III was developed to differentiate the Asian samples. Table 2 shows the PCR conditions and the sequence of primers used.
Figure 1.
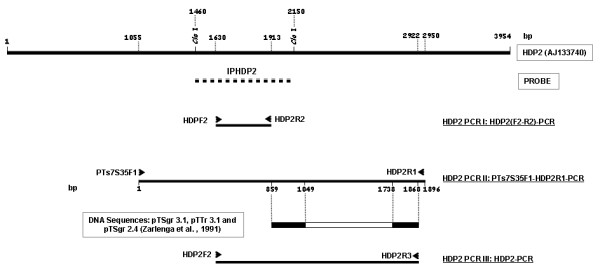
Strategy for amplification and verification of the HDP2 sequences. The location of probe IHDP2 used in Southern-blots is indicated by a discontinuous line below the T.saginata HDP2 sequence [GenBank: AJ133740]. The locations of the oligonucleotide primers within the T. saginata HDP2 DNA sequence [GenBank AJ133740] for the different molecular protocols used, and the directions of these are indicated by arrows: HDP2F2 and HDP2R2 (PCR I: HDP2 (F2-R2)-PCR); PTs7S35F1 and HDP2R1 (PCR II: PTs7S35F1-HDP2R1-PCR); HDP2F2 and HDP2R3 (PCR III: HDP2-PCR). The location of the sequences with similarity to the T. saginata and T. asiatica partial HPD2 DNA fragments (PTs7S35F1-HDP2R1 amplicon), pTSgr 3.1, pTSgr 2.4 and pTTr 3.1, are indicated by the black area of the bar. The restriction enzyme sites are indicated by the lines.
Table 2.
PCR names, primer sequences and amplification conditions used.
| PCR primer name | Primer sequence | Amplification conditions (Taq polymerase; BIOTOOLS , Spain) |
|---|---|---|
| HDP2 PCR I: HDP2F2 HDP2R2 |
5'-GCTGTACCAGCACCTAACCRTCC-3' 5'-GCACACCGCAGCCAATTGGCTG-3' |
94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, 72°C for 7 min (35 cycles) |
| HDP2 PCR II: PTs7S35F1 HDP2R1 |
5'-CAGTGGCATAGCAG AGGAGGAA-3' 5'-GCGGAAAGTGGATCCGACTTCGATGG-3' |
94°C for 30 sec, 56.5°C for 30 sec, 72°C for 2 min, 72°C for 7 min (35 cycles) |
| HDP2 PCR III: HDP2F2 HDP2R3 |
5'-GCTGTACCAGCACCTAACCRTCC-3' 5'-ATCCTGCTCCATGTCGCTTCTAGC-3' |
94°C for 30 sec, 60°C for 30 sec, 72°C for 2 min, 72°C for 7 min (35 cycles) |
To study the HDP2 DNA sequence organization in the 19 Asian T. saginata and T. asiatica isolates, 5 μg of each parasite gDNA was digested to completion with Cla I (Roche, Penzberg, Germany), as recommended by the manufacturer. Electrophoresis, Southern blotting, probe labeling, and hybridization procedures were carried out, with minor modifications, as previously described [12]. The probe used was one fragment derived from the HDP2 sequence, previously named IPHDP2 [12], which is a variable fragment from the mid region of HDP2 (Fig. 1).
In addition, the HDP2 DNA sequence of the Taenia isolates were PCR-amplified with the PTs7S35F1/ HDP2R1 primers set (Fig. 1, Table 2), using 10 ng of gDNA from each parasite sample. After electrophoresis, the amplicons were visualized and purified using a QIAquick Gel Extraction Kit (Qiagen,, Valencia, CA). All the DNA fragments were automatically sequenced by standard Sanger chemistry using a Model 377 ABI PRISM system. We used a primer walking sequencing strategy involving PTs7S35F1, HDP2F2, HDP2R2 forward primers and HDP2R2, HDP2R1 reverse primers (Fig. 1). The DNA sequences obtained and their alignments were analyzed by Seqman II (DNASTAR) and Clustal W2 [34] software packages. DNA sequence comparisons were carried out using the GenBank databases and BLASTn [35]. The nucleotide sequences obtained from amplification of the HDP2 gDNA fragments of the Taenia isolates have been deposited in the EMBL/GenBank databases under the following accession numbers: FM212953-71.
Our analysis showed that the Asian samples could clearly be differentiated using the multiplex PCR described by Yamasaki et al. [6], but not by the HDP2-based multiplex PCR (Fig. 2a/2b) reported by Gonzalez et al. [22], as samples #3, #12,#16,#17 and #19 were incorrectly identified as T. saginata. This finding prompted us to undertake further analysis of the genomic structure and sequence analysis of the HDP2 DNA fragments from the Asian taeniid isolates as now described.
Figure 2.
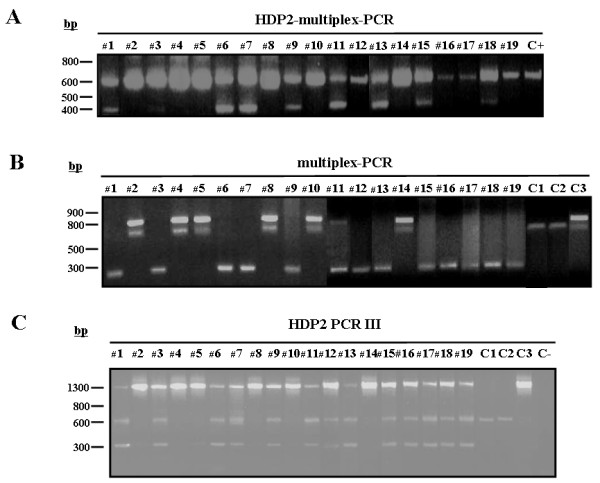
(A) Differential diagnosis of T. saginata and T. asiatica isolates from Asia using the HDP2-multiplex-PCR [22]. Lanes 1, 3, 6, 7, 9, 11, 12, 13, 15, 16, 17, 18, 19, T. asiatica; lanes 2, 4, 5, 8, 10, 14, T. saginata.; C+, T. saginata control sample (Spanish origin). Amplification products were fractionated by electrophoresis in a 1% (w/v) agarose gel and stained with ethidium bromide. The numbers on the left indicate the sizes (in bases pairs, bp) of molecular weight markers. (B) Differential diagnosis of T. saginata and T. asiatica isolates from Asia by multiplex-PCR [6]. Lanes 1, 3, 6, 7, 9, 11, 12, 13, 15, 16, 17, 18, 19, T. asiatica; lanes 2, 4, 5, 8, 10, 14, T. saginata. C1 control sample (T. solium Venezuelan origin); C2 control sample (T. solium Mexican origin); C3 control sample (T. saginata Spanish origin). Amplification products were fractionated by electrophoresis in a 2% (w/v) agarose gel and stained with ethidium bromide. The numbers on the left indicate the sizes (in bases pairs, bp) of molecular weight markers. (C) Differential diagnosis of Taenia spp. DNA samples using the HDP2 PCR III. 10 ng gDNA from Asian isolates of T. asiatica (lanes 1, 3, 6, 7, 9, 11, 12, 13, 15, 16, 17, 18 and 19); Asian isolates of T. saginata (lanes 2, 4, 5, 8, 10 and 14); T. solium (Venezuelan and Mexican origin, C1 and C2); and T. saginata (Spanish origin, C3) were amplified by HDP2-PCR. The amplification products were fractionated by electrophoresis in a 1% (w/v) agarose gel and stained using ethidium bromide. The numbers on the left indicate the sizes (in bases pairs, bp) of molecular weight markers.
After Cla I digestion of gDNAs from the Asian isolates, T. saginata and T. solium controls, and IPHDP2 probe hybridization as described above, different restriction enzyme product profiles were identified (Fig. 3b). With T. saginata, bands of 4.5, 3, 2, 1 and 0.7 kb were produced, whereas with T. asiatica bands of 4, 1 and 0.7 kb were obtained. As an alternative and complementary approach for studying the molecular organization of the HDP2 sequence, T. saginata and T. asiatica gDNA samples were amplified with primers derived from the IHDP2 probe (HDP2 PCR I); three amplicons for T. saginata were observed, but only one for T. asiatica (Fig. 3a). Taking these results together, T. saginata and T. asiatica showed polymorphism in their HDP2 DNA sequences.
Figure 3.
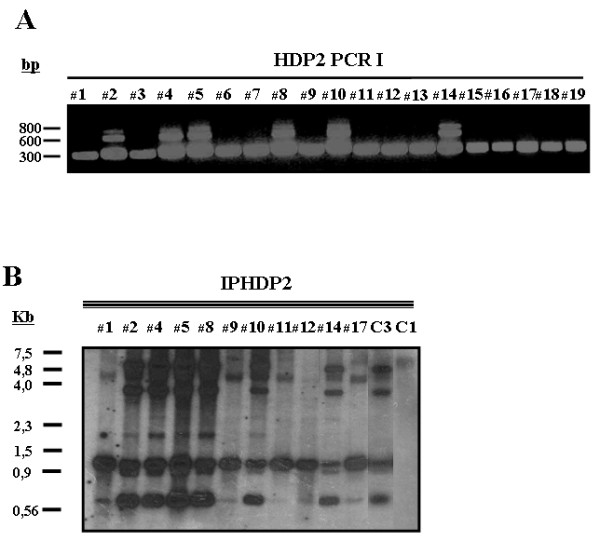
Polymorphism in the HDP2 sequences. (A) HDP2 PCR I using 10 ng gDNA from Asian isolates of T. asiatica (lanes 1, 3, 6, 7, 9, 11, 12, 13, 15, 16, 17, 18 and 19); and Asian isolates of T. saginata (lanes 2, 4, 5, 8, 10 and 14). (B) Southern-blot analysis. 5 μg gDNA from Asian isolates of T. asiatica (lanes 1, 9, 11, 12 and 17); Asian isolates of T. saginata (lanes 2, 4, 5, 8, 10 and 14); T. solium (Venezuelan origin, C1); and T. saginata (Spanish origin, C3) were digested to completion with Cla I and probed with the digoxigenin-labeled T. saginata IPHDP2 probe.
To explore the contribution of nucleotide variability to the observed HDP2 sequence polymorphism, a fragment of 1896 bp from the IHDP2 region was obtained from all 19 Asian taeniid DNA samples using the HDP2 PCR II protocol. A comparison of all the IHDP2 fragments obtained [GenBank: FM212953-FM212971] by Clustal W2, revealed a high degree of homology, particularly in regard to the T. saginata isolates. Thus, alignment of the 1896 bp fragment from the control T. saginata HDP2 sequence [GenBank: AJ133740] with the amplified fragments from the Asian T. saginata samples yielded nucleotide identities from 99.6- 99.7%. Alignment of the control T. saginata HDP2 sequence with the T. asiatica fragments yielded 97.0-98.8% nucleotide identity. This, the Asian T. asiatica HDP2 fragments showed a sequence divergence of 0.1% to 2.1%, compared with divergence of 0.0-0.1% recorded for the Asian T. saginata isolates and the control Spanish T. saginata isolate, thereby confirming low nucleotide variability between the two cestode taxa. Consequently, this structural analysis suggested that the HDP2 sequence polymorphism could be explained by variable representation of partially repeated sequences in the mid region of the HDP2 fragment, IHDP2, rather than due to nucleotide sequence divergence.
Seqman II-alignment of the partial HDP2 DNA fragments for T. saginata and T. asiatica within the NTR of the rDNA sequences of T. saginata (pTSgr 3.1, pTSgr 2.4) and T. asiatica (pTTr 3.1) [21,36], revealed that the sequences were identical. Accordingly, the HDP2 sequence could thus also represent a variable repetitive fragment, being part of the NTR from rDNA. Notably, the identities among the sequences were restricted to nucleotides 859-1049 and 1738-1868 of the HPD2 DNA plus strand; pTSgr 3.1 and pTTr 3.1 has a 0.7 kb internal fragment, whereas pTSgr 2.4 does not (Fig. 1). This genetic variability in the NTR of the rDNA of T. saginata and T. asiatica allowed us to design a PCR protocol (HDP2 PCR III) for the simultaneous differential identification of the two species (Fig. 1). The HDP2F2/HDP2R3 primer set was designed from this region in the mid variable central part of HDP2 (Fig. 1).
Using 10 ng of gDNA from the Asian isolates and control samples, and following the PCR conditions described in Table 2, unambiguous species-specific amplification was obtained for all the samples analyzed (Fig. 2c). Three amplicons were evident with the T. asiatica samples (1300 bp, 600 bp and 300 bp), and two amplicons were generated for T. saginata (a strong band of 1300 bp and a weak band of 300 bp); the control T.solium samples produced one amplicon (600 bp) (Fig. 2c). Therefore, the novel HDP2 PCR III yielded results similar to those of Yamasaki et al. [6] (Fig. 2b). These assays will prove particularly useful in areas such as South East Asia where T. saginata, T. asiatica and T. solium coexist [37,38].
In conclusion, this study showed and defined genetic polymorphism in the HDP2 sequence of the human taeniid cestodes T. saginata and T. asiatica. An unexpected but, nevertheless notable finding, was that HDP2 may be a repetitive fragment of the NTR within taeniid rDNA. This could explain the success of the published PCR protocols derived from this sequence in the sensitive diagnosis of taeniasis and NCC [12,13,17,25,26,39]. These findings were used to design a new HDP2 based PCR protocol, which demonstrated to be useful for the unambiguous discrimination of T. saginata from T. asiatica.
Abbreviations
NCC: neurocysticercosis; ELISA: enzyme-linked immunosorbent assay; PCR: Polymerase chain reaction; ES: excretion-secretion; DNA: Deoxyribonucleic acid; DNAg: genomic DNA; rDNA: ribosomal DNA; NTR: non-translated region; HDP2: a 4- kb polymorphic DNA fragment.
Competing interests
The authors declare that they have no competing interests.
Authors´contributions
LMG, LJSH, RMEP, DPM and TG conceived and designed the experiments.: LMG, BB, EF and MDFG performed the experiments. LMG, EF, DPM and TG analyzed the data. LMG, TG, LJSH, MREP and DPM wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Luis M González, Email: lmgonzal@isciii.es.
Begoña Bailo, Email: begobb@isciii.es.
Elizabeth Ferrer, Email: elizabeth.ferrer@isciii.es.
Maria D Fernandez García, Email: ginebra009@hotmail.com.
Leslie JS Harrison, Email: lharris4@staffmail.ed.ac.uk.
Michael RE Parkhouse, Email: parkhous@igc.gulbenkian.pt.
Donald P McManus, Email: Don.McManus@qimr.edu.au.
Teresa Gárate, Email: tgarate@isciii.es.
Acknowledgements
We would like to thank Prof. PC Fan (now deceased), Dr. J Cross and Prof. P Craig for supplying the Taenia isolates used for analysis. This work was also supported by a grant from FIS (97/0141 and 00/407) and the Spanish Ministry of Science and Innovation and the Instituto de Salud Carlos III within the Network of Tropical Diseases Research (RICET RD06/0021/0019). L.M. Gonzalez was sponsored by fellowships from the ISCIII and RICET.
References
- Flisser A. Neurocysticercosis in Mexico. Parasitol Today. 1998;4:131–137. doi: 10.1016/0169-4758(88)90187-1. [DOI] [PubMed] [Google Scholar]
- García HH, Gonzalez AE, Evans CA, Gilman RH. Cysticercosis Working Group in Peru. Taenia solium cysticercosis. Lancet. 2003;362:547–556. doi: 10.1016/S0140-6736(03)14117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulsby EJL. Helminths, Arthropods and Protozoa of Domesticated Animals. 7. Bailhere and Tyndall: London; 1982. [Google Scholar]
- Schantz PM. In: Taenia solium cysticercosis/Taeniasis is a potentially eradicable disease: developing a strategy for action and obstacles to overcome. García HH, Martínez M, editor. Lima: ICN; 1996. pp. 227–230. Teniasis/Cisticercosis por T. solium. [Google Scholar]
- Proctor BM. Identification of tapeworms. South Afric Med J. 1972;46:234–238. [PubMed] [Google Scholar]
- Yamasaki H, Allan JC, Sato MO, Nakao M, Sako Y, Nakaya K, Qiu D, Mamuti W, Craig PS, Ito A. DNA differential diagnosis of Taeniasis and cysticercosis by multiplex PCR. J Clin Microbiol. 2004;42:548–553. doi: 10.1128/JCM.42.2.548-553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila G, Garcia Noval J, Flisser A, Graig PS. Immunodiagnosis of taen iasis by coproantigen detection. Parasitology. 1990;101:473–447. doi: 10.1017/S0031182000060686. [DOI] [PubMed] [Google Scholar]
- Deplazes P, Eckert J, Pawlowski Z, Machowska L, Gottstein B. An enzyme-linked immunosorbent assay for diagnostic detection of Taenia saginata copro-antigens in humans. Trans Roy Soc Trop Med Hyg. 1991;85:391–396. doi: 10.1016/0035-9203(91)90302-F. [DOI] [PubMed] [Google Scholar]
- Levine MZ, Calderón JC, Wilkins PP, Lane WS, Asara JM, Hancock K, Gonzalez AE, Garcia HH, Gilman RH, Tsang VC. Characterization, cloning, and expression of two diagnostic antigens for Taenia solium tapeworm infection. J Parasitol. 2004;90:631–638. doi: 10.1645/GE-189R. [DOI] [PubMed] [Google Scholar]
- Levine MZ, Lewis MM, Rodriquez S, Jimenez JA, Khan A, Lin S, Garcia HH, Gonzalez AE, Gilman RH, Tsang VC. Development of an enzyme-linked immunoelectrotransfer blot (EITB) assay using two baculovirus expressed recombinant antigens for diagnosis of Taenia solium taeniasis. J Parasitol. 2007;93:409–417. doi: 10.1645/GE-938R.1. [DOI] [PubMed] [Google Scholar]
- Dias AK, Aoki SM, Garcia JF, Nunes CM. Taenia solium and Taenia saginata: identification of sequence characterized amplified region (SCAR) markers. Exp Parasitol. 2007;117:9–12. doi: 10.1016/j.exppara.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Gonzalez LM, Montero E, Harrison LJ, Parkhouse RM, Garate T. Differential diagnosis of Taenia saginata and Taenia solium infection by PCR. J Clin Microbiol. 2000;38:737–744. doi: 10.1128/jcm.38.2.737-744.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez LM, Montero E, Puente S, Lopez-Velez R, Hernández M, Sciutto E, Harrison LJ, Parkhouse RM, Garate T. PCR tools for the differential diagnosis of Taenia saginata and Taenia solium taeniasis/cysticercosis from different geographical locations. Diagn Microbiol Infect Dis. 2002;42:243–249. doi: 10.1016/S0732-8893(01)00356-X. [DOI] [PubMed] [Google Scholar]
- Ito A, Craig PS. Immunodiagnostic and molecular approaches for the detection of taeniid cestode infections. Trends Parasitol. 2003;19:377–381. doi: 10.1016/S1471-4922(03)00200-9. [DOI] [PubMed] [Google Scholar]
- Mayta H, Talley A, Gilman RH, Jiménez J, Verastegui M, Ruiz M, Garcia HH, Gonzalez AE. Differentiating Taenia solium and Taenia saginata infections by simple hematoxylin-eosin staining and PCR-restriction enzyme analysis. J Clin Microbiol. 2000;38:133–137. doi: 10.1128/jcm.38.1.133-137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes CM, Dias AK, Dias FE, Aoki SM, de Paula HB, Lima LG, Garcia JF. Taenia saginata: differential diagnosis of human taeniasis by polymerase chain reaction-restriction fragment length polymorphism assay. Exp Parasitol. 2005;110:412–425. doi: 10.1016/j.exppara.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Nunes CM, Lima LG, Manoel CS, Pereira RN, Nakano MM, Garcia JF. Taenia saginata: polymerase chain reaction for taeniasis diagnosis in human fecal samples. Exp Parasitol. 2003;104:67–69. doi: 10.1016/S0014-4894(03)00111-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Hidalgo R, Geysen D, Benitez-Ortiz W, Geerts S, Brandt J. Comparison of conventional techniques to differentiate between Taenia solium and Taenia saginata and an improved polymerase chain reaction-restriction fragment length polymorphism assay using a mitochondrial 12 S rDNA fragment. J Parasitol. 2002;88:1007–1011. doi: 10.1645/0022-3395(2002)088[1007:COCTTD]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bowles J., McManus DP. Genetic characterization of the Asian Taenia, a newly described Taeniid cestode of Humans. Am J Trop Med Hyg. 1994;50:33–44. [PubMed] [Google Scholar]
- Eom KS, Jeon HK, Kong Y, Hwang UW, Yang Y, Li X, Xu L, Feng Z, Pawlowski ZS, Rim HJ. Identification of Taenia asiatica in China: molecular, morphological, and epidemiological analysis of a Luzhai isolate. J Parasitol. 2002;88:758–764. doi: 10.1645/0022-3395(2002)088[0758:IOTAIC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Zarlenga DS, McManus DP, Fan PC, Cross JH. Characterization and detection of a newly described Asian taeniid using cloned ribosomal DNA fragments and sequence amplification by the polymerase chain reaction. Exp Parasitol. 1991;72:174–183. doi: 10.1016/0014-4894(91)90135-J. [DOI] [PubMed] [Google Scholar]
- Gonzalez LM, Montero E, Morakote N, Puente S, Diaz De Tuesta JL, Serra T, Lopez-Velez R, McManus DP, Harrison LJ, Parkhouse RM, Garate T. Differential diagnosis of Taenia saginata and Taenia saginata asiatica taeniasis through PCR. Diagn Microbiol Infect Dis. 2004;49:183–188. doi: 10.1016/j.diagmicrobio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Jeon HK, Chai JY, Kong Y, Waikagul J, Insisiengmay B, Rim HJ, Eom KS. Differential diagnosis of Taenia asiatica using multiplex PCR. Exp Parasitol. 2009;121:151–156. doi: 10.1016/j.exppara.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Nkouawa A, Sako Y, Nakao M, Nakaya K, Ito A. Loop-Mediated Isothermal Amplification Method for Differentiation and Rapid Detection of Taenia Species. J Clin Microbiol. 2008;47:168–174. doi: 10.1128/JCM.01573-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero E, Gonzalez LM, Puente S, Garate T. Diagnosis of human taeniasis by multiplex-PCR. Med Clin. 2003;120:37. doi: 10.1157/13042005. [DOI] [PubMed] [Google Scholar]
- Villalobos N, Gonzalez LM, Fuentes I, Puente S, Garate T. In: Proceedings IX European Multicolloquium of Parasitology. Bologna, editor. Valencia: Monduzzi; 2004. Differential diagnosis of human taeniasis in the Department of Parasitology, Centro Nacional de Microbiologia, Instituto de Salud Carlos III; pp. 241–244. [Google Scholar]
- Harrison LJS, Delgado J, Parkhouse RME. Differential diagnosis of Taenia saginata and Taenia solium with DNA probes. Parasitology. 1990;100:459–461. doi: 10.1017/S0031182000078768. [DOI] [PubMed] [Google Scholar]
- De Queiroz A, Alkire NL. The phylogenetic placement of Taenia cestodes that parasitize humans. J Parasitol. 1998;84:379–383. doi: 10.2307/3284501. [DOI] [PubMed] [Google Scholar]
- Fan PC, Chung WC. Taenia saginata asiatica: epidemiology, infection, immunological and molecular studies. J Microbiol Immunol Infect. 1998;31:84–89. [PubMed] [Google Scholar]
- McManus DP, Bowles J. Asian (Taiwan) Taenia: species or strain? Parasitol Today. 1994;10:273–275. doi: 10.1016/0169-4758(94)90145-7. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Bessho Y, Kamiya M, Kurosawa T, Horii T. Phylogenetic relationships within Taenia taeniaeformis variants and other taeniid cestodes inferred from the nucleotide sequence of the cytochrome c oxidase subunit I gene. Parasitol Res. 1995;81:451–458. doi: 10.1007/BF00931785. [DOI] [PubMed] [Google Scholar]
- Hoberg EP, Jones A, Rausch RL, Eom KS, Gardner SL. A phylogenetic hypothesis for species of the genus Taenia (Eucestoda: Taeniidae) J Parasitol. 2000;86:89–98. doi: 10.1645/0022-3395(2000)086[0089:APHFSO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory, Cold Spring Harbor: New York; 2003. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Dame JB, Lewis FA, McCutchan TF. The arrangement of ribosomal RNA genes in Schistosoma mansoni. Identification of polymorphic structural variants. Eur J Biochem. 1984;139:41–45. doi: 10.1111/j.1432-1033.1984.tb07973.x. [DOI] [PubMed] [Google Scholar]
- Fan PC, Chung WC, Lin CY, Wu CC. Experimental infection of Thailand Taenia (Chiengmai strain) in domestic animals. Int J Parasitol. 1990;20:121–123. doi: 10.1016/0020-7519(90)90183-N. [DOI] [PubMed] [Google Scholar]
- Ito A, Nakao M, Wandra T. Human Taeniasis and cysticercosis in Asia. Lancet. 2003;362:1918–1920. doi: 10.1016/S0140-6736(03)14965-3. [DOI] [PubMed] [Google Scholar]
- Hernández M, Gonzalez LM, Fleury A, Saenz B, Parkhouse RM, Harrison LJ, Garate T, Sciutto E. Neurocysticercosis: detection of Taenia solium DNA in human cerebrospinal fluid using a semi-nested PCR based on HDP2. Ann Trop Med Parasitol. 2008;102:317–323. doi: 10.1179/136485908X278856. [DOI] [PubMed] [Google Scholar]


