Abstract
Influenza remains a serious public health threat throughout the world. Vaccines and antivirals are available that can provide protection from infection. However, new viral strains emerge continuously because of the plasticity of the influenza genome, which necessitates annual reformulation of vaccine antigens, and resistance to antivirals can appear rapidly and become entrenched in circulating virus populations. In addition, the spread of new pandemic strains is difficult to contain because of the time required to engineer and manufacture effective vaccines. Monoclonal antibodies that target highly conserved viral epitopes might offer an alternative protection paradigm. Herein we describe the isolation of a panel of monoclonal antibodies derived from the IgG+ memory B cells of healthy, human subjects that recognize a previously unknown conformational epitope within the ectodomain of the influenza matrix 2 protein, M2e. This antibody binding region is highly conserved in influenza A viruses, being present in nearly all strains detected to date, including highly pathogenic viruses that infect primarily birds and swine, and the current 2009 swine-origin H1N1 pandemic strain (S-OIV). Furthermore, these human anti-M2e monoclonal antibodies protect mice from lethal challenges with either H5N1 or H1N1 influenza viruses. These results suggest that viral M2e can elicit broadly cross-reactive and protective antibodies in humans. Accordingly, recombinant forms of these human antibodies may provide useful therapeutic agents to protect against infection from a broad spectrum of influenza A strains.
Keywords: influenza matrix 2 protein, monoclonal, pandemic
Seasonal influenza epidemics hospitalize more than 200,000 people each year in the United States and kill an estimated 500,000 people worldwide (1). The immune system affords only partial protection from seasonal strains in most individuals because of constantly arising point mutations in the viral genome, which lead to structural variability known as antigenic drift. Pandemic strains encounter even less immune resistance because of genomic reassortment events among different viruses, which result in more radical shifts in viral antigenic determinants. Consequently, pandemic influenza has the potential to cause widespread illness, death, and economic disruption. Vaccines and antiviral agents are available to counter the threat of influenza epidemics and pandemics. However, the strain composition of influenza vaccines must be determined before the influenza season on an annual basis, and predicting in advance which strains will become dominant is challenging. Moreover, the emergence of strains that evade vaccine-induced, protective immune responses is relatively rapid, which often results in inadequate protection (2). Antiviral drugs include oseltamivir and zanamivir, which inhibit the function of the viral protein neuraminidase (NA), and adamantanes, which inhibit the ion channel function of the viral M2 protein (3, 4). Antiviral agents are effective for sensitive virus strains but viral resistance can develop quickly and has the potential to render these drugs ineffective. In the 2008 to 2009 United States influenza season, nearly 100% of seasonal H1N1 or H3N2 influenza isolates tested were resistant to oseltamivir or adamantane antivirals, respectively (CDC Influenza Survey: http://www.cdc.gov/flu/weekly/weeklyarchives2008-2009/weekly23.htm).
Passive immunotherapy using anti-influenza antibodies represents an alternative paradigm for preventing or treating viral infection. Evidence for the utility of this approach dates back nearly 100 y, when passive serum transfer was used during the 1918 influenza pandemic with some success (5). Although protection provided by anti-influenza mAbs is typically narrow in breadth because of the antigenic heterogeneity of influenza viruses, several groups have recently reported protective mAbs that bind to conserved epitopes within the stem region of viral hemagglutinin (HA) (6–9). However, these epitopes appear to be restricted to a subset of influenza viruses; these anti-HA mAbs would not be expected to provide protection against viruses of the H3 and H7 subtypes. Of these, the former comprises an important component of circulating human strains (10) and the latter includes highly pathogenic avian strains that have caused mortality in humans (11, 12).
Of the three antibody targets present on the surface of the influenza virus, the ectodomain of the viral M2 protein (M2e) is much more highly conserved than either HA or NA, which makes it an attractive target for broadly protective mAbs. Monoclonal antibodies to M2e have been shown to be protective in vivo (13–17), and several groups have demonstrated protection against infection with vaccine strategies based on M2e (18–23). In these cases, purified M2 protein or peptides derived from M2e sequence have been used as immunogens to generate anti-M2e antibodies in animals or as vaccine candidates. In the present study, we have isolated mAbs directly from human B cells that bind to the M2 protein displayed on virus particles and on virus-infected cells. Furthermore, we demonstrate that these antibodies protect mice from a lethal influenza A virus challenge and that they can recognize M2 variants derived from a wide range of human and animal influenza A virus isolates. This combination of properties may enhance the utility of these antibodies to prevent and treat influenza A virus infections.
Results and Discussion
Isolation of a Family of Anti-M2e mAbs from Human B Cells.
To explore the humoral immune response to natural influenza infection in humans, we have isolated antibodies from IgG+ memory B cells of M2e-seropositive subjects. Serum samples from 140 healthy adult, United States-sourced donors were tested for reactivity with M2e expressed on the surface of HEK293 cells that were transfected with a viral M2 gene (derived from A/Fort Worth/1/50 H1N1). IgG+ memory B cells from 5 of the 23 M2e-seropositive subjects were cultured under conditions where they proliferated and differentiated into IgG-secreting plasma cells. B-cell culture wells were screened for IgG reactivity to cell-surface M2e and Ig heavy- and light-chain variable region (VH and VL) genes were rescued by RT-PCR from 17 positive wells and incorporated into a human IgG1 constant region background for recombinant expression and purification. VH and VL sequences of 15 of the 17 anti-M2e mAbs cluster into two related groups (Table S1) (the International ImMunoGeneTics Information System http://www.imgt.org). In group A, assignment of the germ-line VH gene segment is IGHV4-59*01, and in group B, the germ-line gene segment is IGHV3-66*01. The two more distantly related mAbs 62B11 and 41G23 (group C) use the germ-line V gene segment IGHV4-31*03, which has only five amino acid residue differences from the germ-line V gene segment IGHV4-59*01 of group A. All of these mAbs use the same light-chain V gene, IGKV1-39*01 or its allele IGKV1D-39*01, and show evidence of somatic hypermutation from the germ-line heavy- or κ-chain sequence (Fig. S1). Competitive binding experiments showed that all of these human mAbs appear to bind similar sites on native M2e expressed on the surface of CHO cells (Figs. S2 and S3). We selected for further characterization one mAb from each of groups A and B, designated TCN-031 and TCN-032, respectively.
High-Affinity Binding to the Surface of Influenza Virus.
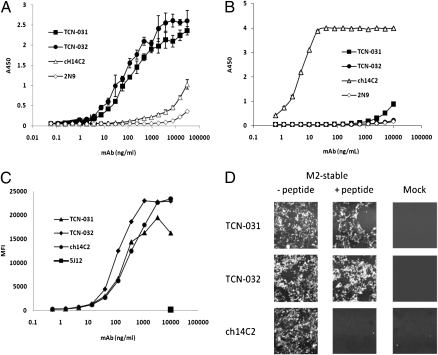
Both TCN-031 and TCN-032 bound directly to an H1N1 virus (A/Puerto Rico/8/34) with high avidity, with half-maximal binding at about 100 ng/mL (Fig. 1A). Fab fragments prepared from TCN-031 and TCN-032 bound virus with affinities (KD) of 14 and 3 nM, respectively, as determined by surface plasmon resonance (Table S2). The human mAbs did not bind appreciably to a 23-amino acid synthetic peptide corresponding to the M2e domain of an H1N1 virus (A/Fort Worth/1/50) (Fig. 1B). A chimeric derivative of the murine anti-M2e mAb 14C2 (ch14C2), which was originally generated by immunization with purified M2 (24), exhibited the opposite behavior to that observed with the human mAbs, with little binding to virus but robust binding to the isolated 23mer M2e peptide with half-maximal binding to peptide at 10 ng/mL (Fig. 1 A and B). Interestingly, both the human mAbs and ch14C2 bound to the surface of Madin-Darby canine kidney (MDCK) cells infected with H1N1 virus (A/Puerto Rico/8/34) with similar avidities (Fig. 1C). It thus appears that viral epitopes recognized by the human anti-M2e mAbs are present and accessible on the surface of both virus and infected cells, although the epitope bound by ch14C2 is accessible only on the surface of infected cells. Our observation that the human anti-M2e mAbs do not bind appreciably to immobilized synthetic peptides derived from M2e, and furthermore that such peptides do not compete for binding of these antibodies to M2e expressed on the surface of mammalian cells (Fig. 1D), supports the idea that secondary structure within the M2e epitope is important for binding by the human antibodies. That ch14C2 binds peptide immobilized on plastic suggests a lesser importance of higher order structure for binding of this mAb.
Fig. 1.
Anti-M2e mAbs TCN-032 and TCN-031 bind virus particles and virus-infected cells but not M2e-derived synthetic peptide. (A) Purified influenza virus (A/Puerto Rico/8/34) was coated at 10 μg/mL on ELISA wells and binding of anti-M2e mAbs TCN-031, TCN-032, ch14C2, and the HCMV mAbs 2N9 was evaluated using HRP-labeled goat anti-human Fc. (B) The 23mer synthetic peptide of M2 derived from A/Fort Worth/1/50 was coated at 1 μg/mL on ELISA wells and binding of mAbs TCN-031, TCN-032, ch14C2, and 2N9 were evaluated as in A. (C) MDCK cells were infected with A/Puerto Rico/8/34 (PR8) and subsequently stained with mAbs TCN-031, TCN-032, ch14C2 and the HCMV mAb 5J12. Binding of antibodies was detected using Alexafluor 647-conjugated goat anti-Human IgG H&L antibody and quantified by flow cytometry. (D) HEK 293 cells stably transfected with the M2 ectodomain of A/Fort Worth/1/50 (D20) were stained with transient transfection supernatant containing mAbs TCN-031, TCN-032, or the control ch14C2 and analyzed by FMAT for binding to M2 in the presence or absence of 5 μg/mL M2e peptide. Mock-transfected cells are 293 cells stably transfected with vector alone. Results shown for A, B, and C are representative of three experiments, and for D, one experiment.
Protection from Lethal Challenges with H5N1 and H1N1 Viruses.
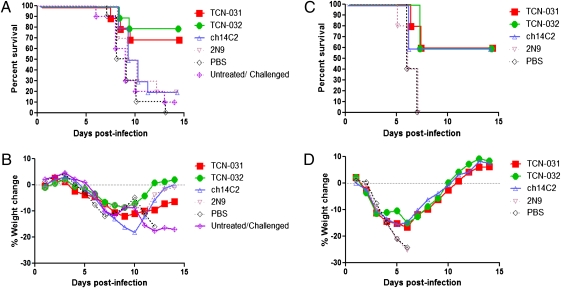
We next examined the protective efficacy of the human anti-M2e mAbs TCN-031 and TCN-032 in a lethal challenge model of influenza infection in mice. Animals were challenged intranasally with 5 × LD50 units of a high-pathogenicity H5N1 virus (A/Vietnam/1203/04) and both human mAbs were protective when treatment was initiated 1 d after viral challenge. In contrast, mice that were subjected to similar treatment regimens with a subclass-matched, irrelevant control mAb 2N9, which targets the AD2 epitope of the gp116 portion of the human cytomegalovirus gB (25), or with a vehicle control were protected to a lesser extent, or not at all, resulting in 70 to 80% survival for mice treated with human mAbs versus 20% survival for control mAb and 0% survival for vehicle (Fig. 2A). The anti-M2e mAb ch14C2 did not confer substantial protection in this model (20% survival) (Fig. 2A), although this mAb has been shown to reduce the titer of virus in the lungs of mice infected with other strains of influenza virus (16). All of the animals, including those in the TCN-031 and TCN-032 treatment groups, exhibited weight loss from days 4 to 8 postinfection, followed by a gradual increase in weight in the surviving animals through the end of the study on day 14 (Fig. 2B), indicating that the human anti-M2e mAbs afforded protection by reducing the severity or extent of infection rather than by completely preventing infection. Indeed, results of immunohistological and viral load analyses of lung, brain, and liver tissue from additional animals in each treatment cohort are consistent with a reduction in the spread of virus beyond the lung to the brain and also possibly liver in animals that were treated with the human anti-M2e mAbs, but not with ch14C2 or the subclass-matched control mAb 2N9. The effect of the human anti-M2e mAbs on viral load in the lung versus the control mAbs was, however, more moderate (Table S3 and Fig. S4, respectively).
Fig. 2.
Therapeutic efficacy of anti-M2 mAbs TCN-031 and TCN-032 in mice. Mice (n = 10) were infected by intranasal inoculation with 5 × LD50 A/Vietnam/1203/04 (H5N1) (A and B) or (n = 5) with 5 × LD50 A/Puerto Rico 8/34 (H1N1) (C and D), followed by 3 i.p. injections with mAbs at 24, 72, and 120 h postinfection (a total of three mAb injections per mouse) and weighed daily for 14 d. Percentage-survival is shown in A and C, whereas percent-weight change of mice is shown in B and D. The results shown for the treatment study of mice infected with A/Vietnam/1203/04 (H5N1) are representative of two experiments.
To test whether protection conferred by the human anti-M2e mAbs mirrors their broad binding behavior, we performed a similar in vivo challenge study with a mouse-adapted isolate of the relatively divergent H1N1 virus A/Puerto Rico/8/34. One-hundred percent of PBS-treated or subclass-matched, control antibody-treated mice were killed by this virus, although a majority of the animals treated with the human anti-M2e mAbs TCN-031 and TCN-032 survived (60%) (Fig. 2C). With this virus, mice treated with ch14C2 provided a similar survival benefit to that of the human anti-M2e mAbs (Fig. 2C). Weight changes in each treatment group throughout the course of infection and its subsequent resolution followed a pattern that was similar to that of mice infected with the H5N1 virus (Fig. 2D).
The human anti-M2e mAbs and ch14C2 bound to cell surface-expressed M2e from A/Vietnam/1203/04 and A/Puerto Rico/8/34 viruses (Fig. 3B and Table S4) and cells infected with A/Puerto Rico/8/34 (Fig. 1C). Mechanisms for antibody-mediated protection could include killing of infected host cells by antibody-dependent cell-mediated cytotoxicity or complement-dependent cytotoxicity (13, 26). We found in vitro evidence for both of these mechanisms with the human anti-M2e mAbs and ch14C2 (Fig. S5 and Fig. S6). An explanation for the enhanced in vivo protection observed with the human anti-M2e mAbs as compared with ch14C2 following challenge by the high-pathogenicity avian virus A/Vietnam/1203/04 as compared with A/Puerto Rico/8/34 could be a result of the unique capability of the human mAbs to bind virus directly, whereas ch14C2 does not appear to bind influenza virions (Fig. 1A). Protective properties of antibodies that bind to virus might be expected to include mechanisms such as antibody-dependent virolysis (27) and clearance via opsonophagocytosis by host cells (28). Some of these mechanisms require efficient interaction between antibodies and host Fc receptors. In our mouse-challenge experiments, all of the mAbs tested had human constant regions; however, other studies have shown that human antibodies can interact productively with murine Fc receptors (29).
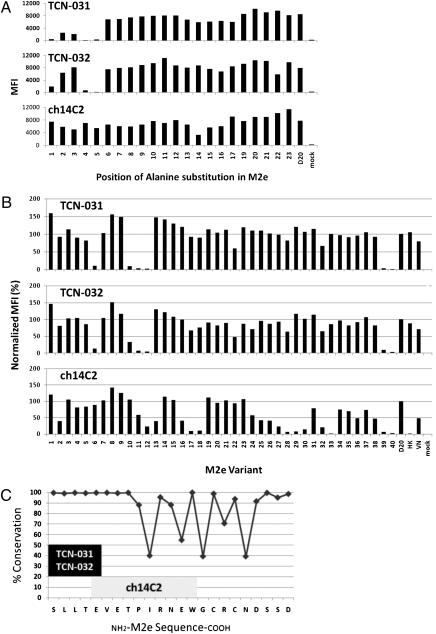
Fig. 3.
Binding of anti-M2e mAbs TCN-031 and TCN-032 to M2 mutants indicates the epitope is located in the highly conserved N terminal of M2e. Mutants with alanine substituted at each position of the M2 ectodomain of A/Fort Worth/1/50 (D20) (A) or 40 wild-type M2 mutants, including A/Vietnam/1203/04 (VN) and A/Hong Kong/483/97 (HK) (B), were transiently transfected into 293 cells. The identity of each wild-type M2 mutant is listed in Table S4. Transfected cells were stained with mAbs TCN-031, TCN-032, or the control ch14C2 and analyzed by FACS for binding to M2 at 24 h posttransfection. The mAbs TCN-031 and TCN-032 do not bind variants with amino acid substitutions at positions 1, 4, or 5 of M2e. (C) The deduced epitope for TCN-031 and TCN-032 occurs in a highly conserved region of M2e and is distinct from that found for ch14C2. Results shown for A and B are representative of three experiments.
Binding to the Highly Conserved N-Terminal Segment of M2e.
To better understand the unique viral binding property of the human anti-M2e mAbs, we mapped their binding sites within the M2e domain. The lack of appreciable binding of the human mAbs to M2e-derived linear peptides precluded a synthetic-peptide approach to fine-structure mapping of their epitopes. Instead, binding of the mAbs to M2e alanine substitution mutants and naturally occurring M2 variants that were expressed on the surface of cDNA-transfected mammalian cells was quantified by flow cytometry. Binding experiments with a panel of M2 mutant proteins where each position in the 23-amino acid M2 ectodomain was substituted with alanine revealed that the first (S), fourth (T), and fifth (E) positions of the mature (methionine-clipped) M2 polypeptide were critical for binding of both TCN-031 and TCN-032 (Fig. 3A). In contrast, the binding of ch14C2 was selectively diminished when alanine was substituted at position 14 of mature M2 (Fig. 3A). These observations were confirmed in studies with a panel of divergent, naturally occurring M2 variants; substitution with proline at position 4 (Table S4: A/Panama /1/1966 H2N2, A/Hong Kong/1144/1999 H3N2, A/Hong Kong/1180/1999 H3N2, and A/chicken/Hong Kong/YU427/2003 H9N2) and glycine at position 5 (Table S4: A/chicken/Hong Kong/SF1/2003 H9N2) correlated with diminished binding of the human anti-M2e mAbs but not ch14C2 (Fig. 3B and Table S4). These results suggest that both TCN-031 and TCN-032 recognize a core sequence of SLLTE at positions 1 to 5 of the N terminus of mature M2e. This theory is supported by data which show that these mAbs compete effectively with each other for binding to M2e expressed on the surface of CHO cells (Fig. S3). In contrast, our results indicate that ch14C2 binds to a site that is spatially distinct and downstream of the SLLTE core that is recognized by the human anti-M2e mAbs. Indeed, previous studies have shown that 14C2 binds a relatively broad, linear epitope with the sequence EVERTPIRNEW at positions 5 to 14 of processed M2e (13).
Although the epitopes recognized by TCN-031 and TCN-032 are likely very similar, there were some differences between these human mAbs in their binding to several of the M2e mutants. For example, TCN-031 appears to have a greater dependence than TCN-032 on residues 2 (L) and 3 (L) of the mature M2e sequence (Fig. 3A). The VH regions of these two human mAbs use different variable, diversity, and joining gene segments, which may explain the minor differences in binding observed between these mAbs. Interestingly, despite the differences in their VH make-up, these human mAbs use the same germ-line κ-chain V gene segments, albeit with distinct κ-chain joining segments.
Localization of the binding region of the human anti-M2e mAbs at the N-terminal region of M2e is especially significant in light of the remarkably high-sequence conservation in this part of the polypeptide among influenza A viruses. The viral M gene segment that encodes M2 also encodes the internal viral protein M1 via differential splicing. However, the splice site is located downstream of the shared N terminus of M2 and M1, resulting in two distinct mature polypeptides with an identical 8-amino acid N-terminal sequence (30). Options for viral escape from host anti-M2e antibodies that bind this region might be limited, as escape mutations in the N-terminal region would result in changes to not just M2 but also the M1 protein. Indeed, this N-terminal 8-amino acid segment of M2e shows nearly complete identity in the 1,364 unique full-length M2 variants cataloged in the National Center for Biotechnology Information (NCBI) Influenza Database (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/multiple.cgi); much lower levels of conservation are seen in M2e sequences downstream of this region (Fig. 3C). In fact, the core human anti-M2e antibody epitope SLLTE is present in ~98% of the 1,364 unique full-length M2e sequences cataloged in the NCBI Influenza Database, including 97, 98, and 98% of the human, swine, and avian viruses, respectively. This finding contrasts to the much lower conservation within the linear binding sites of anti-M2e mAbs elicited by immunization with M2e peptides or proteins. For example, 14C2 and Z3G1 (13) bind sequences that are conserved in less than 40% of influenza A viruses, and conservation within this region is even lower in avian and swine viruses (Table S5).
The linear M2e epitopes recognized by peptide-elicited antibodies may be more sensitive to escape mutations and natural substitutions that are present in some viral isolates. For example, P10L and P10H escape mutations to mAb 14C2 have been mapped to the central portion of M2e (31) and those same substitutions also occur in M2e variants from some highly pathogenic H5N1 strains. We have found that the human mAbs TCN-031 and TCN-032 but not ch14C2 bind to the M2 variant from the H5N1 virus A/Hong Kong/483/97 (HK), which contains the P10L substitution (Fig. 3B and Table S4). Thus, monoclonal antibodies with specificities similar to that of 14C2 are likely to have limited utility as broad spectrum therapeutic agents.
In the examination of five human subjects, we found 17 unique anti-M2e antibodies that bind the conserved N-terminal region of M2e, but did not observe IgG-reactivity with M2e-derived peptides that contain the linear epitopes recognized by 14C2 and other peptide-elicited antibodies. In contrast to the apparently uniform antibody response to M2e in naturally infected or vaccinated humans, mice immunized with M2e-derived peptides produced antibodies with a range of specificities within M2e, including the conserved N terminus and also downstream regions (15). It is tempting to speculate that the human immune system has evolved a humoral response that exclusively targets the highly conserved N-terminal segment of M2e rather than the more divergent, and thus less sustainably protective, downstream sites. Despite the lack of evidence for human antibodies that recognize this internal region of M2e, analysis of the evolution of the M gene suggests that this region of M2e is under strong positive selection in human influenza viruses (32). One explanation for this finding is that selective pressure is being directed at this internal region by immune mechanisms other than antibodies. For example, human T-cell epitopes have been mapped to these internal M2e sites (33).
Recognition of 2009 H1N1 S-OIV.
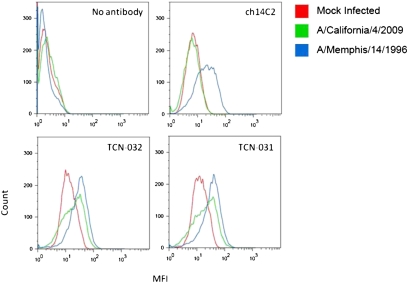
Broadly protective anti-influenza mAbs could be used in passive immunotherapy to protect or treat humans in the event of outbreaks from highly pathogenic, pandemic viral strains. A critical test of the potential for such mAbs as immunotherapeutic agents is whether they are capable of recognizing virus strains that may evolve from future viral reassortment events. As a case in point, the human anti-M2e mAbs TCN-031 and TCN-032 were tested for their ability to recognize the current H1N1 swine-origin pandemic strain (S-OIV). These mAbs were derived from human blood samples taken in 2007 or earlier, before the time that this strain is thought to have emerged in humans (34). Both human mAbs bound to MDCK cells infected with A/California/4/2009 (S-OIV H1N1, pandemic) and A/Memphis/14/1996 (H1N1, seasonal), whereas ch14C2 bound only to cells infected with the seasonal virus (Fig. 4). If this broad binding behavior proves to correlate with protection, as was the case with A/Vietnam/1203/2004 and A/Puerto Rico/8/34, then these human mAbs might be useful to prevent or treat the S-OIV pandemic strain or possibly other pandemic strains that might emerge in the future.
Fig. 4.
Anti-M2e mAbs TCN-031 and TCN-032 bind cells that have been infected with H1N1 A/California/4/09. MDCK cells were infected with Influenza A strain H1N1 A/Memphis/14/96, H1N1 A/California/4/09, or mock infected. Twenty-four hours postinfection cells were stained with mAbs TCN-031, TCN-032, or the control ch14C2 and analyzed by FACS for binding to M2. Results shown are for one of three experiments.
Although it is remarkable that humans have the capability to make antibodies that may confer nearly universal protection against influenza infection, the discovery of this heretofore undescribed class of antibodies raises the question of why this virus is able to mount a productive infection in immunocompetent individuals at all. This apparent paradox may be explained by the nature of the protective M2e epitope and its relative immunogenicity. It has been noted by others that M2e appears to exhibit low immunogenicity in humans (35, 36), especially when compared with the immunodominant virus glycoproteins HA and NA. Therefore, protective anti-M2e antibodies may exist in many individuals but at suboptimal titers. In support of this notion is our observation that most individuals did not display a detectable humoral response to M2e. We observed that fewer than 20% (23/140) of the individuals that we sampled in our cohort of healthy subjects had detectable serum levels of anti-M2e antibodies. The reasons for this phenomenon are not clear, but a similar situation exists in human CMV, where only a minority of human CMV-seropositive subjects has measurable antibodies to the broadly conserved, neutralizing AD2 epitope within the gB complex of human CMV (25, 37, 38).
An important requirement for an immunotherapeutic solution to the influenza threat will be the identification of protective epitopes that are conserved in preexisting and emerging viruses. Using large-scale sampling of the human immune response to native influenza M2, we have identified a naturally immunogenic and protective epitope within the highly conserved N-terminal region of M2e. Human antibodies directed to this epitope, including those described in the present study, may be useful for the prevention and treatment of pandemic and seasonal influenza.
Materials and Methods
A detailed description of all of the experimental procedures is provided in SI Materials and Methods. Briefly, human peripheral blood mononuclear cells (PBMC), or memory B cells that were enriched from PBMC, were collected and setup in a short-term culture in the presence of feeder cells before the culture supernatants were screened for antibody binding to M2 protein expressed on HEK 293 cells stably transfected with influenza virus M2 (H1N1 A/Fort Worth/50). M2e-specific monoclonal antibodies were reconstituted by isolating the mRNA from lysed B-cell cultures followed by reverse transcription of heavy chain, κ- or λ-light chain, variable domain genes with gene-specific primers and PCR amplification using VH, Vκ, and Vλ family-specific primers with flanking restriction sites for cloning into expression vectors.
M2e-specific monoclonal antibodies (mAbs), TCN-031 and TCN-032, were analyzed for binding M2e peptide or whole virus by ELISA for binding to M2 on the surface of cells that were transfected with M2 (H1N1 A/Fort Worth/50) in the presence or absence of competing M2e peptide, for binding to cells that were transfected with M2 from 43 different strains of influenza A, and for binding M2 on the surface of cells that were infected with H1N1 A/Puerto Rico/8/34, A/Memphis/14/96, or A/California/4/09. Therapeutic animal studies were performed to demonstrate the protective capacity of TCN-031 and TCN-032 against infection with H5N1 A/Vietnam/1203/04 and H1N1 A/Puerto Rico/8/34.
Supplementary Material
Acknowledgments
In memory of David Fanning, we would like to thank him for his support and guidance in the development of the anti-M2e antibodies. We thank Gord King, Mark Branum, Ray Fox, Doug Spicer, Courtney Ward, Lee Adams, Roxanne Grondin, Sarmila Basnet, and Anthony Hanson for technical assistance. We also thank Robert Lamb and Michael Katze for their helpful suggestions with the study and the manuscript. Biacore affinity determinations were done by Jason Schuman at GE Healthcare (Seattle, WA) and large-scale preparation of recombinant mAbs was provided by Gregory Bleck at Gala Biotech (Middleton, WI). K.S. was supported by a grant-in-aid for Specially Promoted Research, by the G-COE programme, and by the Program of Funding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministries of Education, Culture, Sports, Science, and Technology and PREST (Japan Science and Technology Agency).
Footnotes
Conflict of interest statement: A.G.G., O.A.O., P.W.H., P.-Y.C.-H., J.M., W.C., Y.K., and M.M. hold stock options in Theraclone Sciences, Inc. M.H. and Y.K. have received consulting fees from Theraclone Sciences for performing the work described in this article.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. TCN-032-VH HM451458, TCN-032-VL HM451459, TCN-031-VH HM451460, TCN-031-VL HM451461, 41_G23-VH HM451462, 41_G23-VL HM451463, 44_I10-VH HM451464, 44_I10-VL HM451465, 43_J07-VH HM451466, 43_J07-VL HM451467, 59_J21-VH HM451468, 59_J21-VL HM451469, 45_O19-VH HM451470, 45_O19-VL HM451471, 44_H04-VH HM451472, 44_H04-VL HM451473, 36_G05-VH HM451474, 36_G05-VL HM451475, 52_C13-VH HM451476, 52_C13-VL HM451477, 55_J06-VH HM451478, 55_J06-VL HM451479, 20_I23-VH HM451480, 20_I23-VL HM451481, 39_P23- VH HM451482, 39_P23-VLHM451483, 48_P18-VH HM451484, 48_P18-VL HM451485, 53_P10-VH HM451486, 53_P10-VL HM451487, 60_D19-VH HM451488, 60_D19-VL HM451489, 62_B11-VH HM451490, and 62_B11-VL HM451491).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911806107/-/DCSupplemental.
References
- 1.Thompson WW, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 2.Carrat F, Flahault A. Influenza vaccine: The challenge of antigenic drift. Vaccine. 2007;25:6852–6862. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Gubareva LV, Kaiser L, Hayden FG. Influenza virus neuraminidase inhibitors. Lancet. 2000;355:827–835. doi: 10.1016/S0140-6736(99)11433-8. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: Characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luke TC, Kilbane EM, Jackson JL, Hoffman SL. Meta-analysis: Convalescent blood products for Spanish influenza pneumonia: A future H5N1 treatment? Ann Intern Med. 2006;145:599–609. doi: 10.7326/0003-4819-145-8-200610170-00139. [DOI] [PubMed] [Google Scholar]
- 6.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell CA, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 11.Fouchier RA, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci USA. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belser JA, Bridges CB, Katz JM, Tumpey TM. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009;15:859–865. doi: 10.3201/eid1506.090072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, et al. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Res. 2008;80:168–177. doi: 10.1016/j.antiviral.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Zou P, Chen YH. Monoclonal antibodies recognizing EVETPIRN epitope of influenza A virus M2 protein could protect mice from lethal influenza A virus challenge. Immunol Lett. 2004;93:131–136. doi: 10.1016/j.imlet.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Fu TM, et al. Characterizations of four monoclonal antibodies against M2 protein ectodomain of influenza A virus. Virology. 2009;385:218–226. doi: 10.1016/j.virol.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 16.Treanor JJ, Tierney EL, Zebedee SL, Lamb RA, Murphy BR. Passively transferred monoclonal antibody to the M2 protein inhibits influenza A virus replication in mice. J Virol. 1990;64:1375–1377. doi: 10.1128/jvi.64.3.1375-1377.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beerli RR, et al. Prophylactic and therapeutic activity of fully human monoclonal antibodies directed against influenza A M2 protein. Virol J. 2009;6:224–234. doi: 10.1186/1743-422X-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu TM, et al. Comparative immunogenicity evaluations of influenza A virus M2 peptide as recombinant virus like particle or conjugate vaccines in mice and monkeys. Vaccine. 2009;27:1440–1447. doi: 10.1016/j.vaccine.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 19.Fan J, et al. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine. 2004;22:2993–3003. doi: 10.1016/j.vaccine.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Slepushkin VA, et al. Protection of mice against influenza A virus challenge by vaccination with baculovirus-expressed M2 protein. Vaccine. 1995;13:1399–1402. doi: 10.1016/0264-410x(95)92777-y. [DOI] [PubMed] [Google Scholar]
- 21.Neirynck S, et al. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med. 1999;5:1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 22.Tompkins SM, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis. 2007;13:426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mozdzanowska K, et al. Induction of influenza type A virus-specific resistance by immunization of mice with a synthetic multiple antigenic peptide vaccine that contains ectodomains of matrix protein 2. Vaccine. 2003;21:2616–2626. doi: 10.1016/s0264-410x(03)00040-9. [DOI] [PubMed] [Google Scholar]
- 24.Zebedee SL, Lamb RA. Influenza A virus M2 protein: Monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer H, Sundqvist VA, Pereira L, Mach M. Glycoprotein gp116 of human cytomegalovirus contains epitopes for strain-common and strain-specific antibodies. J Gen Virol. 1992;73:2375–2383. doi: 10.1099/0022-1317-73-9-2375. [DOI] [PubMed] [Google Scholar]
- 26.Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol. 2004;172:5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura M, Terada M, Sasaki H, Kamada M, Ohno T. Virolysis and in vitro neutralization of HIV-1 by humanized monoclonal antibody hNM-01. Hybridoma. 2000;19:427–434. doi: 10.1089/027245700750053913. [DOI] [PubMed] [Google Scholar]
- 28.Huber VC, Lynch JM, Bucher DJ, Le J, Metzger DW. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J Immunol. 2001;166:7381–7388. doi: 10.4049/jimmunol.166.12.7381. [DOI] [PubMed] [Google Scholar]
- 29.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 30.Lamb RA, Choppin PW. Identification of a second protein (M2) encoded by RNA segment 7 of influenza virus. Virology. 1981;112:729–737. doi: 10.1016/0042-6822(81)90317-2. [DOI] [PubMed] [Google Scholar]
- 31.Zharikova D, Mozdzanowska K, Feng J, Zhang M, Gerhard W. Influenza type A virus escape mutants emerge in vivo in the presence of antibodies to the ectodomain of matrix protein 2. J Virol. 2005;79:6644–6654. doi: 10.1128/JVI.79.11.6644-6654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuse Y, Suzuki A, Kamigaki T, Oshitani H. Evolution of the M gene of the influenza A virus in different host species: Large-scale sequence analysis. Virol J. 2009;6:67. doi: 10.1186/1743-422X-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jameson J, Cruz J, Ennis FA. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J Virol. 1998;72:8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng J, et al. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol J. 2006;3:102. doi: 10.1186/1743-422X-3-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W, Li H, Chen YH. N-terminus of M2 protein could induce antibodies with inhibitory activity against influenza virus replication. FEMS Immunol Med Microbiol. 2003;35:141–146. doi: 10.1016/S0928-8244(03)00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayata M, et al. Different antibody response to a neutralizing epitope of human cytomegalovirus glycoprotein B among seropositive individuals. J Med Virol. 1994;43:386–392. doi: 10.1002/jmv.1890430412. [DOI] [PubMed] [Google Scholar]
- 38.Navarro D, Lennette E, Tugizov S, Pereira L. Humoral immune response to functional regions of human cytomegalovirus glycoprotein B. J Med Virol. 1997;52:451–459. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.