Abstract
Expansions of CTG/CAG trinucleotide repeats, thought to involve slipped DNAs at the repeats, cause numerous diseases including myotonic dystrophy and Huntington's disease. By unknown mechanisms, further repeat expansions in transgenic mice carrying expanded CTG/CAG tracts require the mismatch repair (MMR) proteins MSH2 and MSH3, forming the MutSβ complex. Using an in vitro repair assay, we investigated the effect of slip-out size, with lengths of 1, 3, or 20 excess CTG repeats, as well as the effect of the number of slip-outs per molecule, on the requirement for human MMR. Long slip-outs escaped repair, whereas short slip-outs were repaired efficiently, much greater than a G-T mismatch, but required hMutSβ. Higher or lower levels of hMutSβ or its complete absence were detrimental to proper repair of short slip-outs. Surprisingly, clusters of as many as 62 short slip-outs (one to three repeat units each) along a single DNA molecule with (CTG)50•(CAG)50 repeats were refractory to repair, and repair efficiency was reduced further without MMR. Consistent with the MutSβ requirement for instability, hMutSβ is required to process isolated short slip-outs; however, multiple adjacent short slip-outs block each other's repair, possibly acting as roadblocks to progression of repair and allowing error-prone repair. Results suggest that expansions can arise by escaped repair of long slip-outs, tandem short slip-outs, or isolated short slip-outs; the latter two types are sensitive to hMutSβ. Poor repair of clustered DNA lesions has previously been associated only with ionizing radiation damage. Our results extend this interference in repair to neurodegenerative disease-causing mutations in which clustered slip-outs escape proper repair and lead to expansions.
Keywords: mismatch repair, slipped DNAs, trinucleotide repeat instability, clustered DNA damage, microsatellite instability
Expansions of CTG/CAG trinucleotide repeats (TNRs) cause 14 neurodegenerative and neuromuscular diseases (1, 2). The nonaffected population has stable tracts of 5–24 repeats, whereas unstable disease-causing alleles can have 35–6,550 repeats. Repeats can continue to expand in somatic tissues. The mechanism of TNR instability is unknown, but all models explaining TNR expansions involve slipped-DNAs (1–3).
The role of mismatch repair (MMR) in TNR expansions is an area of intense focus (3). MMR typically maintains genomic integrity by correcting mispaired nucleotides (4) recognized by two eukaryotic protein complexes. The Msh2–Msh6 MutSα complex is required to repair base–base mismatches and nonrepetitive insertion/deletion loops (ID) of 1 to 3 nucleotides or to enhance the repair of IDs of 5, 8, 12, and 27 excess nucleotides (5, 6). The Msh2–Msh3 MutSβ complex can repair some but not all base–base mismatches (refs. 6 and 7 and references therein) but can support or enhance repair of the same 1- to 12-nucleotide IDs repaired by MutSα (5, 6), making it functionally redundant to MutSα (8). MSH2 or MSH6 deficiencies in mice or humans cause a strong mutator phenotype and cancer (3, 4). MSH3 deficiencies cause a weak mutator phenotype and no cancer predisposition (3, 4). msh3/msh6 double mutants recapitulate the phenotype of msh2−/− mutants (3, 4). Characterizing the roles of MutSβ has been difficult because the effects of MSH3 deficiencies may be masked by MutSα. Except for TNR instability, in vivo studies have failed to reveal a function predominantly managed by MutSβ (2, 3).
Curiously, MMR proteins are required for CTG/CAG repeat expansion mutations (3). TNR expansions in mice require MSH2 (9–11), MSH3 (12, 13), and to a lesser degree PMS2 (14). The absence of either MSH2 or MSH3 shifts mutations from 90% expansions to 90% contractions (9, 15). MSH6 deficiency has indirect effects, probably because of competition with Msh3 and Msh6 for binding to Msh2 (12, 13, 16). Absence of one Msh3 allele decreased the frequency of TNR expansions, indicating that Msh3 is rate limiting (13). MMR must be functional, because mice with a defective MSH2 ATPase did not incur TNR expansions (17). It is possible that the mutagenic role of MMR proteins is the result of CTG/CAG repeat structures.
DNA structure plays an important role in MMR; however, some mispaired DNAs are repaired independent of MMR (5, 6, 18,19–20). Unpaired DNA loops formed at nonrepetitive sequences can be mutagenic precursors to deletion or insertion mutations. The length of the DNA loop determines the repair path (5, 6, 18,19–20). Short loops with 1–13 excess nucleotides are repaired in an MMR-dependent manner, but larger loops of up to 216 nucleotides are repaired independent of MMR (5, 6, 18,19–20). Because the size of a nonrepetitive loop affects the selection of the repair path, and because repeat-containing slip-outs differ structurally from nonrepeat loops, it is of interest to study the size of slip-outs formed by disease-relevant CTG/CAG repeats.
Slipped-DNAs probably are mutagenic intermediates of repeat instability (21). To understand the processing of slipped-DNAs, it is critical to appreciate their structural features. Slipped-DNAs form by out-of-register pairings between complementary repeat strands at sites of replication, DNA damage, repair, or recombination. In vitro slipped-intermediate DNAs (SI-DNAs) are heteroduplexes of (CTG)x•(CAG)y where x ≠ y. When x > y or x < y, only a single slip-out containing all the excess repeats is formed, extruding from the complementary CTG/CAG duplex at a unique point (21, 22). Slip-outs cannot branch migrate and once formed do not interconvert to other slipped isoforms (21, 23). CTG slip-outs form intrastrand hairpins with T-T mismatches. CAG slip-outs predominantly assume unpaired loops and hairpins with A-A mismatches. As few as one or two excess CTG or CAG repeats can form intra-strand hairpins (24). In contrast to the distinct structures formed by SI-DNAs, slipped homoduplex DNAs (S-DNAs), having an identical number of repeats in complementary strands (x = y), form a diverse series of slipped-isomers, and each molecule contains multiple short slip-outs. Each molecule of S-DNA formed by (CTG)50•(CAG)50 contained 2–62 short slip-outs (1–31 on each strand), each composed of one to three repeat units. S-DNAs and fully duplexed DNA do not readily interconvert (21). The propensity to form S-DNA and the complexity of slipped species vary with repeat length, tract purity, and flanking nonrepetitive sequences, all factors that contribute to instability in humans suffering diseases caused by TNR repeats (21,22–23, 25,26–27). It is unknown if the structural features of SI-DNAs and S-DNAs determine the path by which they may be processed.
Mechanisms of mammalian MMR have been derived largely from in vitro repair of circular heteroduplexes by human cell extracts (4). We established a similar in vitro assay to process circular slipped-DNAs, which model mutagenic intermediates of instability (28), to elucidate the mechanism of CTG/CAG instability. Previously, we studied SI-DNAs with slip-outs of 20 excess repeats [(CTG)30•(CAG)50 or (CTG)50•(CAG)30] and observed three repair outcomes: (i) correct repair; (ii) escaped repair; or (iii) error-prone repair in which some but not all of the excess repeats are excised. Repair outcome was highly sensitive to the structural features of the slipped-DNA, depending on slip-out sequence (CAG or CTG), on whether the slip-out was in the nicked or continuous strand, and on whether the nick was located upstream or downstream of the slip-out. Outcome of repair for slip-outs of 20 repeats was independent of MMR and nucleotide excision repair proteins (28, 29). Similar results were reported subsequently by others (30, 31). Here we considered whether varying structural features of slipped-DNAs (i.e., slip-out size and slip-out number) could affect repair path selection, repair outcome, and the requirement for MMR proteins.
Results
Length of Slip-Outs and Nick Location.
We focused on isolated slip-outs of CTG repeats, because long slip-outs (20 excess repeats of this sequence) were observed previously to escape repair (28). The ability to escape repair is a means by which expansions could occur: The excess slipped-out repeats would be integrated as expansions. Moreover, in all replication studies, expansions occur when CTG repeat slip-outs form in the newly replicated Okazaki fragments (1). In contrast to CTG slip-outs, long CAG slip-outs were repaired efficiently, regardless of the presence or absence of MMR proteins (28). This ability to be repaired with fidelity reveals that genome maintenance can protect against changes in the length of CAG slip-outs.
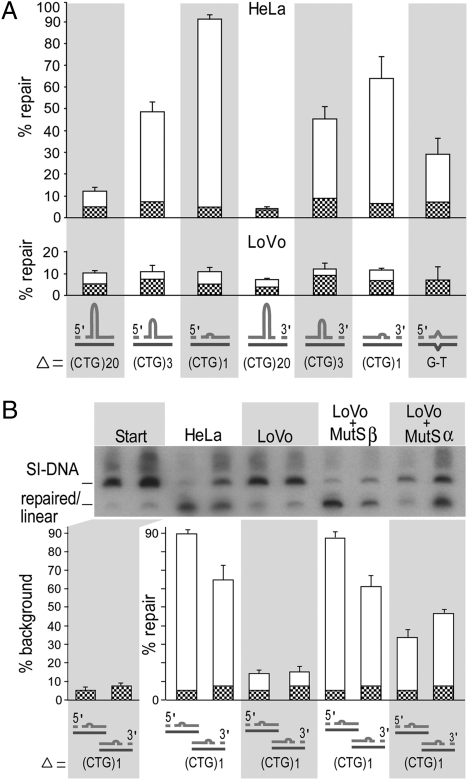
To determine how the size of a TNR slip-out affects repair, we made a series of circular slipped heteroduplex substrates with an excess of 1, 3, or 20 repeats with a nick located either 5′ or 3′ of the slip-out (SI Appendix, Fig. S1). As described in detail (28, 29), substrates were prepared by hybridizing single-stranded circular plasmids with their complementary linearized strand of differing repeat lengths, resulting in nicks located at the sites of linearization (SI Appendix, Fig S1). Substrates were identical except for a single slip-out that varied only in the number of excess repeats. In all substrates, the slip-out extruded at a unique point from a fully base-paired backbone of (CTG)n•(CAG)n, where n could equal 30, 47, 48, or 50 repeats. These substrates were processed in vitro by HeLa extracts and then were assessed for repair. Analysis of repair products by Southern blotting compared with starting material permitted quantitative assessment of repair efficiency at a molar level. Slip-out repair appeared to depend on slip-out size, so that repair efficiency for 1 repeat > 3 repeats > 20 repeats (Fig. 1A). As previously found for repair of SI-DNAs with 20 excess repeats, repair was nick-directed (using the continuous strand as the template for repair) and bidirectional, and 5′-nicked substrates were repaired more efficiently than 3′-nicked substrates (Fig. 1A: Compare substrates 1, 3, and 5 with substrates 2, 4, and 6). Interestingly, substrates with either one or three repeat slip-outs were repaired more efficiently than a G-T mismatch substrate under similar conditions (Fig. 1A). In contrast to the short slip-outs, the slipped-DNA with an excess of 20 CTG repeats escaped repair, yielding levels similar to background of the G-T mismatch incubated in the hMSH2-deficient LoVo extract (Fig. 1A: Compare upper and lower panels).
Fig. 1.
Repair depends on slip-out size and MMR. Circular hybrids with slipped (CTG)x•(CAG)y repeats (x and y are 30, 47, 48, or 50 repeats, and Δ = x – y = 20, 3, or 1 repeats) modeling intermediates of expansions with nicks in slipped-strand (SI Appendix, Fig. S1). (A) MMR-proficient (HeLa) and MMR-deficient (LoVo) extracts (90 μg) in 50-μL reactions, 22 fmol of circular substrate, were incubated for 30 min. Repair efficiencies were calculated for three to six replicates (Materials and Methods). G-T mismatch is an MMR-dependent control. Graph shows starting background (chequered bars) and repair (white bars). (B) (Upper) Southern blot analysis of repair of a single CTG slip-out, processed by 90-μg LoVo extracts with and without 250 ng recombinant MutSβ or MutSα. (Lower) Repair efficiencies were calculated from three to six replicates. Graph shows starting background (chequered bars) and repair (white bars).
Repair of Short Slip-Outs Requires hMutSβ Over hMutSα.
To determine whether MMR proteins are required in the repair of short slip-outs, we processed each of the CTG slipped-DNAs with hMSH2-null (LoVo) extracts, which are devoid of hMSH2, hMSH3, and hMSH6 proteins (32). LoVo extracts processed the long slip-out of 20 CTG repeats as poorly as did the repair-proficient HeLa extract, indicating that hMSH2, hMSH3, and hMSH6 are not required to stabilize or protect this long slip-out from being repaired (Fig. 1A: Compare upper and lower panels). In contrast, the shorter slip-outs were poorly repaired compared with their efficient processing by MMR-proficient HeLa extracts (Fig. 1A). The limited amount of product appearing as repaired for all slip-out substrates by LoVo extracts was similar to the nonspecific repair of a G-T mismatch by this MMR-deficient extract (Fig. 1A, Lower). These low-level repair products by LoVo extracts arise by random incorporation of radionucleotides into both strands (28, 29), implicating them as nonspecific repair. The observation that the short-CTG slip-outs could be repaired by HeLa but not by LoVo extracts suggests that hMutSα or hMutSβ MMR proteins are required in their processing.
To learn if the repair of short CTG slip-outs is dependent upon hMutSα or hMutSβ, repair assays were performed on the single-repeat slip-out using LoVo extracts supplemented with exogenous recombinant human hMutSα or hMutSβ proteins (Fig. 1B). Addition of hMutSβ to LoVo restored repair to levels similar to those seen with HeLa extracts. However, hMutSα was able to restore repair only partially. The ability of hMutSβ to repair single CTG slip-outs in the absence of hMutSα was confirmed by the efficient repair mediated by hMSH6-deficient HCT8 cell extracts (SI Appendix, Fig. S2), indicating that the repair of short slip-outs requires MutSβ rather than MutSα.
Over- or Underexpression of hMutSβ Diminishes Repair.
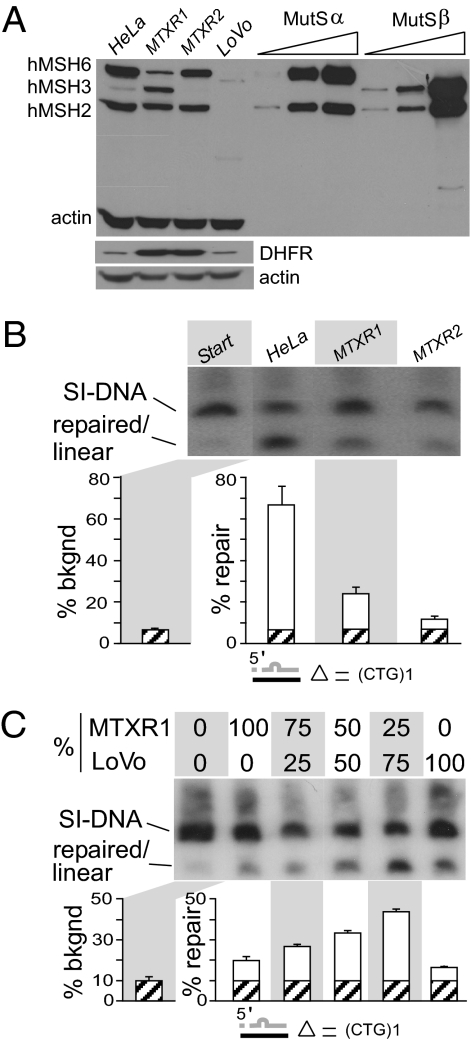
Because it has been suggested that MutSβ binding to CAG/CTG hairpins could misdirect DNA repair (16) and that the levels of MSH3 are rate limiting to CTG/CAG expansions (13), we tested the effect of overexpression of hMSH3 on slip-out repair. We obtained independent single-cell clones of methotrexate-resistant HeLa cells that had undergone >100-fold amplification of the divergently transcribed DHFR-hMSH3 genic region of chromosome 5 (33). In these HeLa variants, the relative levels of hMSH3 and hMSH6 proteins, but not of hMSH2, varied, thereby altering the molar ratios of hMutSα:hMutSβ. In HeLaMTXR1, hMSH3 protein levels are highly elevated (Fig. 2A and SI Appendix, Fig. S3). Levels of hMSH6, and thus hMutSα, are reduced in this line, presumably because the excess hMSH3 sequesters the pool of hMSH2 into hMutSβ, essentially destabilizing hMSH6 (34, 35). In HeLaMTXR2, the levels of hMSH3 are decreased, probably because the initial DNA breakpoint during DHFR gene amplification in this cell clone occurs within the hMSH3 gene, thereby disrupting it but not the DHFR gene (33). DHFR protein is highly amplified in HeLaMTXR1 and HeLaMTXR2. Levels of hMSH6 and presumably hMutSα are unaltered in HeLaMTXR2. Thus, the progenitor HeLa and its methotrexate-resistant derivatives have variable levels of hMSH3 and varied hMutSα:hMutSβ ratios, approximating MutSα >> MutSβ, MutSα < MutSβ, and MutSα >>> MutSβ in HeLa, HeLaMTXR1, and HeLaMTXR2 cells, respectively (Fig. 2A and SI Appendix, Fig. S3B). These attributes make these cells useful tools for studying the effects of varied molar ratios of hMutSα:hMutSβ on repair.
Fig. 2.
Short slip-out repair is sensitive to MutSβ concentration. (A) hMSH3, hMSH6, and hMSH2 proteins in cell extracts. In lanes 1–4, 50 μg of HeLa, HeLaMTXR1, HeLaMTXR2, or LoVo cell extracts, respectively, were separated by SDS/PAGE. In lanes 5–10, 15, 120, or 240 ng of purified hMutSα or 7, 30, or 455 ng of purified hMutSβ were loaded to demonstrate the similar immune responses of the proteins and the lower detection limits of the experiment. Western blotting was simultaneous for hMSH3, hMSH6, hMSH2, and actin (SI Appendix, Fig. S3). DHFR was blotted and probed separately. (B) (Upper) Southern blot analysis of repair of a circular hybrid with a single CTG slip-out, processed by HeLa, HeLaMTXR1, and HeLaMTXR2 with the starting material shown on the left. Autorad is from a single gel, identical exposure time, with intervening lanes excised for presentation. (Lower) Repair efficiencies of corresponding reactions were calculated on three to six replicates. Graph shows starting background (hatched bars) and repair (white bars). (C) (Upper) Southern blot analysis of repair of a circular hybrid with a single CTG slip-out, processed with mixtures of hMSH3-overexpressed HeLaMTXR1, and MMR-deficient LoVo extracts to dilute hMSH3 levels. (Lower) Repair efficiencies were calculated on three to six replicates. Graph shows starting background (hatched bars) and repair (white bars).
To test the effect of altering amounts of hMutSβ on slipped-DNA repair, we processed the single (CTG)1 slip-out by these HeLa variant extracts. At high levels of hMutSβ (HeLaMTXR1), repair of a single-repeat slip-out was reduced (Fig. 2B). It had been shown previously that increasing levels of hMSH3 decreases repair of base–base mismatches requiring hMutSα because of the degradation of hMSH6 (34, 35). However, the decreased repair of the short CTG slip-out probably is the result of high levels of hMSH3 rather than decreased levels of hMSH6, because the complete absence of hMSH6 had no effect on repair efficiency (Fig. 1B and SI Appendix, Fig. S2). Reduced levels of hMSH3, as in HeLaMTXR2 extracts, also diminished repair (Fig. 2B) similar to the complete absence of MMR proteins (LoVo, Fig. 1A). To test further the effect of varying levels of hMSH3 on short slip-out repair, the hMSH3-amplified HeLaMTXR1 extracts were diluted with LoVo extracts, which do not contain hMSH2, hMSH3, or hMSH6. With increasing dilutions (with hMutSβ approaching nonamplified levels), efficiency of repair increased toward levels found in the isogenic HeLa line (Fig. 2C). Further decreasing the levels of hMSH3, as in the HeLaMTXR2 extracts, led to diminished repair (Fig. 2B) and ultimately ablated repair in the absence of hMSH3, as in the LoVo extracts (Fig. 1A). Overexpression of hMSH3 in these cells did not alter CTG/CAG instability at endogenous loci harboring lengths shorter than the instability threshold, suggesting that the effect of hMutSβ may be length dependent (SI Appendix, Fig. S4). Together, these results suggest that the repair of short slip-outs requires hMutSβ rather than hMutSα; however, too much or too little hMutSβ can partially reduce repair.
Multiple Short Slip-Outs in S-DNA Are Not Repaired.
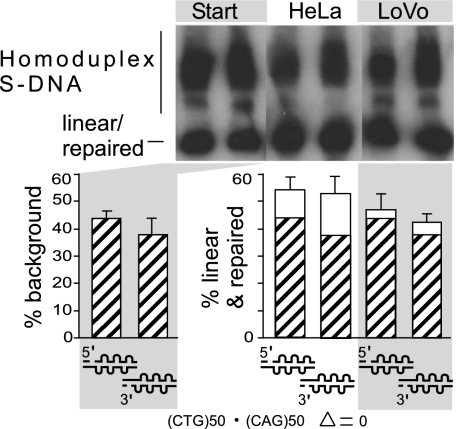
The results described above reveal that structural features of slipped-DNAs can influence repair outcome and the involvement of MMR repair proteins. It has been suggested that the density of A-A or T-T mismatches within CAG or CTG hairpins correlates directly with MutSβ ATPase inhibition (16). As described above, we investigated the effect of the length of an isolated single slip-out in an SI-DNA that extruded at a unique point from a Watson–Crick backbone of around (CTG)50•(CAG)50; longer slip-outs had increased numbers of intrastrand mismatches. Here we consider whether the density of slip-out numbers on a DNA molecule may affect repair. In contrast to the single slip-outs present in heteroduplex SI-DNAs, homoduplex S-DNAs form a series of isomers with numerous slip-outs along the repeat tract, most of which are 1 to 3 repeat units long (21,22–23, 25,26–27). To determine whether S-DNA is processed in the same manner as SI-DNA, we made circular S-DNA substrates with (CTG)50•(CAG)50 repeats, which have been characterized structurally by electrophoresis, chemical and enzymatic probing, and electron microscopy to have from 2 to as many as 62 short slip-outs per molecule distributed on both strands, with each slip-out comprising 1 to 3 repeat units (21,22–23, 25,26–27). The majority of molecules (70%) contained up to 26 slip-outs evenly distributed along the tract length between both strands (26). (Details are shown in SI Appendix, Fig. S5.) Slip-outs cannot branch migrate and once formed do not interconvert readily to fully duplexes, nor does the fully duplexed form convert readily to S-DNA (21). Circular nicked preparations of S-DNA harbored both slipped and fully duplexed DNAs, where the latter was background (Fig. 3, left lanes and hatched bars). S-DNA substrates were incubated with HeLa or LoVo extracts, and repair was assessed. Interestingly, the presence of multiple slip-outs on either strand greatly inhibited but did not completely ablate repair (Fig. 3, white bars). Only a small portion (16–26%) of the S-DNA could be repaired to the fully duplexed form by HeLa extracts (Fig. 3), regardless of nick location (SI Appendix, Fig. S6). The poor repair of S-DNAs does not represent a facile interconversion of S-DNA to and from the fully duplexed form (SI Appendix, Fig. S7). S-DNA repair was reduced further (6–9%) for the hMSH2-deficient LoVo extracts. The limited S-DNA repair and its reduction without MMR is similar to G-T repair and its reduction to background levels without MMR (28% to 7%, Fig 1A). Considering that short isolated CTG slip-outs are repaired efficiently in a MutSβ-dependent manner (approaching 90%; Fig. 1), and our previously observed MMR-independent repair of long CAG slip-outs (20 repeats) (28), the small portion of S-DNA repaired by the HeLa extracts probably represents those slipped isomers that have only a few slip-outs (large or small). The further reduction in repair of S-DNA mediated by the MMR-deficient LoVo extracts (Fig. 3) probably represents the inability to repair any substrates containing slip-outs with <3 repeat units, whereas those with a few large slip-outs may have been repaired independent of MMR. The ability to process S-DNAs was affected only mildly by varying the levels of hMSH3 (SI Appendix, Fig. S8), consistent with the small proportion of reparable S-DNAs. Taken together, repair efficiency depends on both the size of the slip-out, which determines the involvement of hMutSβ, and on the number of slip-outs per DNA molecule, which determines whether repair can proceed (Fig. 4A and SI Appendix, Fig. S9).
Fig. 3.
Multiple slip-outs interfere with repair. Circular hybrids with homoduplex slipped (CTG)50•(CAG)50 S-DNAs with a unique nick, copurified with fully duplexed forms (background) (SI Appendix, Fig. S1). (Upper) Southern blot analysis of S-DNA repair products using MMR-proficient HeLa and MMR-deficient LoVo extracts. Autorad is from a single gel, identical exposure time, with intervening lanes excised for presentation. (Lower) Repair efficiencies were calculated from three to six replicates. Graph shows starting background (hatched bars) and repair (white bars). See also Figs. S6 and S9 in SI Appendix.
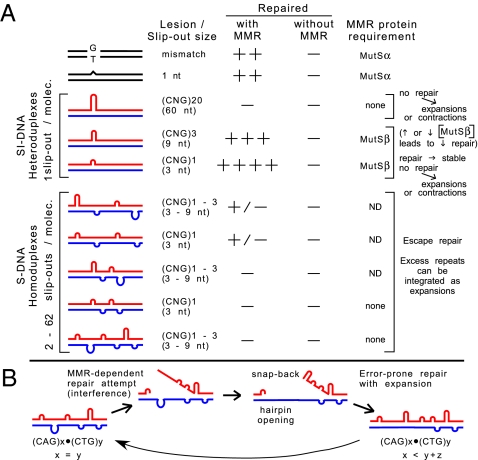
Fig. 4.
MMR and slip-out structure-dependent repair outcomes. (A) Summary of reparability data and MMR dependence of SI-DNA and S-DNA conformations. See also Fig. S9 in SI Appendix. (B) Proposed model (see text).
Discussion
Elucidating the role of MMR proteins in TNR expansions has been challenging (1–3). A widely cited model to explain how MSH2 and MSH3 generate expansions proposes that MutSβ binds to CAG/CTG slip-outs and protects them from repair. This hypothesis is based on the reduced ability of the human MutSβ complex to hydrolyze ATP when bound to a (CAG)13 or (CTG)13 hairpin (16). Therefore the protein complex would be unable to translocate along the helix and unable to signal or interact with downstream repair proteins, leaving the excess repeats unrepaired. However, this model is unlikely, because the ATPase activity of MutSβ generally is decreased when bound to ID DNA, which can be repaired efficiently (8, 16), and, more importantly, the ATPase activity of MSH2 is required to generate repeat expansions in CTG/CAG mice (17). Further support for a functional MMR system comes from the requirement for PMS2 in TNR expansions (14). Furthermore, MMR proteins are not required for either the escaped repair of long slip-outs of (CTG)20 or the efficient repair of (CAG)20 slip-outs (28, 29), an observation confirmed for (CAG)25 (30, 31). Data revealing how MMR is involved in TNR stability or instability have been lacking. The data presented here support processes through which MMR can both prevent and drive repeat expansions and contractions (Fig. 4).
The involvement of MMR depends on slip-out size and the number of slip-outs per molecule. Long, isolated slip-outs of (CTG)20 will escape repair, regardless of MMR, such that these slip-outs can be integrated as expansions or contractions (28). Consistent with this finding, Tian et al. (31) did not observe a significant change in repair of a long (CAG)25 slip-out in the presence of increased MutSβ. For isolated, short (CTG)1–3 slip-outs, repair efficiency is affected by the level of MutSβ. At typical hMutSα:hMutSβ ratios, MutSβ prevents instability by efficiently repairing short, isolated slip-outs. Perturbed levels of MutSβ decreased repair of short CTG slip-outs, allowing them to be integrated as expansions. One possibility is that too much MutSβ may bind and saturate the DNA, thereby impairing downstream events, whereas too little MutSβ is insufficient for the multiple protein complexes required to process a single heteroduplex (36). The sensitivity of short TNR slip-out repair to MutSβ concentration is similar to other reports of repair protein levels affecting repeat instability (29, 37).
Repair of CTG slip-outs depends on slip-out length: Whereas slip-outs of (CTG)20 escape repair, repair of (CTG)3 is highly efficient, and repair of (CTG)1 is most efficient. The efficiency of short slip-out repair exceeds that of a G-T mismatch (the best repaired of all base–base mismatches), indicating a robust protection against mutations by isolated CTG slip-outs. The role of MMR proteins depends on the length of the CTG slip-out: The escaped repair of a long slip-out does not require MMR proteins to protect it from repair by other repair pathways, and shorter slip-outs require MutSβ for repair. The effect of length on the reparability of CTG slip-outs is distinct from the efficient repair of nonrepetitive DNA loops: Whereas shortening the size of the CTG slip-out from 20 to 3 to 1 excess repeats increased repair, shortening nonrepetitive loops from 62 to 27 to 12 to 2 excess nucleotides decreased repair (5). However, in contrast to CTG slip-outs, the efficiency of the repair of long (CAG)20 loops was similar to that of long nonrepetitive loops: Neither required MMR (5, 28). Also, the preferential repair of short CTG slip-outs by MutSβ rather than MutSα is similar to the ability of these complexes to repair small heterologies of nonrepetitive sequences of two to eight nucleotides (6). The repair of short CTG slip-outs and short nonrepetitive loops was nick-directed and bidirectional, indicating that their repair occurs by the MMR pathway. The greater effect of MutSβ on repair of short CTG slip-outs parallels the greater requirement of MSH3 and MSH2 for TNR instability in transgenic mice, relative to the limited requirement of MSH6 (12, 13, 15, 17).
In contrast to the highly efficient repair of isolated short slip-outs, clusters of multiple short slip-outs along a single repeat tract almost escaped repair. The poor repair of clustered slip-outs in S-DNAs was unaffected by the presence of MutSβ, even at high levels (SI Appendix, Fig. S8). We suggest that closely spaced adjacent short slip-outs interfere with each other's repair, possibly serving as DNA structural blockades. Interference has been observed in the repair of adjacent G-T and U-G mispairs (38) and also in palindromic hairpins with adjacent G-T mispairs by human cell extracts (39–41). Notably, the degree of interference increased with diminished distance between the DNA lesions. In bacteria, when two adjacent base–base mismatches are separated by 1 to 3 base pairs, repair of both mismatches can be depressed (42). Similarly, repair was severely inhibited when nonrepetitive 24–32 nucleotide loop heterologies were placed 33 or 46 base pairs from a base–base mismatch, but repair was efficient when spacing was increased to 325 base pairs (41). MMR involves excision tracts that can extend to hundreds of nucleotides (4), possibly allowing efficient corepair of a nonrepairable mismatch or ID loop when separated from another mismatch by 107 or 1,448 base pairs (43). Together, these results reveal proximity-sensitive repair interference between adjacent DNA lesions. Although proximal mispaired lesions are probably rare, high density clustered DNA damage is incurred by ionizing radiation (44, 45). Repair of radiation-induced clustered DNA damage is severely impaired, primarily because of the proximity of the lesions (45). Our results now extend interference with the repair of clustered DNA lesions to neurodegenerative disease-causing mutations in which the clustered lesions are multiple slip-outs on an expanded CTG/CAG tract.
We suggest that the poor repair of S-DNAs is caused by proximal adjacent slip-outs interfering with repair when initiation of repair at one lesion may block the repair of adjacent slip-outs. S-DNAs comprise a mixture of slipped isomers, each containing 2–62 short slip-outs of 1 to 3 repeat units each, extruding at various locations along the tract of (CTG)50•(CAG)50 (21,22–23, 25,26–27). Most molecules (70%) contained up to 26 slip-outs evenly distributed along the tract length between both strands (26). The inability of slip-outs to branch migrate prevents their translocation along the helix away from other slip-outs and thus may act as an immovable block to repair. It is possible that the very limited number of S-DNAs that can be repaired by the MMR-proficient extract represents a subset of slipped isomers in which the adjacent slip-outs are spaced sufficiently far from each other to permit repair (Fig. 3). The further reduced repair of S-DNAs by MMR-deficient extracts might result from an inability to repair the same subset of slipped-isomers, because repair of short slip-outs requires MutSβ. One might expect that in S-DNA the high level of slip-outs creates competition for the repair factors and thus overwhelms the repair machinery. However, we do not favor this interpretation, because the hMSH3-amplified HeLaMTXR1 cell extract did not increase S-DNA repair (SI Appendix, Fig. S8). This result supports the notion that either the DNA lesion or the lesion bound by a protein complex may be a barrier to the efficient signaling of repair, which seems to occur by tracking along the DNA helix (46).
The poor repair of S-DNAs suggests that slipped-DNAs may be retained in cells over long periods, and attempts to repair them may be error prone and lead to expansions (Fig. 4B). We speculate that some attempted repair events on slipped homoduplexes may be arrested, allowing strand displacement, intrastrand slippage, reannealing by further out-of-register mispairing, and leaving a gap that, when filled, results in increased repeats in that strand, essentially producing heteroduplexes with an excess of repeats on one or both strands (Fig. 4B). Reiterations of such events, in the absence of proper repair, would lead to instability (Fig. 4B). Such attempted but error-prone repair, coupled with the dependence of MMR on slip-out size, is consistent with the requirement for MutSβ in TNR expansions in mice (9,10,11,12–13, 15). Interference by adjacent slip-outs in the repair of oxidative DNA damage may have similar effects, explaining the requirement for 8-oxoguanine-DNA glycosylase 1 in CTG/CAG expansions (47).
It is unclear whether TNR expansions occur by multiple stepwise increases of short slip-outs or saltatory jumps of larger slip-outs. The observations that slip-outs of one to three excess repeats require MutSβ for repair and that perturbation of MutSβ levels can reduce repair suggest that the increment of change during a single mutagenic step would be one to three excess repeats. Limited evidence from patient tissues supports incremental changes of 1 to 3 CTG/CAG units per mutation event (48–51), similar to that occurring at other simple repeats such as (CA)n and (A)n (52, 53).
The results presented here support roles for MMR both in preventing and in driving TNR repeat instability. Path selection depends on structural features of slipped-DNAs. Long CTG slip-outs and clustered slip-outs escape repair and can be integrated as expansions, whereas the repair of isolated short slip-outs depends on the presence and concentration of MutSβ.
Materials and Methods
Slipped-DNA Substrates.
Substrates containing pure (CTG)n•(CAG)n repeats, in which n = 30, 47, 48, or 50, and flanking human DM1 sequences were prepared as described (28, 29). G-T reagents were gifts of P. Modrich (Duke University, Durham, NC).
Cells and Extracts.
LoVo (ATCC), HeLa (ATCC), and its clonal methotrexate-resistant variants HeLaMTXR1 and HeLaMTXR2 (33) (gifts of J. L. Hamlin and L. D. Mesner, University of Virginia School of Medicine, Charlottesville, VA) were grown and extracts were prepared as described (28, 29, 33). Only preparations functional in SV40 in vitro replication were used. Western blots were performed by simultaneous blotting (SI Appendix, Fig. S3). Antibody to DHFR was a gift of J. R. Bertino (University of Medicine & Dentistry of New Jersey).
Recombinant hMutSα and hMutSβ Expression.
Baculoviruses expressing his-tagged hMSH2, hMSH3, and hMSH6 were gifts of Guo-Min Li (University of Kentucky, Lexington, KY). Expression and purification were performed as outlined (27, 31, 36, 54) (SI Appendix, Fig. S10).
Repair Reactions and Efficiencies.
Slipped-strand and G-T repair reactions were conducted and efficiencies determined as outlined (refs. 28 and 29 and SI Appendix).
Supplementary Material
Acknowledgments
We thank P. Mahajan (of the Gileadi laboratory) for help in purifying proteins. The Pearson laboratory is supported by the Muscular Dystrophy Association, the Canadian Institutes of Health Research, the Paul Wellstone Muscular Dystrophy Cooperative Research Center, and by Grant U54NS48843 from the National Institutes of Health. M.M.S. was supported by studentships from The Hospital for Sick Children Research Training Competition and the Canadian Institutes of Health Research Collaborative Graduate Training Program in Molecular Medicine.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0909087107/-/DCSupplemental.
References
- 1.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: Mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 2.López Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 3.Slean MM, Panigrahi GB, Ranum LP, Pearson CE. Mutagenic roles of DNA “repair” proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA Repair (Amst) 2008;7:1135–1154. doi: 10.1016/j.dnarep.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 5.Littman SJ, Fang WH, Modrich P. Repair of large insertion/deletion heterologies in human nuclear extracts is directed by a 5′ single-strand break and is independent of the mismatch repair system. J Biol Chem. 1999;274:7474–7481. doi: 10.1074/jbc.274.11.7474. [DOI] [PubMed] [Google Scholar]
- 6.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 7.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian L, Gu L, Li GM. Distinct nucleotide binding/hydrolysis properties and molar ratio of MutSalpha and MutSbeta determine their differential mismatch binding activities. J Biol Chem. 2009;284:11557–11562. doi: 10.1074/jbc.M900908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler VC, et al. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 10.Savouret C, et al. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol Cell Biol. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 12.van den Broek WJ, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 13.Foiry L, et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 14.Gomes-Pereira M, Fortune MT, Ingram L, McAbney JP, Monckton DG. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: Implications for the mechanism of triplet repeat expansion. Hum Mol Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 15.Savouret C, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owen BA, et al. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 17.Tomé S, et al. MSH2 ATPase domain mutation affects CTG•CAG repeat instability in transgenic mice. PLoS Genet. 2009;5:e1000482. doi: 10.1371/journal.pgen.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCulloch SD, Gu L, Li GM. Nick-dependent and -independent processing of large DNA loops in human cells. J Biol Chem. 2003;278:50803–50809. doi: 10.1074/jbc.M309025200. [DOI] [PubMed] [Google Scholar]
- 19.Corrette-Bennett SE, et al. Efficient repair of large DNA loops in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:4134–4143. doi: 10.1093/nar/29.20.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umar A, Boyer JC, Kunkel TA. DNA loop repair by human cell extracts. Science. 1994;266:814–816. doi: 10.1126/science.7973637. [DOI] [PubMed] [Google Scholar]
- 21.Pearson CE, Sinden RR. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 22.Tam M, et al. Slipped (CTG)•(CAG) repeats of the myotonic dystrophy locus: Surface probing with anti-DNA antibodies. J Mol Biol. 2003;332:585–600. doi: 10.1016/s0022-2836(03)00880-5. [DOI] [PubMed] [Google Scholar]
- 23.Pearson CE, et al. Slipped-strand DNAs formed by long (CAG)•(CTG) repeats: Slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng M, Huang X, Smith GK, Yang X, Gao X. Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding. An NMR and UV spectroscopic study. J Mol Biol. 1996;264:323–336. doi: 10.1006/jmbi.1996.0643. [DOI] [PubMed] [Google Scholar]
- 25.Pearson CE, et al. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- 26.Pearson CE, Wang YH, Griffith JD, Sinden RR. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n•(CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson CE, Ewel A, Acharya S, Fishel RA, Sinden RR. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum Mol Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 28.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)•(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12:635–637. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 29.López Castel A, Tomkinson AE, Pearson CE. CTG/CAG repeat instability is modulated by the levels of human DNA ligase I and its interaction with proliferating cell nuclear antigen: A distinction between replication and slipped-DNA repair. J Biol Chem. 2009;284:26631–26645. doi: 10.1074/jbc.M109.034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou C, Chan NL, Gu L, Li GM. Incision-dependent and error-free repair of (CAG)(n)/(CTG)(n) hairpins in human cell extracts. Nat Struct Mol Biol. 2009;16:869–875. doi: 10.1038/nsmb.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian L, et al. Mismatch recognition protein MutSbeta does not hijack (CAG)n hairpin repair in vitro. J Biol Chem. 2009;284:20452–20456. doi: 10.1074/jbc.C109.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang DK, Ricciardiello L, Goel A, Chang CL, Boland CR. Steady-state regulation of the human DNA mismatch repair system. J Biol Chem. 2000;275:18424–18431. doi: 10.1074/jbc.M001140200. [DOI] [PubMed] [Google Scholar]
- 33.Singer MJ, Mesner LD, Friedman CL, Trask BJ, Hamlin JL. Amplification of the human dihydrofolate reductase gene via double minutes is initiated by chromosome breaks. Proc Natl Acad Sci USA. 2000;97:7921–7926. doi: 10.1073/pnas.130194897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci USA. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marra G, et al. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci USA. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 37.Goula AV, et al. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington's disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schanz S, Castor D, Fischer F, Jiricny J. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc Natl Acad Sci USA. 2009;106:5593–5598. doi: 10.1073/pnas.0901726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Hays JB. Mismatch repair in human nuclear extracts: Effects of internal DNA-hairpin structures between mismatches and excision-initiation nicks on mismatch correction and mismatch-provoked excision. J Biol Chem. 2003;278:28686–28693. doi: 10.1074/jbc.M302844200. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Hays JB. Signaling from DNA mispairs to mismatch-repair excision sites despite intervening blockades. EMBO J. 2004;23:2126–2133. doi: 10.1038/sj.emboj.7600153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang YM, et al. Interaction of nick-directed DNA mismatch repair and loop repair in human cells. J Biol Chem. 2004;279:30228–30235. doi: 10.1074/jbc.M401675200. [DOI] [PubMed] [Google Scholar]
- 42.Gasc AM, Garcia P, Sicard M. Inhibition of DNA repair by neighbouring mismatched bases in Streptococcus pneumoniae. J Gen Microbiol. 1988;134:3019–3024. doi: 10.1099/00221287-134-11-3019. [DOI] [PubMed] [Google Scholar]
- 43.Carraway M, Marinus MG. Repair of heteroduplex DNA molecules with multibase loops in Escherichia coli. J Bacteriol. 1993;175:3972–3980. doi: 10.1128/jb.175.13.3972-3980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodhead DT, Thacker J, Cox R. Weiss Lecture. Effects of radiations of different qualities on cells: Molecular mechanisms of damage and repair. Int J Radiat Biol. 1993;63:543–556. doi: 10.1080/09553009314450721. [DOI] [PubMed] [Google Scholar]
- 45.Shikazono N, Noguchi M, Fujii K, Urushibara A, Yokoya A. The yield, processing, and biological consequences of clustered DNA damage induced by ionizing radiation. J Radiat Res (Tokyo) 2009;50:27–36. doi: 10.1269/jrr.08086. [DOI] [PubMed] [Google Scholar]
- 46.Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proc Natl Acad Sci USA. 2007;104:12709–12713. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, Lau R, Marcadier JL, Chitayat D, Pearson CE. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am J Hum Genet. 2003;73:1092–1105. doi: 10.1086/379523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe H, et al. Differential somatic CAG repeat instability in variable brain cell lineage in dentatorubral pallidoluysian atrophy (DRPLA): A laser-captured microdissection (LCM)-based analysis. Hum Genet. 2000;107:452–457. doi: 10.1007/s004390000400. [DOI] [PubMed] [Google Scholar]
- 50.Hashida H, et al. Single cell analysis of CAG repeat in brains of dentatorubral-pallidoluysian atrophy (DRPLA) J Neurol Sci. 2001;190:87–93. doi: 10.1016/s0022-510x(01)00596-2. [DOI] [PubMed] [Google Scholar]
- 51.Shelbourne PF, et al. US-Venezuela Collaborative Research Group Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Genet. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- 52.Weber JL. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990;7:524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- 53.Blake C, Tsao JL, Wu A, Shibata D. Stepwise deletions of polyA sequences in mismatch repair-deficient colorectal cancers. Am J Pathol. 2001;158:1867–1870. doi: 10.1016/S0002-9440(10)64143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson T, Guerrette S, Fishel R. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J Biol Chem. 1999;274:21659–21664. doi: 10.1074/jbc.274.31.21659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.