Abstract
Retroviral vectors offer benefits of efficient delivery and stable gene expression; however, their clinical use raises the concerns of insertional mutagenesis and potential oncogenesis due to genomic integration preferences in transcriptional start sites (TSS). We have shifted the integration preferences of retroviral vectors by generating a library of viral variants with a DNA-binding domain inserted at random positions throughout murine leukemia virus Gag-Pol, then selecting for variants that are viable and exhibit altered integration properties. We found seven permissive zinc finger domain (ZFD) insertion sites throughout Gag-Pol, including within p12, reverse transcriptase, and integrase. Comprehensive genome integration analysis showed that several ZFD insertions yielded retroviral vector variants with shifted integration patterns that did not favor TSS. Furthermore, integration site analysis revealed selective integration for numerous mutants. For example, two retroviral variants with a given ZFD at appropriate positions in Gag-Pol strikingly integrated primarily into four common sites out of 3.1 × 109 possible human genome locations (P = 4.6 × 10-29). Our findings demonstrate that insertion of DNA-binding motifs into multiple locations in Gag-Pol can make considerable progress toward engineering safer retroviral vectors that integrate into a significantly narrowed pool of sites on human genome and overcome the preference for TSS.
Keywords: gene therapy, retroviral integration, random insertion retroviral library
The promising therapeutic success in gene therapy clinical trials for X-linked severe combined immunodeficiency has unfortunately been accompanied by the incidence of leukemia-like clonal T cell expansion in several patients, primarily arising from activation of the LMO2 prooncogene due to nearby retroviral vector integrations (1–3). While immune system function was fully rescued in the unaffected patients, the well-established preference for murine leukemia virus (MLV) integration at the start sites of transcribed regions, with the associated potential genotoxicity (2, 4, 5), represents a general risk that can offset key advantages of using these retroviruses as vectors. An alternative, lentiviruses, preferentially integrates throughout transcriptional units, rather than being concentrated near start sites (6, 7). Lentiviral infections could thus also potentially contribute to oncogenesis, though there has been no experimental evidence of this possibility to date. Various studies have suggested that viral components in the preintegration complex (PIC) in conjunction with host factors, which likely tether the complex to specific chromatin features within the host nucleus, determine retroviral and lentiviral integration patterns (8–10); however, the associated mechanisms are incompletely understood.
There have been several efforts to redirect retroviral integration via fusing sequence-specific DNA-binding domains—including the Sp1 zinc finger domain (ZFD), the DNA-binding domain (DBD) of λ phage repressor, and an engineered ZFD—to the C or N terminus of retroviral integrase (11–14), a critical determinant of integration patterns. The resulting integration behavior was monitored in vitro (11, 12) or in vivo (13, 14) using agarose-gel-based and PCR-based assays. However, likely due to the need to coincorporate wild-type Gag-Pol polyprotein to compensate for viral infectivity completely deprived by the engineered integrase fusions, as well as potential off-target binding of DNA-binding motifs, only modest increases in integration at the intended target site were observed.
In this study we attempted to develop safer retroviral vector systems with high infectivity that do not favor transcriptional start sites (TSS) for integration via inserting an engineered DBD into numerous permissive locations identified in MLV Gag-Pol. Given the incomplete knowledge of the composition of the PIC, and the regions within Gag-Pol that steer integration directly or by association with host factors, the optimal insertion sites for an exogenous DBD to direct integration and/or disrupt viral domains that contribute to wild-type integration preferences is not clear. Accordingly, in this study we have applied a high-throughput protein engineering approach by generating a library of viruses with DBDs inserted into random locations throughout Gag and Pol, without coincorporation of wild-type Gag-Pol polyprotein, and selecting for variants that are viable and avoid integration into TSS. Engineered zinc finger domains (ZFDs) were chosen as the DBD for the modular binding properties of their zinc finger subunits, which enables the engineering of ZFDs with selectivity for a number of DNA sequences (15–17), as well as for their considerable albeit imperfect selectivity for such target sequences (18, 19). Our genome-wide analysis indicates that when inserted into key regions of Gag-Pol, such DBDs can override the intrinsic properties of MLV vectors to shift integration patterns toward safer regions of the genome that lack TSS.
Results and Discussion
Library Construction and Selection Results in Numerous Viable MLV Variants with ZFD Insertions in Gag and Pol.
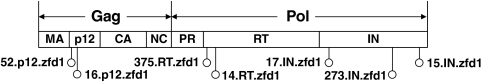
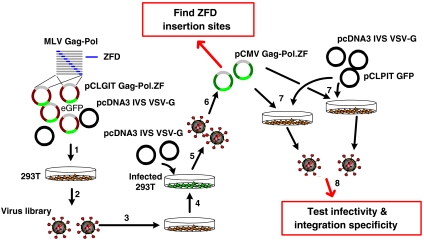
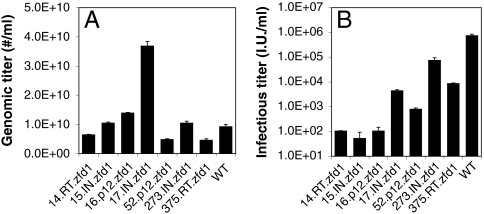
We first constructed a large (4.3 × 105) retroviral library where a 186 amino acid polydactyl zinc finger domain ZFD1—a six zinc finger domain previously designed to recognize an 18-bp sequence (each finger binds a 3-bp sequence) that appears proximal to the γ-globin locus in the human genome (15)—was randomly inserted through the use of a transposon system (20) into likely every position of the MLV Gag and Pol proteins (Figs. 1 and 2 and Fig. S1A). ZFD sequence optimization and a low copy number plasmid were required to avoid plasmid recombination issues. We then selected the library for Gag-Pol.ZF mutant clones that could package in the absence of wild-type Gag-Pol coexpression (Fig. 2). From the library selection and infectivity analysis, we isolated seven viable clones with in-frame ZFD1 insertions (Table 1 and Fig. 1), a relatively surprising number given the substantial size of the polydactyl ZFD (Fig. S2). Specifically, the ZFD was inserted within p12 for two clones (the 16th and 52th sequenced clones, 16.p12.zfd1 and 52.p12.zfd1), within reverse transcriptase (RT) for two variants (14.RT.zfd1 and 375.RT.zfd1), and within integrase (IN) for three clones (15.IN.zfd1, 17.IN.zfd1, and 273.IN.zfd1). Mutant vector genomic titers were equivalent to that of wild-type vector, despite the large ZFD insertions (Fig. 3A), suggesting that key sites within the viral capsid can tolerate large insertions into both Gag and Pol proteins without a severe reduction in assembly efficiency. However, the clones 14.RT.zfd1, 15.IN.zfd1, and 16.p12.zfd1 showed significantly reduced infectious titers, though 17.IN.zfd1, 273.IN.zfd1, and 375.RT.zfd1 exhibited reduced but substantial infectious titers varying from 0.6 to 10% of the wild-type titer (Fig. 3B). These reduced titers could conceivably be offset by the major benefit of integration patterns shifted away from TSS, and further protein engineering could be conducted to enhance titer if necessary.
Fig. 1.
Organization of MLV gag-pol gene. The random insertion library size was 4.3 × 105, estimated by colony counting after transformation of electrocompetent E. coli. Based on the 5,214 nucleotide positions in MLV gag-pol, the library likely covered all possible insertion sites. The ZFD insertion sites of viable clones are indicated by open circles and labels. In general, the name of each clone includes the clone number, the viral protein into which the ZFD is inserted, and the identity of the ZFD. MA, matrix; CA, capsid; NC, nucleocapsid; PR, protease; RT, reverse transcriptase; IN, integrase.
Fig. 2.
Schematic of library selection and clonal analysis. (i) Packaging of library viruses via transfection of plasmid library and VSV-G helper plasmid into 293T cells. (ii) Harvest and purification of viruses. (iii) Infection of 293T cells with virion library at a multiplicity of infection (moi) of < 0.1. (iv) Sorting and subsequent expansion of GFP-positive cells. (v) Rescue of packageable mutants by 293T cell transfection with helper plasmid encoding the VSV-G envelope protein. (vi) Subcloning Gag-Pol.ZF cDNA from packageable mutants into pCMV Gag-Pol helper plasmid. (vii) Packaging virus clones via transfection of 293T cells with pCLPIT GFP and helper plasmids. (viii) Analysis of infectivity and genome-wide integration analysis.
Table 1.
Permissive ZFD insertion sites within MLV Gag and Pol proteins
| Clones | Sequence near inserted zinc fingers | Region of insertion |
| 16.p12.zfd1 | VLSD18APZF_domainAGPHSDSGGP | p12 |
| 16.p12.zfd2 | VLSD18SAZF_domainASSDSGGP | p12 |
| 16.p12.zfd3 | VLSD18SAZF_domainASSDSGGP | p12 |
| 52.p12.zfd1 | TPSL7VRPZF_domainAGPHLGAKP | p12 |
| 52.p12.zfd2 | TPSL7VSAZF_domainASLGAKP | p12 |
| 52.p12.zfd3 | TPSL7VSAZF_domainASLGAKP | p12 |
| 14.RT.zfd1 | EARL71RPZF_domainAGPHRLGIKP | RT |
| 14.RT.zfd2 | EARL71SAZF_domainASLGIKP | RT |
| 14.RT.zfd3 | EARL71SAZF_domainASLGIKP | RT |
| 375.RT.zfd1 | DVSL20VRPZF_domainAGPHLGSTW | RT |
| 375.RT.zfd2 | DVSL20VSAZF_domainASLGSTW | RT |
| 375.RT.zfd3 | DVSL20VSAZF_domainASLGSTW | RT |
| 15.IN.zfd1 | HVKA376VRPZF_domainAGPQAADPG | IN |
| 15.IN.zfd2 | HVKA376VSAZF_domainASGADPG | IN |
| 15.IN.zfd3 | HVKA376VSAZF_domainASGADPG | IN |
| 17.IN.zfd1 | KYWV36RPZF_domainAGPHWVYQGK | IN |
| 17.IN.zfd2 | KYWV36SAZF_domainASVYQGK | IN |
| 17.IN.zfd3 | KYWV36SAZF_domainASVYQGK | IN |
| 273.IN.zfd1 | GAPP272LRPZF_domainAGPHPPLVN | IN |
| 273.IN.zfd2 | GAPP272LSAZF_domainASPPLVN | IN |
| 273.IN.zfd3 | GAPP272LSAZF_domainASPPLVN | IN |
The clone number is followed by the viral protein into which the ZFD was inserted, and the identity of the ZFD. The notations zfd1, zfd2, and zfd3 indicate clones containing a ZFD originally engineered to bind to a 18-bp stretch that appears in the promoter regions of γ-globin, PEDF, and CHK2 genes, respectively (15–17). The positions of amino acid residues flanking the ZFD insertion are also indicated, with the residues of wild-type MLV Gag-Pol proteins, and amino acids additionally introduced via the transposon reaction and cloning steps (underlined).
Fig. 3.
Genomic (A) and infectious (B) titers of wild-type virus and ZFD insertion mutants. (A) A standard retroviral vector that expresses the tetracycline-OFF transcription factor, and drives GFP expression from a tetracycline-responsive promoter (Fig. S1A), was packaged using pCMV Gag-Pol helper plasmids containing wild-type and ZFD variant gag-pol genes. Vector genomic titers and standard errors of the mean are shown. (B) Infectious titers of viruses were measured via duplicate infections of 293T cells, and standard deviations of infectious titer are shown by bars.
Structural Context for Permissive ZFD Insertion Sites.
The exact protein stoichiometry within retroviral particles is not known, but approximately 2,000 copies of each Gag protein (and considerably fewer of each Pol protein) are known to be present in a virion (9), such that the ZFDs in some cases are present in high copy numbers. For example, the ZFD insertions for 16.p12.zfd1 and 52.p12.zfd1—within the early region of p12 (starting at amino acid positions 19 and 8, respectively, Table 1) but distant from the PPPY motif (residues 31–34) that is critical for efficient virus release from the cells (21)—would interestingly be present at very high copy numbers per virion. Because p12 is composed only of random coils (22), this unstructured protein may have sufficient flexibility to accommodate large protein domains such as the ZFD. Based on the roles of p12 in integration (22), the ZFD-p12 fusion presumably remains associated with the PIC and is thus ultimately in a position to affect viral integration patterns. The 14.RT.zfd1 and 375.RT.zfd1 clones harbored insertions within the structured αC chain of the RT fingers domain and within the unstructured region adjacent to the start of the short αA chain of the palm domain (Fig. S3) (23), respectively. Both the finger and palm domains are known to be critical for reverse transcription via their interactions with the primer-template duplex (9), but the residues adjacent to the insertions are not involved in direct DNA contacts (Fig. S3) (24).
Three clones—15.IN.zfd1, 17.IN.zfd1, and 273.IN.zfd1—contained the ZFD next to the second putative α helix near the C terminus of IN, within the first putative β-sheet of the N terminus of IN, and within the sixth α-helix of the catalytic core domain, respectively (25). All three insertions were near IN regions that are relatively variable among eight different MLV strains, while no insertions were observed within the three most conserved domains (residues 254–270, 346–362, and 368–375) (25). In addition, 17.IN.zfd1 harbored the ZFD 19 amino acids upstream of the HHCC domain that binds to a zinc ion, which stabilizes the N-terminal domain and is important for IN multimerization. The functional form of IN has been suggested to be a multimer (perhaps a tetramer or even higher order) (9), and it is thus surprising that the ZFD insertion near the HHCC domain still permitted packaging of infectious particles. Finally, the C-terminal region of IN is known to have nonspecific DNA-binding properties (9), and the incorporation of targeting ZFD into the region may thus modulate the integration specificity.
ZFD Insertion Variant Did Not Favor Transcriptional Start Sites for Integration.
We next adapted a high-throughput method to analyze genomic integration patterns of wild-type and mutant MLV vectors, originally developed for analysis of HIV-1 integration (SI Text). This method, whose results were previously validated by statistical comparison to conventional integration analysis (26), uses the type IIS restriction enzyme Mme I to generate short fragments containing virus-host genome junctions to enable efficient analysis of viral integration patterns. We analyzed a total of 809 sequenced virus-host genome junctions for wild-type and mutant vectors in this study (Table S1). Consistent with prior reports (5), MLV vector with wild-type Gag-Pol exhibited a strong preference in the human embryonic kidney 293 cell genome for TSS and CpG islands that are known to be enriched in promoters (27) (P = 1.392 × 10-49 and P = 4.926 × 10-151 compared to the hypothetical case of random integration, respectively) (Table 2). In stark contrast, none of the 76 sequenced junctions for a representative clone 17.IN.zfd1 (Table S1) was located within 5 kb of a TSS (Table 2), a statistically significant shift from wild-type MLV (P = 0.0214). This mutant also showed integration frequencies in CpG islands that were intermediate between wild-type virus and random integration, but were only marginally statistically distinct from either (P = 0.0817 and P = 0.0606 for comparison to wild-type and random, respectively) (Table 2). In addition, while vector with wild-type Gag-Pol disfavored repeat elements for integration (25.2% vs. 44.6% for hypothetical random integration), the mutant did not disfavor these elements (50.0%) (Table 2). For several other ZFD1 mutants, low numbers of sequenced virus-host genome junctions (Table S1) precluded statistical analysis.
Table 2.
Global integration patterns of wild-type MLV vector and mutants with ZFD insertion
| Genomic feature | WT (n = 107) | 17.IN.zfd1 (n = 10) | 273.IN.zfd2 (n = 22) | 273.IN.zfd3 (n = 9) | 375.RT.zfd2 (n = 14) | Hypothetical random integration |
| Within genes | 39.3% (0.0843) | 10.0% (0.1433/0.0578) | 54.6% (0.2000/0.1432) | 44.4% (0.4032/0.7520) | 50.0% (0.1362/0.4124) | 22.4%–34.3% (31.5%) |
| Within ±5 kb of the transcriptional start | 34.6% (1.392 × 10-49) | 0.0% (0.4874/0.0214) | 9.1% (0.3146/0.0120) | 11.1% (0.3511/0.1385) | 7.1% (0.6497/0.0308) | 3.0%–5.9% (4.6%) |
| Inside or within 1 kb of CpG | 36.5% (4.926 × 10-151) | 10.0% (0.0606/0.0817) | 0.0% (0.5139/0.0004) | 11.1% (0.0430/0.1136) | 0.0% (0.6026/0.0046) | 1.0%–2.6% (1.9%) |
| LINE | 10.3% (0.0127) | 10.0% (0.4330/0.9751) | 4.6% (0.0713/0.3745) | 11.1% (0.5090/0.9362) | 28.6% (0.4164/0.0245) | 19.7%–20.0% (19.9%) |
| SINE | 11.2% (0.5441) | 0.0% (0.2175/0.2614) | 0.0% (0.0674/0.0958) | 0.0% (0.2420/0.2867) | 21.4% (0.3630/0.2249) | 12.9%–13.5% (13.2%) |
| DNA | 0.9% (0.2104) | 20.0% (0.0016/1.600 × 10-10) | 13.6% (0.0034/2.5232 × 10-10) | 0.0% (0.5978/0.7750) | 0.0% (0.5105/0.7214) | 2.8%–3.2% (3.0%) |
| LTR | 2.8% (0.0325) | 20.0% (0.1985/9.773 × 10-4) | 4.6% (0.4976/0.6197) | 33.3% (0.0081/2.8172 × 10-8) | 7.1% (0.8458/0.3246) | 8.4%–8.8% (8.6%) |
| Total repeats (LINE + SINE + DNA + LTR) | 25.2% (5.575 × 10-5) | 50.0% (0.7312/0.0709) | 22.7% (0.0390/0.7894) | 44.4% (0.9925/0.1836) | 57.1% (0.3451/0.0059) | 44.6% |
The frequencies (%) of integrations within featured genomic regions are shown. The relevant P values (chi-square test), calculated by comparison to the data of hypothetical random integration, are shown in parentheses. Additionally, for the mutants the P values (chi-square test) obtained by comparison of the mutant and wild-type integration patterns are shown after a slash (/) in parentheses. The ranges of frequencies for hypothetical random integration, shown in the right-most column, are based on prior work (5, 8, 28–30), and the averages of these reported frequencies are shown in parentheses. For wild-type MLV Gag-Pol, 17.IN.zfd1, 273.IN.zfd2, 273.IN.zfd3, and 375.RT.zfd2, 294, 76, 197, 101, and 93 virus-host genome junctions were sequenced (Table S1), respectively, and only the junctions that matched a single location on the human genome are considered in this table (n in the top row indicates the number of unique junctions identified and statistically considered for each variant). Multiple junctions with the same sequence were counted once for the analysis in this table, and junctions that matched multiple locations on the human genome were not considered for this analysis. RefSeq genes (http://www.ncbi.nlm.nih.gov/RefSeq/) were considered in our analysis. LINE and SINE are long and short interspersed nuclear elements, respectively. DNA and LTR are DNA transposon and long terminal repeat retrotransposon elements, respectively.
Overall, these data indicate that a DBD insertion into a key location of Gag-Pol can significantly shift the overall integration patterns of MLV toward potentially safer regions where TSS are relatively rare. That said, the identified integration locations for the mutant were not near the two addresses (5,271,202 and 5,276,126 on chromosome 11 of human genome) that the ZFD1 was originally designed to target (15), which may be consistent with the potential for off-target binding of ZFDs (18, 19). Regardless, the leukemia cases observed in the X-linked severe combined immunodeficiency trial were attributed to retroviral integrations 3 kb upstream and 2 kb downstream of the LMO2 prooncogene (2); therefore, a significant reduction in the integration preference for TSS as shown in the case of 17.IN.zfd1 likely yields a safer vector.
Two ZFD Variants Integrated with High Frequency into the Same Location in the Human Genome.
In addition to the global changes in the genomic integration patterns for 17.IN.zfd1, analysis of specific integrations intriguingly revealed that variants with ZFD1 inserted into different sites of Gag-Pol mediated integration into the same location in the human genome. That is, 83% and 91% of sequenced junctions for 16.p12.zfd1 and 375.RT.zfd1, respectively, strikingly mapped to the same location, nucleotide 141,457,970 on chromosome 3 (Table 3). Such a rare coincidence of integrations at the same site out of 3.1 × 109 possible human genome locations revealed that insertion of polydactyl ZFDs into the appropriate locations in Gag-Pol can narrow the range of retroviral integrations dramatically (P = 2.6 × 10-9, SI Materials and Methods). In strong contrast, none of the 294 sequenced virus-host genome junctions from three independent infections with MLV vector containing wild-type Gag-Pol (Table S1) mapped to this site on chromosome 3.
Table 3.
Common virus-host genome junction sequences identified for sets of related mutants
| Clones | Virus-host junction sequence | The number of junctions with the shown sequence | Total number of sequenced junctions for each mutant | Percent out of the total number of sequenced junctions | Integration position on the human genome |
|
| Chromosome | Position | |||||
| 16.p12.zfd1 | ACAACCCGTCTTAAATCAATCCA | 5 | 6 | 83.3 | 3 | 141457970 |
| 375.RT.zfd1 | ACAACCCGTCTTAAATCAATCCA | 31 | 34 | 91.2 | ||
| 273.IN.zfd2 | ACATTACAATTACTAATAGTAT | 8 | 197 | 4.1 | 2 | 13496337 |
| 375.RT.zfd2 | ACATTACAATTACTAATAGTAT | 5 | 93 | 5.4 | ||
| 273.IN.zfd2 | ACATTTGGGGGCCTGGACCACT | 1 | 197 | 0.5 | 18 | 43408004 |
| 375.RT.zfd2 | ACATTTGGGGGCCTGGACCACT | 2 | 93 | 2.2 | ||
| 273.IN.zfd2 | ACATTTGGTAGCTGGGATGTTAG | 22 | 197 | 11.2 | X | 153357666 |
| 375.RT.zfd2 | ACATTTGGTAGCTGGGATGTTAG | 7 | 93 | 7.5 | ||
| 273.IN.zfd2 | ACATTGAGTCAAAACTAGAGCCT | 1 | 197 | 0.5 | 15 | 88175538 |
| 375.RT.zfd2 | ACATTGAGTCAAAACTAGAGCCT | 2 | 93 | 2.2 | ||
| 273.IN.zfd2 | ACATTTGGAGACTAAATAAAAT | 47 | 197 | 23.9 | 1 | 58386380 |
| 375.RT.zfd2 | ACATTTGGAGACTAAATAAAAT | 29 | 93 | 31.2 | 9 | 77720767 |
The common junction sequences found among mutants with the same ZFD inserted into different Gag-Pol positions are shown, along with the fraction of total junctions corresponding to that integration position for each mutant. While most junction sequences matched a single location in the human genome, the junction sequence in the bottom two rows matched two locations. The underlined sequences are viral, and the others are from the host genome. During 3′ processing after reverse transcription the viral dinucleotides TT are deleted (9), producing a 5′ overhang that is often but not always filled post integration.
Identified Permissive ZFD Insertion Sites Tolerated Other ZFDs of Similar Length.
To analyze whether the Gag-Pol sites that permitted insertion of the ZFD1 could also tolerate other ZFDs, and whether new ZFDs in these positions could shift integration to different locations in a modular fashion, we inserted ZFDs previously designed to bind to 18-bp sequences that appear in the pigment epithelium-derived factor (PEDF) or CHK2 checkpoint homolog (CHK2) gene promoters (16, 17) (ZFD2 and ZFD3, respectively) (Table 1). We first packaged eight mutants with ZFD2 or ZFD3 grafted into the insertion positions of clones 17, 52, 273, and 375 (Table 1), which previously showed the highest infectivities (Fig. 3B). All of the new Gag-Pol.ZF clones allowed the production of infectious virus particles (Fig. S4), importantly indicating that the insertion sites discovered within Gag-Pol proteins could be generally permissive to other polydactyl ZFDs of similar length (∼180 amino acids).
Genome-wide integration analysis (SI Materials and Methods) was conducted on three of the new mutants: 273.IN.zfd2, 273.IN.zfd3, and 375.RT.zfd2. In contrast to vector with wild-type Gag-Pol, none of the three mutants showed significant integration preferences for TSS (P = 0.3146, P = 0.3511, and P = 0.6497, respectively) (Table 2). In addition, 273.IN.zfd2 and 375.RT.zfd2 did not show significant integration preferences for CpG islands (P = 0.5139 and P = 0.6026, respectively) (Table 2). Therefore, 273.IN.zfd2 and 375.RT.zfd2 exhibited considerable reductions in integration preferences for both TSS and CpG islands relative to wild-type virus (P = 0.0120 and 0.0004 and P = 0.0308 and 0.0046, respectively, Table 2). The consistent observation of substantial reductions in integration preference for TSS and CpG islands for 17.IN.zfd1, 273.IN.zfd2, and 375.RT.zfd2 (Table 2) suggests that judicious insertions of large DBD may override the intrinsic properties of Gag-Pol proteins in governing integration patterns. Furthermore, as shown in the cases of 273.IN.zfd2 and 273.IN.zfd3 (Table 2), insertion of different ZFDs into the same Gag-Pol site can result in statistically different shifts in integration preference for TSS and CpG (P = 0.0120 and 0.0004 and P = 0.1385 and 0.1136, respectively, Table 2), indicating a ZFD sequence-specific effect. Future study may elucidate the extent to which the ZFDs actively tether PIC to the specific regions of host genome, sterically hinder key but unknown regions of Gag-Pol proteins interacting with host factors such as chromatin-associated proteins or both.
Two Variants with ZFD2 Integrated with High Frequency into the Same Location in the Human Genome.
ZFDs were chosen as the DBD for their modularity and relative ease of engineering, though their reportedly very high affinities for their designed DNA targets lead to still high affinity binding to off-target sites (18, 19, 31). For example, a multitarget ELISA titration assay indicated that most ZF subunits within the ZFD1 used in this study can bind two to four off-target triplet sequences with very high binding affinities up to 70% of that for the case of intended target (32–34). This property may be even more a factor when multiple copies of a ZFD are present, as in the multivalent retroviral PIC, and chromatin may further modulate affinities for DNA targets. Consistent with these observations, our retroviral variants succeeded in fundamentally shifting integration patterns; however, as with the ZFD1, the new ZFDs did not find the addresses that the DBDs were originally designed to target.
That said, sequence analysis of individual virus-host genome junctions for new ZFDs strikingly revealed that the two mutants with ZFD2 in IN and RT integrated into a small set of common locations in the human genome. That is, 48% (of 93) and 40% (of 197) analyzed junctions for 375.RT.zfd2 and 273.IN.zfd2 (Table S1), respectively, intriguingly corresponded to a set of five common integration junction sequences despite the ZFD insertion into different viral proteins (four of these five sequences corresponded to unique positions in the human genome, Table 3). The chance that random integration could account for this high degree of coincidence in integration events at these common sites is statistically improbable (P = 4.6 × 10-29, SI Materials and Methods). However, infections of 293 cells with 273.IN.zfd2 and 273.IN.zfd3 did not result in integrations into common sites (Table S2) indicating that different ZFDs at the same site within Gag-Pol direct integrations into different sites of the genome. These observations further illustrate that insertion of ZFDs into RT and IN dramatically shrinks the pool of sites on human genome for the retroviral integrations, thereby potentially creating safer vectors.
Conclusions
In this study, we demonstrated that insertion of DBDs into key sites within Gag-Pol can engineer likely safer retroviral vectors by modulating and shifting integration patterns toward regions where TSS are relatively rare, as well as in many cases considerably narrowing the range of integration positions in the genome. This high-throughput engineering approach can also be extended to engineer other retroviruses, including lentiviruses. Therefore, in general library-based protein engineering of vectors to modulate their integration or other processes is a powerful approach to enhance the properties of viruses for clinical application.
Materials and Methods
Cell Lines and Plasmids.
Human embryonic kidney 293T cells were cultured in Iscove’s modified Dulbecco’s medium with 10% fetal bovine serum at 37 °C and 5% CO2. The retroviral vector plasmids pCLGIT Gag-Pol.ZF and pCLPIT GFP (Fig. S1A) were used for virus packaging during library production and virus clonal analysis, respectively. Three point mutations were made to introduce a Mme I site into the U5 region of these retroviral vector plasmids (Fig. S1B), which enabled adaptation of a prior method (26) to identify the sites of retroviral integration into human genome using a high-throughput linear amplification mediated PCR. The helper plasmids pCMV Gag-Pol and pcDNA3 IVS VSV-G express MLV gag-pol and the vesicular stomatitis virus glycoprotein (VSV-G), respectively, from the cytomegalovirus (CMV) immediate-early promoter. To avoid recombination problems, the ColE replication origin of pCMV Gag-Pol was replaced with a low copy number origin, pRB322, before the wild-type gag-pol was swapped with gag-pol.ZF sequences for clonal analysis.
Construction of pCLGIT Gag-Pol.ZF Library.
DNA encoding a zinc finger domain (ZFD1), with six zinc fingers in tandem, was synthesized (DNA2.0). The domain had been previously designed to recognize two 18-bp addresses near the γ-globin promoter on chromosome 11 of the human genome (15). Sequence repeats within the DNA encoding the ZFD were minimized during the gene synthesis to avoid recombination problems, and codon optimization was performed to maximize expression in human cells, while preserving the amino acid sequence. The gag-pol.ZF insertion library was constructed by replacing the kanR gene of pCLGIT Gag-Pol-kanR plasmid library, where the kanR gene was previously randomly inserted into MLV gag-pol using the Mutation Generation System kit (Finnzymes) (20), with a PCR product containing the ZFD sequence. The resulting pCLGIT Gag-Pol.ZF plasmid library thus expressed gag-pol variants with the ZFD incorporated in random positions. The library size was 4.3 × 105, estimated by colony counting after transformation of electro-competent DH10B Escherichia coli (Invitrogen). Based on the 5,214 nucleotide positions in MLV gag-pol, the library likely covered all possible insertion sites, as our prior work has indicated (20).
Library Production and Packageable Clone Selection.
The gag-pol.ZF library was packaged into retroviral vectors via calcium phosphate transfection of pCLGIT Gag-Pol.ZF library plasmid and pcDNA3 IVS VSV-G plasmids into 293T cells (Fig. 2). Library vector supernatant was twice harvested, two and three days posttransfection, and concentrated by ultracentrifugation. These viral vectors were used to infect 293T cells at a moi of < 0.1. GFP-expressing, infected cells were sorted on an EPICS Elite ESP sorter (Beckman-Coulter). Packageable virus variants were rescued through transfection of pcDNA3 IVS VSV-G into the expanded 293T cells and concentrated by ultracentrifugation. Viral RNA genomes were extracted from the rescued virus particles using the Qiamp viral RNA mini kit (QIAGEN). Following reverse transcription, the gag-pol.ZF cDNAs were amplified by nested PCR using Phusion high-fidelity polymerase (Finnzymes) and cloned into pCMV Gag-Pol to replace the wild-type gag-pol gene with the cDNA encoding the gag-pol.ZF variants. ZFD insertion sites within gag-pol were identified by sequencing the resulting pCMV Gag-Pol.ZF plasmids (UC Berkeley Sequencing Core). For comparison with the ZFD1 mutants, two fragments encoding new domains with six tandem zinc fingers (binding the PEDF promoter region or the CHK2 promoter region, ZFD2 and ZFD3, respectively) (16, 17) were synthesized and swapped for the ZFD1 within the pCMV Gag-Pol.ZF plasmids of packageable clones. The two new ZFDs were also designed and synthesized to minimize repeats and optimize codon usage in human cells (DNA2.0).
Clonal Analysis.
Each virus clone was packaged by transient transfection of pCMV Gag-Pol.ZF, pcDNA3 IVIS VSV-G, and pCLPIT GFP plasmids into 293T cells. Viral supernatant was used for infection either just after filtration with 0.45-μm syringe filters or after concentration via ultracentrifugation following filtration. Vector genomic titers were measured by real-time qPCR (35) using the iCycler iQ Real Time Detection System (Bio-Rad) and SYBR Green I (Invitrogen) with primers 5′- ATTGACTGAGTCGCCCGG-3′ (forward) and 5′- AGCGAGACCACAAGTCGGAT-3′ (reverse). Packaged viruses were analyzed in six serial log dilutions via qPCR. Infectious titers of viruses were measured in duplicate by counting GFP-positive, infected cells via flow cytometry on an EPICS XL-MCL cytometer (Beckman-Coulter).
Supplementary Material
Acknowledgments.
We thank J. T. Koerber and Sung-kuk Lee for technical advice. This work was supported by National Science Foundation BES-0629202 and National Institutes of Health R01 GM073058.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001402107/-/DCSupplemental.
References
- 1.Hacein-Bey-Abina S, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 3.Cavazzana-Calvo M, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 4.Kohn DB, Sadelain M, Glorioso JC. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat Rev Cancer. 2003;3:477–488. doi: 10.1038/nrc1122. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 6.Schroder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell RS, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewinski MK, et al. Retroviral DNA integration: Viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin JM, Hughes SH, Varmus HE. Retroviruses. 1st Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. [PubMed] [Google Scholar]
- 10.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushman FD. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc Natl Acad Sci USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes-Son ML, Appa RS, Chow SA. Molecular genetics and target site specificity of retroviral integration. Adv Genet. 2001;43:33–69. doi: 10.1016/s0065-2660(01)43003-3. [DOI] [PubMed] [Google Scholar]
- 13.Tan W, Dong Z, Wilkinson TA, Barbas CF, III, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng WJ, Chang CM, Lin TH. Target integration by a chimeric Sp1 zinc finger domain-Moloney murine leukemia virus integrase in vivo. J Biomed Sci. 2002;9:171–184. doi: 10.1007/BF02256029. [DOI] [PubMed] [Google Scholar]
- 15.Graslund T, Li X, Magnenat L, Popkov M, Barbas CF., 3rd Exploring strategies for the design of artificial transcription factors: Targeting sites proximal to known regulatory regions for the induction of gamma-globin expression and the treatment of sickle cell disease. J Biol Chem. 2005;280:3707–3714. doi: 10.1074/jbc.M406809200. [DOI] [PubMed] [Google Scholar]
- 16.Tan S, et al. Zinc-finger protein-targeted gene regulation: Genomewide single-gene specificity. Proc Natl Acad Sci USA. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoi K, et al. Gene transfer of an engineered zinc finger protein enhances the anti-angiogenic defense system. Mol Ther. 2007;15:1917–1923. doi: 10.1038/sj.mt.6300280. [DOI] [PubMed] [Google Scholar]
- 18.Cathomen T, Joung JK. Zinc-finger nucleases: The next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 19.Szczepek M, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 20.Yu JH, Schaffer DV. High-throughput, library-based selection of a murine leukemia virus variant to infect nondividing cells. J Virol. 2006;80:8981–8988. doi: 10.1128/JVI.00615-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan B, Li X, Goff SP. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyere SK, Joseph PR, Summers MF. The p12 domain is unstructured in a murine leukemia virus p12-CA(N) Gag construct. PLoS One. 2008;3:e1902. doi: 10.1371/journal.pone.0001902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das D, Georgiadis MM. The crystal structure of the monomeric reverse transcriptase from Moloney murine leukemia virus. Structure. 2004;12:819–829. doi: 10.1016/j.str.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 24.Najmudin S, et al. Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: Functional implications for template-primer binding to the fingers domain. J Mol Biol. 2000;296:613–632. doi: 10.1006/jmbi.1999.3477. [DOI] [PubMed] [Google Scholar]
- 25.Ey PL, Freeman NL, Bela B, Li P, McInnes JL. Nucleotide sequence of the murine leukaemia virus amphotropic strain 4070A integrase (IN) coding region and comparative structural analysis of the inferred polypeptide. Arch Virol. 1997;142:1757–1770. doi: 10.1007/s007050050195. [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Kim Y, Liang T, Sinsheimer JS, Chow SA. A high-throughput method for cloning and sequencing human immunodeficiency virus type 1 integration sites. J Virol. 2006;80:11313–11321. doi: 10.1128/JVI.01737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- 28.Trobridge GD, et al. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci USA. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moalic Y, Blanchard Y, Felix H, Jestin A. Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus. J Virol. 2006;80:10980–10988. doi: 10.1128/JVI.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crise B, et al. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J Virol. 2005;79:12199–12204. doi: 10.1128/JVI.79.19.12199-12204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornu TI, et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 32.Dreier B, et al. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 33.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 34.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towers GJ, et al. One step screening of retroviral producer clones by real time quantitative PCR. J Gene Med. 1999;1:352–359. doi: 10.1002/(SICI)1521-2254(199909/10)1:5<352::AID-JGM57>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.