Abstract
Inherited loss-of-function mutations in the tumor suppressor genes BRCA1, BRCA2, and multiple other genes predispose to high risks of breast and/or ovarian cancer. Cancer-associated inherited mutations in these genes are collectively quite common, but individually rare or even private. Genetic testing for BRCA1 and BRCA2 mutations has become an integral part of clinical practice, but testing is generally limited to these two genes and to women with severe family histories of breast or ovarian cancer. To determine whether massively parallel, “next-generation” sequencing would enable accurate, thorough, and cost-effective identification of inherited mutations for breast and ovarian cancer, we developed a genomic assay to capture, sequence, and detect all mutations in 21 genes, including BRCA1 and BRCA2, with inherited mutations that predispose to breast or ovarian cancer. Constitutional genomic DNA from subjects with known inherited mutations, ranging in size from 1 to >100,000 bp, was hybridized to custom oligonucleotides and then sequenced using a genome analyzer. Analysis was carried out blind to the mutation in each sample. Average coverage was >1200 reads per base pair. After filtering sequences for quality and number of reads, all single-nucleotide substitutions, small insertion and deletion mutations, and large genomic duplications and deletions were detected. There were zero false-positive calls of nonsense mutations, frameshift mutations, or genomic rearrangements for any gene in any of the test samples. This approach enables widespread genetic testing and personalized risk assessment for breast and ovarian cancer.
Keywords: BRCA1, BRCA2, genomics, next-generation sequencing, genetic testing
Inherited mutations in BRCA1 and BRCA2 predispose to high risks of breast and ovarian cancer. Lifetime risks of breast cancer are as high as 80% among women with mutations in these genes, and lifetime risks of ovarian cancer are greater than 40% for carriers of BRCA1 mutations and greater than 20% for carriers of BRCA2 mutations (1). Inherited mutations in the Fanconi anemia genes BRIP1 (FANCJ) and PALB2 (FANCN) are associated with 20–50% lifetime risks of breast cancer (2, 3). Inherited mutations in TP53, PTEN, STK11, and CDH1 are associated with moderate to very high risks of breast cancer in the context of Li-Fraumeni syndrome, Cowden syndrome, Peutz-Jeughers syndrome, and hereditary diffuse gastric cancer syndrome, respectively (4, 5, 6, 7). Inherited mutations in several of the genes responsible for hereditary nonpolyposis colon cancer and endometrial cancer are also associated with elevated risks of ovarian cancer (8).
Genetic testing for BRCA1 and BRCA2 mutations has become an integral part of clinical practice for women with severe family histories of breast or ovarian cancer, whether newly diagnosed or still clinically asymptomatic. However, as many as 50% of breast cancer patients with inherited mutations in BRCA1 and BRCA2 do not have close relatives with breast or ovarian cancer because their mutation is paternally inherited, the family is small, and by chance no sisters or paternal aunts have inherited the mutation of the family (1). Women in such families who carry BRCA1 or BRCA2 mutations have the same high risks of breast and ovarian cancer as women from high-incidence families. At present, women from such families rarely use genetic services.
In the United States, genetic testing of BRCA1 and BRCA2 is carried out almost exclusively by a single commercial company, whose protocol is based on PCR amplification of individual exons and Sanger sequencing of the products (9). In 2007, a quantitative DNA measurement assay (BART) was added as a supplementary test to detect large exonic deletions and duplications that are not detectable by PCR amplification approaches (BRACAnalysis Technical Specifications (updated February 2009) http://www.myriadtests.com/provider/doc/BRACAnalysis-Technical-Specifications.pdf). In Europe, genetic testing of BRCA1 and BRCA2 is more widely available (10, 11). Sequencing of the more moderate-risk breast cancer genes is available in various research or commercial diagnostic laboratories (GeneClinics http://www.ncbi.nlm.nih.gov/sites/GeneTests/?db=GeneTests), but is not routinely performed.
Recent advances in sequencing technologies have dramatically increased the speed and efficiency of DNA testing (12–16). Medical screening of genes responsible for disease generally requires an enrichment step before sequencing (17). This enrichment improves accuracy of mutation detection and reduces cost per sequenced nucleotide. To identify as many mutations as possible that are responsible for inherited predisposition to breast and ovarian cancer, it is useful to analyze multiple genes, not only BRCA1 and BRCA2. The mutational spectra of these genes include single-nucleotide variants, small insertions and deletions, and large genomic rearrangements spanning multiple kilobases. An approach to mutation detection based on next-generation sequencing must be able to accurately and cost-effectively detect all these classes of mutations before it can be used in a clinical diagnostic setting. This project was proof of principle for the application of solution capture and next-generation sequencing to mutation detection for patients at high risk of breast or ovarian cancer.
Results
Our goal was to evaluate the accuracy of DNA capture followed by massive parallel sequencing for identification of inherited mutations in breast and ovarian cancer genes. To carry out DNA capture, we designed oligonucleotides to target complete genomic sequence of 21 genes responsible for inherited risk of these cancers (Table 1). Oligonucleotides were designed to cover coding regions, noncoding intronic sequences, and 10-kb genomic sequence flanking each gene. After repetitive DNA elements were masked, total DNA targeted was ≈1 megabase.
Table 1.
Genomic regions targeted for breast and ovarian cancer genes
| Captured genomic region | |||
| Gene | Chromosome | Start | End |
| BRCA1 | 17 | 41,186,313 | 41,347,712 |
| BRCA2 | 13 | 32,879,617 | 32,983,809 |
| CHEK2 | 22 | 29,073,731 | 29,147,822 |
| PALB2 | 16 | 23,604,483 | 23,662,678 |
| BRIP1 | 17 | 59,759,985 | 59,940,755 |
| p53 | 17 | 7,561,720 | 7,600,863 |
| PTEN | 10 | 89,613,195 | 89,738,532 |
| STK11 | 19 | 1,195,798 | 1,238,434 |
| CDH1 | 16 | 68,761,195 | 68,879,444 |
| ATM | 11 | 108,083,559 | 108,249,826 |
| BARD1 | 2 | 215,583,275 | 215,684,428 |
| MLH1 | 3 | 37,024,979 | 37,102,337 |
| MRE11 | 11 | 94,140,467 | 94,237,040 |
| MSH2 | 2 | 47,620,263 | 47,720,360 |
| MSH6 | 2 | 48,000,221 | 48,044,092 |
| MUTYH | 1 | 45,784,914 | 45,816,142 |
| NBN | 8 | 90,935,565 | 91,006,899 |
| PMS1 | 2 | 190,638,811 | 190,752,355 |
| PMS2 | 7 | 6,002,870 | 6,058,737 |
| RAD50 | 5 | 131,882,630 | 131,989,595 |
| RAD51C | 17 | 56,759,963 | 56,821,692 |
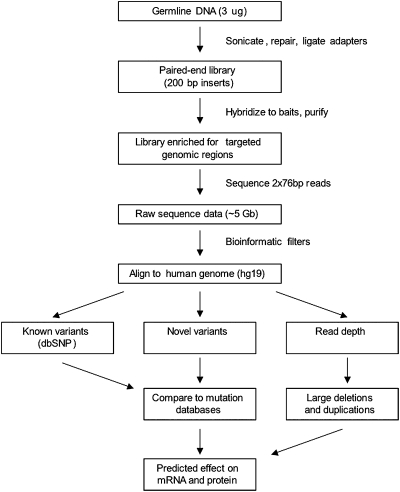
The mutation screening process is outlined in Fig. 1. DNA was extracted from blood and sonicated, and libraries were prepared with a mean insert size of 200 bp. Libraries were hybridized in solution to the custom oligonucleotides and then sequenced on an Illumina Genome Analyzer IIX to generate 2- × 76-bp paired-end reads. An average of 2.4 gigabase (Gb) per sample (range 1.8–4.4 Gb per sample) of high-quality sequence was obtained at targeted sites, representing an average 1,286-fold coverage per nucleotide (range across all samples was 781- to 1854-fold average coverage per nucleotide). DNA sequences were aligned to the human reference genome. Nucleotide coordinates of rare variants were identified as described in Materials and Methods. We required that a potential variant be present on both sequenced DNA strands and represent ≥15% of total reads at that site to be further evaluated. The 15% threshold was chosen because the mutation CHEK2_1100delC was present on only 15% of reads at this site (Table 2) as the result of the existence of CHEK2 pseudogenes on chromosomes 15 and 16. Common polymorphisms were excluded by comparison with dbSNP130. However, because dbSNP erroneously includes some severe disease-associated mutations as benign polymorphisms (e.g., p53_721G > A) (18), we also compared all variants to gene-specific mutation databases. Candidate variants were categorized by gene location (intergenic, intronic, or exonic) and by predicted effect (frameshift, in-frame insertion or deletion, synonymous substitution, nonsynonymous substitution, splice site alteration, or nonsense). Each candidate variant was evaluated by conventional Sanger sequencing. All samples were tested blind to their mutation.
Fig. 1.
Screening for mutations in breast and ovarian cancer genes using targeted DNA capture and next-generation sequencing.
Table 2.
Point mutations and small insertions and deletions identified by the assay
| Mutant sites identified | No. of reads | |||||||||
| Gene | Nucleotide | Effect | Type | Size (bp) | Chromosome | Start | End | Wild type | Variant | % Variant |
| BRCA1 | 4510 del3ins2 | 1465 stop | Deletion-insertion | 1 | 17 | 41,228,596 | 41,228,597 | 525 | 596 | 0.53 |
| BRCA1 | 5083 del19 | 1657 stop | Deletion | 19 | 17 | 41,222,949 | 41,222,968 | 700 | 644 | 0.48 |
| BRCA1 | 5382 insC | 1829 stop | Insertion | 1 | 17 | 41,209,080 | 41,209,081 | 606 | 596 | 0.50 |
| BRCA2 | 999 del5 | 273 stop | Deletion | 5 | 13 | 32,905,141 | 32,905,146 | 363 | 229 | 0.39 |
| BRCA2 | 1983 del5 | 585 stop | Deletion | 5 | 13 | 32,907,366 | 32,907,371 | 304 | 258 | 0.46 |
| BRCA2 | 6174 delT | 2003 stop | Deletion | 1 | 13 | 32,914,438 | 32,914,439 | 565 | 661 | 0.54 |
| BRCA2 | 9179 C > G | 2984 stop | Nonsense | 1 | 13 | 32,953,650 | 391 | 361 | 0.48 | |
| BRIP1 | 3401 delC | 1149 stop | Deletion | 1 | 17 | 59,761,006 | 59,761,007 | 651 | 486 | 0.43 |
| CDH1 | 591 G > A | 157 stop | Nonsense | 1 | 16 | 68,842,406 | 421 | 359 | 0.46 | |
| CHEK2 | 1100 delC | 381 stop | Deletion | 1 | 22 | 29,091,857 | 29,091,858 | 3,293 | 586 | 0.15 |
| MLH1 | ivs14(-1) G > A | 568 stop | Splice | 1 | 3 | 37,083,758 | 1,024 | 683 | 0.40 | |
| MSH2 | 1677 T > A | 537 stop | Nonsense | 1 | 2 | 47,693,895 | 575 | 552 | 0.49 | |
| p53 | 721 G > A | R175H | Missense | 1 | 17 | 7,578,406 | 449 | 306 | 0.41 | |
| PALB2 | 509 delGA | 183 stop | Deletion | 2 | 16 | 23,647,357 | 23,647,359 | 1,283 | 1,233 | 0.49 |
| STK11 | ivs6(-1) G > A | 316 stop | Splice | 1 | 19 | 1,221,947 | 722 | 572 | 0.44 | |
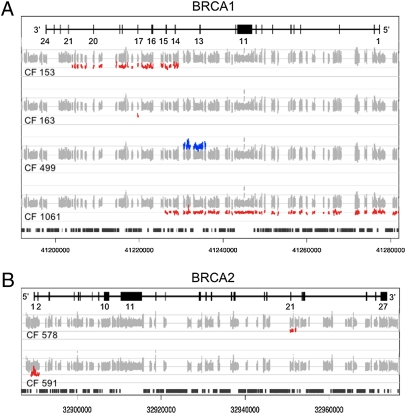
All mutations in the test series were accurately identified and there were zero false-positive calls of mutations in any gene in any of the samples. Point mutations and small insertions and deletions in BRCA1, BRCA2, BRIP1, CDH1, CHEK2, MLH1, MSH2, p53, PALB2, and STK11 ranged in size from 1 to 19 bp (Table 2). The genomic base pairs of each were correctly identified. In addition, by comparing the number of sequence reads at each base pair for each sample to the number of reads at the same base pair for all other samples in the experiment, we screened for large deletions and duplications at each of the 21 loci. Deviations from diploidy were defined as sites at which a test sample yielded <60% or >140% the average number of reads of the other samples in the experiment. We accurately identified the five genomic deletions and one genomic duplication (Table 3, Fig. 2), determining breakpoints on the targeted sequence within 1 kb. Each large deletion and duplication is flanked by Alu sequences that mediate the mutation. Because Alu repeats are not targeted by the oligonucleotides in the capture pool, the exact breakpoints within flanking Alu repeats are not determinable. There was complete concordance between deletions and duplications identified by our read-depth algorithm and by the multiple ligation probe assay (19).
Table 3.
Genomic deletions and duplication identified by the assay
| Mutant sites identified by assay | ||||||
| Gene | Genomic event | Chromosome | Start* | End* | Size (bp) | Ratio† |
| BRCA1 | Deletion exons 1–15 | 17 | 41,226,145 | 41,327,157 | 101,013 | 0.509 |
| BRCA1 | Duplication exon 13 | 17 | 41,230,562 | 41,235,836 | 5,275 | 1.578 |
| BRCA1 | Deletion exons 14–20 | 17 | 41,203,975 | 41,229,297 | 25,323 | 0.519 |
| BRCA1 | Deletion exon 17 | 17 | 41,219,596 | 41,219,755 | 160 | 0.495 |
| BRCA2 | Deletion exons 1–2 | 13 | 32,889,020 | 32,890,900 | 1,881 | 0.489 |
| BRCA2 | Deletion exon 21 | 13 | 32,950,734 | 32,952,070 | 1,337 | 0.544 |
*Breakpoints are flanked by Alu and other repeats, which are not captured.
†Reads per base pair for deletion or duplication/reads per base pair for wild-type genotype.
Fig. 2.
Large genomic deletions and duplications in BRCA1 and BRCA2 identified by analysis of the read depth of sequencing data. Normalized numbers of sequencing reads are indicated for each gene. Exons are indicated by black vertical lines and intervening introns by horizontal lines. Numbers of reads for each base pair are represented in gray. Numbers of reads for each sample deviating from the median of all samples for that base pair occur at deletions (red) and at duplications (blue). DNA repetitive elements are indicated by black vertical bars (bottom of A and B). (A) The BRCA1 locus in DNA from families 153, 163, 499, and 1061. (B) The BRCA2 locus in DNA from families 578 and 591.
Discussion
The landscape of genetic testing in the United States was changed on March 29, 2010, by the decision of Judge Robert Sweet of the Federal District Court in Manhattan, which invalidated Myriad Genetics’ patents on the BRCA1 and BRCA2 genes (20). By declaring that genes are products of nature and therefore not subject to patent, he called into question patents filed on thousands of human genes. In this context, it may be that tools for more efficient genetic testing for cancer susceptibility genes will be developed and clinically applied.
The availability of more rapid and cost-effective testing for multiple breast and ovarian cancer susceptibility genes has major clinical implications. BRCA1 and BRCA2 are the genes most commonly implicated in hereditary breast or ovarian cancer and were patented by Myriad Genetics and the University of Utah after their identification in 1994 and 1995, respectively. Since then, Myriad Genetics has been the sole source in the United States for commercial testing for inherited mutations in BRCA1 and BRCA2. The standard test consists of DNA sequencing of both genes and screening for five large deletions and duplications in BRCA1 and costs $3,340. Comprehensive testing for gene rearrangements is offered as a separate test by Myriad Genetics at an additional cost of $650. If BRCA1 and BRCA2 testing is negative (i.e., wild type), testing for other breast or ovarian cancer genes is typically done selectively, when mutations in certain other genes are suspected on the basis of family history, personal history, or findings from physical examination (21). Sequencing additional genes can add thousands more dollars to the cost of genetic testing. It is possible to identify mutations in the 21 known breast and ovarian cancer genes in one sample for a cost of reagents and consumables less than $1,500. Given the massive redundancy of read depth that we achieved by running a single sample in one flow cell lane, it is likely that a barcoding or indexing strategy would be feasible and reduce the cost to less than $500 per sample.
Other applications of next-generation sequencing to genetic testing of BRCA1 and BRCA2 have been suggested (22–24). The approach described here differs in that we evaluated multiple genes, in addition to BRCA1 and BRCA2, and we evaluated all classes of mutation. Our next-generation sequencing approach identified a wide range of mutations in a variety of genes in 100% of our test cases with zero spurious mutations called. Target mutations included single base substitutions and deletions and insertions of varying sizes. Importantly, we easily identified six large deletions and duplications, all of which would have been missed by standard sequencing (19). This approach thus obviates the need for separate testing for gene rearrangements after Sanger DNA sequencing (25).
Because testing for mutations in genes other than BRCA1 and BRCA2 is done only selectively, the proportion of breast and ovarian cancers attributable to inherited mutations in these other genes is not known. By allowing comprehensive parallel testing of multiple cancer susceptibility genes, we will be able to confidently identify the fraction of women with breast or ovarian cancer who carry a germline alteration in a cancer susceptibility allele and the characteristics of the tumors of patients’ inherited mutations in various genes.
The cost savings in applying such technologies will allow the application of genetic testing to a wider range of individuals than is the current standard. Presently, patients who have a relatively high pretest probability of having a mutation in a given cancer susceptibility allele are chosen for testing (26, 27). However, many breast and ovarian cancer patients with BRCA1 or BRCA2 mutations have a negative family history for cancer (1). Since the advent of inhibitors of the Poly (ADP ribose) polymerase (PARP) enzyme, which effectively kill BRCA1- and BRCA2-mutated carcinomas, understanding the genetic basis of human cancers has therapeutic as well as preventive implications (28, 29). The availability of PARP inhibitors has increased the clinical incentive to identify BRCA1 and BRCA2 mutations in women with breast or ovarian cancer.
Being able to test for multiple cancer susceptibility genes simultaneously will add complexity to the clinical interpretation of results. More variants of uncertain significance will be identified, and clinical recommendations may not be standardized for mutations in some cancer susceptibility genes. As the complexity of genetic testing for cancer risk increases, we emphasize the importance of including a medical geneticist or certified genetic counselor in the testing process. Consequently, these types of tests may not be appropriate to order in a primary practice setting or directly from the company as a product of direct-to-consumer marketing.
As more next-generation sequencing technologies become available for genetic testing, results on sensitivity and specificity should be made freely accessible to those who order the test. Hopefully, comparisons of various technologies will also become available. Guidelines for application of next-generation sequencing to genetic diagnostics are currently under development (30). At present, we believe that apparently positive tests should be validated by standard Sanger sequencing of the patient's DNA before results are reported to the patient. Sanger sequencing would verify the mutation and provide the basis for a simplified test for at-risk relatives. For mutation detection, our goal was to extend genetic testing for inherited risk of breast and ovarian cancer to more women by providing an approach that offers substantially lower cost while maintaining very high sensitivity and specificity.
Materials and Methods
Study Subjects.
Participants were 20 women diagnosed with breast or ovarian cancer and with a known mutation in one of the genes responsible for inherited predisposition to these diseases. The critical mutation for each patient had been previously identified by Sanger sequencing of PCR amplicons or by multiplex ligation-dependant probe amplification, as described (19). The study was approved by the institutional review board at the University of Washington. All participants provided informed consent.
Capture and Sequencing of Genomic DNA.
Genomic DNA was captured by hybridization in solution to custom-designed cRNA oligonucleotide baits (31) following the manufacturer's protocols (Agilent Technologies). BED files of genomic locations of all cRNA oligonucleotide probes are freely available on request to the authors. Captured library DNA (9 pM) was denatured and subjected to cluster amplification on a Paired End Flow Cell v4 with a cBot instrument (Illumina) to generate raw cluster intensity of ∼700,000 mm2. Sequencing was performed on a Genome Analyzer GAIIX for 2 × 76 cycles using Cycle Sequencing v4 reagents (Illumina).
Average coverage for the captured regions ranged from 781- to 1,854-fold per site. To determine if coverage was substantially lower for any region, we calculated the proportion of base pairs for each locus that were captured by <100 reads. All regions were covered by >20 reads. Proportions of base pairs covered by <100 reads were 0.082 for PMS2, 0.067 for STK11, 0.023 for PTEN, and 0.002 for MRE11 and MUTY. Content of guanine and cytosine (GC) basepairs at these poorly covered regions was high; most were CpG islands. Such regions are also refractive to PCR-based sequencing methods. All base pairs of BRCA1, BRCA2, CHEK2, PALB2, BRIP1, p53, CDH1, ATM, BARD1, MLH1, MSH2, NBN, PMS1, RAD50, and RAD51C were captured by >100 reads.
Bioinformatic Analysis of DNA Variants.
Sequencing data were processed through Illumina pipeline v1.6 using default parameters. Reads of high quality were mapped to the reference human genome sequence (GRCh37, UCSC hg19) using MAQ 0.7.1 with default parameters (32). Reads outside the targeted sequences were discarded and statistics on coverage were collected from the remaining reads. Across samples evaluated with this oligonucleotide pool, an average of 69% of reads were on target. Potential single-base-pair variants and small insertions and deletions were identified using the MAQ Perl-based filter after alignment (-map), assembly (-assembly), and consensus calling (-cns2snp) with default parameters.
To detect large genomic deletion and duplication mutations, we developed a script to count sequence reads on captured DNA and then converted total read depth at a given genomic location into deletion and duplication calls. Read depth was corrected for oligonucleotide probe coverage and local GC content. After testing different window sizes, we found that a 100-bp window gave the strongest relationship between GC content and base coverage. We normalized to median coverage across all samples using invariant set methods (33) and then compared each individual's coverage to the median coverage. The derived coverage ratio was then subjected to our sliding-window deletion and duplication-calling method. The minimum accepted call length for deletions and duplications was 100 bp, with threshold ratios <0.6 for deletions and >1.4 for duplications. Using these methods, we identified all six known deletions and duplications. We compared the difference between the number of reads for the sample carrying the putative deletion or duplication with the median number of reads at the corresponding base pair for all samples. Differences were compared by t tests for each event defined by this method. For all deletions and duplications, differences in read depth were significant at P < 10−10.
Acknowledgments
We thank Michael Jacobs, Brian Fritz, and Erica Jonlin for technical advice. This work was supported by the Breast Cancer Research Foundation, the Susan G. Komen Foundation for the Cure, and the Department of Defense Congressionally Directed Medical Research Programs for Breast Cancer and Ovarian Cancer. M.-C.K. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
References
- 1.King MC, Marks JH, Mandell JB New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 2.Seal S, et al. Breast Cancer Susceptibility Collaboration (UK) Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 3.Rahman N, et al. Breast Cancer Susceptibility Collaboration (UK) PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez KD, et al. Beyond Li Fraumeni syndrome: Clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27:1250–1256. doi: 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- 5.FitzGerald MG, et al. Germline mutations in PTEN are an infrequent cause of genetic predisposition to breast cancer. Oncogene. 1998;17:727–731. doi: 10.1038/sj.onc.1201984. [DOI] [PubMed] [Google Scholar]
- 6.Hearle N, et al. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res. 2006;12:3209–3215. doi: 10.1158/1078-0432.CCR-06-0083. [DOI] [PubMed] [Google Scholar]
- 7.Schrader KA, et al. Hereditary diffuse gastric cancer: Association with lobular breast cancer. Fam Cancer. 2008;7:73–82. doi: 10.1007/s10689-007-9172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aarnio M, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer. 1999;81:214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 9.Frank TS, et al. Sequence analysis of BRCA1 and BRCA2: Correlation of mutations with family history and ovarian cancer risk. J Clin Oncol. 1998;16:2417–2425. doi: 10.1200/JCO.1998.16.7.2417. [DOI] [PubMed] [Google Scholar]
- 10.Aymé S, Matthijs G, Soini S ESHG Working Party on Patenting and Licensing. Patenting and licensing in genetic testing. Eur J Hum Genet. 2008;16(Suppl 1) doi: 10.1038/sj.ejhg.5201929. [DOI] [PubMed] [Google Scholar]
- 11.Matthijs G, Hodgson S. The impact of patenting on DNA diagnostic practice. Clin Med. 2008;8:58–60. doi: 10.7861/clinmedicine.8-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lifton RP. Individual genomes on the horizon. N Engl J Med. 2010;362:1235–1236. doi: 10.1056/NEJMe1001090. [DOI] [PubMed] [Google Scholar]
- 13.Goossens D, et al. Simultaneous mutation and copy number variation (CNV) detection by multiplex PCR-based GS-FLX sequencing. Hum Mutat. 2009;30:472–476. doi: 10.1002/humu.20873. [DOI] [PubMed] [Google Scholar]
- 14.Daiger SP, et al. Targeted high-throughput DNA sequencing for gene discovery in retinitis pigmentosa. Adv Exp Med Biol. 2010;664:325–331. doi: 10.1007/978-1-4419-1399-9_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoischen A, et al. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum Mutat. 2010;31:494–499. doi: 10.1002/humu.21221. [DOI] [PubMed] [Google Scholar]
- 16.Chou LS, Liu CS, Boese B, Zhang X, Mao R. DNA sequence capture and enrichment by microarray followed by next-generation sequencing for targeted resequencing: Neurofibromatosis type 1 gene as a model. Clin Chem. 2010;56:62–72. doi: 10.1373/clinchem.2009.132639. [DOI] [PubMed] [Google Scholar]
- 17.Mamanova L, et al. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 18.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 19.Walsh T, et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379–1388. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 20.Kesselheim AS, Mello MM. Gene patenting: Is the pendulum swinging back? N Engl J Med. 2010;362:1855–1858. doi: 10.1056/NEJMp1004026. [DOI] [PubMed] [Google Scholar]
- 21.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Summerer D, et al. Microarray-based multicycle-enrichment of genomic subsets for targeted next-generation sequencing. Genome Res. 2009;19:1616–1621. doi: 10.1101/gr.091942.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder C, Stutzmann F, Weber BH, Riess O, Bonin M. High-throughput resequencing in the diagnosis of BRCA1/2 mutations using oligonucleotide resequencing microarrays. Breast Cancer Res Treat. 2010;122:287–297. doi: 10.1007/s10549-009-0639-z. [DOI] [PubMed] [Google Scholar]
- 24.Morgan JE, et al. Genetic diagnosis of familial breast cancer using clonal sequencing. Hum Mutat. 2010;31:484–491. doi: 10.1002/humu.21216. [DOI] [PubMed] [Google Scholar]
- 25.Hogervorst FB, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- 26.Euhus DM, et al. Pretest prediction of BRCA1 or BRCA2 mutation by risk counselors and the computer model BRCAPRO. J Natl Cancer Inst. 2002;94:844–851. doi: 10.1093/jnci/94.11.844. [DOI] [PubMed] [Google Scholar]
- 27.Berry DA, et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol. 2002;20:2701–2712. doi: 10.1200/JCO.2002.05.121. [DOI] [PubMed] [Google Scholar]
- 28.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 29.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 30.Mattocks CJ for the EuroGentest Validation Group. A standardized framework for the validation and verification of clinical molecular genetic tests. Eur J Hum Genet. 2010 doi: 10.1038/ejhg.2010.101. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tewhey R, et al. Enrichment of sequencing targets from the human genome by solution hybridization. Genome Biol. 2009;10:R116. doi: 10.1186/gb-2009-10-10-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol. 2001;2:0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]