Abstract
Although mammalian polypyrimidine tract-binding (PTB) protein functions in most or all cell types to regulate a wide spectrum of transcripts, Drosophila PTB encodes an abundant male germline-specific mRNA isoform (dmPTB) whose expression correlates with male fertility. The biological function of this isoform is unknown. Using selection–amplification, we show that mammalian and Drosophila PTB have similar RNA sequence preference, suggesting that cell-specific expression rather than unique RNA-binding properties account for the sex-specific function of dmPTB. We also show that the dmPTB protein isoform expressed in the male germline is by far the most abundant isoform, and reduction of its levels correlates with male sterility. Finally, we show that dmPTB expression is necessary for proper spermatid individualization, the terminal step necessary for production of motile sperm. Loss of dmPTB results in severe disruption of the actin cones of the spermatid individualization complex. This represents a cytological defect resulting from PTB loss. We discuss the basis for functional differences between mammalian and Drosophila PTB orthologs.
Keywords: individualization complex, spermatogenesis, male germline, selection–amplification, RNA binding protein
RNA-binding proteins play an important role in posttranscriptional regulation. Among hundreds of known RNA-binding proteins, the biological function is known for only a few. RNA-binding proteins typically bind short, degenerate sequences, which occur frequently by chance throughout the genome and make functional analysis harder. The mammalian polypyrimidine tract-binding protein or heterogeneous nuclear ribonucleoprotein I (PTB/hnRNP I) is one of the well-studied RNA-binding proteins. The hnRNP proteins are ubiquitously expressed, associate with nascent transcripts, and play various roles in RNA metabolism. PTB is known to affect mRNA splicing, polyadenylation, translation, mRNA stability/degradation, and mRNA localization (reviewed in refs. 1 and 2).
Mammalian PTB, which is considered a general splice site repressor, regulates the tissue-specific alternative splicing or localization of a wide spectrum of premRNAs (e.g., CT/CGRP α-tropomyosin, c-src, α-actinin, and fibronectin) (3–6) involving tissue-specific expression of corepressors or PTB antagonists (7). PTB contains four RNA recognition motifs (RRMs) and binds to pyrimidine-rich sequences with (short, degenerate) motifs, such as UCUUC and CUCUCU (8); RRM3 and RRM4 provide the major contribution to specific RNA recognition (9). Diverse mechanisms have been proposed for how PTB binding regulates splice site choice. These include direct competition with the splicing factor U2AF65 (8, 10); multimerization across an exon to create a zone of silencing or cause exon looping (3, 9, 11–13); interference with exon definition (14); and competition with tissue-specific paralogs (nPTB/brPTB) (15) or with PTB antagonists (ETR-3, RBM4, and CELF) (16–18).
Previously, we found that in Drosophila a major PTB transcript is male germline specific, and its presence correlates with fertility (19). However, what cellular role(s) PTB plays during spermatogenesis remained unknown. Spermatogenesis is remarkably similar between Drosophila and mammals, including maintenance of male germ cells, mitotic divisions before meiosis, and spermatid differentiation involving significant morphological changes in almost all cellular components (reviewed in ref. 20). In Drosophila, extensive genetic analysis has identified numerous mutations that produce specific cytological blocks during spermatogenesis, for example, stem cell renewal, mitotic and meiotic amplification, and spermatid differentiation (20, 21). Despite enormous progress from cytological and genetic studies, how relevant mutations influence specific stages during spermatogenesis remains largely unknown.
Relevant to defining the sex-specific biological role of dmPTB, two specific questions arise: (i) What are the specific RNA sequence(s) that the male germline dmPTB isoform binds to? (ii) What is its male germline-specific cellular function? Here we characterize the RNA-binding properties of this PTB isoform and compare it with that of mammalian PTB. Furthermore, we determine that the cellular defect that results in male sterility in the dmPTB loss-of-function mutant heph2 is the disruption of the spermatid individualization complex (IC). These findings highlight that an hnRNP protein (dmPTB) can have a male-specific function that affects a specific step during spermatogenesis.
Results
Mammalian and Drosophila PTB Have Similar RNA-Binding Properties.
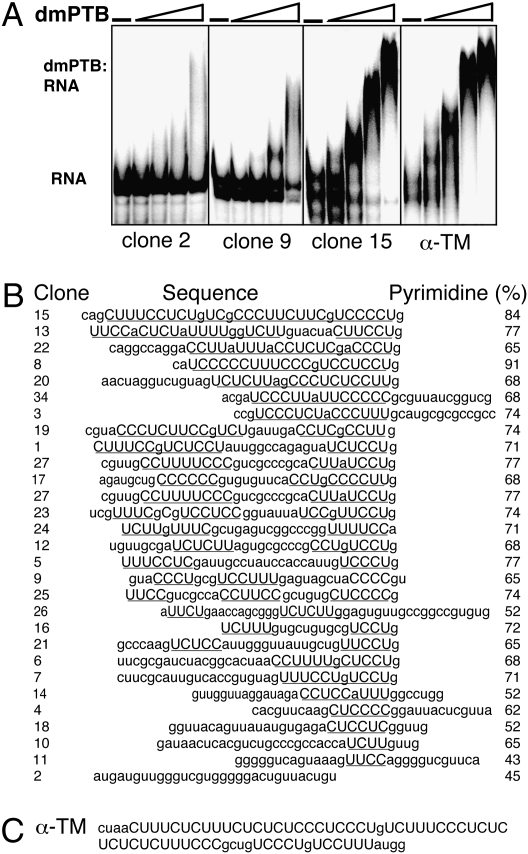
As a first step toward identification of the male-germline function of dmPTB, we characterized its RNA binding properties. To determine the consensus binding sequence of dmPTB, selection–amplification (SELEX) (22) was performed on a random pool of RNA (8) to select sequences that bound to the RNA-binding portion of dmPTB (RRM3 and RRM4). Although there was no binding of dmPTB to the random pool of RNA, significant RNA binding was observed after six cycles of enrichment. Twenty-nine RNAs from Pool 6 were cloned and sequenced (Fig. 1B). In general, the selected sequences had long uninterrupted pyrimidine stretches bounded on the 3′ end by a guanine residue. The average pyrimidine content of the selected sequences was 65%, and all but one had at least one stretch of four or more consecutive pyrimidine nucleotides, for a total of 63 such stretches in the 29 sequences. Whereas some sequences had one long stretch of pyrimidines (e.g., clone 15), others had two shorter stretches (e.g., clones 1 and 19), or just one stretch (e.g., clone 6).
Fig. 1.
SELEX shows that dmPTB binds pyrimidine-rich sequences. (A) Gel mobility shift assay shows that dmPTB binds with high affinity and specificity. (B) Alignment of dmPTB-selected sequences. Pyrimidine tracts are in uppercase and underlined. (C) Sequence of the PTB binding site in the mammalian α-tropomyosin premRNA intron upstream of exon 3.
To determine the binding affinity of the recombinant dmPTB fragment to the individual clones, electrophoretic gel mobility shift assays were performed with several representative clones (Fig. 1A). Of the clones tested, clone 15, which contained the longest pyrimidine stretch (interrupted by guanines) bound with the highest affinity (25 nM). Clone 2, which lacked a definable polypyrimidine tract, showed barely detectable or background level binding. Clone 9, which contained a short polypyrimidine tract, exhibited weak but detectable binding. Furthermore, we found that the binding affinities of dmPTB for clone 15 and for the α-tropomyosin pyrimidine tract, which is known to bind the mammalian PTB (8), were comparable (15 nM). We conclude that dmPTB exhibits a high degree of sequence specificity, that dmPTB binds to long uninterrupted pyrimidine stretches with high affinity, and that the binding affinity of dmPTB for its own selected sequences is similar to that observed for a sequence known to bind the mammalian PTB.
Expression of dmPTB Protein Is Sexually Dimorphic.
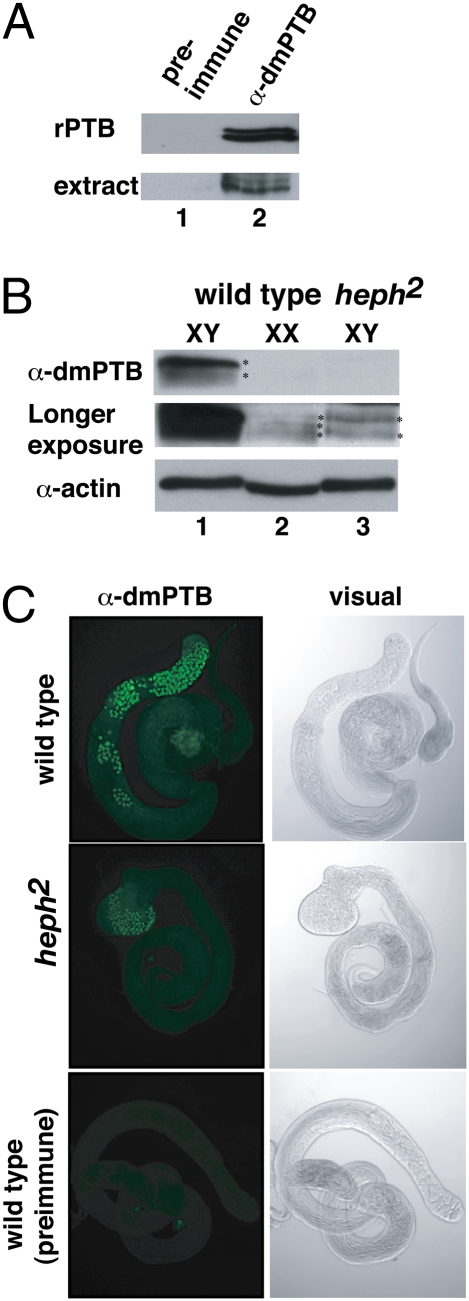
To define dmPTB function in the male germline, we analyzed the dmPTB protein in vivo. Because polyclonal antibodies made against the mammalian PTB (6) did not cross-react with dmPTB, we generated antibodies (α-dmPTB) against the recombinant dmPTB(RRM3,4) protein. These antibodies recognized recombinant dmPTB as well as the endogenous protein from Drosophila extracts (Fig. 2A). There was no detectable signal with preimmune serum, indicating that these antibodies specifically recognize dmPTB.
Fig. 2.
dmPTB protein exhibits sexually dimorphic expression and is predominantly expressed in primary spermatocytes. (A) dmPTB antibodies specifically recognize dmPTB protein (recombinant and endogenous). (B) (Top) Western blot analysis reveals that the two major isoforms of dmPTB are predominantly expressed in wild-type males but not females or heph2 males. (Middle) Overexposure reveals low-level expression of dmPTB isoforms in wild-type females and heph2 males. (Bottom) Anti-actin is loading control. (C) Antibody staining reveals that dmPTB is predominantly expressed in the nuclei of primary spermatocytes of wild-type males. Low-level expression of dmPTB is observed in the nuclei (at the bulged tip testis) of primary spermatocytes in heph2 males.
Next, we analyzed the protein expression pattern in male and female flies and found that expression of the dmPTB protein is sexually dimorphic. One major isoform and one minor isoform were present at high levels in wild-type males (Fig. 2B, Top, lane 1); neither of the isoforms was initially detectable in females (Fig. 2B, Top, lane 2). However, upon longer exposures, we found that females had bands comigrating with these isoforms, but their abundance was much lower [Fig. 2B, Middle (longer exposure), lane 2]. The number and ratio of the isoforms differed between males and females: in males, the longer isoform was more abundant, whereas in females all isoforms were relatively equally expressed at low levels. We conclude that dmPTB protein expression in Drosophila is sexually dimorphic, with an abundant isoform expressed specifically in males.
Loss of dmPTB Correlates with Male Sterility.
The heph2 mutant results from insertion of a transposable P-element into the dmPTB locus. Heph2 flies lack the abundant male-specific dmPTB transcript and are male-sterile (19). Analysis of dmPTB protein expression levels in heph2 males by Western blot showed that dmPTB expression levels were severely reduced in these flies, detectable only upon significantly longer exposure (Fig. 2B, Middle, lane 3). Expression of the more abundant longer isoform seemed to be reduced to a greater extent than that of the shorter isoform. These results indicate that loss of the dmPTB protein correlates with male-sterility.
Given that the abundant male-specific transcript is expressed in the germline, we next determined whether the dmPTB protein observed in wild-type males is expressed in the germline. To determine the expression pattern of dmPTB, we stained testes from wild-type and heph2 males with the anti-dmPTB antibody and a fluorescently labeled secondary antibody. In wild-type testes dmPTB was highly expressed in the nuclei of primary spermatocytes, but not during the later stages of spermatocyte development (Fig. 2C). In heph2 testes dmPTB was observed in the nuclei of primary spermatocytes at the tip of the swollen testis, but at greatly reduced levels.
Generation of a Unique dmPTB Allele.
To provide additional tools with which to examine the function of dmPTB, we generated a new allele (heph2-d1) of dmPTB. The heph2-d1 allele resulted from incomplete excision of the P-element, whereas other generated alleles likely resulted from excision of surrounding genomic DNA along with the P-element.
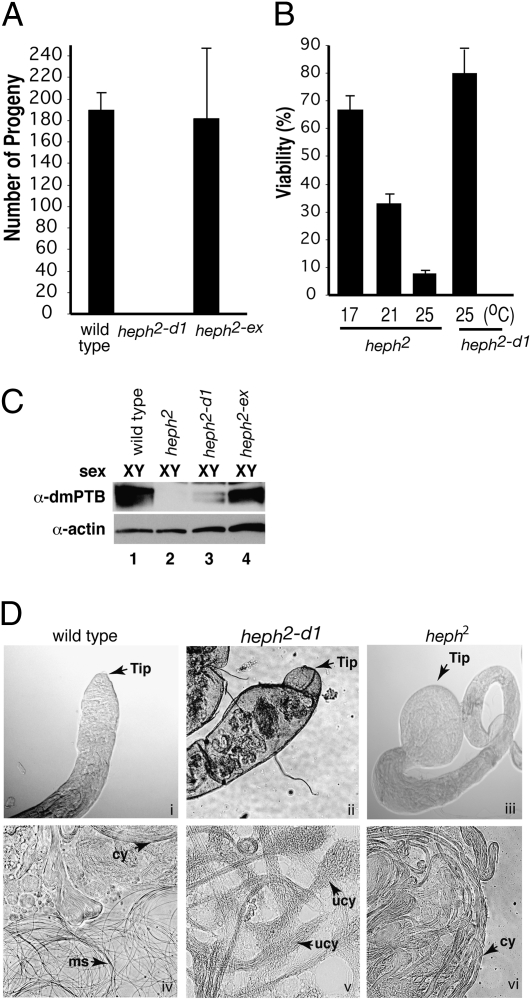
Given that the heph2 homozygous males were sterile, we obtained homozygous lines of the heph2-d1 allele, and males were tested in a standard fertility assay. Wild-type flies and the functionally revertant heph2-ex flies (19) produced similar numbers of progeny, whereas the heph2-d1 flies were male-sterile (Fig. 3A). Similar to heph2, heph2-d1 female flies were fertile. In addition, one of the generated alleles was non–sex-specific lethal, consistent with other known dmPTB alleles, indicating that dmPTB plays a vital non–sex-specific role in addition to its role in male fertility.
Fig. 3.
Male fertility correlates with dmPTB protein expression. (A) Fertility of dmPTB mutants compared with wild-type males. The heph2-d1 allele is sterile. (B) heph2-d1 mutants do not exhibit the same temperature-sensitive, non–sex-specific lethality as heph2 mutants. (C) Male fertility correlates with dmPTB protein expression. Sterile (heph2-d1) mutant has low levels expression of the minor isoforms of dmPTB protein. (D) Testis tip and individualization defects in dmPTB mutants. Loss of dmPTB results in testis tip defects, from minor swelling and formation of knobs (ii) in the heph2-d1 mutant to severe bulging in the heph2 mutant (iii) compared with wild-type testis (i). Loss of dmPTB also results in individualization defects, including partially unraveled unindividualized cysts (v). Cy, cyst; ms, motile sperm; ucy, unraveled, unindividualized cyst.
The original heph2 allele exhibited temperature-sensitive lethality in both sexes (Fig. 3B). At the normal fly-rearing temperature of 25 °C, only 8% of homozygous heph2/heph2 flies reached adulthood. The survival frequency increased to ≈33% and ≈67% when the flies were reared at 21 °C and 17 °C, respectively. The male-sterile heph2-d1 flies were viable at the normal fly-rearing temperature and did not exhibit a temperature-sensitive phenotype. These results indicate that the heph2-d1 mutant affects primarily the male germline function, whereas the heph2 mutant affects the non–sex-specific function(s) as well.
We next tested the effect of the new dmPTB alleles on dmPTB transcript and protein expression. Northern blot analysis revealed that the heph2-d1 allele drastically reduced (barely detectable) the amount of dmPTB transcript as compared with the revertant heph2-ex allele, supporting the correlation between loss of dmPTB and male sterility. However, unlike the heph2 allele, the heph2-d1 allele did not completely abolish the expression of the dmPTB transcript. Similarly, Western blot analysis on protein extracts from male flies (Fig. 3C) revealed a large decrease (barely detectable) in dmPTB protein levels in heph2-d1 flies compared with wild-type and heph2-ex flies.
Loss of dmPTB Results in Defects in Testis Morphology and Spermatid Individualization.
The loss of dmPTB associated with the heph2 allele results in morphological defects to the tip of the testis. In heph2 homozygotes the testis tips were swollen to many times their normal diameter (Fig. 3D, iii vs. i). Examination of the testis tips from heph2-d1 homozygous flies showed that they exhibited a high frequency of testis tip defects (Fig. 3D, ii vs. i), including partially swollen tips with large bumps or knobs.
To characterize spermatogenesis defects in the dmPTB mutants, we dissected testis from flies and prepared testis squashes by compressing the testes between a microscope slide and coverslip to rupture the testis wall and expose all of the stages of spermatogenesis that were present. Heph2 testes showed a lack of motile sperm and a pronounced accumulation of cysts of elongated spermatids (Fig. 3D, vi vs. iv). Testis squashes of heph2-d1 homozygotes revealed unique phenotypes associated with loss of dmPTB. In heph2-d1 testes there was also a lack of individualized nonmotile sperm, which is consistent with its sterility. In addition, many of the cysts (cy) of elongated spermatids appeared to be less tightly bundled and exhibit localized unraveling (Fig. 3D, v vs. iv; ucy). These types of defects are typically found in individualization defective mutants (23), suggesting that loss of dmPTB prevents proper individualization of spermatids.
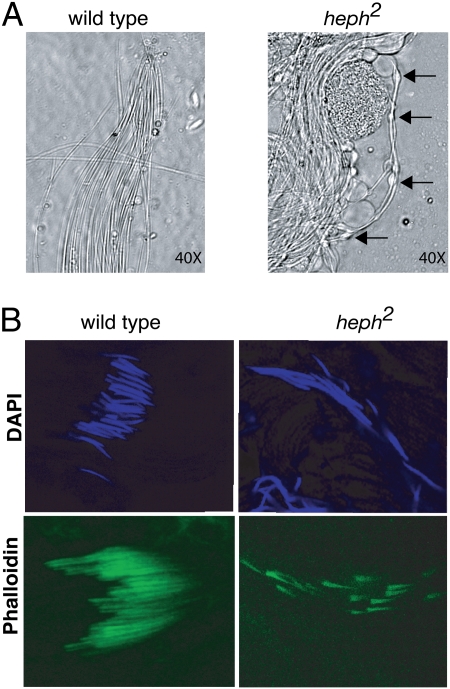
Heph2 testis squashes also exhibited an additional phenotypes characteristic of defects in the spermatid individualization process. A few cysts of elongated spermatids appeared to be partially individualized. Spermatids in cysts often had numerous bulges along their length (Fig. 4A), suggesting aberrant individualization. In contrast, individualized spermatids from wild-type testes exhibited no such bulges (Fig. 4A).
Fig. 4.
dmPTB is necessary for proper functioning of the spermatid IC. (A) Loss of dmPTB in the heph2 mutant results in numerous bulges (arrows) along the length of partially individualized spermatids, a phenotype characteristic of individualization defects. (B) Loss of dmPTB results in interruption of the actin cones of the IC. Anti-phalloidin staining in heph2 males shows that the register of the actin cones is disrupted compared with wild-type actin cones. Anti-DAPI staining indicates that the nuclei of individualizing spermatids in heph2 mutants are disrupted and scattered along the cyst.
Loss of dmPTB Disrupts the Individualization Complex.
To determine how loss of dmPTB affects spermatid individualization, we next examined whether loss of dmPTB affected the spermatid individualization complex (IC), which is composed of highly cross-linked actin cones that surround each individualizing spermatid (23, 24). The IC moves from the basal end of the cyst to the apical end, severing links between spermatids and removing most of the cytoplasm while encasing each spermatid in its own membrane. We stained the actin cones of wild-type and heph2 testes with fluorescently labeled phalloidin, which binds to actin, and with DAPI, which binds to DNA. In wild-type testes, the actin cones were well organized, moved in register (Fig. 4B), and showed regularly organized nuclei of the individualizing spermatids (DAPI). In contrast, in heph2 testes the organization of the actin cones was completely disrupted (Fig. 4B). The cones no longer moved in register, and the nuclei of >90% of the individualizing spermatids became scattered along the cyst (DAPI). These results indicate that dmPTB is necessary for the correct formation and function of the IC in the male germline.
Discussion
The important findings of our present study are that we have characterized the RNA-binding sequence for the Drosophila PTB and that loss of dmPTB specifically results in severe disruption of the actin cones of the spermatid IC. The latter observation represents an identifiable cytological defect resulting from PTB loss. Below we discuss the significance of these findings and reconcile the differences in the functions of the Drosophila and mammalian PTB orthologs, as well as the sex-specific vs. non–sex-specific functions of dmPTB.
Mammalian and Drosophila PTBs Have Similar Sequence Preference.
The functions of mammalian and Drosophila PTBs seem to differ significantly. Mammalian PTB is ubiquitously expressed and regulates (via splicing, polyadenylation, mRNA stability, and translation) a large array of transcripts. In contrast, the predominant mRNA isoform of Drosophila PTB is male germline specific and correlates with male fertility, which is indicative of a highly cell-specific role. This unexpected finding raises an intriguing question as to whether the observed differences between the two organisms relate to differential regulation of expression, or differences in preferred RNA-binding sequences. Whereas three alternative splice variants of PTB (PTB1, -2, and -4) and a neuronal homolog of PTB (nPTB) exist in humans/mammals, there is only one gene (dmPTB) in Drosophila (Fig. 5A). Although there are functional differences between PTB1 and nPTB (2, 3) and between PTB1 and PTB4 (25), our phylogenetic sequence comparison indicates that PTB1 and nPTB are more similar to each other than to dmPTB and that dmPTB has diverged equally from the two human isoforms (Fig. 5B). Our combined results show that the mammalian and Drosophila PTB proteins have similar RNA binding preferences for pyrimidine-rich sequences with long uninterrupted pyrimidine tracts (Fig. 1B) (8, 26, 27). Thus, we favor that the observed differences in function between Drosophila and mammalian PTB are likely due to differential regulation of expression rather than observable differences in RNA-binding function. According to the expressed sequence tags (EST) database (www.flybase.org) there are two promoters for the dmPTB gene (Fig. 5A). The male germline-specific dmPTB uses the distal promoter (Fig. 5A) (19), resulting in abundant expression of dmPTB protein in primary spermatocytes in males (Fig. 2). Abundant expression of the male germline-specific dmPTB transcript and protein accounts for the male-specific expression of dmPTB. It will be interesting to determine whether there is a role for PTB in mammalian/human testis.
Fig. 5.
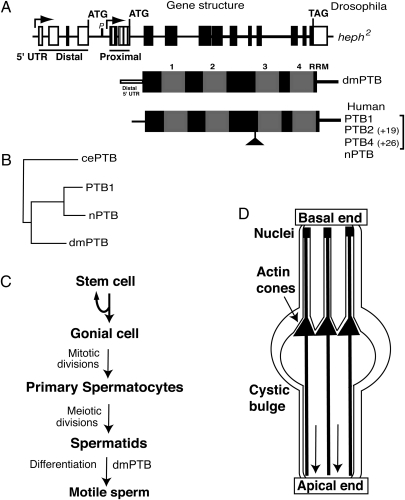
Schematic representations of the dmPTB gene, PTB protein structures, and spermatogenesis. (A) dmPTB gene structure. Arrows represent two transcription start sites. Distal and proximal 5′ UTRs are shown. Exons are rectangles, and lines are introns. The RNA-binding domain of dmPTB is identical for both transcript sets. Protein variants for Drosophila (dmPTB) and human PTB (PTB1, -2, -4, and nPTB) are shown; the PTB variants are also referred to as PTBP1, where 1, 2, and 4 isoforms are labeled as c, b, and a, respectively, and nPTB is referred to as PTBP2 (National Center for Biotechnology Information database). (B) Phylogenetic sequence divergence between dmPTB and the Caenorhabditis elegans (cePTB) and human PTB variants (PTB1 and nPTB) using ClustalW alignment. (C) Schematic of spermatogenesis. The first identifiable defect (spermatid individualization) in the dmPTB mutants is indicated. (D) Spermatid individualization. The IC of actin cones (solid triangles) migrates from the basal end to the apical end (shown by the direction of the downward arrows). Cystic bulge near the migrating actin cone of the IC is shown. Only 3 of the 64 spermatids are shown, for clarity. During the individualization process each axoneme (solid lines from top to bottom) is encased in its own membrane, removing the organelles and cytoplasm and severing interspermatid connections. Sperm nuclei are indicated. Modified from refs. 20 and 21.
Male Germline-Specific vs. Non–Sex-Specific Functions of dmPTB.
Given that the mammalian PTB protein shows ubiquitous expression and binds a wide spectrum of transcripts, it is intriguing why the heph2 mutant shows a highly cell-specific, rather than an expected pleiotropic phenotype. On the basis of our combined results and the EST database, the simplest explanation is that the dmPTB protein is produced from the distal promoter (which would be highly active only in the primary spermatocytes) and provides the sex-specific function in the male germline, whereas the protein from the proximal promoter transcript provides the non–sex-specific function in somatic cells; the RNA-binding portion of dmPTB is identical for transcripts from distal and proximal promoters. The major dmPTB isoform found predominantly in males may be necessary for the male germline function, whereas the dmPTB protein isoforms expressed at much lower levels may be important for non–sex-specific function(s). This scenario would also reconcile the discrepancy between the male sterility of the heph2 and heph2-d1 mutants (Fig. 3A) and non–sex-specific lethality of other dmPTB alleles (28). Given the location of the P-element insertion the heph2 and heph2-d1 alleles could disrupt the abundant distal transcript in the male germline but not the minor proximal transcript, whereas other alleles of dmPTB may disrupt the proximal transcript (Figs. 3A and 5A).
The significance of the much higher expression levels of the male germline-specific isoform of dmPTB remains unresolved. It is possible that a need for higher levels of regulatory protein in the male germline may result from a requirement for higher amounts of the downstream mRNA target(s), for a need to regulate multiple targets, and/or from lower affinity of the binding site or need for efficient regulation. Alternatively, dmPTB may play a structural rather than a regulatory role, requiring stoichiometric amounts of dmPTB. Future studies should distinguish between these possibilities.
Loss of dmPTB Disrupts the IC.
Spermatid individualization is an important cell biological process involving a discrete actin structure that carries out an extraordinary amount of membrane remodeling (24). The dmPTB mutants exhibit numerous defects that are characteristic of disruption of spermatid individualization during spermatogenesis (Figs. 3 and 4) (23, 29, 30). These include cysts of elongated but nonindividualized spermatids, less tight bundling of spermatids, localized unraveling of cysts, accumulation of debris in the testes, numerous bulges along the lengths of spermatids, and disruption of the actin cones of the ICs.
Although several mutants in other genes exhibit defects during spermatid individualization (e.g., purity of essence, jaguar, fuzzy onions, and cytochrome C) (23, 30, 31), the phenotypes associated with loss of dmPTB closely resemble the “expressway” group of IC mutants, which includes the genes crossbronx (cbx), long island expressway (lie), and clathrin heavy chain (Chc) (23). The expressway mutants are characterized by disrupted ICs and spermatid nuclei scattered along the cyst. In addition, expressway mutants [e.g., Chc4 and lie ms(2)42A] are often semilethal, indicating that like dmPTB they are required for vital functions outside of spermatogenesis. Two hypotheses for the disrupted IC and scattered nuclei phenotype of the expressway mutants have been postulated (23) and may be relevant to dmPTB. First, a defective IC may assemble around an intact nuclear bundle, and translocation of the IC then drags the nuclei along the cysts. Second, spermatid nuclei may initially be arranged all along the cyst, and actin cones assemble around each nucleus.
Although dmPTB has been to shown play a role in Notch signaling during wing development (28) and repression of translation of the oskar mRNA during oogenesis (32), the identity or regulation of germline target(s) of dmPTB is currently unknown. Given the shared phenotype with the expressway mutants, it is tempting to speculate that genes relevant for the structure or function of the IC may be potential targets of PTB in Drosophila. Because the mammalian PTB regulates members of the actin cytoskeleton apparatus, such as α- and β-tropomyosin and the actin crosslinker α-actinin, such targets may explain the loss of register observed in the highly cross-linked actin cones of the heph2 IC in Drosophila. Alternatively, mammalian PTB2 plays a role in mRNA stabilization (33). Because most transcription in spermatogenesis occurs during premeiotic stages but translation also occurs at postmeiotic stages (29), it is possible that dmPTB may act in primary spermatocytes to preserve mRNAs until they are translated at later meiotic stages.
In conclusion, we have characterized the RNA-binding site for the dmPTB and have discovered a direct link between dmPTB and specific cytological defects (spermatid individualization) during spermatogenesis. This not only connects an RNA-binding protein to an important cellular process but also provides an important framework for defining the molecular function of dmPTB. These findings have significantly advanced our understanding of the basis for the male germline-specific function of dmPTB and should facilitate future identification of its male germline-specific targets.
Materials and Methods
Fly Strains, Northern, Western, and SELEX.
The P-element excision alleles were generated by using the standard protocol (34). The heph2 flies were from the Bloomington Stock Center and raised (35). For fertility assay, progeny from three matings of relevant males and virgin females were counted. Northern analysis using dmPTB and rp49 probes was performed as previously described (19). Western blots were probed with anti-dmPTB antibodies (1:1,000 dilution) and goat anti-rabbit IgG secondary coupled to HRP and developed using Chemiluminescent Substrate (Pierce). Antibodies were generated against recombinant dmPTB-RRM3 and -4 fragment in rabbits by Covance. Random library SELEX was performed as previously described (8).
Testes Squashes and Testis Staining with Anti-dmPTB-3/4.
Testes squashes were performed as previously described (19). For immunostaining, testes were fixed in paraformaldehyde, incubated with anti-dmPTB antibodies [1:2,500 dilution in 2% BSA in phosphate buffered saline with 1% triton (PBST)], washed with PBST several times, incubated with goat anti-rabbit antibodies (at 1:500 dilution) coupled to Alexa fluor, and viewed using a Leica Confocal microscope.
Visualization of the ICs.
As described previously (24), testes were fixed with 4% formaldehyde, incubated with glycine (20 mg/mL of PBST), permeabilized in 1% TritonX-100 in PBST, stained with 5 μL of Alexa Fluor 488 phalloidin stock or with DAPI solution, mounted, and viewed using a Leica TCS SP2 AOBS confocal microscope.
Acknowledgments
We thank Drs. Tom Blumenthal, Brian DeDecker, Cedric Wesley, and members of the Singh laboratory, especially Joseph Heimiller, for helpful discussions and critical reading of the manuscript. Dr. Robert Boswell (University of Colorado, Boulder, CO) generously provided anti-actin antibodies. This work was supported in part by a grant from the American Cancer Society (to R.S.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 2.Coutinho-Mansfield GC, Xue Y, Zhang Y, Fu XD. PTB/nPTB switch: A post-transcriptional mechanism for programming neuronal differentiation. Genes Dev. 2007;21:1573–1577. doi: 10.1101/gad.1575607. [DOI] [PubMed] [Google Scholar]
- 3.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babic I, Sharma S, Black DL. A role for polypyrimidine tract binding protein in the establishment of focal adhesions. Mol Cell Biol. 2009;29:5564–5577. doi: 10.1128/MCB.00590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gromak N, et al. The PTB interacting protein raver1 regulates alpha-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R, Valcárcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 9.Oberstrass FC, et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 10.Lin CH, Patton JG. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 11.Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monie TP, et al. The polypyrimidine tract binding protein is a monomer. RNA. 2005;11:1803–1808. doi: 10.1261/rna.2214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Y, et al. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izquierdo JM, et al. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci USA. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlet-B N, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 17.Lin JC, Tarn WY. Exon selection in alpha-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol Cell Biol. 2005;25:10111–10121. doi: 10.1128/MCB.25.22.10111-10121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromak N, Matlin AJ, Cooper TA, Smith CW. Antagonistic regulation of alpha-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA. 2003;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robida MD, Singh R. Drosophila polypyrimidine-tract binding protein (PTB) functions specifically in the male germline. EMBO J. 2003;22:2924–2933. doi: 10.1093/emboj/cdg301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller MT. Genetic control of cell proliferation and differentiation in Drosophila speratogenesis. Semin Cell Dev Biol. 1998;9:433–444. doi: 10.1006/scdb.1998.0227. [DOI] [PubMed] [Google Scholar]
- 21.Castrillon DH, et al. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: Characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 23.Fabrizio JJ, Hime G, Lemmon SK, Bazinet C. Genetic dissection of sperm individualization in Drosophila melanogaster. Development. 1998;125:1833–1843. doi: 10.1242/dev.125.10.1833. [DOI] [PubMed] [Google Scholar]
- 24.Noguchi T, Miller KG. A role for actin dynamics in individualization during spermatogenesis in Drosophila melanogaster. Development. 2003;130:1805–1816. doi: 10.1242/dev.00406. [DOI] [PubMed] [Google Scholar]
- 25.Wollerton MC, et al. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolupaeva VG, Hellen CU, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 27.Clerte C, Hall KB. The domains of polypyrimidine tract binding protein have distinct RNA structural preferences. Biochemistry. 2009;48:2063–2074. doi: 10.1021/bi8016872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dansereau DA, Lunke MD, Finkielsztein A, Russell MA, Brook WJ. Hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster. Development. 2002;129:5553–5566. doi: 10.1242/dev.00153. [DOI] [PubMed] [Google Scholar]
- 29.Fuller MT. Spermatogenesis. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1993. pp. 71–147. [Google Scholar]
- 30.Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 31.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 32.Besse F, López de Quinto S, Marchand V, Trucco A, Ephrussi A. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 2009;23:195–207. doi: 10.1101/gad.505709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M, Hecht NB. MSY2 and polypyrimidine tract binding protein 2 stabilize mRNAs in the mammalian testis. Int J Androl. 2008;31:457–461. doi: 10.1111/j.1365-2605.2008.00885.x. [DOI] [PubMed] [Google Scholar]
- 34.Robertson HM, et al. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M. Drosophila. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. p. 2v. [Google Scholar]