Abstract
The emergence of systems biology has reemphasized the advantages of understanding biological processes with a global perspective. One biological process amenable to global approaches is microbial metabolism. This review describes a model system that contributes to the goals of systems biology by experimentally defining metabolic integration found in a bacterial cell and thus providing data needed for implementation and interpretation of systems approaches. We have taken a largely unbiased in vivo approach centered on thiamine biosynthesis to identify new metabolic components and connections and explore uncharacterized paradigms of the integration amongst them. This article summarizes recent results from this approach that include the identification of the function of unknown genes, connections between cofactors biosynthesis and thiamine biosynthesis, and how metabolites from one biosynthetic pathway can be used in thiamine biosynthesis.
From single genes to complex systems: the need to unravel metabolic integration
The interest in understanding microbial metabolism has a long history. In the early years of bacterial genetics, it was proposed that the phenotypes of microbial mutants could facilitate the identification of the function of individual genes. The idea that mutants lacking a particular enzyme could be identified by phenotypic analysis and provide information about the missing reaction was particularly attractive. For instance, if a mutant that required exogenous histidine for growth lacked one of the biochemical activities required for the synthesis of histidine, one could learn a great deal about the relevant gene or protein. Through the decades these types of analyses have contributed to a valuable, if incomplete, catalog of pathways and processes present in microbial cells. Strains in which a mutant phenotype could not be explained by the loss of a single biochemical activity were noted in these early studies, but rarely characterized further. In fact, the characterization of such mutants was not feasible without the technical advances (e.g. PCR, sequencing, etc.) that now facilitate the rapid identification of a mutation.
The advent of global approaches to probe metabolism has partially displaced the one-gene, one-function approach to piecing together metabolism. Progress in transcriptional profiling, computational biology, metabolomics and other ‘-omics’ approaches has allowed scientists to obtain global snapshots of specific features of the metabolic network. While these approaches are increasing our understanding of the metabolic system as a whole, in vivo genetic analysis remains a crucial component of efforts to define new functions and unexpected paradigms that can then be incorporated into and validate the global approaches.
This article focuses on the use of thiamine biosynthesis in Salmonella enterica as a model system to expand our understanding of the metabolic network in the cell (Box 1). The topics discussed here highlight the benefits of using this pathway to uncover, (i) function of uncharacterized genes, (ii) unpredicted metabolic connections, and (iii) inherent potential for robustness in the metabolic network. Several characteristics of this metabolic pathway have made it ideal as a model system for this purpose [1]. Below we detail some recent findings and explore how this approach has provided unpredicted results that increase knowledge of bacterial metabolism and are distinct from those obtained through other efforts.
Box 1. Thiamine is an essential cofactor.
Thiamine was the first vitamin to be discovered and was isolated in 1932 from yeast [52]. This vitamin is an essential component of the human diet and a deficiency in humans causes the disease Beri-beri. Thiamine can synthesized de novo by bacteria, archaea, yeast and plants. The synthesis of thiamine involves the separate formation of a thiazole moiety (4-methyl-5-β-hydroxyethylthiazole phosphate [THX-P]) and a pyrmidine motiey (4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate [HMP-PP]). In bacteria, these two moieties are combined to form thiamine monophosphate by thiamine phosphate synthase. The active form of the cofactor, thiamine pyrophosphate (TPP) is then generated by a single phosphorylation catalyzed by thiamine monophosphate kinase [53]. A schematic of the thiamine biosynthetic pathway from S. enterica is shown in Figure 1.
TPP is a key cofactor for several enzymes in central carbon metabolism, including transketolase, α-ketoglutarate dehydrogenase, pyruvate dehydrogenase, and acetolactate synthase. The role of thiamine involves the stabilization of acyl carbanion intermediates through dissociation of the proton at C2 of the thiazolium ring [54].
Recent studies have identified other forms of thiamine that do not function as cofactors in the cell. Thiamine triphosphate has been identified in Escherichai coli [55] and rat brain cells where it was found to activate chloride channels [56] and be a phosphate donor in the phosphorylation of a synaptic protein [57]. The synthesis of thiamine triphosphate is performed in the mitochondira of rat brain cells and is driven by energy from the respiratory chain [58]. Additionally, adenylated thiamine pyrophosphate has been identified in E. coli and found to act as a signal molecule in response to carbon starvation [59].
Metabolic integration is a component of systems biology
The field of systems biology seeks to integrate data generated by decades of experimental (often focused) studies using mathematical approaches that describe a biological process as a complex system. Several recent reviews describe the approaches and technologies inherent in these approaches [2–6]. One prominent effort in systems biology is the design and implementation of mathematical models to predict the metabolic capacity of an organism from genome content. Such models strive to predict the full capacity of a system based on knowledge of the components and connections within the system. Because of the breadth of organisms involved, these predictions often rely extensively on sequence similarity to experimentally defined systems. Thus, experimental data are often the components that limit the ability of the mathematical models to fully and realistically predict the capacity of a system. Current network reconstructions and analysis used in the creation of the mathematical models cannot be used to identify unknown metabolic reactions or connections [7]. For this reason elucidation of components and connections and the rules that govern their interactions by experimental approaches are essential to facilitate continuing application of mathematical models to metabolism. In vivo analysis in microbial systems provides a powerful way to dissect metabolic integration and thus assist in identifying the metabolic information that is needed for mathematical models to be more complete.
Metabolic integration can be defined as the connections between biochemical processes and pathways mediated by metabolites. Efforts to dissect metabolic integration start with the premise that beyond the enzymatic framework of metabolism, there is a network of interactions mediated by metabolites. Further, it is argued that these interactions contribute to the robustness responsible for adapting to perturbations caused by temporary and/or subtle environmental changes. This inherent network robustness is distinct from the evolutionary changes required to generate a new pathway or to adapt to a constant environmental stress. A complete understanding of metabolic integration will require knowledge of the roles and concentrations of each metabolite in the cell. This understanding will allow modeling approaches to reach their potential in describing metabolism.
The potential of a systems approach for exploiting metabolism is emphasized by the fact that the mathematical model of metabolism in Escherichia coli has been used to address a variety of basic and practical applications (reviewed in Ref. [8]). The model has been used for in silico strain design for optimized production of desired products for industrial use, such as amino acids [9], lycopene [10] ethanol [11] and hydrogen [12]. Additionally, the use of this model allowed for the quantitative interpretation of the physiology of both wild-type and genetically perturbed E. coli strains [13]. Thus, despite the fact that only a subset of the functions encoded by the E. coli genome are included, this model has allowed useful observations and generated significant metabolic data [8]. While a full understanding of metabolism as a complex system is far from complete, progress to date emphasizes the potential of systems approaches that will be achieved as our understanding of the components and connections continues to increase.
Thiamine biosynthesis provides one model system to dissect metabolic integration
Thiamine biosynthesis in Salmonella enterica has served as a model system for biosynthetic pathways that can be exploited to probe metabolic integration in vivo. The use of this system is predicated on the fact that growth phenotypes provide a means to assess the function of the complete system. When caused by a single lesion, a mutant phenotype can be used to dissect the biochemistry underpinning this behavior. This analysis in turn provides information about how the reactions affected by the mutation are integrated in the metabolic network of the wild-type cell, hence broadening our understanding of the system [1].
Thiamine biosynthesis has two characteristics that make it a productive model for analysis of metabolic integration. First, thiamine is an essential nutrient and growth in the absence of exogenous thiamine implies endogenous synthesis is occurring. This truism allows conclusions about the adjustments that can be made in the metabolic network to compensate for a perturbation. Second, the carbon flux required to generate the quantity of thiamine required for growth is low. Thus, this system is sensitive enough to detect slight changes in metabolic flux - an important feature of the system needed to assess inherent or generated robustness.
Conditional requirements provide a portal for dissecting metabolic integration
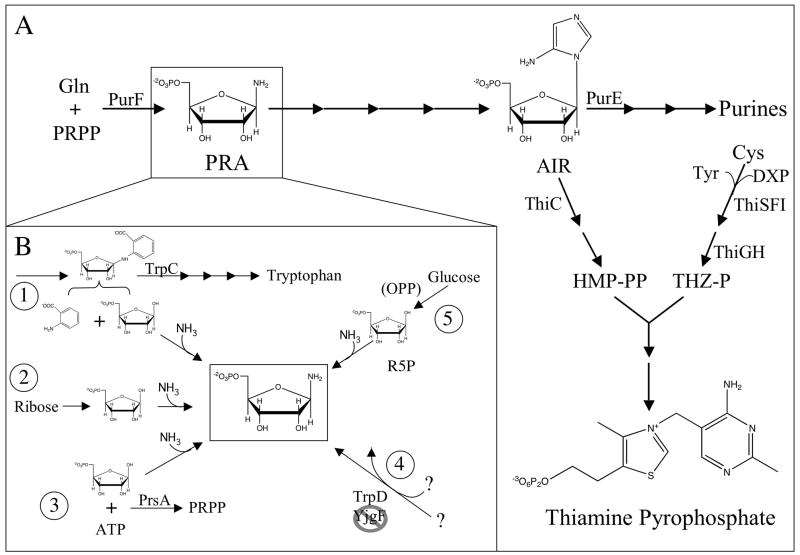
Figure 1 is a schematic representation of thiamine biosynthesis. Salmonella enterica strains completely lacking glutamine phosphoribosylpyrophosphate (PRPP) amidotransferase (EC 2.4.2.14), the product of the purF gene, generate sufficient thiamine for growth under specific conditions [14, 15]. The conditional requirement for thiamine provides a means to identify mutations that increase or decrease accumulation of phosphoribosylamine (PRA), the product of PurF. Characterization of such mutations has been a powerful strategy to identify proteins and processes that affect the synthesis of PRA and/or the efficiency of the subsequent biosynthetic steps. In the absence of PurF, flux to thiamine synthesis is constrained in such a way that under some growth conditions, inherent alternative synthesis of PRA is sufficient for growth, but only if the remaining enzymatic steps function at optimal efficiency. In a sense, thiamine synthesis in a purF mutant is at a tipping point, any increase in synthesis or accumulation of PRA facilitates synthesis, and a change that compromises any of the subsequent biosynthetic steps prevents growth. Characterization of mutations that impact the thiamine biosynthetic process positively or negatively has identified numerous interactions between thiamine synthesis and other areas in metabolism. In some cases the mechanism of the connection is understood, whereas in others investigation is ongoing (Box 2).
Figure 1. The joint thiamine-purine biosynthesis pathway.
(a) Schematic representation of the purine-thiamine biosynthetic pathways in S. enterica. Relevant gene products are noted above the step they catalyze. Not all steps are shown. (b) Schematic representation of the mechanisms for PRA formation that are distinct from PurF. (i) The non-enzymatic breakdown of PR-anthranilate to anthranilate and R5P allows for the non-enzymatic formation of PRA [21]. (ii) High levels of R5P generated when ribose is the sole carbon source allows for non-enzymatic formation of PRA [19]. (iii) R5P can be diverted for non-enzymatic PRA formation when PrsA activity is reduced [19]. (iv) TrpD (bifunctional glutamine amidotransferase/anthranilate phosphoribosyltransferase) is necessary and sufficient for PRA formation in the absence of YjgF [23]. (v) The contribution of R5P from the OPP pathway allows weak non-enzymatic formation of PRA [15]. Abbreviations: AIR, aminoimidazole ribotide; Cys, cysteine; DXP, 1-deoxyxylulose-5-phosphate; Gln, glutamine; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine-pyrophosphate; OPP, oxidative pentose phosphate pathway; PRA, phosphoribosylamine; PRPP, phosphoribosylpyrophosphate; R5P, ribose-5-phosphate THZ-P, thiazole phosphate; TrpD, bifunctional glutamine amidotransferase/anthranilate phosphoribosyltransferase; Tyr, tyrosine.
Box 2. Loci that indirectly impact thiamine biosynthesis.
In the past 15 years lesions in numerous loci have been isolated through their positive or negative affects on thiamine biosynthesis. In some cases the affected loci were of known function and a model could be proposed to describe the mechanism of integration between the relevant locus and thiamine synthesis. In other cases the loci were partially or completely uncharacterized. In these cases, the impact of mutations in these loci on thiamine synthesis provided a tool to pursue the functional role of these gene products in the context of the metabolic network. In all cases, the loci were identified by exploiting a conditional requirement for thiamine that is generated by a lack of the PurF enzyme. On gluconate minimal medium supplemented with adenine, a purF mutant does grow, indicating that there is endogenous synthesis of phosphoribosylamine (PRA) that is sufficient for thiamine synthesis in the absence of PurF. Mutations that eliminate this thiamine-independent growth but are not in known thiamine biosynthetic enzymes can be identified (Table 1). By contrast, the same purF mutant strain fails to grow in the absence of thiamine if the carbon source is glucose. Mutations that allow thiamine-independent growth in this situation have modified the metabolic network to increase the flux to thiamine formation (Table 1). Each of these classes of mutations have been characterized and continue to be characterized to elucidate the mechanism of integration with thiamine synthesis and the functional role of the relevant gene product. Together these analyses have expanded our knowledge of the metabolic network anchored in thiamine biosynthesis.
Increased accumulation of PRA identifies new metabolic links
Evidence that residual PRA is formed in the absence of PurF was obtained by blocking the branchpoint enzyme PurE (Figure 1). In a purF purE double mutant thiamine is not required for growth [15]. This result demonstrated that there was endogenous generation of PRA and it was sufficient for thiamine synthesis if its diversion to purine biosynthesis (via aminoimidazole ribotide [AIR]) was prevented. Introduction of a block in the oxidative pentose phosphate (OPP) pathway prevented this growth and allowed the conclusion that residual PRA was derived from the ribose-5-P (R5P) intermediate of this pathway [15, 16]. Thus, selections for mutations that allowed new PRA synthesis were subsequently preformed in a purF gnd double mutant (gnd encodes 6-phosphogluconate dehydrogenase).
Non-enzymatic PRA formation can be physiologically relevant
Recent results have shown that the non-enzymatic formation of PRA can be significant for growth under some conditions. With ribose as a carbon source in minimal medium [17, 18], a purF gnd strain produces sufficient PRA for thiamine biosynthesis. Subsequent results supported the conclusion that a combination of the 50 mM ammonia present in the medium and the elevated level of R5P caused by ribose catabolism allowed non-enzymatic synthesis of PRA sufficient for thiamine synthesis [19]. These experiments led to a phenotype diagnostic for non-enzymatic PRA synthesis that has been used to identify the mechanism of growth suppression in a number of mutants (Figure 1b, Table 1).
Table 1.
Lesions in numerous loci impact the thiamine biosynthetic pathway
| Lesions that allow thiamine-independent growth of a purF mutant on glucose minimal media | |||
|---|---|---|---|
| Locus | Function | Mechanism of integration | Refs |
| purE | Phosphoribosylaminoimidazole carboxylase catalytic subunit (EC 4.1.1.21) | Diverts all PurF-independent PRA formed to thiamine synthesis | [15] |
| prsA | Phosphoribosylpyrophosphate synthetase (EC 2.7.6.1), essential enzyme | Compromised PrsA activity results in accumulation of substrate R5P and allows non-enzymatic PRA synthesis | [19] |
| trpC | Phosphoribosyl-anthranilate isomerase (EC 5.3.1.24) and indole-3-glycerol-phosphate synthase (EC 4.1.1.48) | Compromised TrpC activity: substrate accumulates, breaks down to increased R5P and allows non-enzymatic PRA synthesis | [21] |
| yjgF | Unknown, member of the YjgF/YER057c/UK114 family of proteins that exist in all three domains of life | Unknown, TrpD is necessary and sufficient for synthesis of PRA. Direct formation hypothesized | [42] |
| pykAF | Pyruvate kinase isozymes I and II (EC 2.7.1.40) | Unknown, allows for thiamine synthesis in gluconate media in the presence of additional mutations | [60] |
| Lesions that prevent thiamine independent growth of a purF mutant on gluconate minimal media | |||
| gnd | 6-phosphogluconate dehydrogenase (EC 1.1.1.44) | Decreased cellular R5P levels on glucose or gluconate media | [16] |
| zwf | Glucose-6-phosphate 1-dehydrogenase (EC 1.1.1.49) | Decreased cellular R5P levels on glucose media | [15] |
| panE | 2-dehydropantoate 2-reductase (EC 1.1.1.169) | Decreased cellular CoA levels due to a defect in pantothenate biosynthesis | [61, 62] |
| nuo | NADH dehydrogenase complex I | Unknown | [63] |
| iscRSUA | Housekeeping [Fe-S] cluster assembly proteins | Disrupts Fe-S cluster metabolism | [64] |
| apbC | [Fe-S] cluster carrier protein | Disrupts Fe-S cluster metabolism | [65, 66] [27] |
| abpE | Unknown lipoprotein, shares homology with the C-terminus of RnfF, a putative membrane complex protein involved in electron transport to nitrogenase, from Rhodobacter capsulatus | Disrupts Fe-S cluster metabolism | [26, 67] [68] |
| rseC | SoxR reducing system protein | Disrupts Fe-S cluster metabolism | [30] [35] |
| gshA | Glutamate-cysteine ligase (EC 6.3.2.2) | Disrupts Fe-S cluster metabolism, unable to produce glutathione | [26] |
The conditional thiamine auxotrophy in derivatives of purF mutant strains offered the means to identify connections to thiamine biosynthesis using suppressor analysis. Mutations that restore growth in the absence of thiamine might be expected to either uncover functional redundancy with PurF, or alter the metabolic network in a way that satisfies the thiamine requirement some other way. The data have shown that thiamine-independent growth was restored most often by changes in metabolite distribution rather than uncovering functional redundancy. This finding was informative as the system was constrained such that one pathway could not be crippled to restore another. Rather, a balance had to be met that provided sufficient function of each of the pathways involved to allow growth (Box 3). The examples uncovered and described below suggest that the equilibrium of the metabolic system can be set in several ways depending on the specific set of requirements for growth. This finding raises the question of what the selection for the ‘normal’ equilibrium setting is. In other words, is there more than one set-point that can result in equally fit organisms? Or does altering pathway flux diminish fitness in a way not apparent in the laboratory setting? Mutations in these analyses might represent natural variants that exist in nature and thus reflect potential flux balances that can arise. While a phenotypic change might only be detectable when a mutation has caused altered flux, the situation represents the potential for dissecting the metabolite cross-talk that can occur in natural organisms or settings not yet described.
Box 3. Positive selections for loss-of-function mutations.
Mutations that restored thiamine synthesis in a purF mutant strain often did so by causing reduction or loss of activity of a relevant gene product (Table 1). This result was surprising as an altered gene product with a new or enhanced activity would be expected to generate a gain-of-function phenotype. Further analyses showed that thiamine biosynthesis provides a system to identify ways to satisfy a metabolic need by altering the production capacity of an enzyme or pathway. This system allowed selections of mutations that constrained flux through various pathways. These mutations resulted in variants with reduced activity that allowed accumulation of the relevant metabolites for re-routing, while maintaining enough activity to produce a necessary product. While there is no obvious way to predict the positive effect a decrease in activity will have, this finding illustrates an additional benefit from an unbiased genetic approach. After the target of a positive selection is identified, the selection can be exploited to study the structure and function of the key, possibly essential, enzyme in the cellular setting.
Protein variants with decreased activity have been isolated in metabolic pathways through their ability to restore growth in a various selections. Variants of threonine dehydratase (EC 4.3.1.19), IlvA [51], PRPP synthetase (EC 2.4.2.14), PrsA [19], phosphoribosyl-anthranilate isomerase (EC 5.3.1.24) and indole-3-glycerol-phosphate synthase (EC 4.1.1.48; TrpC) [21] were isolated with positive selections that demanded restoration of growth. In some cases the specific activity of the enzymes was decreased 45-fold [19].
Studies addressing metabolic integration with in vivo approaches identified two instances where restoration of growth was caused by global changes in regulation that could not be anticipated and were too complex to dissect easily. A mutant allele of the essential gene gyrA, which encodes part of the DNA gyrase complex, was identified by a positive selection for metabolic function. Further analysis showed that the variant protein resulted in reduced negative supercoiling [69] and changed expression of almost every gene tested. No single gene could be identified as the cause of the phenotype. Additionally, a S506F variant of RNA polymerase sigma factor RpoD restored thiamine synthesis in a specific purF mutant background [70]. Analysis of this variant RpoD showed that is had been previously described in the class of ppGpp-independent mutants [71]. We confirmed that other members of this class of rpoD mutants have altered transcription on a global level and restored PurF-independent thiamine synthesis in specific situations. These findings have provided a means of positive selection that can be utilized by researchers interested in dissecting the role of ppGpp and/or supercoiling in gene regulation.
Metabolites can be diverted and used for PRA synthesis
In vivo suppressor analysis allowed the identification of flux changes in two different pathways that caused diversion of metabolites to be used for PurF-independent PRA formation. The trpC gene encodes the bifunctional enzyme N-(5′-phosphoribosyl)anthranilate (PR-anthranilate) isomerase (EC 5.3.1.24) and indole-3-glycerol-phosphate synthase (EC 4.1.1.48) required for tryptophan biosynthesis. Alleles of trpC that compromise the enzymatic activity of TrpC allow PurF-independent PRA synthesis (Figure 1b). In such a strain, PRA synthesis is non-enzymatic and results from the accumulation of PR-anthranilate, the substrate of TrpC, which breaks down to R5P and anthranilate [20, 21]. The increased levels R5P combine with ammonium in the medium to generate sufficient PRA for thiamine biosynthesis [19, 21].
A similar mechanism for PRA formation was a consequence of alleles of prsA, the gene encoding phosphoribosylpyrophosphate synthetase (PrsA, EC 2.4.2.14). PrsA is an essential enzyme that uses R5P and ATP to form PRPP, that is subsequently used in the de novo synthesis of purines, pyrimidines, histidine, tryptophan and nicotinamide coenzymes [22]. The relevant suppressing alleles resulted in variants of PrsA with reduced activity. Consequently, the level of R5P increased, and was used for the non-enzymatic formation of PRA when ammonium was present [19].
Further, in a yjgF mutant strain (see below), the PurF-independent PRA formation requires the presence of TrpD, bifunctional glutamine amidotransferase and anthranilate phosphoribosyltransferase (EC 4.1.1.48, 2.4.2.18) [23]. In the absence of YjgF, PRA is not being generated by a non-enzymatic mechanism [19]. The current model for the function of YjgF to explain this result is described below.
Indirect effects on thiamine synthesis lead to new insights into genes of unknown function
The work described above involved primarily pathways and gene products that had been previously characterized and showed them to be integrated with thiamine synthesis. However, the thiamine system has been equally productive in identifying the function of gene products whose physiological role was unknown.
Compromised thiamine synthesis results from defects in [Fe-S] metabolism
A number of loci with unassigned function were identified involving lesions that resulted in a unique thiamine requirement. Unlike lesions in characterized biosynthetic genes that disrupt one of the two biosynthetic branches, these mutations caused a requirement for either thiamine or both the HMP and THZ moieties. This result indicated that these single lesions led to defects in two distinct pathways (Figure 1a). Further analysis showed that these requirements were suppressed by anaerobic growth and that a lesion in either gshA or the isc operon produced similar phenotypes. Based on the known role of the latter two loci, it was hypothesized that the new loci (apbC, apbE, rseC, etc) were involved in some aspect of [Fe-S] cluster metabolism. Biochemical data in support of this prediction have been reported [24–30]. Subsequent to the identification of ApbC as a protein that was involved in [Fe-S] cluster metabolism in S. enterica, researchers identified ApbC homologs in multiple organisms (including eukaryotes). Studies with several of these proteins, in addition to ApbC, have led to the in vitro functional characterization of the ApbC/Nbp35 family of proteins as [Fe-S] cluster carriers [27, 31–34]. Similarly, RseC was shown to be involved in the re-reduction of the [Fe-S] cluster in the redox-sensitive transcriptional activator SoxR, confirming a role for this protein in [Fe-S] cluster metabolism [35]. Thus the phenotypic analysis of indirect effects led to a general functional assignment for several previously uncharacterized genes and made significant contributions to the field of [Fe-S] cluster metabolism that were distinct from those generated by the researchers focused on this field. In vivo phenotypes continue to inform the efforts in the biochemical characterization of these and other proteins connected with these findings.
Biochemical insights gleaned from in vivo connections
The assignment of apbC, apbE, rseC and yggX genes to the general function of [Fe-S] cluster metabolism provided information on one half of an integrated network. The next question was how the disruption of [Fe-S] metabolism compromised not one, but two branches of thiamine synthesis. Nutritional studies led to the suggestion that the target in the THZ branch was ThiH [26]. This prediction has been verified by the finding that ThiH is a member of the SAM radical superfamily of proteins, characterized by oxygen-labile [Fe-S] clusters [36, 37]. The target in the HMP branch was more problematic as a single gene product (ThiC) had been implicated in the conversion of AIR to HMP. ThiC does not have a canonical [Fe-S] binding motif and had been reported not to have an [FeS] cluster [38]. Encouraged by strong in vivo data, ThiC was purified anoxically and found to be active for the conversion of AIR to HMP [39, 40]. Analysis of this protein showed that ThiC contains a [4Fe-4S] cluster essential for activity, and that interaction with S-adenosylmethionine (AdoMet) generates a novel backbone radical presumed to be on a histidine residue [41]. Based on these and additional studies, ThiC was suggested to define a new class in the AdoMet radical superfamily of proteins [39, 41]. Significantly, results from the in vivo studies predicted integration between [Fe-S] cluster metabolism and ThiH-ThiC before the relevant proteins were considered to be cluster-containing proteins based on other precedents. The fact that each of these proteins has now been found to have the biochemical and physical properties predicted based simply on phenotypic analysis and genetic logic provides a powerful proof of concept and emphasizes a benefit of the approach to studying metabolism that we favor.
Identification of a broadly conserved gene of unknown function by its impact on thiamine synthesis
The only insertional mutations that allow the thiamine-independent growth of a purF gnd double mutant are lesions in yjgF [42]. This gene is a founding member of the YjgF/YER057c/UK114 family of proteins that is conserved in genomes from microbes to human [43]. Multiple high-resolution structures of this protein have been published and homologs have been implicated in diverse cellular processes [43–49]. However, thus far no unifying in vitro biochemical activity has been defined for this protein. Based on several studies it has been proposed that YjgF is involved in eliminating toxic metabolites of the keto-acid class [50, 51] and it was suggested that a yjgF mutant has a metabolite profile that is distinct from a wild-type strain. This model is supported by in vivo work and two structural studies documenting the binding of keto-acid metabolites to YjgF homologs [43, 44, 50, 51]. Additionally, the loss of YjgF results in PurF-independent PRA formation. It has been shown that TrpD (bifunctional glutamine amidotransferase/anthranilate phosphoribosyltransferase [EC 4.1.1.48, 2.4.2.18]) is both necessary and sufficient for this formation from as yet unknown substrates. It has been hypothesized that a metabolite accumulating in a yjgF mutant is one of the substrates [23].
yjgF forms a node for expansion of the metabolic interactions with thiamine synthesis
Beyond the involvement of YjgF function in PRA formation, loss of this protein results in multiple metabolic defects that can be dissected in the effort to understand its biochemical function. Strains lacking YjgF fail to grow on pyruvate, are sensitive to serine, and have reduced specific activity of transaminase b [42, 50]. Each of these phenotypes reflect distinct consequences caused by the absence of YjgF function. These findings provide a functional (phenotypic) context for future analyses of the cellular role of YjgF that was not generated by structural studies. The broad conservation of YjgF across the three domains of life indicates that it is a key component of one or more cellular processes. Perhaps more than any results to date, the identification of this protein from a simple screen for metabolic redundancy emphasizes the potential for new knowledge that comes from general, in vivo genetic studies on cellular metabolism and physiology.
Concluding remarks
The model system described herein provides a largely unbiased approach that can be taken to identify unanticipated metabolic connections and provide a functional context for genes of unknown function. Other, more global approaches can identify potential connections and functions in the cell based on clustering algorithms, sequence similarity and precedent. However an in vivo approach, such as that described herein, can identify relevant (although often subtle) metabolic connections in the cell. Significantly, the connections will be found based on their impact on cellular function, not the quantitative level of flux change. In many cases the connections would not be predicted by current techniques dependent on precedent, because they will represent new paradigms.
Ultimately this model system, and others like it (Box 4) provide a means to generate insights into metabolic network structure and robustness, allowing us to address relevant new questions about metabolic integration. Significantly, this approach identifies new directions of discovery and often provides a new perspective to consider previous results. The data generated from these studies contribute to the base of knowledge needed to increase the rigor and realism of the mathematical models of metabolism and cell function. As this model and others continue to evolve and improve, we come closer to the goal of understanding metabolism at a level that will allow metabolic systems to be productively manipulated.
Box 4. Other nodes are needed to study metabolic integration.
A full understanding of metabolism in a microbial cell will require both high-throughput and focused approaches. Continued study of the model system described here will provide additional metabolic connections that can be anchored in the thiamine biosynthetic pathway. To achieve a complete understanding of the integrated system of metabolism it would be valuable to have similar studies initiated at additional metabolic nodes. Such an approach by multiple investigators will define numerous sub-networks and ultimately uncover the intersections between these networks that together define the integrated system.
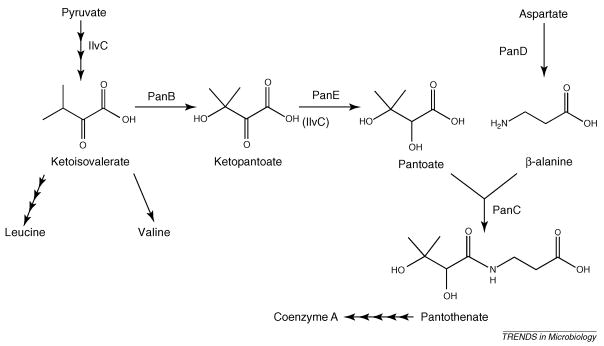
The biosynthetic pathway for pantothenate (Figure I) shares several characteristics with that of thiamine and defines a model system to study a distinct metabolic sub-network that will contribute to our understanding of metabolic integration. Pantothenate is an essential vitamin that is used in the synthesis of coenzyme A (CoA). Previous research determined that eliminating ketopantoate reductase (PanE, EC 1.1.1.169) resulted in an eightfold reduction in CoA levels [72]. A low level of keto-pantothenate is synthesized in this strain by a promiscuous activity of acetohydroxy acid isomeroreductase (IlvC, EC 1.1.1.86). Significantly, a panE mutant grows on minimal medium. This fact indicates that ~10% of the normal levels of CoA are sufficient for growth under these conditions. As might be expected, the lower CoA levels make this strain more sensitive to perturbations of the system that affect CoA metabolism. Thus, the panE mutant is a conditional pantothenate auxotroph that can be exploited with suppressor analysis to probe metabolic integration, much like the strategy that has been successful with the thiamine system.
Figure I. The pantothenate biosynthesis pathway.
A schematic representation of the pantothenate biosynthetic pathway in S. enterica.
Acknowledgments
This work was supported by competitive grant GM47296 from the NIH to D.M.D. Funds were also provided from a 21st Century Scientist Scholars Award from the J.S. McDonnell Foundation to DMD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Downs DM. Understanding microbial metabolism. Annual Review of Microbiology. 2006;60:561–558. doi: 10.1146/annurev.micro.60.080805.142308. [DOI] [PubMed] [Google Scholar]
- 2.VerBerkmoes NC, et al. Systems biology: Functional analysis of natural microbial consortia using community proteomics. Nat Rev Microbiol. 2009;7(3):196–205. doi: 10.1038/nrmicro2080. [DOI] [PubMed] [Google Scholar]
- 3.Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mapelli V, Olsson L, Nielsen J. Metabolic footprinting in microbiology: methods and applications in functional genomics and biotechnology. Trends Biotechnol. 2008;26(9):490–7. doi: 10.1016/j.tibtech.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33(1):191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feist AM, et al. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7(2):129–43. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitling R, Vitkup D, Barrett MP. New surveyor tools for charting microbial metabolic maps. Nat Rev Microbiol. 2008;6(2):156–61. doi: 10.1038/nrmicro1797. [DOI] [PubMed] [Google Scholar]
- 8.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol. 2008;26(6):659–67. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pharkya P, Burgard AP, Maranas CD. Exploring the overproduction of amino acids using the bilevel optimization framework OptKnock. Biotechnol Bioeng. 2003;84(7):887–99. doi: 10.1002/bit.10857. [DOI] [PubMed] [Google Scholar]
- 10.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005;23(5):612–6. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 11.Pharkya P, Maranas CD. An optimization framework for identifying reaction activation/inhibition or elimination candidates for overproduction in microbial systems. Metab Eng. 2006;8(1):1–13. doi: 10.1016/j.ymben.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Pharkya P, Burgard AP, Maranas CD. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 2004;14(11):2367–76. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes S, et al. Essential genes on metabolic maps. Curr Opin Biotechnol. 2006;17(5):448–56. doi: 10.1016/j.copbio.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Downs DM, Roth JR. Synthesis of thiamine in Salmonella typhimurium independent of the purF function. J Bacteriol. 1991;173(20):6597–604. doi: 10.1128/jb.173.20.6597-6604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen L, Enos-Berlage J, Downs DM. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics. 1996;143(1):37–44. doi: 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enos-Berlage JL, Downs DM. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J Bact. 1996;178(5):1476–9. doi: 10.1128/jb.178.5.1476-1479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel HJ, Bonner DM. Acetylornithase of Escherichia coli: partial purification and some properties. Journal of Biological Chemistry. 1956;218:97–106. [PubMed] [Google Scholar]
- 18.Davis RW, et al. Manual for genetic engineering. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. Advanced bacterial genetics; p. x.p. 254. [Google Scholar]
- 19.Koenigsknecht MJ, Fenlon LA, Downs DM. Variants of phosphoribosylpyrophosphate synthetase (PrsA) alter cellular pools of ribose 5-phosphate and influence thiamine synthesis in S. enterica. Microbiology. 2010;156:950–959. doi: 10.1099/mic.0.033050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Creighton TE. The Nonenzymatic Preparation in Solution of N-(5-Phosphoribosyl)anthranilic Acid, an Intermediate in Tryptophan Biosynthesis. Journal of Biological Chemistry. 1968;243(21):5605–5609. [PubMed] [Google Scholar]
- 21.Ramos I, Vivas EI, Downs DM. Mutations in the tryptophan operon allow PurF-independent thiamine synthesis by altering flux in vivo. J Bact. 2008;190(3):815–822. doi: 10.1128/JB.00582-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen KF. Metabolism of 5-phosphoribosyl 1-pyrophosphate (PRPP) in Escherichia coli and Salmonella typhimurium. In: Munch-Petersen A, editor. Metabolism of nucleotides nucleosides and nucleobases in microorganisms. Academic Press Inc. (London) Ltd; London: 1983. pp. 1–26. [Google Scholar]
- 23.Browne BA, Ramos AI, Downs DM. PurF-independent phosphoribosyl amine formation in yjgF mutants of Salmonella enterica utilizes the tryptophan biosynthetic enzyme complex anthranilate synthase-phosphoribosyltransferase. J Bacteriol. 2006;188(19):6786–92. doi: 10.1128/JB.00745-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skovran E, Lauhon CT, Downs DM. Lack of YggX results in chronic oxidative stress and uncovers subtle defects in Fe-S cluster metabolism in Salmonella enterica. J Bacteriol. 2004;186:7626–7634. doi: 10.1128/JB.186.22.7626-7634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beck BJ, Downs DM. The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J Bacteriol. 1998;180(4):885–891. doi: 10.1128/jb.180.4.885-891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gralnick J, et al. Lesions in gshA (encoding gamma-L-glutamyl-L-cysteine synthetase) prevent aerobic synthesis of thiamine in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2000;182(18):5180–5187. doi: 10.1128/jb.182.18.5180-5187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyd JM, et al. Bacterial ApbC can bind and effectively transfer iron-sulfur clusters. Biochemistry. 2008;47:8195–8202. doi: 10.1021/bi800551y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vivas E, Skovran E, Downs DM. Salmonella enterica strains lacking the frataxin homolog CyaY show defects in Fe-S cluster metabolism in vivo. J Bacteriol. 2006;188(3):1175–1179. doi: 10.1128/JB.188.3.1175-1179.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gralnick J, Downs D. Protection from superoxide damage associated with an increased level of the YggX protein in Salmonella enterica. Proc Natl Acad Sci USA. 2001;98(14):8030–8035. doi: 10.1073/pnas.151243198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck BJ, et al. Evidence that rseC, a gene in the rpoE cluster, has a role in thiamine synthesis in Salmonella typhimurium. Journal of Bacteriology. 1997;179(20):6504–6508. doi: 10.1128/jb.179.20.6504-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyd JM, et al. Salmonella enterica requires ApbC function for growth on tricarballylate: evidence of functional redundancy between ApbC and IscU. J Bacteriol. 2008;190(13):4596–602. doi: 10.1128/JB.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hausmann A, et al. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc Natl Acad Sci U S A. 2005;102(9):3266–71. doi: 10.1073/pnas.0406447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Netz DJ, et al. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat Chem Biol. 2007;3(5):278–86. doi: 10.1038/nchembio872. [DOI] [PubMed] [Google Scholar]
- 34.Roy A, et al. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO Journal. 2003;22(18):4826–35. doi: 10.1093/emboj/cdg455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koo MS, et al. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO Journal. 2003;22(11):2614–22. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonardi R, Roach PL. Thiamine biosynthesis in Escherichia coli: In vitro reconstitution of the thiazole synthase activity. J Biol Chem. 2004;279:17054–17062. doi: 10.1074/jbc.M312714200. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Gomez NC, Robers M, Downs DM. Mutational analysis of ThiH, a member of the radical S-adenosylmethionine (AdoMet) protein superfamily. J Biol Chem. 2004;279:40505–40510. doi: 10.1074/jbc.M403985200. [DOI] [PubMed] [Google Scholar]
- 38.Lawhorn BG, Mehl RA, Begley TP. Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Organ Biomol Chem. 2004;2(17):2538–46. doi: 10.1039/B405429F. [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Gomez NC, Downs DM. ThiC is an [Fe-S] cluster protein that requires AdoMet to generate the 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety in thiamin synthesis. Biochemistry. 2008;47(35):9054–6. doi: 10.1021/bi8010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee A, et al. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat Chem Biol. 2008;4(12):758–65. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Gomez NC, et al. Reaction of AdoMet with ThiC generates a backbone free radical. Biochemistry. 2009;48(2):217–9. doi: 10.1021/bi802154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enos-Berlage JL, Langendorf MJ, Downs DM. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. Journal of Bacteriology. 1998;180(24):6519–6528. doi: 10.1128/jb.180.24.6519-6528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsons L, et al. Solution structure and functional ligand screening of HI0719, a highly conserved protein from bacteria to humans in the YjgF/YER057c/UK114 family. Biochemistry. 2003;42(1):80–89. doi: 10.1021/bi020541w. [DOI] [PubMed] [Google Scholar]
- 44.Burman JD, et al. The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct Biol. 2007;7:30. doi: 10.1186/1472-6807-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyakawa T, et al. Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7. Proteins. 2006;62(2):557–61. doi: 10.1002/prot.20778. [DOI] [PubMed] [Google Scholar]
- 46.Mistiniene E, et al. Structure-based ligand binding sites of protein p14.5, a member of protein family YER057c/YIL051c/YjgF. Int J Biol Macromol. 2005;37(1–2):61–8. doi: 10.1016/j.ijbiomac.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 47.Deriu D, et al. Structure and oligomeric state of the mammalian tumour-associated antigen UK114. Acta Crystallographica Section D, Biological Crystallography. 2003;59(Pt 9):1676–1678. doi: 10.1107/s0907444903014306. [DOI] [PubMed] [Google Scholar]
- 48.Sinha S, et al. Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family. Proceedings of the National Academy of Sciences USA. 1999;96(23):13074–13079. doi: 10.1073/pnas.96.23.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volz K. A test case for structure-based functional assignment: the 1.2 A crystal structure of the yjgF gene product from Escherichia coli. Protein Science. 1999;8(11):2428–2437. doi: 10.1110/ps.8.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitz G, Downs DM. Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium. Journal of Bacteriology. 2004;186(3):803–810. doi: 10.1128/JB.186.3.803-810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christopherson MR, Schmitz GE, Downs DM. YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J Bacteriol. 2008;190(8):3057–62. doi: 10.1128/JB.01700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedrich W. Vitamins. Berlin: Walter De Gruyter; 1988. p. 1060. [Google Scholar]
- 53.Jurgenson CT, Begley TP, Ealick SE. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jordan F. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat Prod Rep. 2003;20(2):184–201. doi: 10.1039/b111348h. [DOI] [PubMed] [Google Scholar]
- 55.Lakaye B, et al. Thiamine triphosphate, a new signal required for optimal growth of Escherichia coli during amino acid starvation. J Biol Chem. 2004;279(17):17142–7. doi: 10.1074/jbc.M313569200. [DOI] [PubMed] [Google Scholar]
- 56.Bettendorff L, Kolb HA, Schoffeniels E. Thiamine triphosphate activates an anion channel of large unit conductance in neuroblastoma cells. J Membr Biol. 1993;136(3):281–8. doi: 10.1007/BF00233667. [DOI] [PubMed] [Google Scholar]
- 57.Nghiem HO, Bettendorff L, Changeux JP. Specific phosphorylation of Torpedo 43K rapsyn by endogenous kinase(s) with thiamine triphosphate as the phosphate donor. Faseb J. 2000;14(3):543–54. doi: 10.1096/fasebj.14.3.543. [DOI] [PubMed] [Google Scholar]
- 58.Gangolf M, et al. Thiamine triphosphate synthesis in rat brain occurs in mitochondria and is coupled to the respiratory chain. J Biol Chem. 2010;285(1):583–94. doi: 10.1074/jbc.M109.054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bettendorff L, et al. Discovery of a natural thiamine adenine nucleotide. Nat Chem Biol. 2007;3(4):211–2. doi: 10.1038/nchembio867. [DOI] [PubMed] [Google Scholar]
- 60.Christian T, Downs DM. Defects in pyruvate kinase cause a conditional increase of thiamine synthesis in Salmonella typhimurium. Canadian Journal of Microbiology. 1999;45:565–572. [Google Scholar]
- 61.Downs DM, Petersen L. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1994;176(16):4858–64. doi: 10.1128/jb.176.16.4858-4864.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frodyma ME, Downs D. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J Bacteriol. 1998;180(17):4757–4759. doi: 10.1128/jb.180.17.4757-4759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Claas K, Weber S, Downs DM. Lesions in the nuo operon, encoding NADH dehydrogenase complex I, prevent PurF-independent thiamine synthesis and reduce flux through the oxidative pentose phosphate pathway in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182(1):228–232. doi: 10.1128/jb.182.1.228-232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skovran E, Downs DM. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J Bacteriol. 2000;182(14):3896–3903. doi: 10.1128/jb.182.14.3896-3903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petersen L, Downs DM. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J Bacteriol. 1996;178(19):5676–82. doi: 10.1128/jb.178.19.5676-5682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skovran E, Downs DM. Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J Bacteriol. 2003;185:98–106. doi: 10.1128/JB.185.1.98-106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beck BJ, Downs DM. A periplasmic location is essential for the role of the ApbE lipoprotein in thiamine synthesis in Salmonella typhimurium. Journal of Bacteriology. 1999;181(23):7285–7290. doi: 10.1128/jb.181.23.7285-7290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmehl M, et al. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Molecular and General Genetics. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 69.Schmitz GE, Downs DM. An allele of gyrA prevents Salmonella enterica serovar Typhimurium from growing with succinate as a carbon source. Journal of Bacteriology. 2006;188:3126–3129. doi: 10.1128/JB.188.8.3126-3129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dougherty MJ, Downs DM. A mutant allele of rpoD results in increased conversion of aminoimidazole ribotide to hydroxymethyl pyrimidine in Salmonella enterica. Journal of Bacteriology. 2004;186(12):4034–4037. doi: 10.1128/JB.186.12.4034-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hernandez VJ, Cashel M. Changes in conserved region 3 of Escherichia coli sigma 70 mediate ppGpp-dependent functions in vivo. Journal of Molecular Biology. 1995;252(5):536–49. doi: 10.1006/jmbi.1995.0518. [DOI] [PubMed] [Google Scholar]
- 72.Frodyma M, Rubio A, Downs DM. Reduced flux through the purine biosynthetic pathway results in an increased requirement for coenzyme A in thiamine synthesis in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182(1):236–240. doi: 10.1128/jb.182.1.236-240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]