Abstract
We utilize the recent successful overexpression of recombinant Plasmodium falciparum multidrug resistance transporter, purification and reconstitution of the protein, and a novel high affinity chloroquine analogue to probe hypothesized interaction between the transporter and quinoline drugs. Results suggest that PfMDR1 binding sites for chloroquine, mefloquine, and quinine overlap, that P. falciparum chloroquine resistance transporter has intrinsically higher affinity for chloroquine relative to P. falciparum multidrug resistance transporter, and that there is an isoform specific competition between the two transporters for binding of quinoline antimalarial drugs.
P. falciparum multidrug resistance transporter (PfMDR1) is an ATP-Binding Cassette (ABC) transporter, many of which (e.g. human P-glycoprotein; HsMDR1) have been implicated in drug resistance phenomena. PfMDR1 is composed of two homologous halves, each containing 6 transmembrane (TM) helices and 1 nucleotide binding domain (NBD). Precisely how PfMDR1 contributes to a variety of antimalarial multi-drug resistance phenotypes has been a source of considerable controversy for some time [1 – 5]. PfMDR1 is an integral digestive vacuolar (DV) membrane protein [1], with NBDs cytosolically oriented. If traditional ABC transporter drug-pumping models are invoked, PfMDR1 would paradoxically translocate drugs into the DV, where the principal target for CQ and other drugs is believed to reside (e.g., ferriprotoporphyrin IX [FPIX] released upon hemoglobin catabolism, see [6,7]). How concentrating drugs at the site of action would promote resistance is a puzzle. Also, genetics and a plethora of field studies now show that pfcrt mutations are the main molecular determinants of P. falciparum CQ resistance (CQR) [4,8]. It has therefore been suggested that PfMDR1 only modulates the level of CQR and resistance to other antimalarials in a PfCRT-dependent manner [5,9].
We have recently reported the successful heterologous overexpression of several PfMDR1 isoforms (allelic forms) found in either CQS or CQR parasites [10], have characterized unusual ATPase activity for these [10,11], and have identified residues in one PfMDR1 isoform that confer a specific drug-stimulated ATPase phenotype [11]. The 3D7 isoform is found within CQS strain 3D7 and has a single amino acid difference (N86) relative to the Dd2 isoform found in CQR strain Dd2 (Y86), whereas the 7G8 isoform has 4 amino acid substitutions relative to 3D7 (Y184F, S1034C, N1042D, D1246Y). Strain Dd2 exhibits a CQR phenotype that is partially reversible by verapamil (VPL) whereas CQR observed in strain 7G8 is less VPL sensitive. Taken together, these data suggest that PfMDR1 protein interacts with quinoline-based drugs in some fashion and that amino acid differences among the isoforms may alter drug affinities.
To test interaction between PfMDR1 and CQ more directly, we first combined well-established, previously optimized techniques for purifying and reconstituting ABC transporters [12] as well as PfCRT [13,14], to generate proteoliposomes (PL) harboring PfMDR1. Similar to our earlier results with PfCRT and HsMDR1 [13,12] multiple PfMDR1 isoforms (e.g. 3D7, Dd2 and 7G8 isoforms, see [10]) are easily purified by hexa his chromatography to an approximately equal extent and yield virtually identical protein : lipid ratios in the purified PLs (Fig. 1E, left). Following a similar tag-cleavage approach that we used for PfCRT [13], we find PfMDR1 protein exists in these PLs primarily with C-termini and ATP-binding sites externally disposed (Fig S1, supplemental).
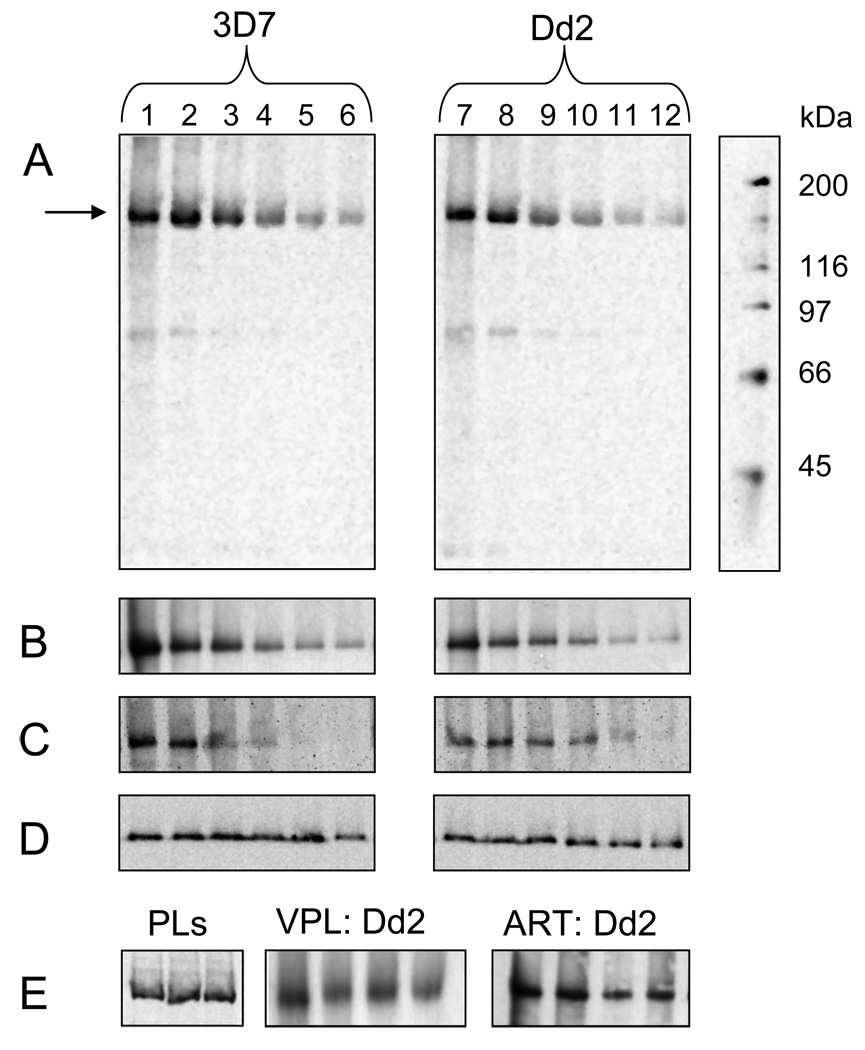
Fig. 1.
Characterization of AzBCQ photolabeling to partially purified 3D7 and Dd2 PfMDR1 isoforms reconstituted into proteoliposomes (PLs). Reaction conditions were as previously reported for PfCRT (13) with a few modifications. Briefly, equivalent amounts of purified PLs were diluted into 0.050 M Mes-Tris buffer at pH 5.2 and aliquots distributed to individual wells of a 96 well plate. A probe to protein molar ratio of approximately 100:1 was utilized in each experiment. 2.5 nmoles (50 µM final concentration) AzBCQ were added, the mixture was incubated at 37°C for 10 min, and then reacted under 254 nM UV light for 10 min. UV illumination was from a Spectroline 8W 60Hz bulb emitting 500 µW / cm2 that was positioned approximately 10 cm above the mixture. Photolabeling was quenched by the addition of 1X Laemmli buffer, and samples were incubated again at 37°C for 10 min before loading half the sample onto one of two 7.5% acrylamide Tris-HCl gels. Gels were run at 110V for 100 min., and immediately transferred to PVDF membrane for 16h at 40 mA and 4°C. Membranes were probed either with Anti-PentaHis-HRP (Qiagen) to detect PfMDR1 protein or Streptavidin-HRP (Amersham) to detect bound AzBCQ (see [13]). A. Representative streptavidin-HRP detection of AzBCQ photolabeling of 3D7 (left) and Dd2 (right) PfMDR1 PLs vs. competition with unlabelled CQ: lanes 1 & 7; no CQ competitor; lanes 2 – 6 and 8 – 12; 16-, 32-, 48-, 64- and 80-fold molar excess (800 µM, 1.6 mM, 2.4 mM, 3.2 mM, and 4.0 mM final concentration) of CQ relative to AzBCQ respectively. B. Representative streptavidin-HRP detection of AzBCQ photolabeling of 3D7 (left) and Dd2 (right) PfMDR1 PLs vs. QN competition: lanes 1 & 7- no QN; lanes 2 – 6 and 8 – 12; 5-, 10-, 20-, 40- and 80-fold excess of QN (250 µM, 500 µM, 1 mM, 2 mM and 4 mM final concentration) relative to AzBCQ, respectively. C. Representative streptavidin-HRP detection of AzBCQ photolabeling of 3D7 (left) and Dd2 (right) PfMDR1 PLs vs MQ competition: lanes 1 & 7- no MQ; lanes 2–6 and 8–12; 0.5-, 1-, 2.5-, 5- and 10-fold excess of MQ (25 µM, 50µM, 125 µM, 250 µM and 500 µM final concentration) relative to AzBCQ respectively. D. Representative PentaHis-HRP detection of 3D7 (left) and Dd2 (right) PfMDR1 PLs used in the CQ drug competition assay (Fig. 1A) showing approximately equal amounts of each protein isoform in all lanes. E, left. PentaHis HRP detection of 3 representative PL preparations. Equal volumes of 3D7 (lane 1), Dd2 (lane 2) and 7G8 (lane 3) PfMDR1 PLs show approximately equal protein:lipid ratio during reconstitution. E, center. Streptavidin-HRP detection of AzBCQ photolabeling of Dd2 PfMDR1 PLs with verapamil (VPL) competition. Lane 1 no VPL; lanes 2–4, 20-, 40- and 80-fold molar excess of VPL (1 mM, 2 mM, and 4 mM final concentration) relative to AzBCQ respectively. E, right. Streptavidin-HRP detection of AzBCQ photolabeling of Dd2 PfMDR1 PLs with artemisinin (ART) competition. Lane 1 no ART; lanes 2–4, 20-, 40- and 80-fold excesses of ART (1 mM, 2 mM, and 4 mM final concentration) relative to AzBCQ respectively.
Earlier we reported on the design, synthesis, and biochemical utilization of a high affinity CQ photolabel analogue, [16-perfluoroazido, 16’-biotinyl] chloroquine (AzBCQ) [13]. This probe combines per fluoro azido C–H bond insertion chemistry with convenient avidin – biotin detection, and binds with high affinity to the CQ transporter PfCRT [13]. Half-maximal inhibition of AzBCQ photolabeling of PfCRT was achieved at 6–25 fold molar excess of unlabelled CQ, depending on the isoform (allelic form) examined, demonstrating that the probe is highly specific for the PfCRT CQ binding site. Correspondingly, a single CQ binding site was mapped by proteolysis and subsequent mass spectroscopy [13]. Also, various levels of competition for binding of other drugs (e.g. quinine [QN], verapamil [VPL]) to the CQ binding site were quantified [13]. We found that AzBCQ binding to the Dd2 PfCRT isoform is well-competed by VPL, whereas binding to the 7G8 PfCRT isoform is not. This presumably reflects in vitro parasite behavior whereby strains harboring the Dd2 PfCRT isoform are CQR and VPL-sensitive, but strains harboring 7G8 PfCRT are CQR and less VPL sensitive.
Fig. 1 shows that using these methods [13] 3D7, Dd2, and 7G8 isoforms of PfMDR1 are also efficiently photolabeled with AzBCQ, and that photolabelling is strongly inhibited by unlabelled CQ (Fig. 1A). The results are surprising since PfMDR1 is not thought to directly confer CQR (as mentioned above, PfCRT mutations confer CQR). Kinetic experiments show that all PfMDR1 isoforms are saturated in labeling in < 1 min (see Fig. S2 in supplemental information), similar to the case previously observed for PfCRT [13]. Importantly, all isoforms of PfMDR1 examined show high specificity in photolabeling, since very low levels of unlabelled CQ effectively compete (e.g. 44-, 64-, and 92-fold molar excess of unlabelled CQ for 3D7, Dd2, and 7G8 isoforms, respectively, see Table 1). These IC50 values are higher than those found earlier for PfCRT isoforms [13], suggesting that under these conditions affinity of CQ for PfMDR1 isoforms is lower than affinity of CQ for PfCRT isoforms. Photolabeling of PfMDR1 is efficiently and selectively competed by QN and MQ as well (Fig. 1B, 1C, respectively), but to varying degrees, depending on the isoform.
Table 1.
Fold molar excess of 50% maximal inhibition of AzBCQ photolabeling of PfMDR1 or PfCRT isoforms [13] via competition with different drugs. In all cases transporters were assayed individually as in Fig. 1. Values in the table are the mean of three independent experiments (3 independent SDS PAGE gel analyses of 3 independent photolabelling reactions), with densitometry of the bands in each experiment quantified twice using Kodak 1D v 3.6.5 K2 software (6 densitometry determinations for each). Standard error of the mean for each value in the table is < 20% in each case.
| PfMDR | PfCRT | |||||
|---|---|---|---|---|---|---|
| 3D7 | Dd2 | 7G8 | HB3 | Dd2 | 7G8 | |
| CQ | 44 | 64 | 92 | 22 | 25 | 6 |
| QN | 11 | 11 | 17 | 19 | 16 | 10 |
| MQ | 4.9 | 3.2 | 6.1 | 2.6 | 2.8 | 2.8 |
| VPL | >80 | >100 | >100 | 35 | 40 | >100 |
However, in contrast to results with PfCRT [13], QN and MQ competition for AzBCQ binding to PfMDR1 is stronger than vs. CQ. Table 1 reports the concentrations of CQ, QN and MQ at which half-maximal inhibition of photolabeling is obtained for the three isoforms and the much smaller values for QN and MQ suggest that affinity of PfMDR1 for QN and MQ is higher than PfMDR1 affinity for CQ.
Virtually no competition is seen vs artemisinin (ART) (Fig. 1E), and unlike the case previously observed for Dd2 and 3D7 PfCRT isoforms [13], no photolabeling inhibition was found for either the Dd2 or 3D7 PfMDR1 isoforms vs. VPL (Fig. 1E, Table 1). Neither the 7G8 PfCRT [13] nor 7G8 PfMDR1 isoforms (Table 1) exhibit AzBCQ photolabeling that is effectively competed by VPL. Taken together then, these data suggest that VPL chemoreversal of the CQR phenotype is due to VPL interaction with Dd2 PfCRT, but not interaction with any PfMDR1 isoform.
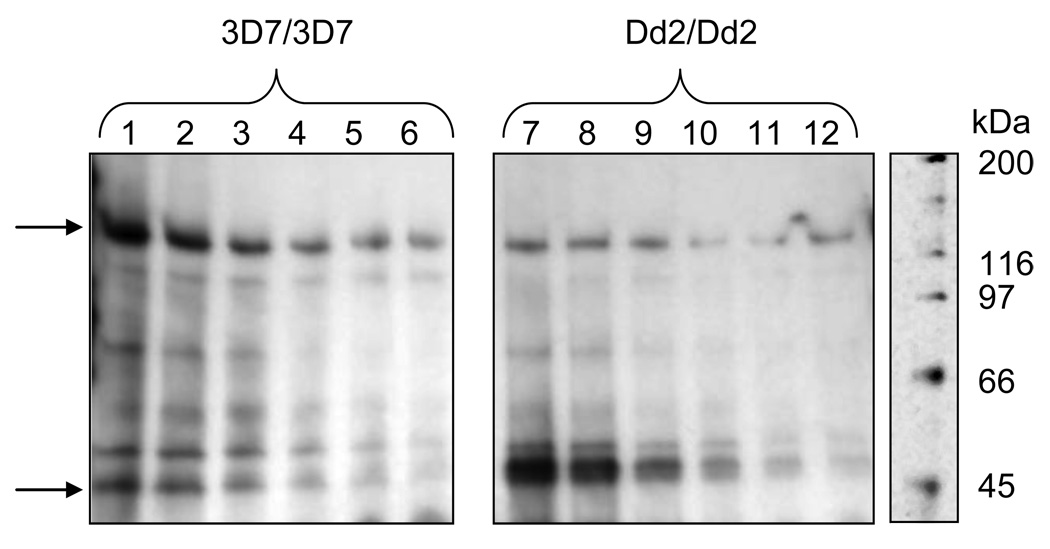
To further analyze how photolabeling of PfMDR1 compares to previous drug-probe interactions elucidated for PfCRT, we mixed equal amounts of PfCRT and PfMDR1 PLs and performed AzBCQ photolabeling vs. increasing concentrations of unlabelled CQ (Fig. 2; top arrow is PfMDR1, bottom arrow is PfCRT). Mixing PLs under these conditions likely promotes PL fusion and randomization of the two proteins across the mixture. We compared photolabel preference and CQ competition for photolabeling for CQS isoforms of the transporters (e.g. 3D7 PfCRT vs. 3D7 PfMDR1, left, lanes 1 – 6) as well as CQR isoforms (Dd2 PfCRT vs. Dd2 PfMDR1, lanes 7 – 12). When CQR isoforms are mixed in equal amounts (equal molar amounts determined by titration as described [10]), then PfCRT photolabeling dominates (Fig. 2 lanes 7 – 12). This is not unexpected, since photolabeling competition with unlabelled CQ for the individual transporters suggests apparent CQ affinity for Dd2 PfCRT is higher than for Dd2 PfMDR1 (half-maximal inhibition at 25-fold [13] vs. 44-fold excess cold CQ [Table 1], respectively). Interestingly however, when CQS isoforms are mixed, PfMDR1 photolabeling dominates. For both PfCRT isoforms, as well as the Dd2 PfMDR1 isoform, relative competition of photolabeling vs. unlabelled CQ remains similar when the transporters are mixed together, showing that the apparent affinity of CQ for the drug binding sites on these three transporters does not change significantly. However, the reduced level of photolabeling for the 3D7 PfCRT isoform in the presence of 3D7 PfMDR1, relative to what has been observed previously for the same 3D7 PfCRT in the absence of PfMDR1 [13], suggests either that the relative affinity of CQ for 3D7 PfMDR1 is increased when 3D7 PfMDR1 is in the presence of equimolar 3D7 PfCRT, or that CQ affinity for 3D7 PfCRT is reduced. The former hypothesis is supported when we plot the densitometry of AzBCQ labeled PfMDR1 and PfCRT bands (e.g. Fig. 2 top and bottom arrows) vs. increasing cold [CQ] and then calculate IC50 from sigmoidal fits to these data (plots not shown, see [13] for methods). That is, half-maximal CQ inhibition of photolabeling for 3D7 PfMDR1 shifts from 44-fold molar excess [CQ : AzBCQ] when it is the only transporter present (Table 1), to 29-fold molar excess when it is mixed with an approximately equimolar amount of 3D7 PfCRT, but IC50 for 3D7 PfCRT stays constant (average of two densitometry plots for two mixed 3D7 PfMDR1 / 3D7 PfCRT experiments using four different PL preparations, data not shown, see Table 1 caption). Many more such mixing studies remain to be done, but these initial data suggest an isoform specific interplay between PfCRT and PfMDR1 proteins that manipulates accessibility and / or affinity of drug binding sites. Another possibility is that the presence of one transporter changes cross linking efficiency for the other (meaning, activated perfluoro azido bond insertion efficiency changes without a change in drug binding affinity) via a non specific conformational change, however, this seems unlikely due to the very low level of quinoline drug needed for selective competition (Fig. 1A–C vs. Fig 1E middle and right).
Fig. 2.
PfMDR1 and PfCRT compete for AzBCQ photolabeling in isoform specific fashion. Equivalent amounts of purified 3D7 PfCRT and 3D7 PfMDR1 PL protein (left panel, lanes 1 – 6) or Dd2 PfCRT and Dd2 PfMDR1 PL protein (right panel, lanes 7 – 12) were mixed and the drug competition assay was performed exactly as in Fig. 1, but using 5.0 nmoles AzBCQ (100 µM final concentration). Note the CQ IC50 does not change for either Dd2 PfMDR1 or Dd2 PfCRT when they are mixed together (Fig. 2) but that the CQ IC50 for 3D7 PfMDR1 drops by nearly a factor of 2 when it is mixed with equimolar 3D7 PfCRT (see text). The reactions were run on a 4–15% Tris-HCl gradient gel (Bio-Rad), to better resolve both high molecular mass protein (PfMDR1, upper arrow) and medium molecular mass protein (PfCRT, lower arrow) on the same gel. Gels were run, transferred, and blotted the same as in Fig 1. Streptavidin-HRP detection of 3D7/3D7 (left) and Dd2/Dd2 (right) AzBCQ-labelled PfMDR1/PfCRT PL proteins vs. CQ competition: lanes 1 & 7- no CQ; lanes 2 – 6 (left) and 8 – 12 (right); 16-, 32-, 48-, 64- and 80-fold molar excess (1.6 mM, 3.2 mM, 4.8 mM, 6.4 mM, and 8.0 mM final concentration) of CQ relative to AzBCQ respectively.
Prior to these data, it was quite surprising to us that CQ was found to have the greatest effect on PfMDR1 ATPase activity [10] relative to other quinoline drugs. Table 1 shows that PfMDR1 affinity for MQ and QN is likely higher than for CQ, so the higher effect of CQ on PfMDR1 ATPase activity still remains a bit puzzling. Another conclusion from our ATPase data was that 7G8 PfMDR1 consistently showed lower levels of endogenous ATPase activity relative to the other two isoforms, with little to no fluctuation in activity upon introduction of quinoline drugs. Correspondingly, the present data show the 7G8 isoform consistently requires higher concentrations of competitor for photolabeling inhibition relative to the other two isoforms (Table 1). Combined with observed effects on ATPase activity then [10,11] these photolabeling results are both the most direct evidence to date for highly isoform-specific interaction between PfMDR1 and quinoline drugs, and suggestive of a complex relationship between PfMDR1 drug binding and drug effects on PfMDR1 ATPase activity.
Our initial efforts to map the location of AzBCQ binding site(s) in PfMDR1 have included trypsinization of labeled 3D7 protein followed by gel extraction and mass spectrometry as previously described for AzBCQ labeled PfCRT [13]. MS peak analysis yields two prominently labeled peptides encompassing residues 610 – 627 and residues 1104 – 1129, corresponding to domains within loop 6 (between putative helices 6,7) and within the C-terminal tail (after putative helix 12). Perhaps not coincidentally, loop 6 and the C terminal tail also harbor the PfMDR ATPase sites, and as mentioned we previously observed quinoline drug effects on PfMDR1 ATPase activity [10]. We note that previous mapping of the covalent attachment site of [125I]-iodoarylazidoprazosin (IAAP) to the PfMDR1 homologue HsMDR1 (human P-glycoprotein or Pgp) revealed two covalent attachment sites as well, the first located immediately C-terminal to putative HsMDR1 helix 6 and the second C-terminal to putative HsMDR1 helix 12 [20, 21]. Thus one or more drug binding site(s) for PfMDR1 and HsMDR1 appear to have similar locations. Assuming the overall structures for PfMDR1 and HsMDR1 are similar to that recently elucidated for MuMDR1 [22], then the loop 6 and C terminal tail domains are rather far apart, suggesting two drug binding sites near each domain. However, ATP hydrolysis could cause a conformational change that brings the two domains closer together such that covalent attachment to either domain occurs from only one drug binding site (see [22]).
The PfMDR1 and PfCRT mutations that are selected for in CQR field isolates illustrate symbiotic relationships between PfCRT and PfMDR1 isoforms. These relationships likely confer either preferred resistance patterns, fitness adaptations, or perhaps both. Variation in Dd2 vs. 7G8 PfMDR1 drug affinity and ATPase parameters (Vmax and Km see [10]) along with variation in Dd2 vs. 7G8 PfCRT drug affinity [13] likely reflect fitness adaptations relevant to the unique resistance phenotypes exhibited by Dd2 and 7G8 strains [18].
Finally, we hypothesize that the recently described distinction between resistance to CQ cytostasis (CQRCS) vs. CQ cytotoxicity (CQRCR) [15] might reconcile the paradoxical cytosolic disposition of PfMDR1 ATPase domains, and also offers a framework from which to better understand data such as that in Fig. 2. As mentioned, at lower levels of drug commensurate with heme-binding induced cytostasis [15], the disposition of PfMDR1 is the wrong orientation to confer resistance, since it would be acting to translocate drugs to the site of action, not away. However, we previously reported that although inhibition of Hz formation through heme binding is very time-dependent [16], resistance to cytotoxic effects of CQ is much less time-dependent [15, 17]. In fact, quite interestingly, for CQR ring stage parasites, the duration of bolus dose cytotoxic exposure does not seem to correlate at all with the degree of cell death [17]. We suggest then that it is possible that other (non-DV localized) targets are more relevant for quinoline drug cytotoxicity, that parasites evolved to be resistant to both quinoline cytostatic and cytotoxic effects [15], and that PfCRT could be more important for the former whereas PfMDR1 may be more important for the latter. If so, PfCRT could confer CQRCS [15] by acting to both alter DV ionic environment and remove CQ from the DV where targets particularly relevant for CQ cytostasis reside, whereas PfMDR1 could contribute to, for example, resistance to quinine cytotoxicity (QNRCT), by acting to move drug the other way, to sequester much higher (toxic) levels of drug within the DV, away from extra-DV drug targets that are more relevant for QN cytotoxicity [15]. It is underappreciated perhaps, but QN and CQ are very different drugs in terms of their concentration and phase-dependent heme reactivities [19], and QN vs. CQ effects on PfMDR1 ATPase are very strongly dose-dependent and in opposite directions [10].
More detailed comparison between QN and CQ pharmacology and cell biology, as well as continued studies such as those reported herein, will ultimately elucidate the molecular basis of quinoline antimalarial multi-drug resistance.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIAID / NIH (RO1 AI056312) and a DoD predoctoral awards to PP (W81XWH-06-1-0454)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- 1.Cowman AF, Karcz S, Galatis D, Culvenor JG. A P-glycoprotein homologue of Plasmodium falciparum is localized on the digestive vacuole. J Cell Biol. 1991;113(5):1033–1042. doi: 10.1083/jcb.113.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 3.Wilson CM, Serrano AE, Wasley A, Bogenschutz MP, Shankar AH, Wirth DF. Amplification of a gene related to mammalian mdr genes in drug-resistant Plasmodium falciparum. Science. 1989;244(4909):1184–1186. doi: 10.1126/science.2658061. 9. [DOI] [PubMed] [Google Scholar]
- 4.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6(4):861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403(6772):906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 6.de Dios AC, Tycko R, Ursos LMB, Roepe PD. NMR Studies of Chloroquine – Ferriprotoporphyrin IX Complex. J Phys Chem A. 2003;107:5821–5825. [Google Scholar]
- 7.Pisciotta JM, Sullivan D. Hemozoin: oil versus water. Parasitol Int. 2008;57(2):89–96. doi: 10.1016/j.parint.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184(6):770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 9.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57(4):913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 10.Amoah LE, Lekostaj JK, Roepe PD. Heterologous expression and ATPase activity of wildtype vs mutant PfMDR1 protein. Biochemistry. 2007;46(20):6060–6073. doi: 10.1021/bi7002026. [DOI] [PubMed] [Google Scholar]
- 11.Lekostaj JK, Amoah LE, Roepe PD. A single S1034C mutation confers altered drug sensitivity to PfMDR1 ATPase activity that is characteristic of the 7G8 isoform. Mol Biochem Parasitol. 2008;157(1):107–111. doi: 10.1016/j.molbiopara.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard EM, Roepe PD. Purified human MDR 1 modulates membrane potential in reconstituted proteoliposomes. Biochemistry. 2003;42(12):3544–3555. doi: 10.1021/bi026706i. [DOI] [PubMed] [Google Scholar]
- 13.Lekostaj J, Natarajan JK, Paguio M, Wolf C, Roepe PD. Photoaffinity Labeling of the Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) with a Novel Perfluorophenylazido Chloroquine. Biochemistry. 2008;47(39):10394–10406. doi: 10.1021/bi8010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paguio M, Cabrera M, Roepe PD. Chloroquine transport in Plasmodium falciparum II: Analysis of PfCRT mediated Drug Transport Using Proteoliposomes and a Fluorescent Chloroquine Probe. Biochemistry. 2009;48:9482–9491. doi: 10.1021/bi901035j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabrera M, Paguio MF, Xie C, Roepe PD. Reduced digestive vacuolar accumulation of chloroquine is not linked to resistance to chloroquine toxicity. Biochemistry. 2009;48:11152–11154. doi: 10.1021/bi901765v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glikoreijevic B, McAllister R, Urbach J, Roepe PD. Spinning disk confocal microscopy of intraerythrocytic malarial parasites I: quantification of hemozoin development. Biochemistry. 2006;45:12400–12410. doi: 10.1021/bi061033f. [DOI] [PubMed] [Google Scholar]
- 17.Glikoreijevic B, Purdy K, Elliot DA, Cooper RA, Roepe PD. Stage Independent Chloroquine Resistance and Chloroquine Toxicity Revealed via Spinning Disk Confocal Microscopy. Mol Biochem Parasitol. 2008;159:7–23. doi: 10.1016/j.molbiopara.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel J, Thacker D, Tan JC, Checkley L, Gonzales JM, Deng B, Pleeter P, Roepe PD, Cooper R, Ferdig M. Chloroquine Susceptibility and Reversibility in a Plasmodium falciparum Genetic Cross. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2010.07366.x. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casabianca LB, An D, Natarajan J, Alumasa J, Roepe PD, Wolf C, de Dios AC. Quinine and chloroquine differentially perturb heme monomer-dimer equilibrium. J Inorg Chem. 2008;47:6077–6081. doi: 10.1021/ic800440d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruggemann EP, Currier SJ, Gottesman MM, Pastan I. Characterization of the azidopine and vinblastine binding site of P-glycoprotein. J Biol Chem. 1992;267:21020–21026. [PubMed] [Google Scholar]
- 21.Greenberger LM. Major photoaffinity drug labeling sites for iodoaryl azidoprazosin in P-glycoprotein are within, or immediately C-terminal to, transmembrane domains 6 and 12. J Biol Chem. 1993;268:11417–11425. [PubMed] [Google Scholar]
- 22.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;5922:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.