Abstract
Genetic analysis of mammalian color variation has provided fundamental insight into human biology and disease. In most vertebrates, two key genes, Agouti and Melanocortin 1 receptor (Mc1r), encode a ligand-receptor system that controls pigment type-switching, but in domestic dogs, a third gene is implicated, the K locus, whose genetic characteristics predict a previously unrecognized component of the melanocortin pathway. We identify the K locus as β-defensin 103 (CBD103) and show that its protein product binds with high affinity to the Mc1r and has a simple and strong effect on pigment type-switching in domestic dogs and transgenic mice. These results expand the functional role of β-defensins, a protein family previously implicated in innate immunity, and identify an additional class of ligands for signaling through melanocortin receptors.
AUTHORS’ SUMMARY.
The marked spectrum of color and diversity of patterns that we see in mammals arises, unexpectedly, from variation in the quantity, quality, and regional distribution of just two types of pigment—black eumelanin and yellow pheomelanin. The appeal of unusual coat colors and patterns has motivated their selection in domestic animals, providing geneticists with a model for studying gene action and interaction that began a century ago and continues today. Most of the work has been carried out in laboratory mice, where studies of more than 100 different coat-color mutations have provided insight into stem cell biology (hair graying), biogenesis of intracellular organelles (pigmentary dilution), and hormone-receptor interactions (switching between the synthesis of eumelanin and pheomelanin).
The latter process—commonly known as pigment “type-switching”—is controlled primarily by the melanocortin system, in which a family of G protein–coupled receptors (identified by virtue of their response to α-melanocyte–stimulating hormone or adrenocorticotrophic hormone) has been implicated not only in pigmentation but also in cortisol production, body weight regulation, and exocrine gland secretion. In most mammals, pigment type-switching is controlled by two genes, the Melanocortin 1 receptor (Mc1r) and Agouti, which encode a seven transmembrane–domain receptor and its extracellular ligand, respectively. Indeed, our current understanding of melanocortin biology stems from the identification in laboratory mice of Mc1r mutations as the cause of recessive yellow and Agouti mutations as the cause of lethal yellow.
Clarence Cook Little, who developed many of the original laboratory mouse strains and founded The Jackson Laboratory, was also one of the first dog geneticists. He recognized that dominant inheritance of a black coat was mediated differently in dogs than in other animals (1). Using classical linkage analysis, we realized that the dominant black gene represented a previously unrecognized component of the melanocortin pathway (2). Unexpectedly, we found the responsible gene to encode a β-defensin, a secreted protein previously studied for its role in immunity.
The identification of dominant black (formally, an allele of the “K locus”) relied on two major advances in dog genetics: the sequencing of the dog genome and recognition that the distinctive genetic structure of dog breeds allows for efficient gene mapping (3). Dogs were domesticated from wolves more than 15,000 years ago and expanded into a diverse population until the recent establishment of dog breeds. This population history is well-suited for high-resolution genetic mapping of old traits, like black coat color, that are found in multiple modern breeds. Using a combination of pedigree analysis and association studies within and among dog breeds, we identified a mutation in a β-defensin gene, CBD103, that correlates with black coat color in 38 different breeds. We confirmed the role of CBD103 in pigment types-witching by demonstrating that the dog gene causes a black coat in transgenic mice. CBD103 is a member of a large family of secreted peptides with structures similar to that of Agouti and is highly expressed in dog skin.
We used biochemical and cell-based assays to show that CBD103, like Agouti, binds competitively to the Mc1r, leading to an updated model of the pigment type-switching pathway (see figure). Moreover, studies with another β-defensin and additional melanocortin receptors reveal the potential for extensive cross-talk between β-defensins and the melanocortin system. In humans and other animals, β-defensins are highly polymorphic in sequence and copy number. Current β-defensin research is focused primarily on the immune system. This stems from the early discovery of defensins in phagocytic cells and their antimicrobial properties in vitro, together with more recent work demonstrating that defensins can act as receptor-specific chemotactic agents. Our work indicates that β-defensins do more than defend and suggests that the marked molecular variation in this family supplies a diverse and rapidly evolving family of ligands for G protein–coupled receptors in many different biologic systems.
Summary References
1. C. C. Little, The Inheritance of Coat Color in Dogs (Comstock, Ithaca, NY, 1957).
2. J. A. Kerns et al., Genetics 176, 1679 (2007).
3. K. Lindblad-Toh et al., Nature 438, 803 (2005).
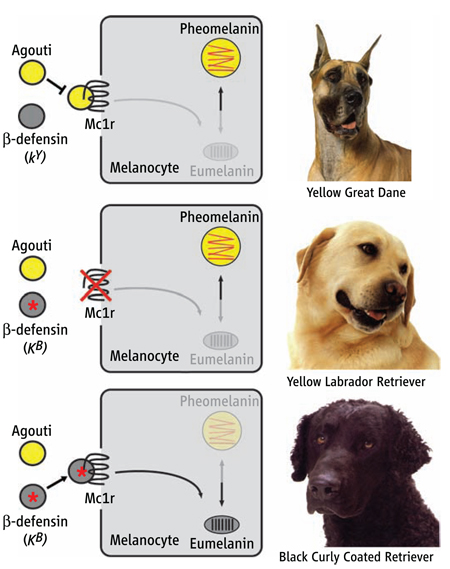
Production of yellow versus black pigment in dogs is controlled by three genes: Mc1r, Agouti, and CBD103. Dogs carrying wild-type alleles for all three genes have a yellow coat resulting from Agouti antagonism of Mc1r signaling in melanocytes (yellow Great Dane, top). Dogs carrying a loss-of-function mutation at Mc1r have a yellow coat, regardless of their genotype at Agouti or CBD103 (yellow Labrador Retriever, middle). Dogs carrying wild-type alleles for Mc1r and Agouti, together with the dominant black allele of CBD103 (KB) have a black coat resulting from the interaction between a β-defensin and Mc1r (black Curly Coated Retriever, bottom).
Genetic analysis of model systems in laboratory animals underlies much of what we know about major signaling pathways in multicellular organisms. In mammals, coat-color mutations have proven especially fruitful, because much of the molecular machinery used by the pigmentary system is either shared by, or homologous to, genes used for other physiologic pathways (1).
This approach has been particularly useful for pigment “type-switching,” a phenomenon in which melanocytes synthesize eumelanin (a black or brown pigment) versus pheomelanin (a red or yellow pigment), depending on the phase of the hair growth cycle, position on the body, and the genotype of several key loci (2). In most mammals, two genes that control pigment type-switching have been recognized: the Mc1r, which encodes a seven transmembrane–spanning domain protein expressed on melanocytes (3–5), and Agouti, which encodes a ligand for the Mc1r that is secreted by specialized dermal cells and which inhibits Mc1r signaling (6–9). Mc1r activation causes exclusive production of eumelanin, whereas Mc1r inhibition causes exclusive production of pheomelanin (5, 10). Thus, gain-of-function Mc1r mutations cause dominant inheritance of a black coat, whereas gain-of-function Agouti mutations cause dominant inheritance of a yellow coat. The Mc1r was first recognized by virtue of its ability to respond to peptides derived from proopiomelanocortin (POMC), such as α-melanocyte–stimulating hormone (α-MSH) (3, 4); however, a null mutation of Pomc has no effect on pigmentation in C57BL/6 mice, leading to the suggestion that the Mc1r has high basal activity and/or additional stimulatory ligands (11).
In a classic series of papers in the early 1900s, Sewall Wright (12) concluded that genetic mechanisms for color variation were largely conserved across mammals. An exception, however, later became apparent from the work of Clarence Cook Little on domestic dogs (13), in which dominant inheritance of a black coat was shown to involve a locus distinct from Mc1r. At the time, Little posited that dominant black was caused by an unusual allele of Agouti; however, using molecular linkage analysis, we recently demonstrated the presence of a third gene in dogs that interacts with Agouti and Mc1r, which we named the K locus (14). We found that the K locus has three alleles with a simple dominance order [Black (KB) > brindle (kbr) > yellow (ky)], that the K locus and Agouti behave similarly in genetic interaction studies (Mc1r is epistatic to both K and Agouti), and that the genetic map position of K does not correspond to the predicted location of any previously known pigmentation gene. We use the distinctive evolutionary history of domestic dogs to show that the K locus encodes a previously unrecognized class of melanocortin receptor ligands.
Linkage and association mapping of the K locus
We showed previously that the KB mutation mapped to a 12-Mb interval on the distal end of dog chromosome 16 (CFA16), between markers REN292N24 and FH3592 (14). We ascertained additional kindreds segregating KB, kbr, and ky to refine the map location [see supporting online material (SOM) text] and defined overlapping critical regions of 3.8 and 7.6 Mb for the KB and kbr mutations, respectively (Fig. 1Afigs. S1 to S4).
Fig 1.
Genetic mapping of the K locus. (A) Initial linkage studies [phase 1, (14)] defined a 12-Mb critical region for KB; ascertainment and characterization of additional kindreds narrowed the interval to 3.8 Mb (phase 2, figs. S1 to S4). Association analysis for 60 markers in brindle (n = 12) versus yellow (n = 10) Boxers, and for 51 markers in black (n = 9) versus yellow (n = 10) Great Danes, was carried out as described in the text. (B) Candidate genes in the 320-kb region of greatest association in Great Danes; this region includes 12 β-defensin genes (shown in red). (C and D) Significance, plotted as −log of P values from a chi-square test of allele counts, is shown as a function of distance along CFA16 (only for SNPs present at greater than 10% frequency and genotyped in at least 75% of the samples). The dashed red line indicates a Bonferroni-corrected 5% significance level; these regions are indicated by hatched and black bars for Boxers and Great Danes, respectively, in (A). Annotation is based on the Non-dog RefSeq track in the UCSC Genome Browser, except for CBD102, identified by Patil et al. (16).
We used an association-based strategy to narrow the critical region. Because most breeds were derived in the past 200 years from small founding populations (15), mutations within a breed are expected to be identical by descent and share extended haplotypes. In Boxers and Great Danes, we identified broad peaks of significant association (Bonferroni-corrected P value < 0.05) that extended over 1.9 Mb and 320 kb, respectively (Fig. 1, B to D). Sixteen genes have been annotated to the region of significant association in Great Danes, including a gene cluster that encodes 12 β-defensins (16): small antimicrobial peptides that are secreted mainly by epithelial cells (17, 18). We sequenced the mature protein-coding regions for nine members of the β-defensin cluster (those known at the time) in dogs carrying KB and/or ky and identified several polymorphisms concordant with the KB allele, including a 3–base pair (bp) deletion in the second exon of CBD103, the ortholog of human DEFB103, that predicts an in-frame glycine deletion (ΔG23).
To evaluate the extent to which the ΔG23 polymorphism distinguishes KB versus ky more broadly, we examined dogs from 38 breeds that could be classified into two categories with regard to their putative K locus genotype (SOM text). Among 454 dogs, there were 13 cases where the ΔG23 polymorphism did not correlate with coat-color phenotype. However, sequencing of Agouti and Mc1r revealed that each discordant case could be explained by known epistatic interactions (19, 20) (table S3). These results indicate that KB alleles in all breeds are probably identical by descent and suggest that the ΔG23 polymorphism or a closely linked variant in complete linkage disequilibrium (LD) is the KB mutation.
Short-range haplotype and resequencing analysis of KB-bearing chromosomes
By contrast to the pattern of LD within breeds, which affords a powerful approach for association mapping with megabase resolution, the pattern of LD across breeds is more fine-grained and therefore provides the opportunity for high-resolution haplotype mapping when mutations in different breeds are identical by descent (21). We identified 28 polymorphisms [22 single-nucleotide polymorphisms (SNPs) and six indels including the ΔG23 polymorphism] in a 20-kb interval surrounding CBD103 that were then used to infer short-range haplotypes for 14 KB-bearing and 16 ky-bearing chromosomes selected from seven breeds (SOM text). We observed six “parental” ky-bearing and five “parental” KB-bearing chromosomes (depicted in yellow and blue, respectively, in Fig. 2A). We also identified eight chromosomes that carried a single ancestral recombination event, which together defined a maximal interval for KB of 9146 bp (Fig. 2A). Complete resequencing of this interval (except for three homopolymer tracts) in five ky/ky animals, one KB/ky animal, and four KB/KB animals from seven breeds revealed two polymorphisms besides ΔG23 that are perfectly concordant with K locus genotype (S104 and S105) (Fig. 2A and table S2).
Fig 2.
Resequencing and recombinant haplotype-based mutation analysis for KB-bearing versus ky-bearing chromosomes. (A) A 20-kb region surrounding CBD103ΔG23 was resequenced (except for repetitive regions) in 10 dogs from 7 breeds, and haplotypes were inferred for 28 high-frequency biallelic polymorphisms. Blue and yellow squares represent the major and minor alleles in KB-bearing chromosomes, respectively, and allow some haplotypes to be designated as “ky-parental,” “KB-parental,” or “proximal recombinant,” as indicated. White squares represent missing data. Genotypes for five Great Danes (denoted by asterisks) were determined in a second resequencing round targeted specifically for distal recombinants as described in the SOM text. Within the 9.1-kb interval defined by recombinant haplotype analysis, three polymorphisms are completely associated with KB versus ky, as indicated in the upper part of the figure. (B) Exon structure of transcripts within the maximal candidate interval and alignment of selected CBD103 orthologs (38).
The 9146-bp interval contains both exons of CBD103, the first exon of dog expressed sequence tag (EST) CX990240, and dog EST CO665262 (Fig. 2). However, several considerations indicate that CBD103 is, indeed, the K locus and that the ΔG23 deletion in CBD103 is the KB mutation. First, the other two transcribed elements in the critical interval are represented in the database by single ESTs and are not known to encode proteins or to be expressed in the skin. Second, S104 and S105 lie in a long terminal repeat element that is 3 kb upstream of the first exon of CBD103 and have no effect on mRNA levels of CBD103 (Fig. 3A). Finally, as discussed further below, CBD103 is highly expressed in skin, the ΔG23 deletion affects CBD103 protein function, and pharmacologic studies reveal that CBD103 can modulate melanocortin signaling.
Fig 3.
Expression of β-defensin mRNA and protein in skin and in cultured keratinocytes. (A) Levels of Agouti, CBD1, or CBD103 mRNA from black or yellow dog skin, as indicated, determined relative to Bactin by quantitative RT-PCR, and expressed as percentage of mRNA present in the yellow samples. Results shown represent the mean ± SEM of four different animals. (B) Expression of epitope (V5)–tagged CBD103 (+) or epitope (V5)–tagged CBD103ΔG23 (ΔG23) in cell layer and media after transfection of mouse keratinocytes as determined by Western blotting with antisera against the V5 epitope. Representative results are shown for one of four experiments; for each experiment, the two constructs were transfected in triplicate or quadruplicate.
The preceding discussion has referred to K locus variation as though ky is ancestral, whereas KB is derived: a hypothesis based on the comparative genetic distribution of coat-color phenotypes and inheritance patterns. Considerations based on sequence alignments confirm this hypothesis: Mammalian CBD103 orthologs that we identified from the available genome sequence are each 67 amino acids in length, and the optimal sequence similarity alignment contains no gaps or insertions (Fig. 2B), indicating that ΔG23 and consequently the KB mutation occurred specifically within the canid lineage.
Expression of dog defensins in skin and in transfected keratinocytes
We isolated RNA from the skin of a ky/ky Doberman Pinscher and a KB/ky mixed-breed dog and surveyed the expression of the 19 β-defensin genes that are clustered on chromosomes 16 or 25 by reverse transcription polymerase chain reaction (RT-PCR) (16). Expression was detectable only for two genes: CBD1 and CBD103 (fig. S6). We then used quantitative RT-PCR to measure levels of skin mRNA from four KB/ky samples and four ky/ky samples, which were all from mixed-breed dogs, and found no effect of K locus genotype on levels of CBD1, CBD103, or Agouti mRNA (Fig. 3A).
Available antisera against human DEFB103 are unable to detect the endogenous dog protein by Western blotting or immunohistochemistry; therefore, we generated epitope-tagged expression constructs for each allele (CBD103V5 and CBD103DG23V5) and studied their patterns and levels of protein expression after transfection of cultured mouse keratinocytes.
In cell extracts analyzed by Western blotting, antisera against the V5 epitope detect a single fragment whose size (about 8 kD) corresponds to the expected molecular mass of the tagged protein after signal peptide cleavage; in media, an additional slightly smaller band is present, which suggests additional processing (Fig. 3B). The relative ratios of the two bands are similar in media from keratinocytes transfected with either construct; however, the total amount of immunodetectable protein in media was significantly greater for CBD103DG23V5 as compared with that for CBD103V5 (P = 0.0021, Cochran-Mantel-Haenszel chi-square test). Thus, loss of the N-terminal glycine from CBD103 does not affect intracellular processing but allows more of the mature protein to accumulate in the media and/or extracellular space.
CBD103 activity in vivo and in vitro
To further explore the function of CBD103 in an experimental genetic system, we generated transgenic mice in which a cDNA encoding either the KB or the ky allele was driven by a strong and widely expressed promoter (22). We chose a genetic background that normally has Agouti-banded hairs and observed that two transgenic founders generated with the CBD103ΔG23 cDNA (the KB allele) displayed a predominantly black coat with small patches of banded hair (Fig. 4). Unexpectedly, the normal CBD103 cDNA (the ky allele) also produced transgenic mice with a black coat in 20 out of 21 founders (Fig. 4). Furthermore, we observed that transgenic animals were smaller than their nontransgenic littermates. By 2 weeks of age, female transgenic animals were easily recognized by their dark coat and small size; in adult mice, reduced body weight persists in both males and females (fig. S7).
Fig 4.
Pigmentary effects of CBD103 in transgenic mice. Photographs of transgenic (Tg) and non-transgenic littermates, representative of 2/2 independent founders for Tg.CBD103ΔG23 and 20/21 independent founders for Tg.CBD103.
These considerations suggest three possible mechanisms by which CBD103 might act to modulate melanocortin signaling: (i) by binding to and activating the Mc1r, (ii) by binding to the Mc1r and preventing its inhibition by Agouti protein, or (iii) by binding to Agouti protein leading to its sequestration and/or degradation. To distinguish among these ideas, we generated synthetic forms of CBD103 and tested their ability to interact with the Mc1r and agouti signaling protein–YY (ASIP-YY), a synthetic version of the C terminus of Agouti protein that behaves as a competitive antagonist of α-MSH at the Mc1r and Mc4r (23).
The CBD103 ky allele predicts a mature peptide (after signal sequence cleavage) of 45 amino acids that contains six cysteine residues and begins with the glycine that is deleted in the KB allele (Fig. 2B). The predicted signal sequence cleavage site is supported by biochemical studies of the orthologous human protein [known as DEFB103 or human β-defensin 3 (HBD3)] purified from human tissues (24). We synthesized the 45-residue ky form and 44-residue KB form of the dog protein (hence referred to as CBD103 and CBD103DG23, respectively), allowed oxidative refolding, and used mass spectrometry and high-performance liquid chromatography to confirm recovery of a single congener with three intrachain disulfide bonds.
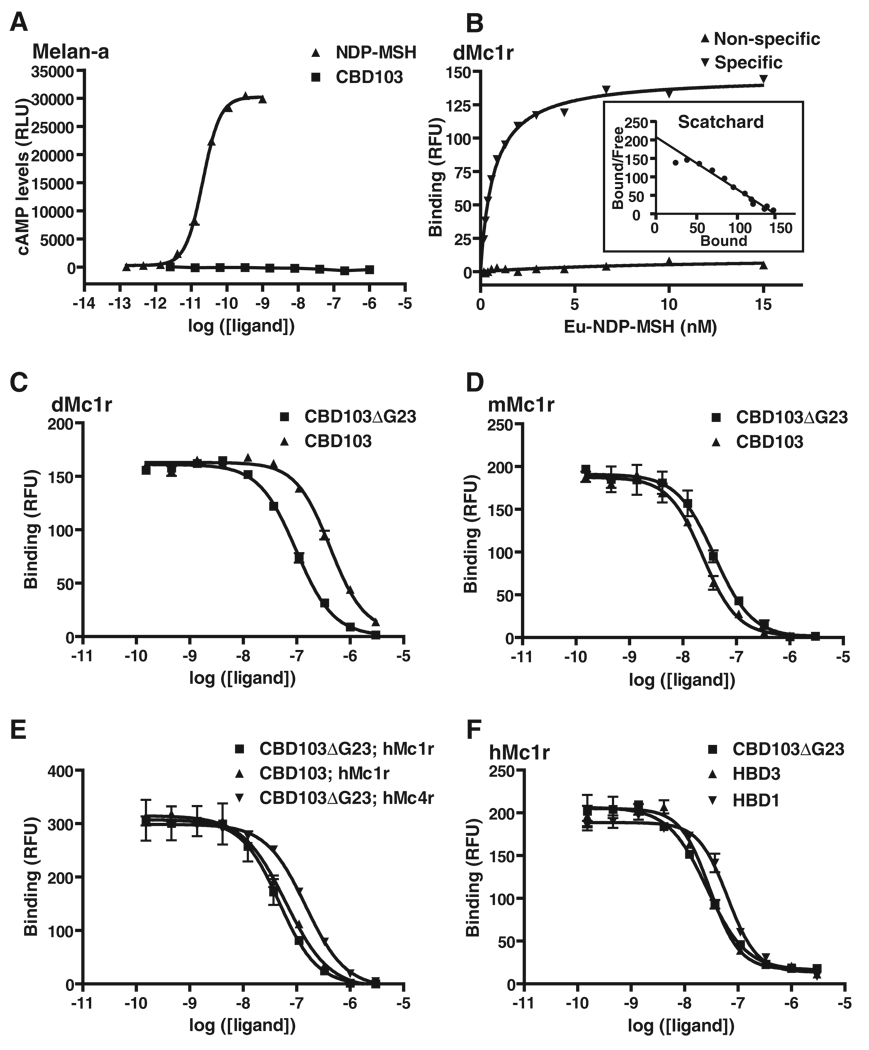
[Nle4, d-Phe7]-α–MSH (NDP-MSH), a potent derivative of α-MSH, stimulates robust accumulation of cyclic adenosine monophosphate (cAMP) in melanocytes (25). However, neither CBD103 nor CBD103ΔG23 has any effect on cAMP accumulation (Fig. 5A), indicating that CBD103 is not a conventional Mc1r agonist. To test for receptor binding, we transiently transfected human embryonic kidney (HEK) 293 cells with Mc1r expression constructs (22) and used europium-labeled NDP-MSH (Eu-NDP-MSH) as a fluorescent tracer. We used saturation binding assays to first calculate the affinity of Eu-NDP-MSH for the dog Mc1r (Fig. 5B) and then carried out displacement assays with progressively increasing concentrations of CBD103, CBD103ΔG23, or ASIP-YY. All three peptides exhibited qualitatively similar profiles characteristic for competitive binding to a single high-affinity site (Fig. 5C and Table 1). Quantitatively, inhibition constant (Ki) values estimated from the data depicted in Fig. 5C were 150.6 nM for CBD103 and 34.2 nM for CBD103ΔG23. These estimates varied according to experimental conditions, but CBD103ΔG23 consistently exhibited higher affinity for the dog Mc1r than did CBD103 (mean of fivefold across four paired experiments; Table 1). In these same experiments, ASIP-YY exhibited Ki values of 0.51 to 0.95 nM for the dog Mc1r, in the same range as reported previously for ASIP-YY at the human Mc1r (23). Using quantitative RT-PCR, we found that the levels of CBD103 mRNA in total skin were ~300-fold greater than that of Agouti mRNA; to the extent that this difference reflects protein abundance, the levels of CBD103ΔG23 in dog skin are likely to be much greater than those of Agouti protein, which is consistent with a model in which CBD103ΔG23 competitively inhibits the ability of Agouti protein to antagonize Mc1r signaling.
Fig 5.
Pharmacology of β-defensin action on melanocortin receptors. (A) Ability of NDP-MSH or CBD103 to stimulate cAMP accumulation in cultured melanocytes. (B) Saturation binding of Eu-NDP-MSH to HEK293 cells transiently transfected with the dog Mc1r. (C to F) Competition binding assays in which varying amounts of unlabeled synthetic β-defensins were added together with Eu-NDP-MSH tracer (at 1.8 to 3 nM) to HEK293 cells transiently transfected with dog (d), mouse (m), or human (h) melanocortin receptors, as indicated. In all panels, ligand concentration {either in nM (B) or log of the molarity [(A) and (C) to (F)]} is plotted on the abscissa; amount of cAMP formed (A) or Eu-NDP-MSH bound [(B) to (F)], measured, respectively, as relative light units (RLUs) or relative fluorescent units (RFUs), is plotted on the ordinate. Each curve represents a single experiment carried out in triplicate; error bars represent SEM.
Table 1.
Affinity constants for melanocortin receptor ligands. In the column for Eu-NDP-MSH, saturation binding assays as depicted in Fig. 5B were used to derive dissociation constant (Kd) values (in nM) by fitting the data to a hyperbolic dose-response curve with the use of nonlinear regression. In the remaining columns, displacement binding assays as depicted in Fig. 5, C to F, were used to derive Ki values (in nM) by fitting the data to a sigmoidal dose-response curve with variable slope. For some ligand-receptor combinations, multiple experiments were carried out, in which case the mean Ki value is given followed by the range and number (in parentheses) of the separate experiments. ND, not determined.
| Receptor | Eu-NDP-MSH | ASIP-YY | CBD103ΔG23 | CBD103 | HBD1 | HBD3 |
|---|---|---|---|---|---|---|
| Dog Mc1r | 0.70 | 0.59 (0.51–0.95) (n = 3) |
37.0 (16.4–61.1) (n = 5) |
221.0 (150.6–398.7) (n = 4) |
ND | ND |
| Mouse Mc1r | 1.38 | 2.30 | 15.1 (12.4–17.0) (n = 4) |
9.7 (8.9–10.4) (n = 2) |
ND | ND |
| Human Mc1r | 2.59 | 0.95 | 19.6 (12.3–26.8) (n = 2) |
35.5 | 30.0 | 13.8 |
| Human Mc4r | 3.17 | 0.82 | 104.5 | ND | ND | ND |
Finally, we investigated a potential interaction between Agouti protein and CBD103 by preincubating ASIP-YY with either CBD103 or CBD103ΔG23 before the binding assay, and we found no evidence for a functional interaction (fig. S8). We also examined the effects of mixing the two peptides on two-dimensional nuclear magnetic resonance spectra and found no evidence for a structural interaction. Taken together, these observations indicate that CBD103 is a high-affinity ligand for the Mc1r and that the ΔG23 mutation may cause a black coat color in dogs via two different mechanisms: increased affinity for the Mc1r and increased availability of the mature protein in vivo.
Other mammals, other melanocortin receptors, and other defensins
Dogs are the only mammals known for which dominant inheritance of black coat color is not caused by a Mc1r mutation. However, we observed that both CBD103 and CBD103ΔG23 are high-affinity ligands for the mouse (Fig. 5D) and the human (Fig. 5E) Mc1r, with estimated Ki values that were, unexpectedly, lower than that observed at the dog Mc1r (Table 1). Furthermore, unlike at the dog Mc1r, the affinity of CBD103 as compared to that of CBD103ΔG23 was similar at both the mouse and the human Mc1r (Table 1), which may explain why expression of either allele caused a black coat in transgenic mice (Fig. 4).
Given the effects of the CBD103 transgenes on body size, we investigated the potential for cross-talk between β-defensins and other melanocortin receptors. We found that CBD103ΔG23 binds to the human Mc4r with an intermediate affinity (Ki = 104.5 nM) between that of CBD103ΔG23 and CBD103 for the dog Mc1r (Table 1 and Fig. 5E). We also synthesized HBD3, the human ortholog of canine CBD103, and observed high-affinity binding (Ki = 13.8 nM) to the human Mc1r (Table 1 and Fig. 5F). Finally, we tested human β-defensin 1 (HBD1), which lies in the same cluster as HBD3 but is more distantly related; HBD1 also exhibited high-affinity binding to the human Mc1r (Ki = 30 nM) (Table 1 and Fig. 5F).
Defensins and melanocortin signaling
β-defensin genes exhibit strong signatures of diversifying selection (26, 27), which is consistent with their proposed role as endogenous antibiotics. Confirming and studying this role in vivo, however, have been challenging; the results of mouse knockout experiments have been reported for only one gene, Defb1, and revealed no obvious effects on viability or innate defense (28). By contrast, the CBD103 KB mutation causes a marked change in hair color that is readily apparent among a large number of dog breeds with diverse genetic backgrounds. We propose that β-defensin genes play a broader role than currently envisaged, in which the ability to modulate melanocortin receptor signaling may have been selected during vertebrate evolution to provide camouflage and/or adaptive visual cues. From this perspective, evolutionary lability of the β-defensins with regard to both diversifying selection and copy number variation (26, 27, 29) may represent a response to a shifting spectrum of environmental challenges that includes microbial pathogens, carnivorous predators, and nutrient availability. More generally, our biochemical studies indicate that at least two human β-defensins, HBD1 and HBD3, may also modulate melanocortin receptor signaling in vivo. Both genes are expressed by a broad range of epithelial and other tissues (18) and could therefore act not only on Mc1r but also on other melanocortin receptors, including Mc5r, for which an endogenous melanocortin ligand has not yet been identified. The range of Ki values that we observed suggests that CBD103ΔG23, and potentially HBD1 or HBD3, modulate pigmentation in the low nanomolar range; however, the antimicrobial effects of β-defensins are typically observed in the low micromolar range (30). Finally, several β-defensins promote chemotaxis of immune cells, and although there is controversy regarding which receptors are involved, there is a growing consensus that β-defensins contribute to adaptive as well as to innate immunity (31–33). From this perspective, several studies have pointed to a role for Mc1r signaling in immune cell function (34), which could be mediated by β-defensins acting as melanocortin receptor ligands.
Modulation of melanocortin receptor signaling by β-defensins may also help explain the apparent paradox that mutations of Pomc have a relatively minor effect on pigmentation (11, 35–37). These observations have been often interpreted as evidence that melanocortin receptors have high levels of constitutive activity, but our results suggest that β-defensins may raise “basal” levels of melanocortin receptor signaling in the absence of melanocortin peptides.
Much of the early work on melanocortin signaling was driven by ideas associated with the way in which Pomc had been discovered and named; additional biological functions of melanocortins in behavior and energy homeostasis did not become apparent for decades. An analogous situation may apply in the case of β-defensins, which underscores the utility of phenotype-driven genetics to provide an agnostic view of gene function.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/1147880/DC1
Materials and Methods
SOM Text
Figs. S1 to S10
Tables S1 to S3
References
References and Notes
- 1.Bennett DC, Lamoreux ML. Pigment Cell Res. 2003;16:333. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 2.Barsh GS. In: The Pigmentary System. Nordlund JJ, et al., editors. Oxford: Blackwell; 2006. pp. 395–410. [Google Scholar]
- 3.Chhajlani V, Wikberg JE. FEBS Lett. 1992;309:417. doi: 10.1016/0014-5793(92)80820-7. [DOI] [PubMed] [Google Scholar]
- 4.Mountjoy KG, Robbins LS, Mortrud MT, Cone RD. Science. 1992;257:1248. doi: 10.1126/science.1325670. [DOI] [PubMed] [Google Scholar]
- 5.Robbins LS, et al. Cell. 1993;72:827. doi: 10.1016/0092-8674(93)90572-8. [DOI] [PubMed] [Google Scholar]
- 6.Millar SE, Miller MW, Stevens ME, Barsh GS. Development. 1995;121:3223. doi: 10.1242/dev.121.10.3223. [DOI] [PubMed] [Google Scholar]
- 7.Bultman SJ, Michaud EJ, Woychik RP. Cell. 1992;71:1195. doi: 10.1016/s0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- 8.Miller MW, et al. Genes Dev. 1993;7:454. doi: 10.1101/gad.7.3.454. [DOI] [PubMed] [Google Scholar]
- 9.Ollmann MM, Lamoreux ML, Wilson BD, Barsh GS. Genes Dev. 1998;12:316. doi: 10.1101/gad.12.3.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klungland H, Vage DI. Ann. N.Y. Acad. Sci. 2003;994:331. doi: 10.1111/j.1749-6632.2003.tb03197.x. [DOI] [PubMed] [Google Scholar]
- 11.Slominski A, et al. Endocrinology. 2005;146:1245. doi: 10.1210/en.2004-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright S. J. Hered. 1917;8:224. [Google Scholar]
- 13.Little CC. The Inheritance of Coat Color in Dogs. Ithaca, NY: Comstock; 1957. [Google Scholar]
- 14.Kerns JA, et al. Genetics. 2007;176:1679. doi: 10.1534/genetics.107.074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sampson J, Binns MM. In: The Dog and Its Genome. Ostrander EA, Giger U, Lindblad-Toh K, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. pp. 19–30. [Google Scholar]
- 16.Patil AA, Cai Y, Sang Y, Blecha F, Zhang G. Physiol. Genomics. 2005;23:5. doi: 10.1152/physiolgenomics.00104.2005. [DOI] [PubMed] [Google Scholar]
- 17.Ganz T. Nat. Rev. Immunol. 2003;3:710. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 18.Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Cell. Mol. Life Sci. 2006;63:1294. doi: 10.1007/s00018-005-5540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton JM, et al. Mamm. Genome. 2000;11:24. doi: 10.1007/s003350010005. [DOI] [PubMed] [Google Scholar]
- 20.Kerns JA, et al. Mamm. Genome. 2004;15:798. doi: 10.1007/s00335-004-2377-1. [DOI] [PubMed] [Google Scholar]
- 21.Lindblad-Toh K, et al. Nature. 2005;438:803. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 22.For further details, see the Supporting Online Material
- 23.McNulty JC, et al. J. Mol. Biol. 2005;346:1059. doi: 10.1016/j.jmb.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 24.Harder J, Bartels J, Christophers E, Schroder JM. J. Biol. Chem. 2001;276:5707. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 25.Sawyer TK, et al. Proc. Natl. Acad. Sci. U.S.A. 1980;77:5754. doi: 10.1073/pnas.77.10.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semple CA, Gautier P, Taylor K, Dorin JR. Mol. Divers. 2006;10:575. doi: 10.1007/s11030-006-9031-7. [DOI] [PubMed] [Google Scholar]
- 27.Hughes AL. Cell. Mol. Life Sci. 1999;56:94. doi: 10.1007/s000180050010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison G, Kilanowski F, Davidson D, Dorin J. Infect. Immun. 2002;70:3053. doi: 10.1128/IAI.70.6.3053-3060.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollox EJ, Armour JA, Barber JC. Am. J. Hum. Genet. 2003;73:591. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, et al. Peptides. 2006;27:931. doi: 10.1016/j.peptides.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Yang D, et al. Science. 1999;286:525. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 32.Soruri A, Grigat J, Forssmann U, Riggert J, Zwirner J. Eur. J. Immunol. 2007;37:2474. doi: 10.1002/eji.200737292. [DOI] [PubMed] [Google Scholar]
- 33.Biragyn A, et al. Science. 2002;298:1025. doi: 10.1126/science.1075565. [DOI] [PubMed] [Google Scholar]
- 34.Luger TA, Scholzen TE, Brzoska T, Bohm M. Ann. N.Y. Acad. Sci. 2003;994:133. doi: 10.1111/j.1749-6632.2003.tb03172.x. [DOI] [PubMed] [Google Scholar]
- 35.Yaswen L, Diehl N, Brennan MB, Hochgeschwender U. Nat. Med. 1999;5:1066. doi: 10.1038/12506. [DOI] [PubMed] [Google Scholar]
- 36.Challis BG, et al. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4695. [Google Scholar]
- 37.Smart JL, Low MJ. Ann. N.Y. Acad. Sci. 2003;994:202. doi: 10.1111/j.1749-6632.2003.tb03181.x. [DOI] [PubMed] [Google Scholar]
- 38.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 39.This work was supported by grants from NIH to G.S.B. and to G.L.M. and by a grant from the Donald E. and Delia B. Baxter Foundation to S.I.C. We thank H. Manuel for technical assistance; Y. Kobayashi and the Stanford Transgenic Facility for advice and help with transgenic experiments; D. Bennett for melan-a cells; Y.-K. Yang for providing the Mcr expression constructs; E. A. Ostrander for providing some of the samples; and the dog owners and breeders who generously submitted DNA samples.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.