Abstract
The effect of desferoxamine (DFO)-induced hypoxia on neuronal human mu-opioid receptor (hMOR) gene expression was investigated using NMB cells. DFO decreased cell viability and increased cellular glutathione levels in a dose- and time-dependent manner. Confocal analysis using annexin-V-fluorescein and propidium iodide staining revealed that surviving/attached cells under DFO challenge were morphologically similar to control (vehicle-treated) cells. RT-PCR analysis demonstrated that the hypoxia inducible factor-1α (HIF-1α) mRNA level was augmented in these surviving neurons. DFO treatment for 8hrs or longer down-regulated the hMOR message, but not that of the delta-opioid receptor. Functional analysis using luciferase reporter assay showed that the hMOR 5′-regulatory region, from −357bp to translational initiation site (+1), contains the active promoter with an inhibitory region located in the −422 to −357bp region. DFO decreased hMOR promoter activity as compared to control. Mutation analysis suggested the existence of both dsDNA and ssDNA elements, located in a CT-rich region of hMOR, mediating the DFO-response. RT-PCR further revealed that DFO exhibited no effect on hMOR mRNA stability. In conclusion, DFO-induced hypoxia specifically affects neuronal hMOR gene expression at the transcriptional, not post-transcriptional, level.
Keywords: desferoxamine, HIF1-α, hMOR promoter, transcription, neurons
Introduction
Hypoxia (low oxygen condition), often caused by such conditions as physical trauma, cardiac arrest, or stroke [1], increases the expression of hypoxia inducible factor-1 (HIF-1) transcription factor. HIF-1, in turn, can up- and down-regulate expressions of various genes against insult [2]. It contains HIF-1α and 1β subunits. Under hypoxic condition, only HIF-1α mRNA is upregulated and it can be used as a cellular hypoxic marker.
Among different cell types, neurons are particularly sensitive to low levels of oxygen. With the reduction of oxygen availability and a subsequent decrease of ATP, neurons are unable to efficiently maintain membrane potential (required ATP), resulting in depolarization, calcium influx, and finally cell death [3]. However, some neurons still survive under the same hypoxic condition, suggesting the development of adaptation processes to overcome the insult [2].
Pain sensation can be triggered in patients suffering from hypoxia. Opioids, such as morphine, are used clinically to treat pain from surgery, trauma or myocardial infarction [4]. Three types of opioid receptors, mu (MOR), delta (DOR) and kappa (KOR), have been reported [5]. Among these, MOR is the key mediator for morphine-induced analgesia, and it is mainly expressed in the central nervous system (CNS) [6]. Therefore, it is important to determine whether hypoxia affects neuronal human MOR (hMOR) gene expression.
Regulation of hMOR gene expression in the neuronal system is not as well understood as that of the mouse MOR (mMOR) gene at the transcriptional level. Three different promoters (proximal, distal, and far upstream) of mMOR gene initiate neuronal mMOR gene expression at different transcriptional initiation sites were reported [7-9]. The proximal promoter, close to the translation initiation site, dominantly drives mMOR transcription in the CNS [10]. However, only one transcription initiation site, also close to the translational initiation site, has been documented for the hMOR gene [11].
Previous analysis [12-13] reported that the mMOR proximal core promoter contains a 26bp CT-rich region (also known as PPy/u region) with an overlapping double-stranded (ds) and single-stranded (ss) DNA element. Based on sequence comparisons, the hMOR gene also possesses a similar CT-rich element located closely to the transcription initiation site.
Here we investigated the effect of deferoxamine (DFO) on neuronal hMOR gene expression at the transcriptional or posttranscriptional level using human neuronal cells, NMB, endogenously expressing hMOR. DFO creates hypoxia by chelating irons [14] and altering the iron status of iron- and O2-dependent hydroxylases, from which HIF receives the cellular O2 level information. DFO has also been shown to increase HIF-1 gene expression in cells and brains from animal models. Thus, we reported the mechanism underlying DFO-induced hypoxia on hMOR gene expression in neuronal cells, which survived under DFO-induced hypoxic stress.
Materials and methods
Cell culture and counting
Human neuroblastoma NMB [15] were cultivated in RPMI 1640 medium with 10% heat-inactivated fetal calf serum in an atmosphere of 5% CO2 and 95% air at 37 °. Cells treated with deferoxamine (DFO) were rinsed gently with PBS, and detached with PBS/EDTA for cell counting using trypan blue staining (Sigma).
Glutathione assay
Cellular glutathione level was determined using GSH-Glo Glutathione Assay (Promega). Briefly, cells were incubated with GSH-Glo reagent for 30min at R.T., and then incubated with luciferin detection reagent for 15min at R.T. Light signal from the reaction was measured using a luminometer (Berthold).
RNA extraction and RT-PCR
Total RNA from cells was isolated using TriReagent (Molecular Research Center). RT-PCR was performed as previously described [16] using human-specific primers: HIF-1α 5′-CCAGCAGACTCAAATACAAGAACC-3′ and 5′-GTATGTGGGTAGGAGATGGAGAT-3′; β-actin 5′-CCTTCCTGGGCATGGAGTCCTG-3′ and 5′-TACAGCGAGGCCAGGATGG-3′; DOR 5′-GTTCACCAGCATCTTCACGCTC-3′ and 5′-CGGTCCTTCTCCTTGGAGCCC-3′; MOR 5′-CTGGAAGGGCAGGGTACTGGTG-3′ and 5′-CTGCCCCCACGAACGCCAGCAAT-3′.
Cell staining and confocal microscopy
Cells grown on coverslips were washed with PBS, and then were stained using Annexin-V-FLUOS Staining Kit following the company’s instructions (Roche). Cell image was taken using a laser scanning confocal microscope (Fluoview 1000, Olympus).
Isolation of genomic DNA from NMB cells and PCR amplification of 5′-regulatory region of hMOR gene
NMB genomic DNA was prepared by following the manufacturer’s procedure (Molecular Research Center). Briefly, cells were lysed using TriReagent and then incubated with chloroform. The mixture was then subjected to centrifugation at 12,000 rpm at 4°C. DNA was precipitated using ethanol from interphase and organic phase. To amplify hMOR 5′-regulatory region (−422 to +1 bp and −357 to +1 bp, respectively) by PCR, two sense primers and one antisense primer were used: −422, 5′-CTCTCTCCCCAACCCTTCTCTCCATCTCC-3′; −357, 5′-GAAGAGACCTACTCCTTGGATCGCTTTG-3′; antisense, 5′-CCATGGTACTGACGGCCGGG-3′.
Plasmid construction
Plasmids, containing various lengths of hMOR 5′-regulatory region, were made using a promoterless vector, pGL3-basic (Promega), containing luciferase gene. The pRh285/325 plasmid contained the reverse orientation of the −325 to −285 bp region. To prepare plasmids containing mutated ss or ds element, the wild type plasmid, ph325/285, was used as the template and PCR reaction was performed with mutant oligonucleotides. ph325/285-ds, 5′-CTTTTCCCTGGACGGGGCGGGGCAGCC-3, ph325/285-ss 5′-CTTTTCCCTCCTGGTTCCCTTCCA-3′. All inserts were examined by sequencing.
Transient transfection and reporter gene assay
Cells were transfected using lipofection method, as described previously [16]. Forty-eight hrs after transfection, cells were harvest and lysed with lysis buffer (Promega). Normalization of samples followed the method described previously [16]. Luciferase activity was determined following the manufacturer procedure (Promega) using a luminometer (Berthold).
Results
Dose-dependent effects of DFO-induced hypoxia on human NMB neuronal cells
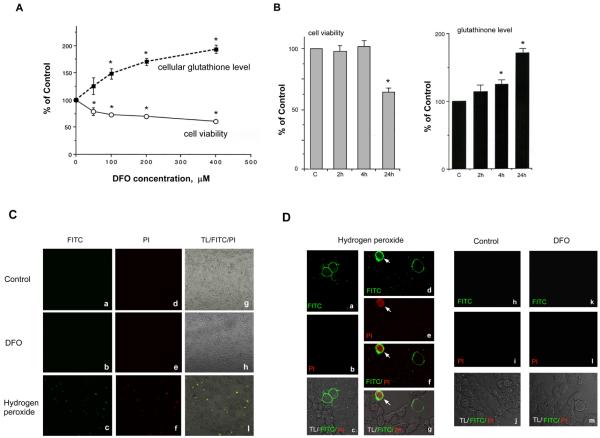
Human NMB cells, endogenously expressing mu-opioid receptors (MOR), were treated with deferoxamine (DFO), to create a DFO-induced hypoxic cell model. DFO can cause various degree of cell death; therefore, cell survival rate under different concentrations (50, 100, 200 and 400 μM) of DFO treatment for 24 hrs was determined first. Total number of surviving/attached cells was counted using non-DFO-treated group (control) as 100%. As shown in Fig. 1A (open circles), the percentage of surviving cells decreased significantly as DFO concentration increased (50 and 100 μM, respectively). The slope of the linear line (% surviving vs. DFO concentration) was −0.42 initially, falling off to −0.09 at the higher concentrations (using 100, 200, and 400μM data points).
Fig. 1.
Effect of DFO on NMB cells
A-B, Cells were treated without (control) or with different concentrations of DFO for 24 hrs (A), or 200μM DFO for 2, 4 or 24 hrs (B). Total number of attached cells was determined. Cell viability (open circles in A; gray bars in B) is presented as a percentage of total cell number from DFO-treated group divided by the number from control (as 100%). Cellular glutathione level (closed squares in A; black bars in B) is present as the percentage of the average amount of glutathione per cell from DFO-treated group divided by control (as 100%). Data is present as mean± S.E. “*” indicates p< 0.01 (student paired t-test). C and D, Surviving/attached cells under DFO, no treatment (Control), or H2O2 treatment (positive control) were stained using annexin-V-FLUOS (as FITC in green color) and propidium iodide (as PI in red color). Cells were imaged using confocal microscope under 10x in C or 40x magnification of object lens in D. Merged images are overlapped images of transmitted light (TL) with FITC and PI images, or an overlapped image (panel f in D) of FITC and PI images. An arrow indicates the cell staining by both annexin-V and PI.
Cells surviving DFO challenge may develop adaptive processes. To examine if this adaptation was developed in surviving/attached cells, glutathione level was measured using the average amount of glutathione per cell obtained from control as 100%. As shown in Fig. 1A (close squares), as DFO concentration increased to 50 and 100 μM, the level of cellular glutathione increased sharply (slope of 1.0), though the rate of increase was less at concentrations up to 200 and 400 μM DFO (0.26 slope).
Collectively, these results showed that the increase of DFO concentration caused a decrease of cell viability and an increase of cellular glutathione level in neurons surviving DFO-induced hypoxia, with the effect near maximum at 200 μM DFO. Therefore, this concentration was chosen for further experiments.
Impact of DFO exposure time on cell survival rate was then examined by treating cells with DFO for a short (2 and 4hrs) or longer (24 hrs) time. As shown in Fig. 1B, left panel, cell viability did not change significantly at 2hr- or 4hr-DFO treatment as compared to that of control (as 100%). Only 24hr treatment displayed a significant difference. Concomitantly, as shown in Fig. 1B, right panel, a minor increase of cellular glutathione level was observed at 2 or 4hr treatment as compared to control (100%), but again only 24hr treatment generated a substantial increase. Thus long-term (24hr), but not short-term, DFO exposure produced extensive changes on cell viability and cellular glutathione level.
We next examined if these surviving/attached cells, under 24hr DFO challenge, were morphologically different from that of control at the stage of apoptosis or necrosis, using annexin-V-FLUOS and propidium iodide (PI) staining method. Annexin-V-FLUOS (green color), interacting with phosphatidylserine on the outer plasma membrane, was used to stain the plasma membrane of an apoptotic cell. PI (red color), entering the cell and interacting with nuclear DNA and RNA molecules in nucleus and cytosol regions, was used to stain nucleus and cytosol of necrotic cells or cells in the late phase of apoptosis. As shown in Fig. 1C, under a low magnification, no significant green fluorescence or PI staining (red color) was observed in control and DFO-treated groups. Merged images (TL/FITC/PI) obtained from the transmitted light (TL) with FITC and PI images are also shown.
In addition, a positive control using NMB treated with 100 μM hydrogen peroxide for 4 hrs was also imaged. Fluorescence-reactive cells are shown in Fig. 1C. These fluorescence-reactive cells were further viewed using higher magnification (Fig. 1D). Annexin-V-FLUOS (green color) labeled the plasma membrane in panels a and d, suggesting fluorescence-reactive cells (panels a-c) were at the apoptotic stage. In panels d-g, one cell was at the apoptotic stage, and the other cell (with green and red labeling; indicated by an arrow) was at the necrosis or the late stage of apoptosis. Again, in both control (panels h-j) and DFO-treated groups (panels k-m), no fluorescence-reactive cells were observed, suggesting that the majority of attached neuronal cells were not at the apoptotic or necrotic stage. In summary, upon DFO challenge, the surviving/attached neurons displayed no apoptosis or necrosis.
Increase of HIF1α levels in survival cells under DFO challenge
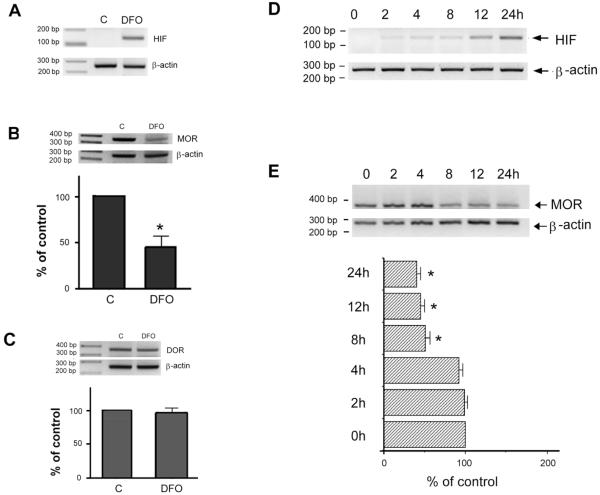
To determine if surviving/attached neuronal cells actually developed the cellular hypoxic response, hypoxia-induced factor 1α (HIF-1α) mRNA level was examined. RNA from NMB cells treated without (control, C) or with DFO for 24 hrs was subjected to semiquantitative RT-PCR. As shown in Fig. 2A, a significant DFO-induced increase of HIF-1α mRNA level was observed as compared to that of control (C), while β-actin levels (as an internal standard) were similar in both C and DFO groups. This augmentation of HIF-1α in surviving cells established that these surviving/attached neurons did develop the adaptation upon DFO treatment.
Fig. 2.
DFO alters endogenous HIF-1α and hMOR message levels
RNA was extracted from cells treated without (control, C) or with DFO (A-C) for 24 hrs, or with DFO for 0, 2, 4, 8, 12 or 24 hrs (D-E). RT-PCR was performed using human HIF-1α (A, D), MOR (B, E) or DOR (C)-specific primers. Human β-actin specific primers were included in every PCR reaction (added at the cycle 19) as an internal control for normalization purpose. The normalized message from control, or from cells at time zero, was arbitrarily defined as 100%. Quantitative analysis of message levels are presented as mean ± S.E. “*” indicates p< 0.01. (student paired t-test).
Reduction of neuronal hMOR gene expression under DFO challenge
Influence of DFO on hMOR gene expression was then investigated using semiquantitative RT-PCR with RNAs from cells treated without (control, C) or with DFO for 24hrs. As shown in Fig. 2B, a significant decrease of hMOR mRNA level (MOR) was observed as compared to control, while β-actin messages (β-actin) remained unaffected. Quantitative data of normalized hMOR message was also shown with normalized hMOR messages from control as 100%. Surviving neurons produced approximately 45% of hMOR messages as compared to control.
Effect of DFO on hDOR gene expression
To investigate if DFO-induced change was specific to hMOR mRNA, human δ-opioid receptor (hDOR) mRNA was also investigated using NMB. Semiquantitative RT-PCR showed (Fig. 2C) that DFO resulted in no significant change of hDOR message as compared to control (C), suggesting that DFO had no effect on hDOR gene expression.
DFO-induced alterations of hMOR and hHIF-1α gene expressions in a time-dependent manner
To determine the time course of DFO effect on hMOR gene expression, RNA from cells treated with DFO for 2, 4, 8, 12 and 24hrs was examined using semiquantitative RT-PCR. As shown in Fig. 2E, a significant decrease of hMOR mRNA level was observed at 8, 12 and 24hr treatment as compared to control (0 hr; as 100%). In addition, DFO also resulted in an increase of HIF-1α expression (Fig. 2D) in a time-dependent manner in these surviving/attached neurons.
Analysis of hMOR gene promoter region
To further examine if the mechanism controlling DFO-induced reduction of hMOR gene expression occurred at the transcription level, we proceeded to analyze hMOR promoter. Since hMOR promoter is not as well understood, the active hMOR promoter region was first determined.
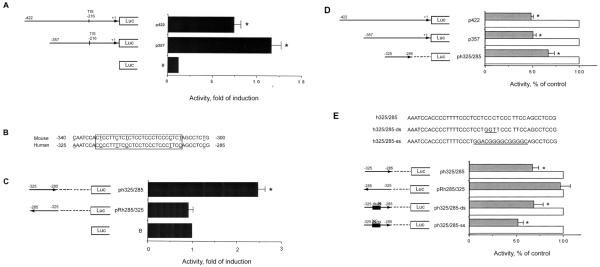
Only one transcriptional initiation site of hMOR gene has been reported [11], implicating the existence of an active promoter. An active promoter region is commonly composed of a region of approximately 200bp above the transcriptional initiation site (TIS), which is at −216bp for the hMOR gene with the translation initiation site ATG designated as +1. We therefore generated sequences of approximately 200 bp and 150 bp of hMOR 5′-flanking region, encompassing of the TIS, by PCR using NMB genomic DNA as the template. PCR products with correct DNA sequences were cloned into the pGL3-basic plasmid (B), containing the luciferase reporter gene. Two resulting plasmids, p422 (−422 to +1 bp) and p357 (−357 to +1 bp), were made (Fig. 3A).
Fig. 3.
Identification of hMOR gene promoter and DFO-effect on promoter activity
Various lengths of hMOR 5′-regulatory regions were cloned into the promoterless luciferase vector, pGL3-basic (B). Numbers on the left and right of each plasmid refer to the nucleotide number at the 5′- and 3′-ends, of each inserted fragment with its orientation indicated by the arrow. Reverse orientation is shown by an arrow pointing toward left. A deletion is indicated by a dashed line. Translation initiation site is designated as +1, and the transcription initiation site (TIS) is −216 bp. Transient transfection and luciferase assays were performed. A and C, Promoter activity of each construct was expressed as n-fold activation of pGL3-basic plasmid activity. Histograms represent mean values of fold activation. Error bars indicate S.E. “*” indicates p< 0.01 (student paired t-test). B, Sequence comparison of hMOR 5′-regulatory and mMOR proximal promoter regions. Open box represents the CT-rich region, with C/T transition underlined. D-E, “X” indicates the mutated ss or ds element. DNA sequences of wild type (h325/285) and mutant (h325/285-ds with GGT mutation; h325/285-ss with the GC box substitution underlined) are shown. Cells, transfected with plasmid, were treated without (control) or with DFO for 24hrs. Luciferase activity from control was defined as 100%. Data are presented as mean ± S.E. “*” indicates p< 0.01.
These plasmids along with pGL3-basic vector were transfected into NMB cells. The hMOR promoter activity was determined via luciferase reporter activity. As shown in Fig. 3A, the p422 and p357 plasmids displayed significant promoter activities (7.4- and 11.6-fold induction, respectively) as compared to that of pGL3-basic (B; defined as 1), suggesting that the p357 plasmid contained hMOR active promoter region and a possible inhibitory site in the −422 to −357bp region.
The p357 plasmid possesses a −325 to −285 bp region of hMOR promoter, containing 83% of DNA sequence homology to that of mMOR proximal core promoter. A 26bp CT-rich region (indicated by a box) with 5 nucleotides C/T transition (underlined) is depicted in Fig. 3B. The PCR product corresponding to the −325 to −285bp region was generated, sequenced and cloned into pGL3-basic plasmid. The pRh285/325 plasmid containing a reversed orientation was also made. As shown in Fig. 3C, ph325/285 plasmid showed a significant activity (2.5-fold), whereas pRh285/325, reversed orientation, displayed no promoter activity as compared to that of pGL3-basic vector (B). These results suggested that the −325 to −285bp region of hMOR gene possessed the promoter activity in an orientation-dependent manner.
DFO affected hMOR gene expression at transcriptional level
Having identified the hMOR active promoter region, the effect of DFO on this region was tested. As shown in Fig. 3D, upon DFO treatment (gray bars), a significant decrease of the promoter activity (p422 or p357 plasmid) was observed as compared to control (open bars; arbitrarily defined as 100%).
We further examined if the −325 to −285 bp region of hMOR was involved in mediating the DFO effect. As shown in Fig. 3E, there was a significant decrease of the promoter activity of ph325/285 plasmid upon DFO challenge (gray bar) as compared to control (open bar; as 100%). In contrast, the reverse orientation pRh285/325 displayed no significant difference between DFO treated group and control.
This −325 to −285 bp (CT-rich) region is known to encompass the overlapping ss and ds DNA element [12-13]. A GGT mutation is known to ablate the dsDNA cis-element only (Fig. 3E, upper panel). Conversely, replacing the right-half of the CT-rich region with a consensus GC box is known to abolish only the ssDNA cis-element. Accordingly, these mutations were constructed, in the form of ph325/285-ds and ph325/285-ss plasmids, containing the intact ds and ss DNA element, respectively. Promoter activities of mutant hMOR were then examined. The ph325/285-ds plasmid (ds element intact) retained 63.6±9 % of promoter activity as compared to that of the WT control (100%), and ph325/285-ss plasmid (ss element intact) possessed 60.3±8.8% activity.
The contribution of ds and/or ssDNA elements on DFO-mediated effect was then examined. As shown in Fig. 3E, lower panel, DFO (gray bar) resulted in a significant decrease of the promoter activities of ph325/285-ds and ph325/285-ss plasmids, as compared to control (open bar; as 100%). These results thus suggested that both ds and ss elements, located at the −325 to −285 bp region, participated in DFO-induced response.
Effect of hypoxia on hMOR message at the posttranscriptional level
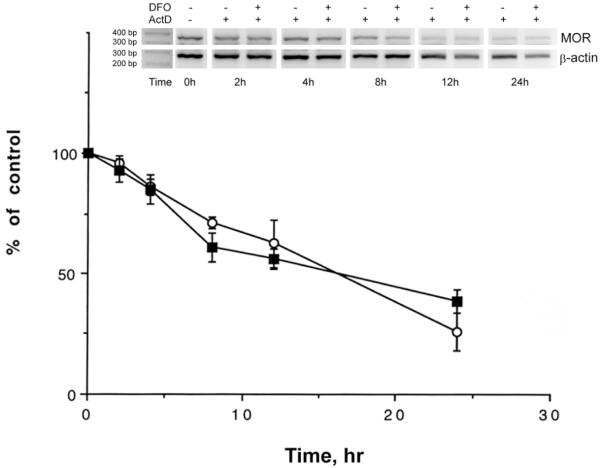
Data above showed that DFO-induced reduction of neuronal hMOR gene expression occurred at transcriptional level. To further determine if DFO also affected the stability of hMOR messages, the influence of DFO on hMOR mRNA degradation rate was investigated using actinomycin D (ActD), which inhibits mRNA synthesis [16]. RNA, extracted from cells treated with ActD (Fig. 4) for various time periods along with or without DFO treatment, was subjected to semiquantitative RT-PCR. As shown in Fig. 4, the degradation rate of hMOR mRNA displayed no significant difference between ActD (circles) and ActD plus DFO (squares) treatment, suggesting that DFO did not affect the stability of hMOR mRNA in neuronal cells.
Fig. 4.
Effect of DFO on hMOR mRNA stability
RT-PCR was performed using hMOR-specific primers with RNA from cells treated with Actinomycin D (ActD) or DFO plus ActD for 0, 2, 4, 8, 12 or 24 hrs. The β-actin was used as an internal control. PCR products were resolved using electrophoresis. Sizes of DNA markers are indicated. Quantitative analysis of hMOR message levels are presented as mean ± S.E., with normalized hMOR message at time zero as 100%. “*” indicates p< 0.01 (student paired t-test).
Discussion
Using a DFO-induced hypoxia model, we found (Figs. 1A and B) that the enhancement of cellular glutathione levels was more sensitive to increasing DFO concentration than to the decrease of cell viability. The faster onset and stronger response of increasing cellular glutathione may provide the neuroprotection and reduce cell death [17], explaining why some neuronal cells can still survive under DFO-induced hypoxia.
Although these surviving/attached neurons appeared morphologically similar to non-DFO treated cells (Figs. 1C and D), they developed a cellular adaptive response by increasing the HIF-1α mRNA level (Figs. 2A and D), in addition to the increase of the cellular glutathione amount.
Interestingly, DFO-induced hypoxia affected opioid receptor gene expressions differentially in these survival/attached neurons (Figs. 2D, C and E). DFO treatment resulted in approximately 50% decrease of hMOR message only as compared to controls, while levels of hDOR were not altered. In fact DOR has been suggested to provide neuroprotection under hypoxia in animal model [19].
Functional analysis with plasmids containing a hMOR promoter-luciferase fusion gene (Fig. 3) demonstrated that the active hMOR promoter, consisting of a region of approximately 150bp upstream from the transcription initiation site, was involved in DFO-induced response. Furthermore, both ss and dsDNA elements, located in the CT-rich region (−318 to −293 bp), participated in the DFO-induced response (Fig. 3E), contributing to about 30% of this reduction. Moreover, MOR mRNA stability was not altered via DFO treatment (Fig. 8), suggesting that no contribution of DFO-induced hMOR message reduction is from the post-transcriptional level. Taken together, these results suggest that another region of hMOR promoter (outside the CT-rich region) and an additional mechanism, such as epigenetic regulation [18], may also contribute to the DFO-mediated reduction of hMOR gene expression.
It is interesting to note that the contribution of the hMOR CT-rich region to hMOR promoter activity was not as vital as that of the mouse CT-rich region to the mMOR proximal promoter. The mMOR CT-rich region is known to be a vital element for mMOR core proximal promoter [12-13], whereas hMOR CT-rich region contributes only about 20% of total hMOR promoter activity (Figs. 3A and C). However, we cannot ignore the involvement of the hMOR CT-rich region in hMOR promoter activity; especially it alone exhibits independent promoter activity in an orientation-dependent manner (Fig. 3C).
DFO-induced hypoxia is a complicated event, which may affect various cellular pathways and gene expressions. Although HIF-1 can influence gene expressions, there is no HIF-1 DNA binding site (hypoxia response element) located in the hMOR CT-rich region, suggesting no direct HIF-1 involvement in the DFO-mediated response in this promoter region.
Acknowledgments
We thank Drs. Hsien-Ching Liu and Andrew Smith for editing the manuscript. Fluoview 1000 confocal microscope was acquired via the MRI fund of NSF. This research was supported by SHU Research Fund, SURF (RJC/JLK) and NIH research grant DA-016673.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Anderson RN, Smith BL. Deaths: Leading Causes for 2002. Natl Vital Stat. Reports. 2005;53:1–90. [PubMed] [Google Scholar]
- [2].Nangaku M, Kojima I, Tanaka T, et al. Novel drugs and the response to hypoxia: HIF stabilizers and prolyl hydroxylase. Recent Pat. Cardiovasc. Drug Discov. 2006;1:129–139. doi: 10.2174/157489006777442522. [DOI] [PubMed] [Google Scholar]
- [3].Won SJ, Kim DY, Gwag BJ. Cellular and molecular pathways of ischemic neuronal death. J. Biochem. Mol. Biol. 2002;35:67–86. doi: 10.5483/bmbrep.2002.35.1.067. [DOI] [PubMed] [Google Scholar]
- [4].Herlitz J, Hjalmarson A, Waagstein F. Treatment of pain in acute myocaridial infarction. Br. Heart J. 1989;61:9–13. doi: 10.1136/hrt.61.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kieffer BL, Gaveriaux-Ruff C. Exploring opioid system by gene knockout. Prog. Neurobiol. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- [6].Mansour A, Fox CA, Akil H, et al. Opioid-receptor mRNA expression in the rat CNS. Trends Neurosci. 1995;18:22–29. doi: 10.1016/0166-2236(95)93946-u. [DOI] [PubMed] [Google Scholar]
- [7].Min BH, Augustin LB, Felsheim JA, et al. Genomic structure and analysis of promoter sequence of a mouse mu opioid receptor gene. Proc. Natl. Acad. Sci. USA. 1994;91:9081–9085. doi: 10.1073/pnas.91.19.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liang Y, Mestek A, Yu L, et al. Cloning and characterization of the promoter region of the mouse mu opioid receptor gene. Brain Res. 1995;679:82–88. doi: 10.1016/0006-8993(95)00222-c. [DOI] [PubMed] [Google Scholar]
- [9].Pan YX, Xu J, Mahurter L, et al. Generation of mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc. Natl. Acad. Sci. USA. 2001;98:14084–14089. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ko JL, Chen HC, Loh HH. Differential promoter usage of mouse mu-opioid receptor gene during development. Mol. Brain. Res. 2002;104:184–193. doi: 10.1016/s0169-328x(02)00357-1. [DOI] [PubMed] [Google Scholar]
- [11].Wendel B, Hoehe MR. Human mu opioid receptor gene: 5′ regulatory and intronic sequences. J. Mol. Med. 1998;76:525–532. doi: 10.1007/s001090050246. [DOI] [PubMed] [Google Scholar]
- [12].Ko JL, Liu HC, Minnerath SR, et al. Transcriptional regulation of mouse mu-opioid receptor gene. J. Biol. Chem. 1998;273:27678–27685. doi: 10.1074/jbc.273.42.27678. [DOI] [PubMed] [Google Scholar]
- [13].Ko JL, Loh HH. Single-stranded DNA-binding complex involved in transcriptional regulation of mouse mu-opioid receptor gene. J. Biol. Chem. 2001;276:788–795. doi: 10.1074/jbc.M004279200. [DOI] [PubMed] [Google Scholar]
- [14].Wang GL, Semenza GL. Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity. Blood. 1993;82:3610–3615. [PubMed] [Google Scholar]
- [15].Baumhaker Y, Ben-Dor T, Bar-Hamburger R, et al. Characterization of a triple opioid system in the human neuroblastoma NMB cell line. Brain Res. 1994;665:94–100. doi: 10.1016/0006-8993(94)91156-8. [DOI] [PubMed] [Google Scholar]
- [16].Lin YC, Flock KE, Cook RJ, et al. Effects of Trichostatin A on Neuronal mu-Opioid Receptor Gene Expression. Brain Res. 2008;1246:1–10. doi: 10.1016/j.brainres.2008.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo S, Miyake M, Liu KJ, et al. Specific inhibition of hypoxia inducible factor 1 exaggerates cell injury induced by in vitro ischemia through deteriorating cellular redox environment. J. Neurochem. 2009;108:1309–1321. doi: 10.1111/j.1471-4159.2009.05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Formisano L, Noh K, Miyawaki T, et al. Ischemic insults promote epigenetic reprogramming of mu opioid receptor expression in hippocampual neurons. Proceed. Nalt. Acad. Sci. USA. 2007;104:4170–4175. doi: 10.1073/pnas.0611704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ma MC, Qian H, Ghassemi F, et al. Oxygen-sensitive {delta}-opioid receptor-regulated survival and death signals: novel insights into neuronal preconditioning and protection. J. Biol. Chem. 2005;280:16208–16218. doi: 10.1074/jbc.M408055200. [DOI] [PubMed] [Google Scholar]