Abstract
The protein tyrosine phosphatase (PTP) Shp2 (PTPN11) is an attractive target for anticancer drug discovery because it mediates growth factor signaling and its gain-of-function mutants are causally linked to leukemias. We previously synthesized SPI-112 from a lead compound of Shp2 inhibitor, NSC-117199. In this study, we demonstrated that SPI-112 bound to Shp2 by surface plasmon resonance (SPR) and displayed competitive inhibitor kinetics to Shp2. Like some other compounds in the PTP inhibitor discovery efforts, SPI-112 was not cell permeable, precluding its use in biological studies. To overcome the cell permeation issue, we prepared a methyl ester SPI-112 analog (SPI-112Me) that is predicted to be hydrolyzed to SPI-112 upon entry into cells. Fluorescence uptake assay and confocal imaging suggested that SPI-112Me was taken up by cells. Incubation of cells with SPI-112Me inhibited epidermal growth factor (EGF)-stimulated Shp2 PTP activity and Shp2-mediated paxillin dephosphorylation, Erk1/2 activation, and cell migration. SPI-112Me treatment also inhibited Erk1/2 activation by a Gab1-Shp2 chimera. Treatment of Shp2E76K mutant-transformed TF-1 myeloid cells with SPI-112Me resulted in inhibition of Shp2E76K-dependent cell survival, which is associated with inhibition of Shp2E76K PTP activity, Shp2E76K-induced Erk1/2 activation, and Bcl-XL expression. Furthermore, SPI-112Me enhanced interferon-γ (IFN-γ)-stimulated STAT1 tyrosine phosphorylation, ISRE-luciferase reporter activity, p21 expression, and the anti-proliferative effect. Thus, the SPI-112 methyl ester analog was able to inhibit cellular Shp2 PTP activity.
Keywords: PTPN11, Shp2, protein tyrosine phosphatase, inhibitor, Erk1/2
1. Introduction
Protein tyrosine phosphorylation affects cellular activities that control tumor growth and progression. Protein tyrosine phosphorylation is regulated by protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs). Many studies in the last three decades have demonstrated the roles of various protein tyrosine kinases (PTKs) in human cancer [1]. PTK inhibitors such as imatinib and gefitinib are now well-recognized as targeted therapy drugs in cancer treatment [2, 3]. While PTPs catalyze the reverse reaction of PTKs, increasing evidence suggests that cell signaling and oncogenesis require coordinated action of both PTKs and PTPs. For instance, PTPs such as Shp2, PTP1B, Cdc25, and PRL-3 have been found to be positively involved in oncogenesis and tumor progression [4–7]. Consequently, the search for PTP inhibitors as a new class of potential drugs for targeted cancer therapy has intensified in recent years [8–13].
Shp2 is a non-receptor PTP encoded by the PTPN11 gene. It contains two SH2 domains, a PTP domain, and a C-terminal region [14]. The N-SH2 domain in the wildtype Shp2 interacts with the PTP domain, resulting in autoinhibition of the Shp2 PTP activity. When the SH2 domains bind to specific phosphotyrosine docking sites in growth factor- or cytokine-stimulated cells, it relieves the autoinhibition and Shp2 is activated [15]. A well-recognized Shp2-regulated signaling pathway is the Ras-Erk1/2 MAP pathway. For instance, Shp2 is positively involved in epidermal growth factor (EGF)-stimulated Erk1/2 activation [15, 16]. Shp2 PTP activity is required for transformation of human glioblastoma cells by EGFRvIII [17] and human mammary epithelial cells by ErbB2 [18]. In the last few years, mutations in the Shp2 gene PTPN11 have been identified in several types of leukemias and in some cases of solid tumors [4, 19]. In particular, PTPN11 is the most frequently mutated gene in juvenile myelomonocytic leukemia (JMML), associating with ~35% of JMML cases. Most of leukemia-associated Shp2 mutations occur in the N-SH2 domain that interacts with the PTP domain [20]. These and other cancer-associated Shp2 mutants are predicted or have been demonstrated to be gain-of-function mutations [4, 21, 22]. Importantly, no loss-of-function Shp2 mutant has ever been found in human cancer. Laboratory experiments have established the oncogenic activity of several leukemia-associated Shp2 mutants [21, 23]. These findings point to Shp2 PTP as a potential target for cancer therapy.
Whereas Shp2 plays a positive role in the Ras-Erk1/2 MAP kinase pathway, several reports have indicated that Shp2 is a negative regulator of interferon (IFN) signaling. Shp2 was able to dephosphorylate STAT1 in vitro, suggesting that STAT1 is a substrate of Shp2 PTP [24]. Consistently, increased IFN-stimulated STAT1 tyrosine phosphorylation was observed in mouse embryonic fibroblast cells lacking a functional Shp2 [24, 25]. The inhibitory effect of Shp2 on STAT1 tyrosine phosphorylation may contribute to modulation of the antiviral effect of IFN [26].
While PTPs have increasingly attracted attention as novel targets of cancer drug discovery, only a few selective PTP inhibitors have been characterized biologically. Among PTP inhibitors identified in recent years, many of them contain one or more negatively-charged functional groups [8, 10, 27, 28]. This property is reminiscent of PTP substrates since phosphotyrosine is negatively charged and negatively charged Asp and Glu residues are frequently present near tyrosine phosphorylation sites. While aryl sulfonic compounds have been found to exert cellular activities in some cases [10, 17, 28–30], compounds containing aryl phosphate or carboxylate groups often require modifications for cell permeation and/or prodrug strategies for delivery into cells [8, 31].
Several Shp2 PTP active site inhibitors with in vitro IC50 < 10 µM have been reported in the last few years. We identified NSC-87877 from the NCI Diversity Set-1 library [28]. NSC-87877 is a potent Shp2 inhibitor (Shp2 IC50: 0.32 µM) but it inhibits Shp1 with a similar potency. NSC-87877 has two aryl sulfonic groups (supplementary Fig. 1). Inhibition of cellular Shp2 activity by NSC-87877 has been reported in certain cells, including epithelial/carcinoma cells, fibroblasts, endothelial cells, muscle cells, and neuronal/glioma cells [17, 28, 29, 32, 33]. Using a biology-oriented synthesis approach, Nören-Müller et al. [34] discovered a tetrazolefurofuran Shp2 inhibitor furanofuran-2a (sFig. 1, Shp2 IC50: 2.5 µM) that has a >40 fold selectivity against PTP1B. It is unclear if furanofuran-2a is cell permeable. Starting with a virtual screening, Hellmuth et al [10] identified phenylhydrazonopyrazolone sulfonate 1 (PHPS1) as a Shp2 inhibitor. PHPS1 has > 10 selectivity against most of other PTPs, including a 14-fold selectivity against Shp1. PHPS1 appears to have broader cell permeability than NSC-87877. However, the activity of this aryl sulfonic acid compound in hematopoietic cells remains to be determined. Wu et al [35] identified 7-deshydroxypyrogallin-4-carboxylic acid (DCA) as a Shp2 inhibitor (IC50: 2.1 µM) from a chemical library screen effort. Similar to NSC-87877, DCA inhibits both Shp1 and Shp2 with the same potency. Recently, Zhang and colleagues synthesized a salicylic acid based Shp2 inhibitor II-B8 (Shp2 IC50: 5.5 µM) [36]. It was reported that II-B8 is cell active. Significantly, a Shp2-IIB8 co-crystal structure has been solved [36]. This may help the further optimization effort to obtain more potent and selective Shp2 inhibitors.
NSC-117199 was the second lead compound that we identified from the NCI Diversity Set-1. In a previous study, we synthesized >100 analogs in our lead optimization effort [9]. SPI-112 (Compound 10m in ref. [9]) was among the best Shp2 inhibitor derived from NSC-117199. However, these Shp2 PTP inhibitors have either a polar –NO2 or a negatively charged –COOH group and have no detectable cellular activity, suggesting that they are not cell permeable. In this study, we performed kinetic analyses of SPI-112 binding and inhibition of Shp2 and showed that SPI-112 is a competitive inhibitor of the Shp2 PTP. To deliver SPI-112 into cells, we prepared a methyl ester prodrug of SPI-112 (SPI-112Me) and demonstrated that SPI-112Me was able to inhibit the Shp2 PTP activity in intact cells.
2. Materials and methods
2.1. Chemical synthesis of SPI-112 and SPI-112Me
SPI-112 [(Z)-3-(2-(5-(N-(4-fluorobenzyl)sulfamoyl)-2-oxoindolin-3-ylidene)hydrazinyl)benzoic acid] and SPI-112Me [(Z)-3-(2-(5-(N-(4-fluorobenzyl)sulfamoyl)-2-oxoindolin-3-ylidene)hydrazinyl)benzoic acid methyl ester] were synthesized using procedure similar to that described previously [9]. The detailed description of SPI-112Me synthesis is provided in the online supplementary information.
2.2. Computer docking
Computer docking was performed with the X-ray crystal structure of human Shp2 (PDB identification code: 2SHP) [37] using the GLIDE (Grid-Based Ligand Docking from Energetics software available from Schrödinger, L.L.C., San Diego, CA) program similar to that reported [28]. The Jorgensen OPLS-2001 force field was applied in the GLIDE program. The optimal binding geometry was obtained by utilization of Monte Carlo sampling techniques coupled with energy minimization.
2.3. Surface Plasmon resonance (SPR) binding assay
An anti-His-tag antibody (Qiagen, Valencia, CA) was immobilized on a CM5 Sensor Chip for Biacore T100 using a protocol similar to that described [38]. His-tagged Shp2 (amino acids 1–525) was captured on this antibody surface for each cycle of compound or running buffer injection. SPI-112 meglumine salt was diluted in the running buffer [50mM Bis-Tris HCl pH 7.0, 150mM NaCl, 2mM dithiothreitol (DTT), 0.01% TritonX-100, 3% dimethyl sulfoxide (DMSO)] to yield final inhibitor concentrations of 10, 2, 3.3, 1.1, 0.37, and 0.12 µM. Duplicate independent kinetics experiments were carried out in a manner similar to that used by Nordin and colleagues [39]. Kinetics analysis was performed using BIAevaluation software (GE Life Sciences, Piscataway, NJ) fitting Langmuir (1:1) binding. Local Rmax calculations were used for our model fitting due to variation between cycles in theoretical Rmax values. Baseline RU drift over time in each experiment was corrected to improve the fitting [40]. The kinetic constant and rates of association and dissociation were from the average of two experiments.
2.4. Cell lines
MDA-MB-468 breast cancer cells and HT-29 colon cancer cells were from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco’s modified Eagle’s medium (DMEM)/10% fetal bovine serum (FBS) and RPMI 1640/10% FBS, respectively. TF-1/Shp2E76K and TF-1/V cells were established by infection of TF-1 cells with a retrovirus encoding a Flag-tagged Shp2E76K mutant or the control virus as reported previously [21]. Flp-In-T-Rex-293 cells (Invitrogen, Carlsbad, CA) containing a doxycycline (dox)-inducible Gab1PH-Shp2 ΔN chimera has been described [28].
2.5. Shp2 PTP activity assays
In vitro Shp2 PTP activity inhibition assay for determination of IC50 was performed with a recombinant GST-Shp2 PTP domain protein using 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP, Invitrogen) as the substrate similar to that described previously [28]. Curve fitting and IC50 were obtained using the GraphPad Prism program (GraphPad Software, San Diego, CA).
Enzyme kinetics analysis was performed with a non-fusion Shp2 PTP protein in which the GST fragment had been removed by PreScission protease and using DiFMUP as the substrate. Reaction was carried out in duplicate at room temperature in black, half-area 96-well plates. Each reaction mixture (75 µl) contained 25 mM Hepes, pH 7.3, 50 mM NaCl, 1 mM DTT, 0.01% Triton-X100, 18.7–280 µM DiFMUP, 3% DMSO (vehicle) or SPI-112 (0.1, 0.25, 0.5, 1.0 µM), and 0.16 µM Shp2 PTP protein. Reaction was initiated by the addition of the enzyme. Fluorescent signal was measured at 5 and 10 min with a Wallac 1420 Victor2 Multilabel Plate Reader (Perkin Elmer; Waltham, MA) at excitation/emission wavelengths of 355 nm/460 nm. Fluorescent signal was converted to the amount of product (nmole) using reference curve generated with 6,8-difluoro-4-methylumbelliferone (DiFMU, Invitrogen). Data obtained at the 5 min time point was used in the kinetics analysis. This is because >5% of substrate had been converted to the fluorescent DiFMU product at the later time point in some reaction wells and thus the steady state enzyme kinetics could no longer be ensured. Kinetics analysis of the data was performed with the VisualEnzymics program (Softymics, Princeton, NJ).
To measure Shp2 PTP activity in immune complexes, cells were lysed in ice-cold PTP lysis buffer [25 mM Hepes, pH 7.4, 150mM NaCl, 1 mM DTT, 2mM EDTA, 0.5% Triton X-100, 1:50 diluted protease inhibitor cocktail (Roche, Indianapolis, IN)]. Cell lysate supernatants (0.2 mg/each) were incubated with an anti-Shp2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or with an anti-Flag antibody M2 (Sigma, St. Louis, MO) plus protein G-Sepharose for 2 h at 4°C. Immunoprecipitates were washed twice with the PTP lysis buffer and twice with the Reaction Buffer A (25 mM Hepes, pH 7.4, 50 mM NaCl, 1 mM DTT, 0.05% Triton X-100). Each Shp2 immune complex was resuspended in 100 µl of reaction buffer containing 50 µM DiFMUP and incubated at room temperature for 20 min. After a brief centrifugation, supernatants were transferred into 96-well plates and the DiFMU fluorescence signal was measured. The remaining immune complex pellets were used for immunoblotting analysis of Shp2 protein.
For in vitro dephosphorylation of STAT1, STAT1 was immunoprecipitated from serum-starved HT-29 cells stimulated with IFN-γ (100 U/ml, 30 min). STAT1 immunoprecipitates were incubated with different amounts of GST-Shp2 PTP protein in Reaction Buffer A at 30 °C for 20 min. After the reaction, the immune complex was washed and analyzed by immunoblotting.
2.6. SPI-112 and SPI-112Me uptake assay
TF-1/Shp2E76K cells (6 × 105 cells) were incubated in 12-well plates with indicated concentrations of SPI-112, SPI-112Me, or solvent (DMSO) for 5 h in 1 ml RPMI 1640/10% FBS at 37 °C/5% CO2. Aliquots of cell culture suspension (0.1 ml/each) were used for measuring the fluorescent signal in cell cultures. The remaining cell suspensions were centrifuged (3,000 rpm, 5 min). Cell pellets were washed 2 times with 1 ml culture medium. Each washed cell pellet was resupended in 0.1 ml culture medium. Fluorescent signal in cell culture, cell-free medium, wash medium, and cell pellet was measured using an Envision 2102 Multilabel Reader (Perkin Elmer) at excitation/emission wavelengths of 485 nm/535 nm, and total fluorescent signal in each fraction was calculated. Values of fluorescent signal in cell-free supernatant and the wash solution were combined and designated as fluorescent signal in the cell-free medium.
For confocal laser scanning microscopy, cells were cultured in glass bottom culture dishes (MatTeck, Ashland, MA). After drug treatment (25 µM, 3 h), Hoechst 33342 (5 µg/ml, Invitrogen) was added. Confocal microscopy was performed using a Leica TCS SP5 AOBS laser scanning confocal microscope with a 63X/1.40NA Plan Apochromat oil immersion objective lens (Leica Microsystems, Germany). Argon 458 nm and 405 nm laser Diode were applied to excite the samples and a tunable emission filters were used to minimize crosstalk between fluorescent signals. Images were captured through photo multiplier tubes and prepared with the LAS AF software version 2.1.0 (Leica Microsystems). Image Z-stacks were acquired at 1.0 µm intervals. Bright field images were captured with an Argon 488 nM laser line and transmitted light photo multiplier tube. A 4× zoom was applied prior to acquiring the image.
2.7. Immunoblotting and immunoprecipitation
Cells were treated as indicated in figure legends. Except experiments involving measurement of Shp2 PTP activity in immune complex, cells were lysed with the lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 25 mM NaF, 5 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mM dithiothreitol, 20 mM p-nitrophenyl phosphate, 1% Triton X-100, 1:50 diluted protease inhibitor cocktail). After incubation on ice for 30 min, cell lysates were centrifuged twice at 16,000 rpm with a microfuge at 4 °C for 15 min. Cleared cell lysate supernatants were used for immunoblotting analysis or incubated with specific antibodies indicated in figure legends for immunoprecipitation. Immune complexes were collected with protein G-agarose or protein A-agarose. Immunoprecipitated proteins were separated on SDS-polyacrylamine gels and transferred to nitrocellulose membranes. Immunoblotting was performed as described previously [16].
2.8 Cell growth and apoptosis assays
TF-1/Shp2E76K and HT-29 Cells (1,000 cells/well in triplicate) were plated in 96-well plates in RPMI160/10% FBS plus test agent(s) in a total volume of 100 µl. The volume of SPI-112Me or SPI-112 was kept ≤0.1% of the total volume. After incubation for 4 days at 37 °C/5% CO2, relative viable cell number was measured using the CelltiterGlo reagent (Promega) following the supplier’s instruction.
Synergism of two agents was calculated based on the Bliss definition [41] as below:
Assume X is the fraction of inhibition caused by the first agent, Y is the fraction of inhibition caused by the second agent, and Z is the fraction of inhibition caused by the combination of both agents. In the presence of the first agent, the remaining possible fraction of inhibition is 1-X. If two agents are Bliss independence, then the fraction of inhibition to the second agent in the presence of the first agent is Y(1−X). Thus, Z = X + Y(1−X) = X + Y− XY.
If Z = X + Y – XY, then the effect of two agents is additive.
If Z > X + Y – XY, then the effect of two agents is synergistic.
If Z < X + Y – XY, then the effect of two agents is antagonistic.
Apoptosis was examined using the ethidium bromide and acridine orange (EB/AO) staining assay [42]. In this assay, AO is taken up by all cells and labels nucleus green whereas EB only enters late apoptotic and necrotic cells that have lost membrane integrity. When a nucleus is stained with both EB and AO, the EB fluorescent signal is dominant and the nucleus appears red. Therefore, cells with green normal appearance nuclei are live cells; cells with condensed and fragmented green nuclei are early apoptotic cells; cells with condensed and fragmented red nuclei are late apoptotic cells, and cells with normal appearance red nuclei are necrotic cells.
TF-1/Shp2E76K cells were incubated in granulocyte-macrophage colony-stimulating factor (GM-CSF)-free RPMI1640/10% FBS medium at 37 °C/5% CO2 in the presence of absence of SPI-112Me as indicated in the figure legend for 3 days. Aliquots (100 µl/each) of cells were then transferred to a 96-well plate and centrifuged at 1,000 rpm. EB/AO dye mix (100 µg/ml EB and 100 µg/ml AO in PBS) were added to each well (8 µl/well). Nuclear staining by EB/AO was examined under a fluorescent microscope (400 × magnification). The EB fluorescence was captured with a rhodamine filter (excitation 540–552 nm, emission 575–640 nm). The DNA bound AO fluorescence was captured with a FITC filter (excitation 450–490 nm, emission 500–550 nm). Live, apoptotic, and necrotic cells were enumerated in at least 150 cells in multiple fields in two wells. Data were from three independent experiments.
2.9. Cell migration assay
Cell migration assay was performed using polycarbonate membrane Transwell cell culture insert (6.5 mm, 8.0 µm pore size tissue culture treated, Costar, Lowell, CA). The membrane was coated with rat tail type I collagen (10 µg/ml) overnight at 4°C and air-dried. MDA-MB-468 cells (2.5 × 104) in 0.2 ml DMEM/0.1% BSA were placed in the upper chamber. The lower chamber contained 0.6 ml DMEM/1% FBS. EGF (10 ng/ml) was included in the media in both chambers where indicated. After incubation at 37 °C/5% CO2 for 16 h, cells on the upper membrane surface were removed with a cotton swab. Cells that had migrated to the lower side of membrane were fixed and stained using the HEMA3 reagents (Fisher Scientific, Pittsburg, PA). Cells were enumerated under a microscope in at least 8 randomly selected fields per well.
2.10. Luciferase reporter assay
HT-29 cells (5 × 105 cells/each in 12-well plate) were co-transfected with 1.6 µg pISRE-Luc (Clontech, Mountain View, CA) and 0.4 µg β-gal plasmids with LipofectAMINE 2000 reagent (Invitrogen). Eighteen hours after transfection, cells were incubated in fresh medium, pretreated with indicated concentrations of SPI-112Me or mocked treated for 2 h, and then stimulated with 50 U/ml IFN-γ (Peprotech, Rocky Hill, NJ) for 6 h or left unstimulated. Luciferase and β-galactosidase activities were determined as described [21]. The luciferase activity was then normalized to β-galactosidase activity as an internal control for transfection efficiency.
3. Results
3.1 The Shp2 PTP inhibitor SPI-112 and its methyl ester prodrug SPI-112Me
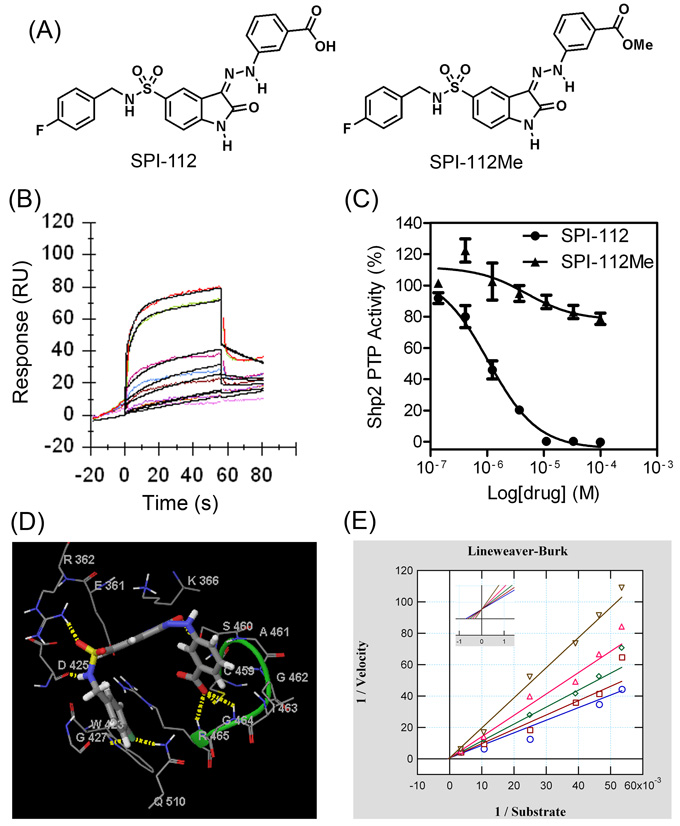
SPI-112 (Fig. 1A) was synthesized based on a hit (NSC-117199) from the NCI Diversity Set-1 in our lead optimization effort [9]. In SPR binding assay, SPI-112 displayed a 1:1 stoichiometric binding kinetics to Shp2 with a kinetic constant (KD) of 1.30±0.14 µM and the association and dissociation rates of Ka = 2.24 × 104/Ms and Kd = 0.029/s (Fig. 1B). However, SPI-112 and other NSC-117199 analogs had no detectable activity in cell-based Shp2 inhibition assays, suggesting that they are not cell permeable. This is likely due to the presence of a negatively charged carboxylic acid on these Shp2 inhibitors.
Fig. 1.
SPI-112 and its methyl ester prodrug SPI-112Me. (A) Chemical structures of SPI-112 and SPI-112Me. (B) A Biacore sensorgram of SPI-112 binding to His-tagged Shp2 at the inhibitor concentrations of 10, 3.3, 2, 1.1, 0.37, and 0.12 µM. The data were locally fit to a 1:1 interaction model (black lines). (C) Comparison of Shp2 PTP inhibition by SPI-112 and SPI-112Me in vitro. IC50s of SPI-112 and SPI-112Me on Shp2 PTP were determined in vitro using a GST-Shp2 PTP protein as the enzyme and DiFMUP as the substrate. The results were from three (SPI-112) and two (SPI-112Me) experiments performed in duplicates. (D) Computer docking of SPI-112 binding to the Shp2 PTP domain. Carbon atoms are colored in grey, oxygen in red, nitrogen in blue, hydrogen in white, and sulfur in yellow. SPI-112 is shown in cylindrical representation. Amino acid residues of Shp2 are shown as lines. Yellow dashed lines are predicted hydrogen bonds between SPI-112 and Shp2, which are shown schematically but not to scale. The hydrogen bonds are defined with a minimum donor angle of 120° and minimum acceptor angle of 90° and maximum length of 2.5 Å. The green loop identifies the catalytic P-loop of Shp2. (D) Lineweaver-Burk plot of enzyme kinetics data of inhibition of the Shp2 PTP with SPI-112. The data were fit to competitive, noncompetitive and uncompetitive linear inhibition models with noncompetitive and uncompetitive model fits calculated with proportional weighting and using the Levenberg-Marquart Robust algorithm to minimize influence of any outliers in the data. All models were compared to the competitive model as the simplest model by Akaike’s Information Criterion (AIC) evidence ratio since most models did not yield a Chi-square value for computing an F-value to run an F-test. The linear competitive model fit the best with a Ki value of 0.8 µM ± 0.9 µM (SEM) (from three experiments) and the lowest AICc values with AIC ratios of 3.0×108 and 6.5 versus linear uncompetitive and noncompetitive, respectively, which are greatly statistically significant (AIC ratio >> 2) in favor of the simpler model.
To circumvent the cell permeation issue, we prepared a methyl ester analog of SPI-112 (SPI-112Me, Fig. 1A) to shield the negative charge of the carboxylic acid group of SPI-112 (Fig 1A). The methyl ester analog is predicted to be hydrolyzed to the corresponding carboxylic acid by cellular esterases upon entry into cells similar to other methyl ester prodrugs [43, 44]. In vitro Shp2 PTP inhibition assay showed that, while SPI-112 potently inhibited Shp2 PTP (IC50: 1.0 µM) as reported previously [9], the methyl ester analog did not inhibit Shp2 PTP (IC50 > 100 µM) (Fig. 1C). Computer modeling (Fig. 1D) suggested that SPI-112 binds to the catalytic pocket. The phenyl carboxylic acid points inwards mimicking a phosphotyrosine residue, forming hydrogen bonds with Gly-464 and Arg-465 of the P-loop, and this suggests that it is essential for binding and consequently the inhibitory activity. In agreement with the computer model, enzyme kinetic data obtained with SPI-112 were best fitted with the competitive inhibition model (Ki: 0.8 µM; Fig. 1E), suggesting that SPI-112 interacts with the catalytic site of Shp2.
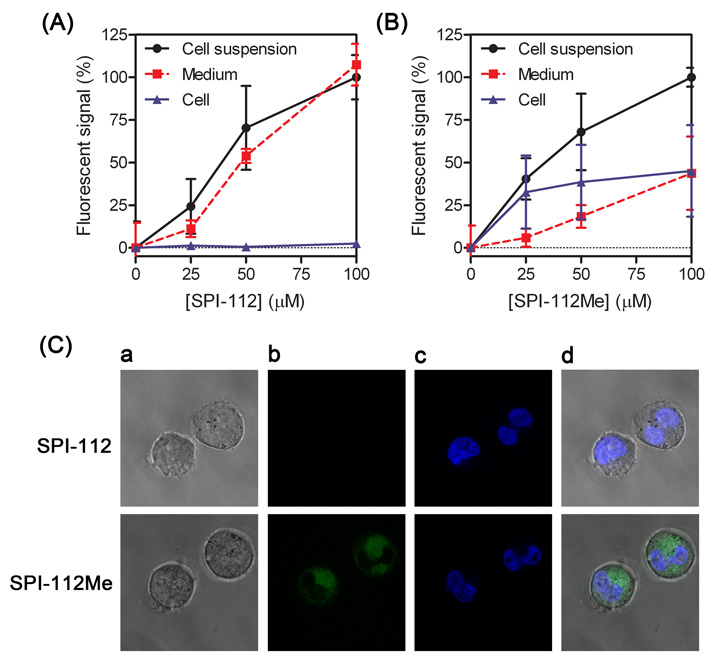
SPI-112 and SPI-112Me emitted concentration-dependent fluorescent signal under a Wallac 2100 EnVision multilable plate reader (PerkinElmer) at excitation/emission wavelengths of 485 nm/535 nm. This property provided a means for us to assess the cellular uptake of SPI-112 and SPI-112Me. We incubated TF-1/Shp2E76K cells with SPI-112 and SPI-112Me at different concentrations and then measured the fluorescent signal in cell culture, cell-free medium, and the cell pellets. When TF-1/Shp2E76K cells were incubated with SPI-112, essentially all fluorescent signal remained in the cell-free supernatants and only trace amount of fluorescent signal was associated with the cell pellets (Fig. 2A). In contrast, when TF-1/Shp2E76K cells were incubated with SPI-112Me, a substantial amount of the fluorescent signal was associated with the cells (Fig. 2B).
Fig. 2.
Comparison of SPI-112 and SPI-112Me uptake into TF-1/Shp2E76K cells. TF-1/Shp2E76K cells were incubated with indicated concentrations of SPI-112 (A) or SPI-112Me (B) for 5 h. Fluorescent signals in cell culture suspension, cell-free medium, and cell pellet were measured. The fluorescent signal of cell suspension with 100 µM compounds in each sample was set as 100%. The data were from two duplicate experiments (n = 4). (C) Cells were examined with a confocal microscope and representative images of internal sections are shown. Magnification: 2,520×. (a) bright field images, (b) SPI-112/SPI-112Me fluorescence, (c) Hoechst 33342 fluorescence, (d) overlay of (b) and (c).
To confirm that the cell-associated compound was present inside the cells, we examined cells treated with SPI-112 and SPI-112Me with a confocal laser scanning microscope. Cells treated with SPI-112 did not have green fluorescence in confocal images representing the internal sections of the cells, as evident by Hoechst dye-stained nucleus (Fig. 2C, upper panels). In comparison, green fluorescence emitted by SPI-112/SPI-112Me was readily detectable in confocal planes of the internal sections of cells that had been treated with SPI-112Me (Fig. 2C, lower panels). The green fluorescent signal was largely present in the cytoplasm. Together, these results suggest that SPI-112Me, but not SPI-112, is able to enter cells. In addition, SPI-112 was detected in the extract of cells treated with SPI-112Me by high performance liquid chromatography (HPLC) analysis and by liquid chromatography/mass spectrometry (LC/MS) analysis (supplementary data), indicating that SPI-112Me was hydrolyzed to SPI-112 in the cells.
3.2. SPI-112Me displays Shp2 inhibition properties in MDA-MB-468 cells
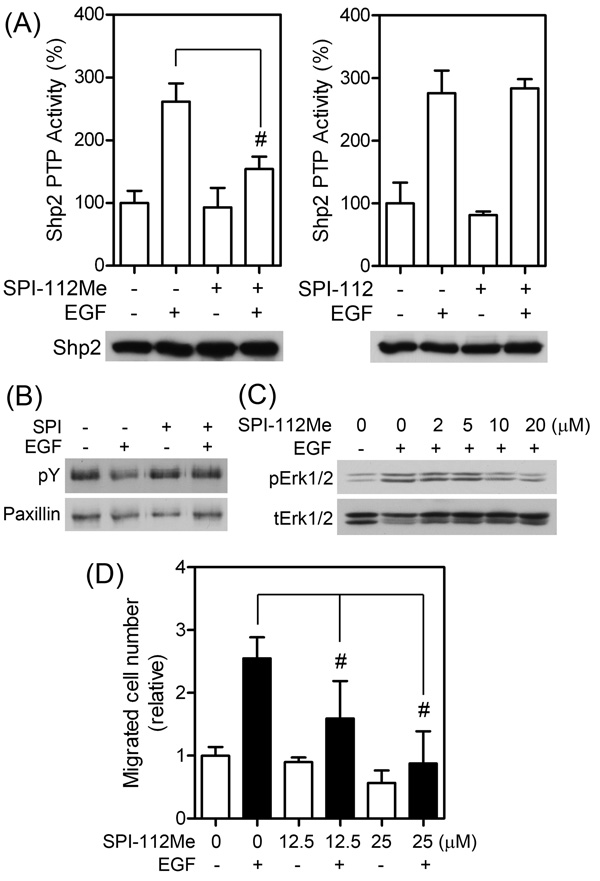
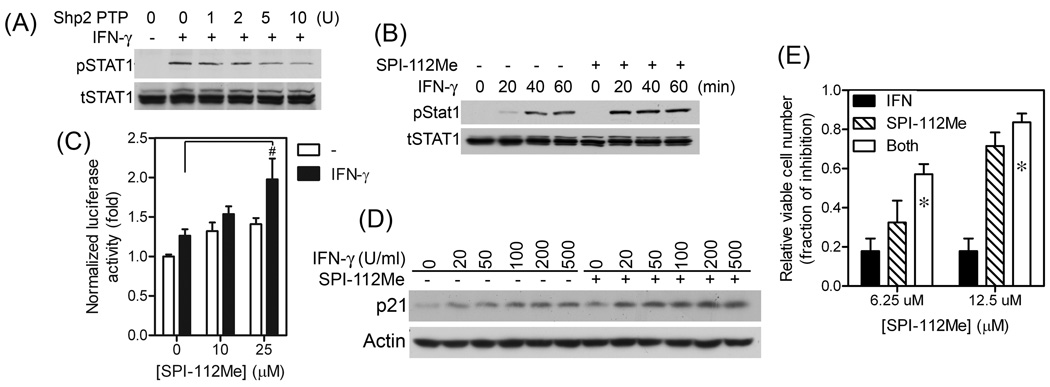
Shp2 is activated by EGF in MDA-MB-468 breast cancer cells. To test if SPI-112Me and SPI-112 are able to inhibit EGF-stimulated Shp2 in the cells, serum-starved MDA-MB-468 cells were pre-treated with or without SPI-112Me or SPI-112 and then stimulated with EGF. Shp2 was immunoprecipitated from cell lysate supernatants and its PTP activity was determined. Shp2 from EGF-stimulated cells had 2.6-fold higher PTP activity (Fig. 3A). SPI-112Me (20 µM) pre-treatment significantly (p = 0.003) reduced the EGF-stimulated Shp2 PTP activity by 77%. In contrast, SPI-112 did not affect the EGF-stimulated Shp2 PTP activity (Fig. 3A).
Fig. 3.
SPI-112Me suppresses Shp2 PTP activity in MDA-MB-468 cells. (A) Sub-confluent MDA-MB-468 cells were serum-starved in DMEM/0.1% BSA for 18 h, pretreated overnight with SPI-112Me (20 µM) or SPI-112 (100 µM) and stimulated with EGF (50 ng/ml, 10 min) or mocked treated as indicated. Shp2 was immunoprecipitated from cell lysate supernatants and Shp2 PTP activity was measured. Data were from two experiments performed in duplicates. After the PTP reaction, a portion of each immunoprecipitate was used for immunoblotting with an anti-Shp2 antibody (lower panel). #, p < 0.05. (B) Paxillin was immunoprecipitated from serum-starved MDA-MB-468 cells treated with or without SPI-112Me (20 µM, 2 h) or EGF (50 ng/ml, 10 min) as indicated and analyzed by immunoblotting with antibody to phosphotyrosine (pY) or paxillin. (C) Serum-starved MDA-MB-468 cells were pretreated with indicated concentrations of SPI-112Me and then stimulated with EGF (2 ng/ml, 5 min). Cell lysate supernatants were analyzed by immunoblotting with antibodies to phospho-Erk1/2 (pErk1/2) or total Erk1/2 (tErk1/2). (D) Transwell cell migration assay of MDA-MB-468 in the presence or absence of indicated concentrations of SPI-112Me and/or EGF (10 ng/ml). The data were from two duplicate experiments (n =4).
Paxillin is a physiological substrate of Shp2 [10, 16, 45, 46]. We demonstrated previously that Shp2 dephosphorylates paxillin in EGF-stimulated MDA-MB-468 cells [16]. Inhibition of EGF-stimulated paxillin dephosphorylation is an established surrogate marker of Shp2 inhibition [10, 28]. As shown in Fig. 3B, paxillin became dephosphorylated in EGF-stimulated cells. Pre-treatment of MDA-MB-468 cells with SPI-112Me prevented EGF-induced paxillin tyrosine dephosphorylation. Shp2 is known to mediate EGF-stimulated Erk1/2 activation [16]. To determine if SPI-112Me is able to inhibit EGF-stimulated Erk1/2 activation, MDA-MB-468 cells were pre-treated with SPI-112Me at various concentrations (0–20 µM) and then stimulated with EGF. Erk1/2 activation was assessed by immunoblotting analysis of cell lysates with an antibody against the active, dual phosphorylated form of Erk1/2. Inhibition of EGF-stimulated pErk1/2 signal was observed at 10 µM SPI-112Me in this assay (Fig. 3C). Shp2 is involved in EGF-stimulated MDA-MB-468 cell migration [16]. We measured EGF-stimulated MDA-MB-468 cell migration by the Transwell cell migration assay in the presence or absence of SPI-112Me. Fig. 3D shows that EGF-increased MDA-MB-468 cell migration 2.5-fold. The EGF-stimulated MDA-MB-468 cells migration was significantly (p = 0.049) reduced 62% by 12.5 µM SPI-112Me and was completed blocked in the presence of 25 µM SPI-112Me. These data indicate that SPI-112Me could inhibit Shp2 PTP activity in the EGF-stimulated MDA-MB-468 cells.
3.3 SPI-112Me inhibits Erk1/2 activation induced by a Gab1-Shp2 chimera
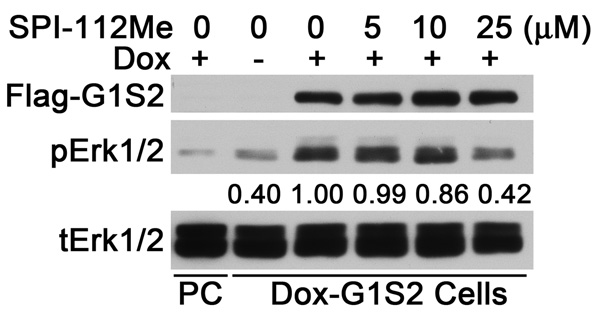
We found previously that expression of a fusion protein consisting of the Gab1 PH domain and an N-SH2 domain deletion mutant of Shp2 (Gab1PH-Shp2ΔN) caused constitutive Erk1/2 activation [28, 47]. The Gab1PH-Shp2ΔN-induced Erk1/2 activation bypasses the up-stream growth factor receptor signaling steps. HEK293 cells containing dox-inducible Gab1PH-Shp2 ΔN were generated using Flp-In-T-Rex-293 cells (Dox-G1S2). As shown in Fig. 4, no Flag-tagged Gab1PH-Shp2ΔN and a low basal level of pErk1/2 were detected in Dox-G1S2 cells in the absence of dox. Incubation of cells with dox induced Gab1PH-Shp2ΔN expression and activation of Erk1/2 (Fig. 4), whereas dox had no effect on Erk1/2 activation in the parental Flp-In-T-Rex-293 cells. Inhibition of Gab1PH-Shp2ΔN-induced Erk1/2 activation was observed in the presence of 10–25 µM SPI-112Me (Fig. 4).
Fig 4.
SPI-112Me inhibits Erk2 activation by a Gab1-Shp2 chimera. Flp-In-T-Rex-293 cells (PC) or Flp-In-T-Rex-293 cells containing a dox-inducible Gab1PH-Shp2DN (Dox-G1S2) were incubated in serum-free medium, pretreated with indicated concentrations of SPI-112Me for 30 min and induced with dox. Cell lysates were prepared and analyzed by immunoblotting with antibodies to Flag-tag, phospho-Erk1/2 (pErk1/2) and total Erk1/2 (tErk12/). Numbers under the pErk1/2 panel indicate the relative signal intensity (average from two experiments).
3.4. SPI-112Me suppresses leukemia-associated Shp2E76K mutant-dependent survival of TF-1 cells
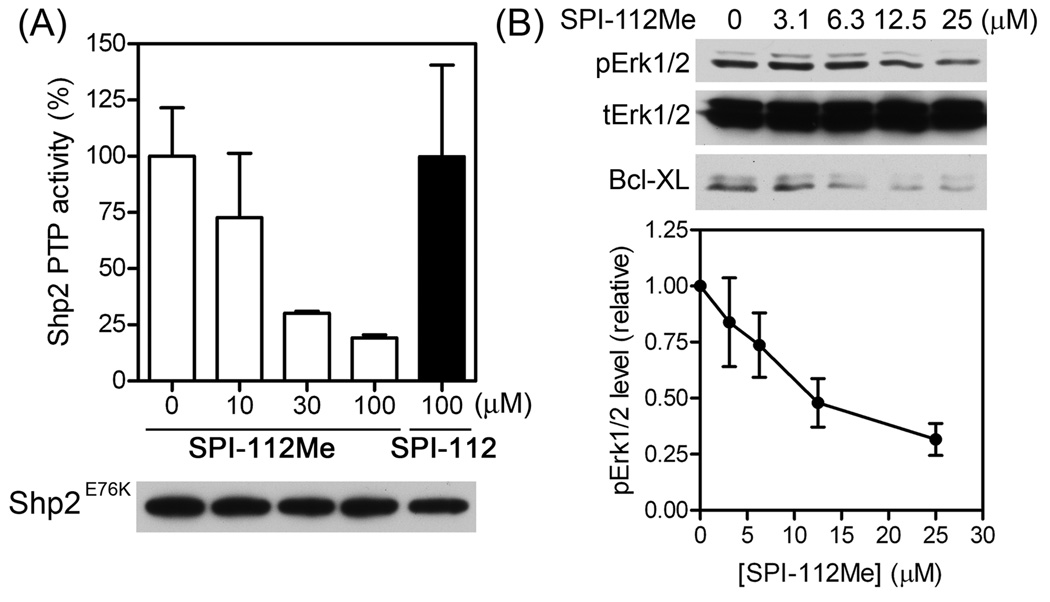
The gain-of-function Shp2E76K mutant has constitutively active PTP activity and is able to convert the cytokine-dependent TF-1 myeloid cells into cytokine-independence by up-regulation of Bcl-XL through the Erk1/2 pathway [21]. To assess if Shp2E76K PTP activity was inhibited in SPI-112Me-treated TF-1/Shp2E76K cells, TF-1/Shp2E76K cells were incubated in GM-CSF-free medium in the presence of different concentrations of SPI-112Me or a high concentration of SPI-112 (negative control). The Flag-tagged Shp2E76K was immunoprecipitated and the PTP activity was measured. Whereas there was no decrease in the Shp2E76K PTP activity in SPI-112-treated TF-1/Shp2E76K cells, lower Shp2E76K PTP activity was detected in SPI-112Me-treated TF-1/Shp2E76K cells (Fig. 5A). Shp2E76K activates Erk1/2 and up-regulates Bcl-XL in TF-1 cells to confer cytokine-independent cell survival [21]. Consistently, immunoblotting analyses of cell lysates showed that SPI-112Me reduced the amounts of pErk1/2 and Bcl-XL in TF-1/Shp2E76K cells in a concentration-dependent manner (Fig. 5B).
Fig. 5.
Inhibition of Shp2E76K and Shp2E76K-induced Bcl-XL expression by SPI-112Me. (A) TF-1/Shp2E76K cells were incubated in RPMI1640/10% FBS and treated with SPI-112Me or SPI-112 at the indicated concentrations. Shp2E76K was immunoprecipitated from cell lysates supernatant with an anti-Flag antibody and the PTP activity was determined. After the PTP reaction, a portion of each immunoprecipitate was used for immunoblotting with an anti-Shp2 antibody. Data were from two experiments. (B) TF-1/Shp2E76K cells were incubated in RPMI1640/10% FBS with the indicated concentrations of SPI-112Me. Cell lysates were analyzed by immunoblotting with antibodies to phospho-Erk1/2, total Erk1/2, or Bcl-XL. The pErk1/2 signal intensities were quantified using the ImageQuant program (n = 4, lower panel).
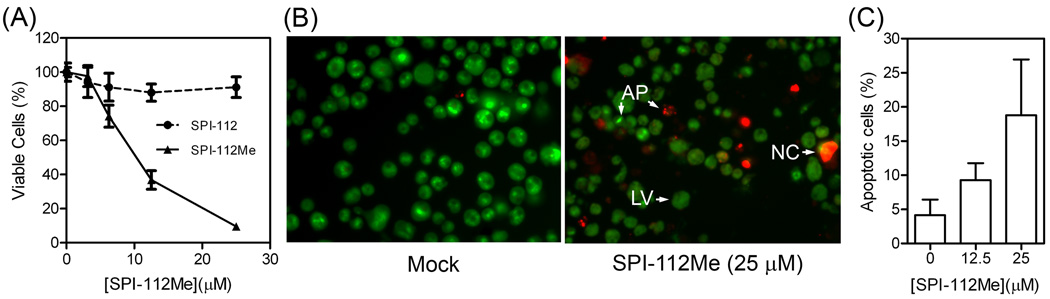
To determine if SPI-112Me and SPI-112 affect Shp2E76K-dependent cell growth, TF-1/Shp2E76K cells were cultured in GM-CSF-free medium in the presence of various concentrations of SPI-112Me or SPI-112 for 4 days and relative viable cell number was measured. SPI-112Me caused a concentration-dependent decrease in viable cells (Fig. 6A). A 50% decrease in viable TF-1/Shp2E76K cells was observed at 10 µM SPI-112Me in this assay. In contrast, SPI-112 did not affect TF-1/Shp2E76K cells, which is consistent with the notion that SPI-112 is not cell permeable.
Fig. 6.
SPI-112Me inhibits a gain-of-function Shp2 mutant-dependent survival of TF-1 cells. (A) TF-1/Shp2E76K cells (1,000 cells/wells) were incubated in RPMI1640/10% FBS plus various concentrations of SPI-112 or SPI-112Me for 4 days and relative number of viable cells was measured with the CellTiterGlo reagent (Promega). The data were from two triplicate experiments. (B) and (C) TF-1/Shp2E76K cells were incubated in RPMI1640/10% FBS plus SPI-112Me as indicated for 3 days and then processed for EB/AO staining assay of apoptosis as described in the Materials and Methods. (B) Representative images of nuclear stainings. LV, live cells, NC, necrotic cells, AP, apoptotic cells. (C) Percentage of apoptotic cells from three experiments.
The decrease in viable cell number could be due to inhibition of cell survival, cell proliferation, or both. An initial testing indicated that SPI-112Me-treated cells were fluorogenic under the laser light of flow cytometer, interfering with flow cytometric analysis of apoptosis. Therefore, a microscopic EB/AO staining assay [42] was employed to examine apoptosis of SPI-112Me-treated TF-1/Shp2E76K cells. Fig. 6B illustrates the nuclear morphology of EB/AO staining cells. As shown in Fig. 6C, in the absence of SPI-112Me, there was an average of 4.2% of apoptotic cells. In the presence of 12.5 and 25 µM SPI-112Me, 9.3% and 18.8% of apoptotic cells were observed, which were significantly increased (p = 0.019 and 0.001, respectively) above the base level. Thus, SPI-112Me is able to inhibit Shp2E76K-dependent survival of TF-1/Shp2E76K cells.
3.5. SPI-112Me enhances IFN-γ-stimulated tyrosine phosphorylation of STAT1
It was reported that STAT1 is a Shp2 substrate [24] and increased IFN-stimulated STAT1 tyrosine phosphorylation was observed in mouse embryonic fibroblasts from Shp2 knockout mice [24, 25]. We immunoprecipitated STAT1 from HT-29 cells treated with or without IFN-γ. As shown in Fig. 7A, STAT1 tyrosine phosphorylation was induced by IFN-γ in HT-29 cells. Incubation of STAT1 isolated from IFN-γ-treated cells with GST-Shp2 PTP protein in vitro resulted in concentration-dependent STAT1 tyrosine dephosphorylation, supporting the notion that pSTAT1 is a Shp2 PTP substrate (Fig. 7A). We reasoned that if SPI-112Me inhibits Shp2 PTP in the cells, it would enhance the IFN-γ-stimulated STAT1 tyrosine phosphorylation. Indeed, comparison of STAT1 tyrosine phosphorylation in HT-29 cells stimulated with IFN-γ alone or with a combination of IFN-γ and SPI-112Me showed increased amount of tyrosine-phosphorylated STAT1 in cells treated with both IFN-γ and SPI-112Me (Fig. 7B). This results show that SPI-112Me was able to enhance the ability of IFN-γ to stimulate STAT1 tyrosine phosphorylation.
Fig. 7.
SPI-112Me enhances the IFN-γ-induced responses. (A) STAT1 was immunoprecipitated from HT-29 cells with or without IFN-γ stimulation and incubated with a recombinant GST-Shp2 PTP as indicated. After the in vitro reaction, immunoprecipitates were washed and analyzed by immunoblotting with antibody to phosphotyrosine or total STAT1. (B) HT-29 cells were pre-treated with or without SpI-112Me (20 µM, 2 h) and stimulated with IFN-γ (20 U/ml) for the indicated time. Cell lysates were analyzed by immunoblotting with antibodies to phospho-STAT1, total STAT1, or β-actin. (C) HT-29 cells co-transfected with pISRE-Luc and β-gal plasmids were pretreated with indicated concentrations of SPI-112Me for 2 h and then stimulated with 50 U/ml IFN-γ for 6 h. Luciferase activity was determined and normalized to β-galactosidase activity. Data were from two duplicate experiments (n = 4). #, p < 0.05. (D) HT-29 cells were treated with or without SPI-1112Me (12.5 µM) and the indicated amounts of IFN-γ for 24 h. Cell lysates were analyzed by immunoblotting for the amounts of p21 and β-actin. (E) HT-29 cells were plated in 96-well plates (1,000 cells/well) and incubated with IFN-γ (100 U/ml) and/or the indicated concentrations of SPI-112Me or vehicle control for 4 days. Viable cell number was measured. * indicates a synergistic effect. The data were from three experiments performed in triplicates (n= 9).
To determine if SPI-112Me could enhance the STAT1-mediated transcription activity in response to a sub-saturated concentration of IFN-γ, HT-29 cells were transfected with pISRE-Luc and β-gal plasmids, stimulated with 50 U/ml IFN-γ in the presence or absence of SPI-112Me. Fig. 7C shows that IFN-γ alone activated the ISRE luciferase activity by 26%, which was further increased to 54% and 98% in the presence of 10 and 25 µM SPI-112Me. It was reported that the cyclin-dependent kinase inhibitor p21 (CDKN1A) is a STAT1 target gene [48]. As shown in Fig. 7D, stimulation of HT-29 cells with IFN-γ increased p21 expression and higher levels of p21 were detected in HT-29 cells treated with both IFN-γ and SPI-112Me. To determine if SPI-112Me could enhance the anti-proliferative activity of IFN-γ, HT-29 cells were cultured in the presence or absence of 100 U/ml IFN-γ, 0–12.5 µM SPI-112Me, or combination of both agents for 4 days. Relative number of viable cells was measured and analyzed as fraction of inhibition (Fig. 7E). IFN-γ alone had a fraction of inhibition of 0.18. Similar to a previous report using a different Shp2 inhibitor [10], HT-29 cell growth was sensitive to inhibition of the Shp2 inhibitor. The fraction of inhibition caused by 6.25 and 12.5 µM SPI-112Me were 0.32 and 0.71, respectively. In the presence of both IFN-γ (100 U/ml) and SPI-112Me (6.25 and 12.5 µM), the fraction of inhibition increased to 0.57 and 0.84, respectively. Thus, IFN-γ and SPI-112Me displayed a synergistic effect on inhibition of HT-29 cell growth.
4. Discussion
Although SPI-112 is among the most potent in vitro Shp2 inhibitors derived from NSC-117199 [9], its lack of activity in intake cells precluded its use in biological studies. In this study, we used the methyl ester prodrug strategy to enhance the cellular uptake of SPI-112. In vitro Shp2 PTP inhibition assay indicated that SPI-112Me had very weak Shp2 PTP inhibitor activity (IC50 >100 µM). This is consistent with the computer docking data, which suggests that the carboxylic acid group is involved in the binding of SPI-112 to the Shp2 PTP catalytic site. Thus, neutralization of the negative charge of the carboxylic acid with the methyl group is predicted to hinder the inhibitory activity.
Several lines of evidence showed that Shp2 PTP activity and function were inhibited in cells treated with SPI-112Me, indicating that SPI-112Me is a cell-active Shp2 PTP inhibitor. These include 1) inhibition of EGF-stimulated Shp2 PTP activity, paxillin dephosphorylation, Erk1/2 activation, and migration; 2) inhibition of Erk1/2 activation induced by an intracellular Gab1-Shp2 chimera; 3) inhibition of PTP activity of the leukemia-associated Shp2E76K mutant, Shp2E76K-dependent Bcl-XL expression and cell survival; and 4) enhancement of IFN-γ-induced STAT1 tyrosine phosphorylation, STAT1-mediated transcription activity, and anti-proliferation activity. Together with the SPI-112Me fluorescent uptake experiment, confocal microscope imaging, HPLC and LC/MS analyses, these data indicate that SPI-112Me is able to enter cells and convert to the potent Shp2 inhibitor SPI-112 in the cells.
Shp2 mutants such as Shp2E76K are linked to several types of leukemias, particularly the deadly JMML. It is important to test if a leukemia-associated Shp2 mutant-induced transformed phenotype in myeloid cells can be suppressed by a Shp2 PTP inhibitor. A transformation property of JMML cells is cytokine-independent growth. However, because of the lack of a suitable Shp2 PTP inhibitor that can efficiently enter hematopoietic cells, the suppression effect of a Shp2 PTP inhibitor on Shp2 mutant-induced cytokine-independent growth has not been shown previously. Using the Shp2E76K-induced cytokine-independent survival of TF-1 cells as the model, we demonstrate here that SPI-112Me effectively suppresses the PTP activity of the leukemia-associated Shp2 mutant and the Shp2 mutant-mediated cytokine-independence activity in hematopoietic cells.
While mutant Shp2 has been demonstrated as an oncogene in hematopoietic progenitor cells, it is not yet known whether mutant Shp2 is an oncogene in carcinomas. Shp2 mutations occur infrequently in carcinomas. Therefore, even if a gain-of-function Shp2 mutant can induce carcinomas, it is an uncommon event. However, the wildtype Shp2 is activated by oncogenic receptor tyrosine kinases such as ErbB and Met and is positively involved in their signaling. It will be very important to evaluate if Shp2 is required for the initiation and maintenance of malignant phenotypes induced by these oncogenic receptor tyrosine kinases and thus functions as a non-oncogene addiction gene [49] in carcinomas. The availability of SPI-112Me and other suitable Shp2 inhibitors should facilitate the evaluation of the role of Shp2 in carcinomas and the suitability of using a PTP inhibitor for cancer therapy.
We demonstrate here that SPI-112Me can enhance IFN-γ signaling. This interesting finding not only provides a line of evidence that SPI-112Me is hitting the intended target (Shp2) in cancer cells but also reinforces the notion that SPI-112Me does not non-specifically impair the cellular signaling machinery or cell functions. This is because a global impairment of cellular functionality would likely prevent an increase in transcription activity. IFNs are noted for their anti-viral and anti-tumor activities but the use of high doses of IFNs is limited by toxicity. The development of SPI-112Me and other novel Shp2 inhibitors should facilitate the exploration of enhancing the anti-viral and anti-tumor efficacies of these biological agents at lower doses with a chemical inhibitor.
Supplementary Material
Acknowledgment
This work was supported by the National Institutes of Health National Cancer Institute (Grants P01CA118210, R01CA077467, P30CA076292, and U56CA118809). We thank Dr. Wesley Brooks of the High Throughput Screening and Chemistry Core, Mark Lloyd and Joseph Johnson of the Analytic Microscopy Core of the Moffitt Cancer Center for their assistance.
Abbreviations
- AO
acridine orange
- DiFMUP
6,8-difluoro-4-methylumbelliferyl phosphate
- DiFMU
6,8-difluoro-4-methylumbelliferone
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethyl sulfoxide
- dox
doxycycline
- DTT
dithiothreitol
- EB
Ethidium bromide
- EGF
epidermal growth factor
- Erk1/2
extracellular signal-regulated kinases 1 and 2
- FBS
fetal bovine serum
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- HPLC
high performance liquid chromatography
- LC/MS
liquid chromatography/mass spectrometry
- JMML
juvenile myelomoncytic leukemia
- IFN
interferon
- MAP kinase
mitogen-activated protein kinase
- PTP
protein tyrosine phosphatase
- SPI-112
(Z)-3-(2-(5-(N-(4-fluorobenzyl)sulfamoyl)-2-oxoindolin-3-ylidene)hydrazinyl)benzoic acid
- SPI-112Me
(Z)-3-(2-(5-(N-(4-fluorobenzyl)sulfamoyl)-2-oxoindolin-3-ylidene)hydrazinyl)benzoic acid methyl ester
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 3.Mulloy R, Ferrand A, Kim Y, Sordella R, Bell DW, Haber DA, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67:2325–2330. doi: 10.1158/0008-5472.CAN-06-4293. [DOI] [PubMed] [Google Scholar]
- 4.Bentires-Alj M, Paez JG, David FS, Keilhack H, Halmos B, Naoki K, et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 5.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 6.Stephens BJ, Han H, Gokhale V, Von Hoff DD. PRL phosphatases as potential molecular targets in cancer. Mol Cancer Ther. 2005;4:1653–1661. doi: 10.1158/1535-7163.MCT-05-0248. [DOI] [PubMed] [Google Scholar]
- 7.Tonks NK, Muthuswamy SK. A brake becomes an accelerator: PTP1B--a new therapeutic target for breast cancer. Cancer Cell. 2007;11:214–216. doi: 10.1016/j.ccr.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 8.Jiang ZX, Zhang ZY. Targeting PTPs with small molecule inhibitors in cancer treatment. Cancer Metastasis Rev. 2008;27:263–272. doi: 10.1007/s10555-008-9113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence HR, Pireddu R, Chen L, Luo Y, Sung SS, Szymanski AM, et al. Inhibitors of Src homology-2 domain containing protein tyrosine phosphatase-2 (Shp2) based on oxindole scaffolds. J Med Chem. 2008;51:4948–4956. doi: 10.1021/jm8002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellmuth K, Grosskopf S, Lum CT, Wurtele M, Roder N, von Kries JP, et al. Specific inhibitors of the protein tyrosine phosphatase Shp2 identified by high-throughput docking. Proc Natl Acad Sci U S A. 2008;105:7275–7280. doi: 10.1073/pnas.0710468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geronikaki A, Eleftheriou P, Vicini P, Alam I, Dixit A, Saxena AK. 2-Thiazolylimino/heteroarylimino-5-arylidene-4-thiazolidinones as new agents with SHP-2 inhibitory action. J Med Chem. 2008;51:5221–5228. doi: 10.1021/jm8004306. [DOI] [PubMed] [Google Scholar]
- 12.Johnston PA, Foster CA, Tierno MB, Shun TY, Shinde SN, Paquette WD, et al. Cdc25B dual-specificity phosphatase inhibitors identified in a high-throughput screen of the NIH compound library. Assay Drug Dev Technol. 2009;7:250–265. doi: 10.1089/adt.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott LM, Lawrence HR, Sebti SM, Lawrence NJ, Wu J. Targeting Protein Tyrosine Phosphatases for Anticancer Drug Discovery. Curr Pharm Des. 2010;16:1843–1862. doi: 10.2174/138161210791209027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neel BG, Gu H, Pao L. The 'Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 15.Cunnick JM, Mei L, Doupnik CA, Wu J. Phosphotyrosines 627 and 659 of Gab1 constitute a bisphosphoryl tyrosine-based activation motif (BTAM) conferring binding and activation of SHP2. J Biol Chem. 2001;276:24380–24387. doi: 10.1074/jbc.M010275200. [DOI] [PubMed] [Google Scholar]
- 16.Ren Y, Meng S, Mei L, Zhao ZJ, Jove R, Wu J. Roles of Gab1 and SHP2 in paxillin tyrosine dephosphorylation and Src activation in response to epidermal growth factor. J Biol Chem. 2004;279:8497–8505. doi: 10.1074/jbc.M312575200. [DOI] [PubMed] [Google Scholar]
- 17.Zhan Y, Counelis GJ, O'Rourke DM. The protein tyrosine phosphatase SHP-2 is required for EGFRvIII oncogenic transformation in human glioblastoma cells. Exp Cell Res. 2009;315:2343–2357. doi: 10.1016/j.yexcr.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Agazie YM. Molecular mechanism for SHP2 in promoting HER2-induced signaling and transformation. J Biol Chem. 2009;284:12226–12234. doi: 10.1074/jbc.M900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tartaglia M, Gelb BD. Germ-line and somatic PTPN11 mutations in human disease. Eur J Med Genet. 2005;48:81–96. doi: 10.1016/j.ejmg.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Kratz CP, Niemeyer CM, Castleberry RP, Cetin M, Bergstrasser E, Emanuel PD, et al. The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood. 2005;106:2183–2185. doi: 10.1182/blood-2005-02-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren Y, Chen Z, Chen L, Woods NT, Reuther GW, Cheng JQ, et al. Shp2E76K mutant confers cytokine-independent survival of TF-1 myeloid cells by up-regulating Bcl-XL. J Biol Chem. 2007;282:36463–36473. doi: 10.1074/jbc.M705789200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J Biol Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 23.Chan G, Kalaitzidis D, Usenko T, Kutok JL, Yang W, Mohi MG, et al. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 2009;113:4414–4424. doi: 10.1182/blood-2008-10-182626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu TR, Hong YK, Wang XD, Ling MY, Dragoi AM, Chung AS, et al. SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem. 2002;277:47572–47580. doi: 10.1074/jbc.M207536200. [DOI] [PubMed] [Google Scholar]
- 25.You M, Yu DH, Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol Cell Biol. 1999;19:2416–2424. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baron M, Davignon JL. Inhibition of IFN-gamma-induced STAT1 tyrosine phosphorylation by human CMV is mediated by SHP2. J Immunol. 2008;181:5530–5536. doi: 10.4049/jimmunol.181.8.5530. [DOI] [PubMed] [Google Scholar]
- 27.Bialy L, Waldmann H. Inhibitors of protein tyrosine phosphatases: next-generation drugs? Angew Chem Int Ed Engl. 2005;44:3814–3839. doi: 10.1002/anie.200461517. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Sung SS, Yip ML, Lawrence HR, Ren Y, Guida WC, et al. Discovery of a novel shp2 protein tyrosine phosphatase inhibitor. Mol Pharmacol. 2006;70:562–570. doi: 10.1124/mol.106.025536. [DOI] [PubMed] [Google Scholar]
- 29.Zhao XT, Qian YK, Chan AW, Madhavan R, Peng HB. Regulation of ACh receptor clustering by the tyrosine phosphatase Shp2. Dev Neurobiol. 2007;67:1789–1801. doi: 10.1002/dneu.20556. [DOI] [PubMed] [Google Scholar]
- 30.Fuchikawa T, Nakamura F, Fukuda N, Takei K, Goshima Y. Protein tyrosine phosphatase SHP2 is involved in Semaphorin 4D-induced axon repulsion. Biochem Biophys Res Commun. 2009;385:6–10. doi: 10.1016/j.bbrc.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 31.Boutselis IG, Yu X, Zhang ZY, Borch RF. Synthesis and cell-based activity of a potent and selective protein tyrosine phosphatase 1B inhibitor prodrug. J Med Chem. 2007;50:856–864. doi: 10.1021/jm061146x. [DOI] [PubMed] [Google Scholar]
- 32.Qian YK, Chan AW, Madhavan R, Peng HB. The function of Shp2 tyrosine phosphatase in the dispersal of acetylcholine receptor clusters. BMC Neurosci. 2008;9:70. doi: 10.1186/1471-2202-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee PK, Al Abed Y, Sherry B, Metz CN. Cholinergic agonists regulate JAK2/STAT3 signaling to suppress endothelial cell activation. Am J Physiol Cell Physiol. 2009 doi: 10.1152/ajpcell.00160.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noren-Muller A, Reis-Correa I, Jr, Prinz H, Rosenbaum C, Saxena K, Schwalbe HJ, et al. Discovery of protein phosphatase inhibitor classes by biology-oriented synthesis. Proc Natl Acad Sci U S A. 2006;103:10606–10611. doi: 10.1073/pnas.0601490103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu D, Pang Y, Ke Y, Yu J, He Z, Tautz L, et al. A conserved mechanism for control of human and mouse embryonic stem cell pluripotency and differentiation by shp2 tyrosine phosphatase. PLoS One. 2009;4:e4914. doi: 10.1371/journal.pone.0004914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, He Y, Liu S, Yu Z, Jiang ZX, Yang Z, et al. Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2) J Med Chem. 2010;53:2482–2493. doi: 10.1021/jm901645u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 38.Muller KM, Arndt KM, Bauer K, Pluckthun A. Tandem immobilized metal-ion affinity chromatography/immunoaffinity purification of His-tagged proteins--evaluation of two anti-His-tag monoclonal antibodies. Anal Biochem. 1998;259:54–61. doi: 10.1006/abio.1998.2606. [DOI] [PubMed] [Google Scholar]
- 39.Nordin H, Jungnelius M, Karlsson R, Karlsson OP. Kinetic studies of small molecule interactions with protein kinases using biosensor technology. Anal Biochem. 2005;340:359–368. doi: 10.1016/j.ab.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 40.Joss L, Morton TA, Doyle ML, Myszka DG. Interpreting kinetic rate constants from optical biosensor data recorded on a decaying surface. Anal Biochem. 1998;261:203–210. doi: 10.1006/abio.1998.2744. [DOI] [PubMed] [Google Scholar]
- 41.Greco WR, Faessel H, Levasseur L. The search for cytotoxic synergy between anticancer agents: a case of Dorothy and the ruby slippers? J Natl Cancer Inst. 1996;88:699–700. doi: 10.1093/jnci/88.11.699. [DOI] [PubMed] [Google Scholar]
- 42.Ribble D, Goldstein NB, Norris DA, Shellman YG. A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol. 2005;5:12. doi: 10.1186/1472-6750-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Streiber M, Picard F, Scherer C, Seidel SB, Hartmann RW. Methyl esters of N-(dicyclohexyl)acetyl-piperidine-4-(benzylidene-4-carboxylic acids) as drugs and prodrugs: a new strategy for dual inhibition of 5 alpha-reductase type 1 and type 2. J Pharm Sci. 2005;94:473–480. doi: 10.1002/jps.20265. [DOI] [PubMed] [Google Scholar]
- 44.Manne V, Yan N, Carboni JM, Tuomari AV, Ricca CS, Brown JG, et al. Bisubstrate inhibitors of farnesyltransferase: a novel class of specific inhibitors of ras transformed cells. Oncogene. 1995;10:1763–1779. [PubMed] [Google Scholar]
- 45.Manes S, Mira E, Gomez-Mouton C, Zhao ZJ, Lacalle RA, Martinez AC. Concerted activity of tyrosine phosphatase SHP-2 and focal adhesion kinase in regulation of cell motility. Mol Cell Biol. 1999;19:3125–3135. doi: 10.1128/mcb.19.4.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vadlamudi RK, Adam L, Nguyen D, Santos M, Kumar R. Differential regulation of components of the focal adhesion complex by heregulin: role of phosphatase SHP-2. J Cell Physiol. 2002;190:189–199. doi: 10.1002/jcp.10054. [DOI] [PubMed] [Google Scholar]
- 47.Cunnick JM, Meng S, Ren Y, Desponts C, Wang HG, Djeu JY, et al. Regulation of the mitogen-activated protein kinase signaling pathway by SHP2. J Biol Chem. 2002;277:9498–9504. doi: 10.1074/jbc.M110547200. [DOI] [PubMed] [Google Scholar]
- 48.Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 49.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.