Abstract
While insights into the molecular processes that specify adoption of the αβ and γδ fates are beginning to emerge, the basis for control of specification remains highly controversial. This review highlights the current models attempting to explain T lineage commitment. Recent observations support the hypothesis that the T cell receptor (TCR) provides instructive cues through differences in TCR signaling intensity and/or longevity. Accordingly, we review evidence addressing the importance of differences in signal strength/longevity, how signals differing in intensity/longevity may be generated, and finally how such signals modulate the activity of downstream effectors to promote the opposing developmental fates.
Keywords: T cell development, lineage commitment, γδTCR, signal strength, signal duration
1. Distinctions between αβ and γδ lineage T cells
T lymphocytes comprise two distinct lineages that express either αβ or γδ TCR complexes and perform non-overlapping roles in immune responses. αβ T cells localize primarily in secondary lymphoid organs, recognize peptide ligands presented by class I and II major histocompatibility complex (MHC) antigens, and respond to infection by facilitating the production of antibodies reactive with the pathogen or by lysing infected target cells. γδ T cells comprise a small percentage of lymphoid cells in the thymus and secondary lymphoid tissues; however, they are quite abundant at epithelial surfaces lining the inside and outside of the body [1, 2]. γδ T cells recognize a much wider variety of antigens, including non-classical MHC molecules, heat shock proteins, and lipids [3]. Although the precise role of γδ T cells in immune responses remains unclear at present, these cells are thought to lie at the interface between the innate and adaptive immune systems and to perform functions that are at least partially distinct from those of αβ T cells. Indeed, certain bacterial infections (e.g., Nocardia asteroides) that are normally cleared in wild type mice are rapidly fatal in mice lacking γδ T cells [4]. γδ T cells have also been implicated in preservation of epithelial barriers and in eradication of cutaneous malignancies [5–7]. Despite the important, distinct functions of these two T lineages, our understanding of the developmental cues responsible for promoting immature T cell progenitors to adopt either the αβ or γδ fate remains limited.
2. Development of αβ and γδ lineage T cells in the thymus
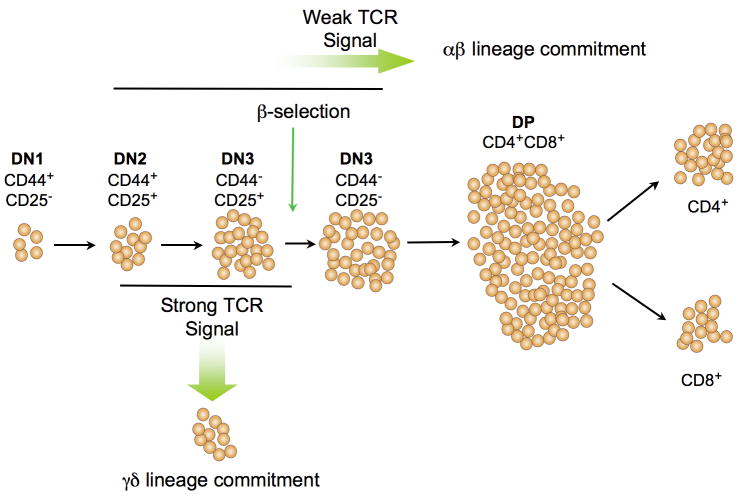
Developmental milestones achieved by T cell precursors are marked by prescribed changes in expression of the differentiation antigens CD4, CD8, CD25, and CD44, which can be used to divide thymocytes into distinct subsets schematized in ascending order of maturity in Figure 1 [8]. CD44+CD25− (DN1) cells upregulate CD25 expression (CD44+CD25+, DN2) coincident with their commitment to the T lineage and begin to rearrange their TCRγ, δ, and β genes [9–11]. Those precursors that productively rearrange their TCRγ and δ genes are eligible to become γδ T cells, which usually remain DN and exit the thymus to populate peripheral lymphoid organs or epithelial surfaces [1]. Generation of γδ T cells in the mouse is more pronounced during fetal life and occurs in waves of cells expressing particular sets of Vγ and Vδ genes [12–14]. The developmental stage at which γδ lineage T cells diverge from αβ T cells has not been precisely defined but recent evidence suggests it is complete upon arrival at the CD44− CD25+ (DN3) stage when β-selection occurs [15–19]. β-selection stipulates that only thymocytes that maintain the translational reading frame of TCRβ will survive and differentiate; those failing to do so die by apoptosis [18, 20, 21]. Expression of TCRβ protein promotes development through assembly with pre-Tα and the CD3 signaling subunits (CD3γ, δ,ε,ζ) to form the pre-T cell receptor (pre-TCR) complex [22,23]. Pre-TCR assembly initiates signaling through a poorly-understood ligand-independent process [24, 25]. Pre-TCR signals, which are required for traversal of the β-selection checkpoint, rescue DN3 thymocytes from apoptosis and induce massive proliferative expansion as αβ lineage cells differentiate to the CD4+CD8+ (DP) stage, a differentiation step that committed, γδ lineage thymocytes do not undergo [20, 26–28]. Therefore, development of γδ lineage T cells requires productive rearrangement of the TCRγ and δ loci and signaling through the γδ TCR, whereas commitment to the αβ lineage requires productive rearrangement of the TCRβ locus and pre-TCR signaling.
Figure 1. αβ/γδ lineage commitment during thymocyte development.
TCR γ, δ, and β gene rearrangement begins in DN2 thymocytes. αβ/γδ lineage commitment is thought to occur between the onset of gene rearrangement and arrival at the DN3 stage. Precursors that have committed to the γδ lineage express the γδ TCR complex, but usually not CD4 or CD8. In contrast, commitment to the αβ lineage usually occurs in response to pre-TCR signaling and is characterized by development of thymocytes to the DP stage. While commitment to the αβ and γδ lineage is most often directed by signals from the pre-TCR and γδ TCR complexes, respectively, these decisions are not irrevocably tied to the receptor isotype. Rather, they are determined by the nature of the TCR signal, with weaker signals favoring adoption of the αβ fate and stronger signals promoting adoption of the γδ fate.
3. Markers of αβ and γδ lineage commitment in early precursors
Analysis of the molecular control of αβ/γδ T lineage commitment continues to be hampered by the lack of definitive lineage markers of the early stages of commitment in DN thymocytes. Through Serial Analysis of Gene Expression (SAGE) performed by the Hayday lab, a γδ biased gene signature was established; however, this profile was more closely linked to function than to lineage commitment [29, 30]. The TCR complexes (i.e., pre-TCR expression for αβ lineage and γδTCR for γδ lineage) have also been employed as lineage markers, but the TCR isotype alone is no longer sufficient to assign lineage fate to developing DN thymocytes because both the pre-TCR and γδTCR are able to support αβ lineage commitment and development to the DP stage [31]. Accordingly, CD4 and CD8 expression must also be taken into consideration in assigning lineage, such that γδTCR expressing cells that remain DN are assigned to the γδ lineage, while those developing to the DP stage in response to TCR signals from any receptor isotype are assigned to the αβ lineage [28]. We and others have utilized downregulation of CD24 (HSA) among DNs as an additional marker of γδ commitment [28, 32–34], but this too has been questioned as many of the γδTCR+ cells exiting the thymus are CD24+ [35–37]. Indeed, γδ cells newly exported from the thymus (recent thymic emigrants; RTE) represent a mixture of CD24− and CD24+ cells; however, while the CD24− population is long-lived, a high proportion of CD24+ RTEs seem to die off soon after reaching the periphery, suggesting they make a minimal contribution to the long-lived peripheral ©™ pool [35–37]. More recent efforts identified the transcription factor Sox13 as being highly enriched in γδ T cells. Nevertheless, it is not clear whether this factor reliably marks all γδ lineage cells [38]. Accordingly, while efforts to gain insight into the molecular control of αβ/γδ lineage commitment would benefit from definitive molecular markers distinguishing DN thymocytes committing to the αβ lineage from those committing to the γδ lineage, such markers remain elusive.
4. Models of αβ/γδ lineage commitment
There is widespread agreement that αβ and γδ T cells arise from a common progenitor, but the respective roles of the pre-TCR and γδ TCR complexes in selection of the αβ and γδ lineages remain controversial [15, 17, 19]. A central question is whether the TCR complexes function to specify fate (instruction model) or alternatively serve to rescue the viability of cells whose fate was pre-determined without input from the TCR complex (stochastic model) [39, 40]. For many years, only these two models were advanced to explain the role of TCR complexes in lineage commitment: however, these models were not adequate to explain the status of TCR gene rearrangements in αβ and γδ lineage cells, nor did they appropriately explain the lineage infidelity observed in TCR transgenic and gene-targeted mice. The stochastic model predicts that the frequency of in-frame rearrangements of the “irrelevant” TCR loci (e.g., the TCRβ locus in γδ cells) should not exhibit signs of selection against in-frame rearrangements whereas the instructional model predicts that in-frame rearrangements of these loci should be depleted [41]. Interestingly, αβ lineage cells were found to be depleted of in-frame TCRγ rearrangements while γδ T cells exhibited no obvious selection against in-frame TCRβ rearrangements [9, 19, 42–47]. These observations are inconsistent with the predictions of both instructional and stochastic models. Analysis of TCR transgenic and gene-targeted mice produced observations that similarly defied explanation [48–51]. One particular example is the finding that the γδ TCR complex was capable of promoting development of αβ lineage DP thymocytes [31]. This observation violates predictions made by strict interpretation of both instructional and stochastic models suggesting that pre-TCR and γδTCR complexes should always give rise to αβ and γδ lineage cells, respectively. However, there are two observations supporting the stochastic model. The first is that CD127hi and CD127lo DN2 cells were reported to exhibit differences in αβ/γδ lineage potential, inferring that commitment occurred prior to TCR gene rearrangement [52]. One caveat tempering interpretation is that because CD127 is the IL-7 receptor (IL-7R) α chain and IL-7R signaling can influence TCRγ rearrangement, the bias in lineage potential might be secondary to alterations in TCRγ rearrangement [53–55]. Another observation supportive of stochastic lineage assignment is that the γδ T cell restricted transcription factor Sox13 is induced in DN2 thymocytes and is required for efficient generation of γδ T cells. However, the blockade of development in Sox-13 deficient mice was incomplete and it was not clear whether Sox13 expression occurred prior to or subsequent to TCR expression [38]. Further, some have raised the possibility that Sox13 may not mark all γδ lineage cells [28, 34].
While there is no question that pre-TCR signaling and γδTCR signaling are most often responsible for development of αβ and γδ lineage cells, respectively, the notable exceptions described above provided clues as to the nature of the differences in TCR signaling involved in these alternate fate choices. In an attempt to reconcile the discrepant observations described above, a signal strength model for αβ/γδ lineage commitment was proposed [56]. This model contends that weak signals promote commitment to the αβ lineage while comparatively strong signals promote commitment to the γδ lineage, irrespective of the TCR complex from which they originate. In support of this model, we demonstrated that the same transgenic γδTCR could both promote development of γδ lineage cells in response to engagement by selecting ligand, T10d, and also divert cells to the αβ fate when signaling was attenuated either by removal of ligand or by ablation of the gene encoding the Src kinase p56lck [33]. Paul Love’s lab reported similar findings using a different γδTCR Tg model [57]. The differences in signal strength regulating fate selection were found to be critically dependent upon the amplitude of activation of the ERK-Early growth response (Egr)-Id3 pathway [33]. These observations cannot be accommodated by strict stochastic or instructional models in which the TCR isotype determines fate with fidelity. Nevertheless, since the aforementioned analysis was performed on bulk populations, it remained unclear whether the differences in signal strength were acting instructionally to dictate fate or stochastically to rescue viability of pre-committed precursors. The Zúñiga-Pflücker lab sought to resolve this issue, first employing a single-cell progenitor assay to demonstrate that fate selection is complete by the DN3 stage, and then ectopically expressing either pre-TCR or γδTCR complexes in TCR-deficient DN3 and demonstrating that the TCR complexes dictated the expected fate [17]. These data provide strong support for the notion that TCR signals act instructively to dictate fate. This topic will be addressed more extensively in two other reviews in this issue (See Narayan et al., and Wong et al).
5. Factors enabling the γδTCR to signal more robustly than the pre-TCR
Receptor characteristics
The association of TCR signals of greater intensity/longevity with γδ lineage commitment engenders questions regarding the molecular basis by which γδTCRs transduce such signals. A number of attributes that differ between the pre-TCR and γδTCR may contribute. Perhaps the most important difference is their surface density. The pre-TCR is expressed at levels (~400 per cell) about 100-fold lower than the αβTCR on mature T cells or the γδTCR on thymocytes [58]. Further, the γδTCR also appears to be a more potent signaling complex than the αβTCR following antibody-stimulation, which may result from the differing complement of CD3 signaling subunits, as the γδTCR complex lacks CD3δ, instead possessing two CD3δε dimers [59]. Finally, it was recently reported that some TCRγδ pairs may be able to signal in a ligand-independent fashion, as was previously shown for the pre-TCR [25, 60]. Nevertheless, when the ligand-independent signaling of a single γδTCR Tg of known antigenic specificity was tested in vivo, it promoted development to the DP stage and αβ lineage commitment, suggesting that ligand-independent signaling by γδTCR complexes may be similar in intensity/duration to pre-TCR signals [33].
Ligand
In contrast to the pre-TCR for which ligands have not been identified, there is clear evidence that at least some γδTCRs recognize endogenous ligands. For these γδTCRs, the intensity/duration of γδTCR signaling may be significantly modulated by intrathymic ligands and there is compelling evidence in support of ligand-involvement in their development. Dendritic epidermal γδ T cells (DETC) are a skin resident subset with a nearly invariant Vγ3Vδ1 γδTCR and development of this subset bears the hallmarks of ligand-selection [61, 62]. Skint1 protein has been proposed as a potential ligand for the DETC γδTCR. Nevertheless, while Skint1 is essential for DETC development, its function as a bona fide ligand has not been formally demonstrated [63]. The most extensive analysis of ligand involvement in γδ development has focused on γδ cells with TCR complexes reactive with T10/T22. T10 and T22 are non-classical MHC class I molecules that require β2M for surface expression. Early analysis in β2M-deficient mice suggested that the development of T10/T22 reactive γδ cells in two TCR transgenic (Tg) models (G8 and KN6) was dependent upon ligand [32, 64]. A subsequent report using the G8 transgenic model reported that ligand caused deletion rather than promoting development [65]. The basis for this discrepancy has never been resolved but has been suggested to result from differences in the background strains of the mice utilized [65]. More recently, the role of ligand in development of T10/22 reactive γδ cells was monitored using T10 tetramer-binding, which showed that tetramer-binding cells developed and exited the thymus even in the absence of their presumptive ligand [60]. However, interpretation of this result is complicated by the fact that T10 tetramers bind to TCRs employing the Dδ2 element with widely varying affinity (differing by at least 15-fold) [66]. Accordingly, it is possible that tetramer binding lacks the precision to identify precursors with the appropriate affinity for positive selection on T10. Consistent with this notion, neither ERK phosphorylation nor CD5 induction among T10 tetramer staining γδTCR+ thymocytes was detectably altered by the presence of the presumptive high-affinity T10b ligand (Meyers et al., this issue); however, both TCR signaling (ERK phosphorylation and CD5 induction) and fate selection were altered by ligand in thymocytes transgenic for the T10d-selected KN6 γδTCR [33]. These data raise the possibility that a substantial fraction of γδTCRs that utilize Dδ2 (and are thus bound by T10 tetramer) are reactive with and perhaps selected upon other intrathymic ligands. Irrespective of these findings, we have demonstrated that commitment of KN6 γδTCR transgenic thymocytes to the γδ lineage is dependent upon β2M (required for surface expression of T10/T22), as KN6 Tg thymocytes are diverted to the αβ fate in its absence [33, 34]. Likewise, we have recently shown that specific targeting of T10/T22 ligand in OP9-DL1 cultures using shRNA-mediated knockdown prevents KN6 γδTCR expressing thymocytes from maturing along the γδ-lineage and instead diverts them to the DP stage of αβ-lineage differentiation [34]. This represents the first example where a specific, defined γδ ligand has been shown to be required for γδ lineage commitment and development. Nevertheless, use of the KN6 γδTCR has been suggested to produce artifactual results because of abnormally early expression; however, we maintain that these concerns are unfounded as the KN6 TCR is expressed under the control of endogenous elements which are not activated until the DN2 stage, coincident with rearrangement of the endogenous TCRγ and δ loci. Whether ligand-involvement in KN6 Tg γδ cell development represents the exception or represents a more common phenomenon awaits the identification and evaluation of additional γδTCR ligands. The controversial role of ligand in γδ development will be addressed in two other articles in this issue (Meyer et al., and Kreslavsky et al).
6. How do signals that control the alternate lineage choices differ?
While support for TCR signal strength/duration as an important determinant of αβ/γδ fate choice is accumulating, our understanding of the key signaling effectors that are differentially regulated during fate choice remains rudimentary. Evidence from gene-targeting approaches suggests that the signaling cascades upon which γδ and αβ lineage development depends are genetically-separable (reviewed in [40, 67]). For example, TCR stimulation induces phosphorylation of the adaptor molecule LAT on multiple tyrosines, each of which exhibits some specialization in the SH2-domain-containing proteins it recruits (reviewed in [68]). Importantly, LAT-deficiency causes a severe blockade of both the αβ and γδ lineages [69]; however, mice expressing LAT molecules selectively defective in PLCγ recruitment exhibit a preferential block in development of αβ lineage cells [70–72]. Gene-targeting experiments have also identified numerous other molecular effectors that are selectively required for either the αβ or γδ lineage development (e.g., our identification of the ribosomal protein Rpl22; [73]), but this information has not yet been integrated into well-defined molecular pathways tied to development of either αβ of γδ cells (reviewed by Hayes et al., in this issue). Nevertheless, our data suggest that differences in activation of the ERK-Egr-Id3 axis are an important manifestation of the differences in signal strength/duration that influence αβ/γδ fate choice [33, 34].
ERK signaling
TCR signaling leads to ERK/MAP kinase (MAPK) activation and is required for normal thymocyte development [74]. We, and others, have shown that ERK is more highly phosphorylated in developing γδ than in αβ lineage cells [33, 57]. ERKs are the terminal enzymes in a cascade of three protein kinases, MAP kinase kinase kinase (MAP3K; Raf), MAP kinase kinase (MAP2K; MEK) and MAPK (ERK), which sequentially activate their downstream targets by phosphorylation at specific amino acid residues. Upon activation, ERK not only phosphorylates regulatory targets in the cytosol, but is also capable of translocating to the nucleus, where it regulates gene expression by activating transcription factors such as Elk-1, c-Fos, or c-Myc [75, 76]. The greater ERK activation associated with adoption of the γδ fate appears to be functionally important as γδ commitment can be impaired in vitro using pharmacologic inhibitors of ERK signaling (S.-Y. Lee, unpublished observation); however, the requirement for ERK in γδ lineage commitment and development has not been rigorously tested genetically.
Egr proteins
ERK/MAPK induces the expression of immediate early gene, zinc-finger transcription factors of the Egr family [77], which we and others have shown to be critical for normal thymocyte development [78–80]. The Egr family of transcription factors contains four members: Egr1, Egr2, Egr3, and Egr4. The zinc-finger DNA-binding domains of the Egr family members are highly homologous, but their N-terminal activation domains are more divergent and are thought to scaffold interactions with distinct transcriptional regulators that influence the target specificity of particular Egr family members [81]. Egr proteins are induced in response to various mitogenic stimuli and regulate genetic programs controlling growth and differentiation of diverse cell types [81]. Importantly, induction of Egr proteins has been demonstrated to be proportional to signal strength [82, 83]. Accordingly, we found that Egr induction correlates with signal strength during αβ/γδ lineage commitment in that γδ lineage choice was associated with greater induction of Egr proteins than was adoption of the αβ fate [33]. Differential induction of Egr proteins appears to play an important role in fate choice as elevating Egr levels through ectopic expression augmented development of γδ lineage cells while causing a commensurate decrease in αβ lineage cells [33, 34]. While these data clearly indicate that the extent of Egr induction is an important component of the signals that promote commitment to the γδ lineage, loss of function analysis has been complicated by functional redundancy among the 4 family members expressed in the thymus and constraints imposed on generation of compound-deficient mice by impaired reproduction and survival of some of the strains.
Id proteins and their targets
An important target of TCR-induced Egr proteins is the helix-loop-helix (HLH) factor, Id3 [84–86]. Like ERK and Egr proteins, Id3 is induced in proportion to signal strength and is more highly expressed in γδ lineage progenitors than in those adopting the αβ fate [33, 34, 87]. Id3 function has also been causally-linked to control of αβ/γδ fate using both gain- and loss-of-function analysis. Enforced expression of Id3 in thymic progenitors blocks development of αβ lineage cells while enabling γδ development to continue unimpaired [88]. Id3 is also epistatic to Egr1 induction, as Id3-deficiency impairs the ability of Egr1 to promote development of γδ lineage cells in fetal thymic organ cultures [33] [34]. Several groups have reported that Id3-deficient mice exhibit perturbations in γδ cell development in that Id3-deficiency results in the selective expansion of innate type Vγ1.1Vδ6.3 γδ cells, although the reports differ in interpretation of this result [34, 89–91]. Our analysis revealed that other γδ subsets were diminished by Id3-deficiency (e.g., Vγ2 and DETC), and we postulate that the relative sensitivity of different γδ subsets reflects differences in TCR signal strength, perhaps resulting from intrathymic ligands of differing affinity [34]. In support of this view, we have shown that Id3-deficiency reduces the number of mature γδ lineage KN6 γδTCR Tg thymocytes generated by positive selection in the presence of moderate affinity T-10d ligand [34]. In contrast, in the presence of T-10b ligand (10-fold higher affinity), Id3-deficiency markedly increased the number of mature γδ lineage, KN6 thymocytes, suggesting that Id3-deficiency enabled them to escape the deletion normally resulting from engagement by high affinity ligand [32, 34]. Altogether, these findings support a dichotomy of Id3 function, with Id3 promoting commitment and development of γδ cells in the context of the stronger signals that typically accompany this process, while inducing deletion (or restraining the expansion) of other sublineages (some of which are PLZF-dependent) when signal strength exceeds a particular threshold. Therefore, we suggest that an important factor contributing to the expansion of Vγ1.1Vδ6.3 cells in Id3-deficient mice is their escape from Id3-mediated restraint, perhaps following encounter with a high affinity ligand.
These results raise the question of how the extent of Id3 induction influences αβ/γδ lineage choice. One possibility is that differences in Id3 induction produce distinct developmental outcomes through graded suppression of E proteins. E proteins are basic helix-loop-helix (bHLH) transcription factors that bind DNA at E-box motifs (CANNTG) either as homodimers, or heterodimers with other bHLH proteins [92]. Of the 4 family members found in mammals, two are expressed in developing T cells, E2A and HEB, and their DNA-binding activity is antagonized by pairing with Id family members like Id3. Consequently, the magnitude of E protein inhibition is likely to be an important manifestation of signal intensity, with very strong signals nearly extinguishing E protein activity during γδ commitment and mimicking the effect of E protein gene-ablation. Consistent with this view, deficiencies in the E proteins, E2A and HEB, appear to differentially affect development of αβ and γδ T cells. E2A deficiency perturbs development of αβ T cells and at least certain subsets of γδ T cells, while HEB deletion selectively impairs αβ T cell development, leaving development of γδ T cells unaffected [93, 94]. Impairment of γδ development is less severe in mice lacking both E2A and HEB than in mice lacking E2A alone, suggesting that HEB expression in the absence of E2A may be responsible for the defect [95, 96]. E proteins play a pivotal role in preventing thymocytes from developing beyond the β-selection checkpoint, as evidenced by the ability of pre-TCR deficient thymocytes to traverse the β-selection checkpoint and differentiate to the DP stage in the absence of E2A [95, 97]. Paradoxically, E protein deficiency blocks development beyond the β-selection checkpoint of pre-TCR expressing cells, suggesting that the induction of αβ-lineage development by pre-TCR signals is dependent upon partial or temporally-restricted suppression of E protein activity [84, 86]. Conversely, the mild impairment of γδ development by E protein deficiency is consistent with the effect of strong TCR signals inducing greater Id3 expression and more profound or sustained reductions of E protein activity. Altogether, these data suggest a model whereby graded reductions in E protein activity mediated by differences in TCR signal strength play an important role in fate adoption and development (Fig. 2).
Figure 2. Model by which strong TCR signals render γδ lineage cells Notch independent.
Development beyond the β-selection checkpoint requires suppression of E protein function. We hypothesize that T lineage fate and developmental characteristics are determined by the extent to which E protein activity is repressed in a model encompassing graded suppression of E protein function by TCR signals of differing strength. Pre-TCR signals are too weak by themselves to suppress E proteins beyond the threshold required for the αβ lineage differentiation program. They require assistance from Notch to do so, providing an explanation for the Notch-dependence of αβ lineage differentiation to the DP stage. γδ lineage commitment, in contrast, is dictated by strong TCR signals capable of suppressing E protein function beyond the threshold required for γδ lineage commitment, and do so without assistance from Notch. Notch is able to contribute to E protein suppression both through Id3 induction and by ERK-dependent degradation of E proteins; however, our data suggest Notch represses E proteins primarily by inducing their degradation. Lightning bolt size denotes signals of differing strength.
Signal strength and Notch-dependence
The extent or mode of E protein suppression following TCR signaling also appears to influence the relative dependence of αβ and γδ precursors on Notch [34]. Notch molecules are surface receptors involved in cell fate decisions in a wide variety of organisms but the participation of Notch in αβ/γδ lineage choice has been controversial until recently [98–102]. The Zúñiga-Pflücker lab recently clarified this issue. They demonstrated that γδ lineage thymocytes become Notch-independent only upon expression of the γδTCR complex, while αβ lineage precursors are dependent upon Notch signaling throughout the entirety of their differentiation to the DP stage, although the molecular basis for this differential dependence was unclear [17]. Recent evidence from the Murre lab (as well as genetic analysis from Drosophila) suggests interplay between E proteins and the Notch pathway may underlie the differential Notch-dependence of αβ and γδ lineage progenitors [103, 104]. Indeed, we recently determined that the Notch-independence of γδ lineage cells requires Id3-mediated suppression of E protein activity, whereas in αβ lineage precursors, E protein activity is also suppressed by Notch signaling [34]. Altogether, these observations suggest that Notch-dependence is determined by graded reductions in E protein activity mediated by differences in TCR signal strength. Specifically this model suggests that pre-TCR signals partially suppress E protein activity by induction of Id3 but require Notch-ligand interactions to further suppress E protein activity to reach the threshold required for αβ-lineage development (Fig. 2). Notch signaling has been reported to suppress E protein function both by promoting ERK-dependent E protein degradation and through induction of Id3 [105–107]. Conversely, the strong signals that confer Notch-independent differentiation upon γδ-lineage cells are dependent upon Id3 induction alone and are sufficient to suppress E protein activity below the threshold required for γδ-lineage development without assistance from Notch.
7. How do differences in ERK activation influence fate selection?
ERK activation plays a critical role in the interpretation of cellular stimuli that regulate proliferation, differentiation, and survival, with the outcome presumably determined by the extent of ERK activation and the constellation of cellular targets that are phosphorylated [108]. Differential ERK signaling has been shown to be involved in many cell fate decisions in lower organisms as well as in the mammalian immune system (e.g., positive vs. negative selection) [109–113]. In most cases within the context of fate decisions in the immune system, it remains to be demonstrated whether these differences in ERK activation reflect differences in intensity, duration, or both. Indeed, while both the Haks and Hayes studies demonstrated that γδ lineage cells exhibit greater ERK activation, it remains unclear whether this reflects differences in the magnitude of ERK induction, the duration, or both [33, 57]. Nevertheless, there is increasing evidence that signal duration plays an important role in other aspects of T cell development. The kinetic signaling model of CD4/8 lineage commitment incorporates this thinking by hypothesizing that sustained signals support CD4 lineage commitment, whereas CD8 commitment is associated with transient signals [114]. This model received strong support from studies by Sarafova et al., demonstrating that CD4/8 lineage commitment could be altered by manipulating the duration of TCR-coreceptor signals [115]. Regarding the longevity of ERK signaling in particular, the Palmer and Hogquist labs have reported that negative selection is associated with strong but transient ERK activation while positive selection involves weaker but more sustained activation [113, 116].
There are two well characterized models in which the duration of ERK activation has been critically linked to distinct biological outcomes: rat PC12 pheochromocytoma cells and Swiss 3T3 fibroblasts [117]. Stimulation of PC12 cells with epithermal growth factor (EGF) induces transient ERK activation, which results in proliferation, while nerve growth factor (NGF) stimulation produces sustained ERK activation that leads to differentiation into cells resembling sympathetic neurons [118, 119]. Prolonging ERK activation following EGF stimulation transforms the proliferative signal into one inducing differentiation to the neural fate [120]. Conversely, truncating ERK activation following NGF stimulation induces proliferation rather than differentiation [121]. Likewise, treatment of Swiss 3T3 fibrobasts with fibroblast growth factor (FGF) results in transient ERK signaling and quiescence while the prolonged activation of ERK following EGF stimulation induces proliferation [122]. These observations clearly attest to the importance of the duration of ERK signals in determining the biological outcome of an inductive stimulus. However, neither the molecular basis whereby differences in the intensity/duration of ERK activation are regulated nor the way such differences might be interpreted by the cell are fully understood [123]
Regulators of ERK signaling
A recent proteomic analysis provided insights into the complex regulatory networks involved in controlling ERK activation, identifying 143 proteins whose association with ERK differed in transient versus sustained signals [124]. The data suggest that the control of ERK signaling is not focused on a single, particularly important step in the cascade, but is instead distributed at multiple steps along the pathway. Examples affecting several distinct control points will be illustrated in this section. Differences in the extent or kinetics of ERK activation have been shown to result from modulation of the G proteins sitting atop the MAPK cascade. For example, Ras activation is differentially controlled by nucleotide exchange factors like Ras-GRP1, which produce modest but prolonged ERK signaling following Ras activation at the Golgi complex, but produce intense yet brief ERK signals following Ras activation (along with SOS) at the plasma membrane during negative selection [116]. Immediately downstream of the G proteins, lie the MAP3Ks, which can produce signals of differing duration depending upon the particular G protein-MAP3K pair employed [108]. In PC12 cells, Ras/Raf causes transient activation of ERK, while Rap1/B-Raf leads to sustained ERK signals [125]. The duration of MAP3K activation can in turn be controlled by a series of negative regulators, including members of the Sprouty family as well as Raf kinase inhibitory protein (RKIP) [126] [127, 128]. Sprouty 1 has been reported to inhibit ERK activation following TCR signaling in Th1 CD4 T cells [129]. Immediately downstream of the MAP3Ks, are the MAP2Ks, Mek1 and 2, whose ability to support signals differing in duration results from distinct susceptibility to negative regulation [130]. The function of the final effector kinase in the signaling cascade, MAPK/ERK, is also modulated by a host of proteins with which ERK proteins directly interact. These molecules function either to modulate the magnitude or duration of ERK activity or to modulate access to substrates. Examples of the former are dual specificity MAPK phosphatases (MKPs or DUSPs) that dephosphorylate and inactivate ERK or the Ras-GTPase activating protein, Neurofibromin 1 (NF1), which decreases the duration of ERK activity by turning off Ras. Both of these effectors have been implicated in controlling TCR-dependent developmental steps in the thymus [131–133]. An example of the latter class is PEA-15, which blocks ERK-dependent transcription and proliferation by preventing ERK translocation into the nucleus [134]. Sustained ERK activation is accompanied by dissociation of ERK from PEA-15 [124]. Many of the molecular effectors listed above have been shown to regulate the duration of ERK activation in other contexts; however, their involvement in regulating TCR signal strength or duration in the context of αβ/γδ lineage commitment has not been explored.
Cellular responses to differences in ERK intensity/duration
Another fundamental question is how differences in the intensity or duration of ERK activation induce alternate fate choices at a molecular level. It has been proposed that prolonged activation of ERK might result in translocation to different subcellular locations, which could produce alternative developmental outcomes through phosphorylation of distinct substrate pools resident at that site. Two interrelated mechanisms have been proposed to control ERK compartmentalization, scaffold targeting and dimerization. Scaffold proteins assemble together all of the components of a particular MAPK signaling cascade (e.g., Raf, Mek, ERK assembled with KSR1), which both facilitates efficient activation of the MAPK pathway and integrates incoming signals in a localized microenvironment [108]. A number of binding proteins that may serve as scaffolds for the ERK pathway have been identified; KSR, MEK partner-1 (MP1), β-arrestins, similar expression to FGF (Sef) and IQGAP [135]. Many of these scaffolds can target the signaling machinery to distinct subcellular locations (e.g., KSR1 to cholesterol rich domains in the plasma membrane, MP1 to endosomes, and Sef to the Golgi complex) [136–138]. KSR is expressed in T cells and has been implicated in controlling the intensity and duration of ERK signaling in thymocytes [139]. KSR1 is targeted to the Golgi complex in thymocytes receiving positive selection signals, but to the plasma membrane in cells undergoing negative selection, and these changes in location are associated with differences in both the intensity and duration of ERK signaling [113, 116]. The basis for these differences in targeting is not understood. Scaffolds are also able to control the subcellular location of ERK by facilitating ERK dimer formation. Interaction of ERK with a number of different scaffolds promotes dimer formation, which is required for retention in the cytosol, interaction with cognate cytosolic substrates, and the induction of proliferation as well as transformation. Conversely, dimer formation does not appear to be necessary for activation of nuclear substrates [140]. It is, therefore, tempting to speculate that ERK dimer-formation might predominate during αβ commitment and the massive proliferative burst that accompanies differentiation to the DP stage. Conversely, γδ commitment may primarily involve ERK monomer signals, as γδ development is believed to involve less extensive proliferation, although this point remains controversial [87, 141].
The Blenis lab has provided additional insights into the molecular basis by which differences in the duration of ERK activation might produce distinct biological outcomes [117]. Using the Swiss 3T3 cell model, it was demonstrated that FGF stimulation resulted in transient ERK signaling and quiescence while the prolonged activation of ERK following EGF stimulation induced proliferation. Using this experimental model system, the Blenis lab has produced evidence in support of the immediate early gene (IEG) sensor model. This model proposes that prolonged ERK activity produces altered biological outcomes by regulating the stability of IEG protein products [122]. The idea is that when ERK activation is transient, it decays prior to synthesis of IEG protein products, which remain unstable and are rapidly degraded (Fig 3A). In contrast, if ERK activation is sustained until IEG protein products are expressed, ERK physically docks with IEG proteins through motifs termed DEF domains (docking site for ERK; FXFP) and stabilizes them, thereby leading to the accumulation of IEG protein products. Accordingly, cells “perceive” a sustained ERK signal as one that results in the accumulation of IEG protein products. Since many of the IEG are in fact transcription factors, their accumulation is able to lead to a second wave of transcriptional activation resulting in induction of “intermediate early genes” such as Fra-1 and Fra-2 [122, 142]. The implication is that this second wave of transcriptional activation is absent or blunted in cells receiving transient signals and this likely plays an important role in dictating the ultimate biological outcome. While the involvement of differential accumulation of IEG in αβ/γδ lineage commitment has not been formally tested, our findings regarding the importance of the IEG, Egr1, and its target Id3 are consistent with this model [33, 34]. However, these finding could be explained by either differences in the amplitude or longevity of ERK induction. The IEG sensor model suggests that the longevity of ERK activation controls the extent of IEG protein accumulation per unit of encoding mRNA and predicts that ERK signals of increased duration should lead to more IEG protein per unit mRNA than transient ERK signals. To test this prediction, we compared the amount of Egr1 protein per unit mRNA in cells adopting the γδ fate to that in cells adopting the αβ fate in our KN6 Tg model. Interestingly, we found that cells committing to the γδ lineage accumulate more Egr1 protein product per unit mRNA than cells committing to the αβ lineage, as predicted if longer ERK signals were stabilizing IEG proteins in γδ lineage cells (Fig. 3B,C). While these findings are consistent with αβ/γδ fate specification being associated with differences in the duration of ERK signaling, this conclusion remains to be rigorously tested.
Figure 3. Signal duration and lineage commitment.
A. ERK signals can differ in amplitude or duration. Transient ERK activation decays prior to synthesis of the protein encoded by immediate early genes (IEG), resulting in their rapid degradation and failure to accumulate. In contrast, during prolonged signals ERK signals persist until IEG protein products are expressed, enabling them to physically dock with ERK. Physical interaction enables ERK to phosphorylate and stabilize the IEG protein so that it accumulates, leading to protein levels disproportionate to the encoding mRNA. B. γδ commitment is associated with disproportionate increases in the IEG protein Egr1. The mRNA and protein levels of Egr1 were measured in thymocytes committing to the γδ lineage (KN6+Lig+) and the αβ lineage (KN6+Lig−) and normalized to control (KN6−). Egr1 protein levels were disproportionately high in cells committing to the γδ lineage, consistent with the notion that γδ lineage commitment involves signals of increased duration. C. γδ lineage commitment is accompanied by ERK signals lasting longer than those occurring in cells committing to the αβ lineage.
8. Concluding remarks
When first proposed, the TCR signal strength model of αβ/γδ lineage commitment provided plausible explanations for the status of TCR gene rearrangements in αβ and γδ lineage cells and the lineage infidelity observed in TCR transgenic and gene-targeted mice. However, many important and controversial questions remain to be addressed, some of which are represented in this issue. Evidence is support of the signal strength model continues to accumulate, and while we think most evidence is consistent with the hypothesis that TCR signals of differing strength are acting instructionally, the possibility that they may be acting stochastically has not been formally eliminated. The basis by which γδTCR complexes are able to transduce signals that are more robust than those of the pre-TCR also remains in question, particularly whether the γδTCR is intrinsically able to transduce stronger signals or requires ligand-engagement to do so. There are notable examples of particular γδTCR specificities requiring ligand, but the determination of whether these examples represent the exception or the rule must wait for identification of more ligands. Our data has implicated the skeletal framework of the ERK-Egr-Id3 pathway as being critical in lineage commitment. Nevertheless, other important pathways remain not only to be identified but also to be integrated in order to provide a comprehensive understanding of how these diverse signaling pathways cooperate to produce differing fates. Signal duration has been shown to be important in fate-determination in a number of other developmental contexts and our early evidence suggests it will be in αβ/γδ lineage commitment as well. Distinguishing these possibilities will require construction of model systems in which the longevity of TCR signaling in general or perhaps ERK in particular can be manipulated and measured at the single cell level in vivo.
Acknowledgments
We thank Dr. Juan-Carlos Zúñiga-Pflücker for reviewing the manuscript and gratefully acknowledge the assistance of the following core facilities of the Fox Chase Cancer Center: Cell Culture, DNA Sequencing, DNA Synthesis, Flow Cytometry, and Laboratory Animal. This work was supported by NIH grants AI081814, AI073920, NIH core grant P01CA06927, Center grant #P30-DK-50306, and an appropriation from the Commonwealth of Pennsylvania. SYL and JS were supported by the Greenwalt Fellowship and NIH T32 CA009035 training grant, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayday AC. [gamma][delta] cells: a right time and a right place for a conserved third way of protection. Annu Rev Immunol. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 2.Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol. 2002;2:336–45. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 3.Salerno A, Dieli F. Role of gamma delta T lymphocytes in immune response in humans and mice. Critical Reviews in Immunology. 1998;18:327–57. doi: 10.1615/critrevimmunol.v18.i4.30. [DOI] [PubMed] [Google Scholar]
- 4.King DP, Hyde DM, Jackson KA, Novosad DM, Ellis TN, Putney L, et al. Cutting edge: protective response to pulmonary injury requires gamma delta T lymphocytes. Journal of Immunology. 1999;162:5033–6. [PubMed] [Google Scholar]
- 5.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, et al. Regulation of Cutaneous Malignancy by {gamma}{delta} T Cells. Science. 2001;20:20. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc Natl Acad Sci U S A. 2002;99:14338–43. doi: 10.1073/pnas.212290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharp LL, Jameson JM, Cauvi G, Havran WL. Dendritic epidermal T cells regulate skin homeostasis through local production of insulin-like growth factor 1. Nat Immunol. 2005;6:73–9. doi: 10.1038/ni1152. [DOI] [PubMed] [Google Scholar]
- 8.Wiest DL, Berger MA, Carleton M. Control of early thymocyte development by the pre-T cell receptor complex: A receptor without a ligand? Seminars in Immunology. 1999;11:251–62. doi: 10.1006/smim.1999.0181. [DOI] [PubMed] [Google Scholar]
- 9.Wilson A, de Villartay JP, MacDonald HR. T cell receptor delta gene rearrangement and T early alpha (TEA) expression in immature alpha beta lineage thymocytes: implications for alpha beta/gamma delta lineage commitment. Immunity. 1996;4:37–45. doi: 10.1016/s1074-7613(00)80296-4. [DOI] [PubMed] [Google Scholar]
- 10.Capone M, Hockett RD, Jr, Zlotnik A. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) Pro-T thymocytes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12522–7. doi: 10.1073/pnas.95.21.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak F, Tourigny M, Schatz DG, Petrie HT. Characterization of TCR gene rearrangements during adult murine T cell development. Journal of Immunology. 1999;162:2575–80. [PubMed] [Google Scholar]
- 12.Havran WL, Allison JP. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. 1988;335:443–5. doi: 10.1038/335443a0. [DOI] [PubMed] [Google Scholar]
- 13.Ito K, Bonneville M, Takagaki Y, Nakanishi N, Kanagawa O, Krecko EG, et al. Different gamma delta T-cell receptors are expressed on thymocytes at different stages of development. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:631–5. doi: 10.1073/pnas.86.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carding SR, Kyes S, Jenkinson EJ, Kingston R, Bottomly K, Owen JJ, et al. Developmentally regulated fetal thymic and extrathymic T-cell receptor gamma delta gene expression. Genes Dev. 1990;4:1304–15. doi: 10.1101/gad.4.8.1304. [DOI] [PubMed] [Google Scholar]
- 15.Shortman K, Wu L, Kelly KA, Scollay R. The beginning and the end of the development of TCR gamma delta cells in the thymus. Current Topics in Microbiology & Immunology. 1991;173:71–80. [PubMed] [Google Scholar]
- 16.Petrie HT, Scollay R, Shortman K. Commitment to the T cell receptor-alpha beta or -gamma delta lineages can occur just prior to the onset of CD4 and CD8 expression among immature thymocytes. European Journal of Immunology. 1992;22:2185–8. doi: 10.1002/eji.1830220836. [DOI] [PubMed] [Google Scholar]
- 17.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity. 2006;25:105–16. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Dudley EC, Petrie HT, Shah LM, Owen MJ, Hayday AC. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1994;1:83–93. doi: 10.1016/1074-7613(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 19.Dudley EC, Girardi M, Owen MJ, Hayday AC. Alpha beta and gamma delta T cells can share a late common precursor. Current Biology. 1995;5:659–69. doi: 10.1016/s0960-9822(95)00131-x. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman ES, Passoni L, Crompton T, Leu TM, Schatz DG, Koff A, et al. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes & Development. 1996;10:948–62. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 21.Fehling HJ, von Boehmer H. Early alpha beta T cell development in the thymus of normal and genetically altered mice. Current Opinion in Immunology. 1997;9:263–75. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- 22.Berger MA, Dave V, Rhodes MR, Bosma GC, Bosma MJ, Kappes DJ, et al. Subunit composition of pre-T cell receptor complexes expressed by primary thymocytes: CD3 delta is physically associated but not functionally required. Journal of Experimental Medicine. 1997;186:1461–7. doi: 10.1084/jem.186.9.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groettrup M, von Boehmer H. A role for a pre-T-cell receptor in T-cell development. Immunology Today. 1993;14:610–4. doi: 10.1016/0167-5699(93)90201-U. [DOI] [PubMed] [Google Scholar]
- 24.Irving BA, Alt FW, Killeen N. Thymocyte development in the absence of pre-T cell receptor extracellular immunoglobulin domains. Science. 1998;280:905–8. doi: 10.1126/science.280.5365.905. [DOI] [PubMed] [Google Scholar]
- 25.Yamasaki S, Ishikawa E, Sakuma M, Ogata K, Sakata-Sogawa K, Hiroshima M, et al. Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat Immunol. 2006;7:67–75. doi: 10.1038/ni1290. [DOI] [PubMed] [Google Scholar]
- 26.Aifantis I, Buer J, von Boehmer H, Azogui O. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus [published erratum appears in Immunity 1997 Dec;7(6):following 895] Immunity. 1997;7:601–7. doi: 10.1016/s1074-7613(00)80381-7. [DOI] [PubMed] [Google Scholar]
- 27.Kruisbeek AM, Haks MC, Carleton M, Michie AM, Zuniga-Pflucker JC, Wiest DL. Branching out to gain control: how the pre-TCR is linked to multiple functions. Immunology Today. 2000;21:637–44. doi: 10.1016/s0167-5699(00)01744-8. [DOI] [PubMed] [Google Scholar]
- 28.Kreslavsky T, Garbe AI, Krueger A, von Boehmer H. T cell receptor-instructed alphabeta versus gammadelta lineage commitment revealed by single-cell analysis. J Exp Med. 2008;205:1173–86. doi: 10.1084/jem.20072425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pennington DJ, Silva-Santos B, Shires J, Theodoridis E, Pollitt C, Wise EL, et al. The inter-relatedness and interdependence of mouse T cell receptor gammadelta+ and alphabeta+ cells. Nat Immunol. 2003;4:991–8. doi: 10.1038/ni979. [DOI] [PubMed] [Google Scholar]
- 30.Silva-Santos B, Pennington DJ, Hayday AC. Lymphotoxin-mediated regulation of gammadelta cell differentiation by alphabeta T cell progenitors. Science. 2005;307:925–8. doi: 10.1126/science.1103978. [DOI] [PubMed] [Google Scholar]
- 31.Passoni L, Hoffman ES, Kim S, Crompton T, Pao W, Dong MQ, et al. Intrathymic delta selection events in gammadelta cell development. Immunity. 1997;7:83–95. doi: 10.1016/s1074-7613(00)80512-9. [DOI] [PubMed] [Google Scholar]
- 32.Pereira P, Zijlstra M, McMaster J, Loring JM, Jaenisch R, Tonegawa S. Blockade of transgenic gamma delta T cell development in beta 2-microglobulin deficient mice. EMBO Journal. 1992;11:25–31. doi: 10.1002/j.1460-2075.1992.tb05023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, et al. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, et al. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–75. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zorbas M, Scollay R. Development of gamma delta T cells in the adult murine thymus. Eur J Immunol. 1993;23:1655–60. doi: 10.1002/eji.1830230739. [DOI] [PubMed] [Google Scholar]
- 36.Kelly KA, Pearse M, Lefrancois L, Scollay R. Emigration of selected subsets of gamma delta + T cells from the adult murine thymus. Int Immunol. 1993;5:331–5. doi: 10.1093/intimm/5.4.331. [DOI] [PubMed] [Google Scholar]
- 37.Tough DF, Sprent J. Lifespan of gamma/delta T cells. J Exp Med. 1998;187:357–65. doi: 10.1084/jem.187.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, et al. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–3. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 39.MacDonald HR, Wilson A. The role of the T-cell receptor (TCR) in alpha beta/gamma delta lineage commitment: clues from intracellular TCR staining. Immunological Reviews. 1998;165:87–94. doi: 10.1111/j.1600-065x.1998.tb01232.x. [DOI] [PubMed] [Google Scholar]
- 40.Lauritsen JP, Haks MC, Lefebvre JM, Kappes DJ, Wiest DL. Recent insights into the signals that control alphabeta/gammadelta-lineage fate. Immunol Rev. 2006;209:176–90. doi: 10.1111/j.0105-2896.2006.00349.x. [DOI] [PubMed] [Google Scholar]
- 41.Hayday AC, Barber DF, Douglas N, Hoffman ES. Signals involved in gamma/delta T cell versus alpha/beta T cell lineage commitment. Semin Immunol. 1999;11:239–49. doi: 10.1006/smim.1999.0180. [DOI] [PubMed] [Google Scholar]
- 42.Kang J, Baker J, Raulet DH. Evidence that productive rearrangements of TCR gamma genes influence the commitment of progenitor cells to differentiate into alpha beta or gamma delta T cells. European Journal of Immunology. 1995;25:2706–9. doi: 10.1002/eji.1830250946. [DOI] [PubMed] [Google Scholar]
- 43.Livak F, Petrie HT, Crispe IN, Schatz DG. In-frame TCR delta gene rearrangements play a critical role in the alpha beta/gamma delta T cell lineage decision. Immunity. 1995;2:617–27. doi: 10.1016/1074-7613(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 44.Burtrum DB, Kim S, Dudley EC, Hayday AC, Petrie HT. TCR gene recombination and alpha beta-gamma delta lineage divergence: productive TCR-beta rearrangement is neither exclusive nor preclusive of gamma delta cell development. Journal of Immunology. 1996;157:4293–6. [PubMed] [Google Scholar]
- 45.Mertsching E, Ceredig R. T cell receptor-gamma, delta-expressing fetal mouse thymocytes are generated without T cell receptor V beta selection. European Journal of Immunology. 1996;26:804–10. doi: 10.1002/eji.1830260412. [DOI] [PubMed] [Google Scholar]
- 46.Mertsching E, Wilson A, MacDonald HR, Ceredig R. T cell receptor alpha gene rearrangement and transcription in adult thymic gamma delta cells. European Journal of Immunology. 1997;27:389–96. doi: 10.1002/eji.1830270208. [DOI] [PubMed] [Google Scholar]
- 47.Aifantis I, Azogui O, Feinberg J, Saint-Ruf C, Buer J, von Boehmer H. On the role of the pre-T cell receptor in alphabeta versus gammadelta T lineage commitment. Immunity. 1998;9:649–55. doi: 10.1016/s1074-7613(00)80662-7. [DOI] [PubMed] [Google Scholar]
- 48.Bonneville M, Ishida I, Mombaerts P, Katsuki M, Verbeek S, Berns A, et al. Blockage of alpha beta T-cell development by TCR gamma delta transgenes. Nature. 1989;342:931–4. doi: 10.1038/342931a0. [DOI] [PubMed] [Google Scholar]
- 49.Dent AL, Matis LA, Hooshmand F, Widacki SM, Bluestone JA, Hedrick SM. Self-reactive gamma delta T cells are eliminated in the thymus. Nature. 1990;343:714–9. doi: 10.1038/343714a0. [DOI] [PubMed] [Google Scholar]
- 50.Bruno L, Fehling HJ, von Boehmer H. The alpha beta T cell receptor can replace the gamma delta receptor in the development of gamma delta lineage cells. Immunity. 1996;5:343–52. doi: 10.1016/s1074-7613(00)80260-5. [DOI] [PubMed] [Google Scholar]
- 51.Terrence K, Pavlovich CP, Matechak EO, Fowlkes BJ. Premature expression of T cell receptor (TCR)alphabeta suppresses TCRgammadelta gene rearrangement but permits development of gammadelta lineage T cells. Journal of Experimental Medicine. 2000;192:537–48. doi: 10.1084/jem.192.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang J, Volkmann A, Raulet DH. Evidence that gammadelta versus alphabeta T cell fate determination is initiated independently of T cell receptor signaling. Journal of Experimental Medicine. 2001;193:689–98. doi: 10.1084/jem.193.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maki K, Sunaga S, Ikuta K. The V-J recombination of T cell receptor-gamma genes is blocked in interleukin-7 receptor-deficient mice. Journal of Experimental Medicine. 1996;184:2423–7. doi: 10.1084/jem.184.6.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perumal NB, Kenniston TW, Jr, Tweardy DJ, Dyer KF, Hoffman R, Peschon J, et al. TCR-gamma genes are rearranged but not transcribed in IL-7R alpha-deficient mice. Journal of Immunology. 1997;158:5744–50. [PubMed] [Google Scholar]
- 55.Kang J, Coles M, Raulet DH. Defective development of gamma/delta T cells in interleukin 7 receptor-deficient mice is due to impaired expression of T cell receptor gamma genes. Journal of Experimental Medicine. 1999;190:973–82. doi: 10.1084/jem.190.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayes SM, Shores EW, Love PE. An architectural perspective on signaling by the pre-, alphabeta and gammadelta T cell receptors. Immunol Rev. 2003;191:28–37. doi: 10.1034/j.1600-065x.2003.00011.x. [DOI] [PubMed] [Google Scholar]
- 57.Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 2005;22:583–93. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 58.Borst J, Jacobs H, Brouns G. Composition and function of T-cell receptor and B-cell receptor complexes on precursor lymphocytes. Current Opinion in Immunology. 1996;8:181–90. doi: 10.1016/s0952-7915(96)80056-2. [DOI] [PubMed] [Google Scholar]
- 59.Hayes SM, Love PE. Distinct structure and signaling potential of the gamma delta TCR complex. Immunity. 2002;16:827–38. doi: 10.1016/s1074-7613(02)00320-5. [DOI] [PubMed] [Google Scholar]
- 60.Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Havran WL, Chien YH, Allison JP. Recognition of self antigens by skin-derived T cells with invariant gamma delta antigen receptors. Science. 1991;252:1430–2. doi: 10.1126/science.1828619. [DOI] [PubMed] [Google Scholar]
- 62.Xiong N, Kang C, Raulet DH. Positive selection of dendritic epidermal gammadelta T cell precursors in the fetal thymus determines expression of skin-homing receptors. Immunity. 2004;21:121–31. doi: 10.1016/j.immuni.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 63.Boyden LM, Lewis JM, Barbee SD, Bas A, Girardi M, Hayday AC, et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nature Genetics. 2008;40:656–62. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wells FB, Gahm SJ, Hedrick SM, Bluestone JA, Dent A, Matis LA. Requirement for positive selection of gamma delta receptor-bearing T cells. Science. 1991;253:903–5. doi: 10.1126/science.1831565. [DOI] [PubMed] [Google Scholar]
- 65.Schweighoffer E, Fowlkes BJ. Positive selection is not required for thymic maturation of transgenic gamma delta T cells. Journal of Experimental Medicine. 1996;183:2033–41. doi: 10.1084/jem.183.5.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams EJ, Strop P, Shin S, Chien YH, Garcia KC. An autonomous CDR3delta is sufficient for recognition of the nonclassical MHC class I molecules T10 and T22 by gammadelta T cells. Nature Immunology. 2008;9:777–84. doi: 10.1038/ni.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayes SM, Love PE. A retrospective on the requirements for gammadelta T-cell development. Immunol Rev. 2007;215:8–14. doi: 10.1111/j.1600-065X.2006.00476.x. [DOI] [PubMed] [Google Scholar]
- 68.Sommers CL, Samelson LE, Love PE. LAT: a T lymphocyte adapter protein that couples the antigen receptor to downstream signaling pathways. Bioessays. 2004;26:61–7. doi: 10.1002/bies.10384. [DOI] [PubMed] [Google Scholar]
- 69.Zhang W, Sommers CL, Burshtyn DN, Stebbins CC, DeJarnette JB, Trible RP, et al. Essential role of LAT in T cell development. Immunity. 1999;10:323–32. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]
- 70.Nunez-Cruz S, Aguado E, Richelme S, Chetaille B, Mura AM, Richelme M, et al. LAT regulates gammadelta T cell homeostasis and differentiation. Nat Immunol. 2003;4:999–1008. doi: 10.1038/ni977. [DOI] [PubMed] [Google Scholar]
- 71.Sommers CL, Park CS, Lee J, Feng C, Fuller CL, Grinberg A, et al. A LAT mutation that inhibits T cell development yet induces lymphoproliferation. Science. 2002;296:2040–3. doi: 10.1126/science.1069066. [DOI] [PubMed] [Google Scholar]
- 72.Aguado E, Richelme S, Nunez-Cruz S, Miazek A, Mura AM, Richelme M, et al. Induction of T helper type 2 immunity by a point mutation in the LAT adaptor. Science. 2002;296:2036–40. doi: 10.1126/science.1069057. [DOI] [PubMed] [Google Scholar]
- 73.Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, et al. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–72. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 74.Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–43. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 75.Lin LL, Wartmann M, Lin AY, Knopf JL, Seth A, Davis RJ. cPLA2 is phosphorylated and activated by MAP kinase. Cell. 1993;72:269–78. doi: 10.1016/0092-8674(93)90666-e. [DOI] [PubMed] [Google Scholar]
- 76.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–44. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelly K, Siebenlist U. Immediate-early genes induced by antigen receptor stimulation. Curr Opin Immunol. 1995;7:327–32. doi: 10.1016/0952-7915(95)80106-5. [DOI] [PubMed] [Google Scholar]
- 78.Miyazaki T. Two distinct steps during thymocyte maturation from CD4−CD8− to CD4+CD8+ distinguished in the early growth response (Egr)-1 transgenic mice with a recombinase-activating gene-deficient background. Journal of Experimental Medicine. 1997;186:877–85. doi: 10.1084/jem.186.6.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carleton M, Haks MC, Smeele SA, Jones A, Belkowski SM, Berger MA, et al. Early growth response transcription factors are required for development of CD4(-)CD8(-) thymocytes to the CD4(+)CD8(+) stage. Journal of Immunology. 2002;168:1649–58. doi: 10.4049/jimmunol.168.4.1649. [DOI] [PubMed] [Google Scholar]
- 80.Xi H, Kersh GJ. Early growth response gene 3 regulates thymocyte proliferation during the transition from CD4−CD8− to CD4+CD8+ J Immunol. 2004;172:964–71. doi: 10.4049/jimmunol.172.2.964. [DOI] [PubMed] [Google Scholar]
- 81.Liu C, Rangnekar VM, Adamson E, Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Therapy. 1998;5:3–28. [PubMed] [Google Scholar]
- 82.Shao H, Kono DH, Chen LY, Rubin EM, Kaye J. Induction of the early growth response (Egr) family of transcription factors during thymic selection. Journal of Experimental Medicine. 1997;185:731–44. doi: 10.1084/jem.185.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–71. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 84.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–26. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 85.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 86.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of e2a protein activity. J Exp Med. 2001;194:733–46. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 88.Blom B, Heemskerk MH, Verschuren MC, van Dongen JJ, Stegmann AP, Bakker AQ, et al. Disruption of alpha beta but not of gamma delta T cell development by overexpression of the helix-loop-helix protein Id3 in committed T cell progenitors. EMBO Journal. 1999;18:2793–802. doi: 10.1093/emboj/18.10.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gammadelta lineage during thymopoiesis. J Immunol. 2009;182:5306–16. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gammadelta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 184:1268–79. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL. Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent “innate” gammadelta T cells. PLoS One. 5:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–86. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 93.Bain G, Romanow WJ, Albers K, Havran WL, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. Journal of Experimental Medicine. 1999;189:289–300. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barndt R, Dai MF, Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. Journal of Immunology. 1999;163:3331–43. [PubMed] [Google Scholar]
- 95.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–26. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–85. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. Embo J. 2004;23:202–11. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–53. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 99.Garcia-Peydro M, de Yebenes VG, Toribio ML. Sustained Notch1 signaling instructs the earliest human intrathymic precursors to adopt a gammadelta T-cell fate in fetal thymus organ culture. Blood. 2003;102:2444–51. doi: 10.1182/blood-2002-10-3261. [DOI] [PubMed] [Google Scholar]
- 100.Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, et al. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–57. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, et al. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997;88:833–43. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 102.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta Lineage Thymocytes. Immunity. 2002;16:869–79. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 103.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–42. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–81. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nie L, Xu M, Vladimirova A, Sun XH. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. Embo J. 2003;22:5780–92. doi: 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ordentlich P, Lin A, Shen CP, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, et al. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–9. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reynaud-Deonauth S, Zhang H, Afouda A, Taillefert S, Beatus P, Kloc M, et al. Notch signaling is involved in the regulation of Id3 gene transcription during Xenopus embryogenesis. Differentiation. 2002;69:198–208. doi: 10.1046/j.1432-0436.2002.690413.x. [DOI] [PubMed] [Google Scholar]
- 108.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–12. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 109.Sharp LL, Schwarz DA, Bott CM, Marshall CJ, Hedrick SM. The influence of the MAPK pathway on T cell lineage commitment. Immunity. 1997;7:609–18. doi: 10.1016/s1074-7613(00)80382-9. [DOI] [PubMed] [Google Scholar]
- 110.Alberola-Ila J, Hernandez-Hoyos G. The Ras/MAPK cascade and the control of positive selection. Immunol Rev. 2003;191:79–96. doi: 10.1034/j.1600-065x.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 111.Alberola-Ila J, Forbush KA, Seger R, Krebs EG, Perlmutter RM. Selective requirement for MAP kinase activation in thymocyte differentiation. Nature. 1995;373:620–3. doi: 10.1038/373620a0. [DOI] [PubMed] [Google Scholar]
- 112.Mariathasan S, Zakarian A, Bouchard D, Michie AM, Zuniga-Pflucker JC, Ohashi PS. Duration and strength of extracellular signal-regulated kinase signals are altered during positive versus negative thymocyte selection. J Immunol. 2001;167:4966–73. doi: 10.4049/jimmunol.167.9.4966. [DOI] [PubMed] [Google Scholar]
- 113.McNeil LK, Starr TK, Hogquist KA. A requirement for sustained ERK signaling during thymocyte positive selection in vivo. Proc Natl Acad Sci U S A. 2005;102:13574–9. doi: 10.1073/pnas.0505110102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singer A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr Opin Immunol. 2002;14:207–15. doi: 10.1016/s0952-7915(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 115.Sarafova SD, Erman B, Yu Q, Van Laethem F, Guinter T, Sharrow SO, et al. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 2005;23:75–87. doi: 10.1016/j.immuni.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 116.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–9. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 117.Murphy LO, Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem Sci. 2006;31:268–75. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 118.Heasley LE, Johnson GL. The beta-PDGF receptor induces neuronal differentiation of PC12 cells. Mol Biol Cell. 1992;3:545–53. doi: 10.1091/mbc.3.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nguyen TT, Scimeca JC, Filloux C, Peraldi P, Carpentier JL, Van Obberghen E. Co-regulation of the mitogen-activated protein kinase, extracellular signal-regulated kinase 1, and the 90-kDa ribosomal S6 kinase in PC12 cells. Distinct effects of the neurotrophic factor, nerve growth factor, and the mitogenic factor, epidermal growth factor. J Biol Chem. 1993;268:9803–10. [PubMed] [Google Scholar]
- 120.Traverse S, Seedorf K, Paterson H, Marshall CJ, Cohen P, Ullrich A. EGF triggers neuronal differentiation of PC12 cells that overexpress the EGF receptor. Curr Biol. 1994;4:694–701. doi: 10.1016/s0960-9822(00)00154-8. [DOI] [PubMed] [Google Scholar]
- 121.Schlessinger J, Bar-Sagi D. Activation of Ras and other signaling pathways by receptor tyrosine kinases. Cold Spring Harb Symp Quant Biol. 1994;59:173–9. doi: 10.1101/sqb.1994.059.01.021. [DOI] [PubMed] [Google Scholar]
- 122.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–64. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 123.Yasuda T, Kurosaki T. Regulation of lymphocyte fate by Ras/ERK signals. Cell Cycle. 2008;7:3634–40. doi: 10.4161/cc.7.23.7103. [DOI] [PubMed] [Google Scholar]
- 124.von Kriegsheim A, Baiocchi D, Birtwistle M, Sumpton D, Bienvenut W, Morrice N, et al. Cell fate decisions are specified by the dynamic ERK interactome. Nat Cell Biol. 2009;11:1458–64. doi: 10.1038/ncb1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kao S, Jaiswal RK, Kolch W, Landreth GE. Identification of the mechanisms regulating the differential activation of the mapk cascade by epidermal growth factor and nerve growth factor in PC12 cells. J Biol Chem. 2001;276:18169–77. doi: 10.1074/jbc.M008870200. [DOI] [PubMed] [Google Scholar]
- 126.Brady SC, Coleman ML, Munro J, Feller SM, Morrice NA, Olson MF. Sprouty2 association with B-Raf is regulated by phosphorylation and kinase conformation. Cancer Res. 2009;69:6773–81. doi: 10.1158/0008-5472.CAN-08-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hanafusa H, Matsumoto K, Nishida E. Regulation of ERK activity duration by Sprouty contributes to dorsoventral patterning. Nat Cell Biol. 2009;11:106–9. doi: 10.1038/ncb1820. [DOI] [PubMed] [Google Scholar]
- 128.Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–30. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 129.Choi H, Cho SY, Schwartz RH, Choi K. Dual effects of Sprouty1 on TCR signaling depending on the differentiation state of the T cell. J Immunol. 2006;176:6034–45. doi: 10.4049/jimmunol.176.10.6034. [DOI] [PubMed] [Google Scholar]
- 130.Catalanotti F, Reyes G, Jesenberger V, Galabova-Kovacs G, de Matos Simoes R, Carugo O, et al. A Mek1-Mek2 heterodimer determines the strength and duration of the Erk signal. Nat Struct Mol Biol. 2009;16:294–303. doi: 10.1038/nsmb.1564. [DOI] [PubMed] [Google Scholar]
- 131.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–13. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 132.Bettini ML, Kersh GJ. MAP kinase phosphatase activity sets the threshold for thymocyte positive selection. Proc Natl Acad Sci U S A. 2007;104:16257–62. doi: 10.1073/pnas.0705321104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ingram DA, Zhang L, McCarthy J, Wenning MJ, Fisher L, Yang FC, et al. Lymphoproliferative defects in mice lacking the expression of neurofibromin: functional and biochemical consequences of Nf1 deficiency in T-cell development and function. Blood. 2002;100:3656–62. doi: 10.1182/blood-2002-03-0734. [DOI] [PubMed] [Google Scholar]
- 134.Formstecher E, Ramos JW, Fauquet M, Calderwood DA, Hsieh JC, Canton B, et al. PEA-15 mediates cytoplasmic sequestration of ERK MAP kinase. Dev Cell. 2001;1:239–50. doi: 10.1016/s1534-5807(01)00035-1. [DOI] [PubMed] [Google Scholar]
- 135.Yao Z, Seger R. The ERK signaling cascade--views from different subcellular compartments. Biofactors. 2009;35:407–16. doi: 10.1002/biof.52. [DOI] [PubMed] [Google Scholar]
- 136.Matheny SA, Chen C, Kortum RL, Razidlo GL, Lewis RE, White MA. Ras regulates assembly of mitogenic signalling complexes through the effector protein IMP. Nature. 2004;427:256–60. doi: 10.1038/nature02237. [DOI] [PubMed] [Google Scholar]
- 137.Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3:803–14. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 138.Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 139.Laurent MN, Ramirez DM, Alberola-Ila J. Kinase suppressor of Ras couples Ras to the ERK cascade during T cell development. J Immunol. 2004;173:986–92. doi: 10.4049/jimmunol.173.2.986. [DOI] [PubMed] [Google Scholar]
- 140.Casar B, Pinto A, Crespo P. Essential role of ERK dimers in the activation of cytoplasmic but not nuclear substrates by ERK-scaffold complexes. Mol Cell. 2008;31:708–21. doi: 10.1016/j.molcel.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 141.Prinz I, Sansoni A, Kissenpfennig A, Ardouin L, Malissen M, Malissen B. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat Immunol. 2006;7:995–1003. doi: 10.1038/ni1371. [DOI] [PubMed] [Google Scholar]
- 142.Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol Cell Biol. 2004;24:144–53. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]