Abstract
Study on molecular mechanism of conformational reorientation of RXR-alpha ligand binding domain is presented. We employed CABS - a reduced model of protein dynamics to model folding pathways of binding 9-cis retinoic acid to apo-RXR molecule and TRAP220 peptide fragment to the holo form. Based on obtained results we also propose a sequential model of RXR activation by 9-cis retinoic acid and TRAP220 coactivator. Methodology presented here may be used for investigation of binding pathways of other NR/hormone/cofactor sets.
Keywords: protein interactions, flexible docking, molecular modeling, nuclear receptors
1. Introduction
Nuclear hormone receptors (NR) are ligand-activated transcription factors regulating the expression of target genes [1]. They play widespread and important roles in development, metabolism, homeostasis and disease [2, 3]. Retinoid X Receptor (RXR) occupies a central position among other nuclear receptors and plays a crucial role as universal heterodimerization partner for many other members of NR superfamily [4]. Its natural ligand - 9-cis reti noic acid activates RXR by binding to a pocket located in RXR’s ligand binding domain (LBD). Investigation of crystallographic structures of holo and apo forms of RXR-alpha’s LBD show significant structural reorientation of the receptor upon ligand binding. Usually nuclear receptors are investigated in respect of their interaction only with primary ligands [5–7], but they also form complexes with other molecules. In holoform NRs bind with various cell-specific co-activators, which link receptor with the RNA polymerase II – a gear in transcriptional mechanism, while in apo-form NRs form complexes with co-repressors and act as transcriptional suppressors. Abundance of functions of different nuclear receptors and structural similarity between them at the same time makes the NR class very promising pharmacological target.
2. Materials and methods
Two crystallographic structures of RXR-alpha LBD were analyzed. 1lbd is a structure of monomeric apo form [8] and 1xdk holds the structure of holo-RXR-alpha/holo-RAR-beta heterodimer, liganded by 9-cis retinoic acid and bound to 13 amino acid long peptide - TRAP220 coactivator fragment containing LXXLL motif [9]. RXR residues were divided into four separate classes: ligand active site, cofactor active site, structurally conserved residues and others (Fig. 1). Residue was marked as ligand active site if any of its heavy atoms was located within 4.5 Å radius from any of ligand’s heavy atoms. Analogues procedure was used to find cofactor active site. Global distance test (GDT) was calculated on 1lbd and 1xdk structures to find fragments that are highly structurally conserved in both structures. GDT cutoff was set to 0.35 Å in order to match the accuracy of casting the structures to the lattice used in CABS model. Residues previously marked as either ligand or cofactor active site were excluded from the conserved residues set. Table 1 presents comparison between these residue classes. CABS is a reduced model of protein dynamics and thermodynamics, which proved to be extremely effective in various modeling tasks, such as comparative modeling [10, 11], protein fragment reconstruction [12], modeling of folding pathways [13–15] and protein docking [16, 17]. CABS stands for Cα(CA), Cβ(B) and united pseudoatom located in the center of mass of the side chain (S), which are the only interaction centers. Positions of Cα atoms are restricted to simple cubic lattice, with spacing equal to 0.61 Å, while remaining atoms are located off the lattice. Sampling is controlled by Replica Exchange Monte Carlo (REMC) scheme [18]. Force field was derived from statistical analysis of regularities found in known protein structures. It consists of generic terms and biases forcing very flexible Cα-trace to behave in protein-like manner and context-dependant pairwise potentials for side chains’ interactions. CABS has been fully automated and integrated with user-friendly interface and data processing/analysis tools [19, 20] in modeling platform SPMP, commercially available from Selvita Life Sciences Solutions (selvita.com. Command-line version of CABS is available for non-commercial users on our website (biocomp.chem.uw.edu.pl). Detailed description of the CABS model was presented earlier [21].
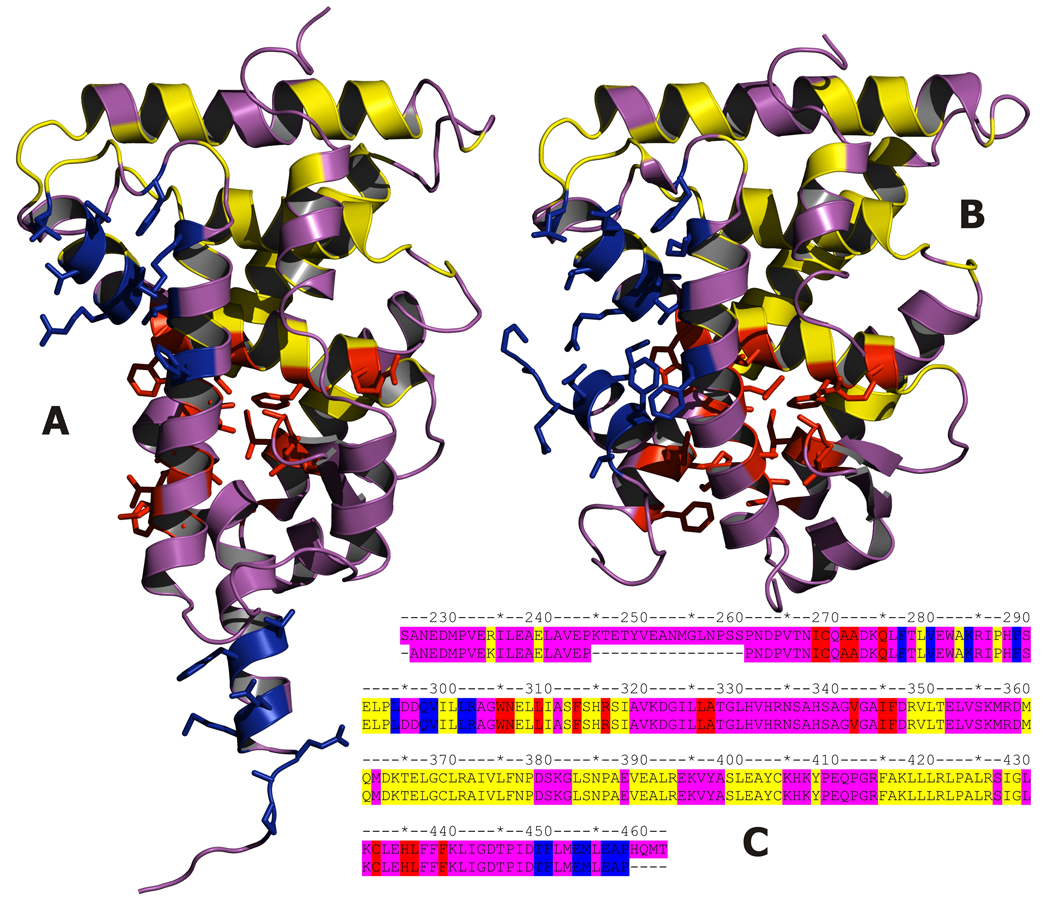
Fig. 1.
Crystal structures of RXR-alpha LBD in apo A) and holo B) forms. Residues in contact with retinoic acid in apo-RXR are presented in red, those in contact with TRAP220 coactivator in apo-RXR are presented in blue. In yellow most structurally conserved residues (rmsd between apo and holo forms less than 0.35Å) C) Alignment of sequences from 1lbd and 1xdk structures, colors as above.
Table 1.
Structural comparison of 1lbd and 1xdk structures.
| RESIDUE SELECTION | NUMBER OF RESIDUES |
RMSD | GDT | |||||
|---|---|---|---|---|---|---|---|---|
| 0.5 | 1.0 | 2.0 | 5.0 | 7.5 | 10.0 | |||
| ALL | 217 | 7.36 | 0.57 | 0.81 | 0.89 | 0.97 | 1.00 | 1.00 |
| CONSERVED REGIONS | 71 | 0.34 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| LIGAND ACTIVE SITE | 19 | 2.97 | 0.53 | 0.74 | 0.84 | 1.00 | 1.00 | 1.00 |
| COFACTOR ACTIVE SITE | 16 | 14.79 | 0.56 | 0.56 | 0.56 | 0.56 | 0.63 | 0.75 |
| OTHER | 111 | 6.28 | 0.28 | 0.68 | 0.83 | 0.97 | 1.00 | 1.00 |
RMSD (root mean square deviation) is defined as square root of averaged distances between corresponding atoms in two sets.
GDT (global distance test) is defined as a number of residues in a subset, for which rmsd is below given threshold, divided by total number of residues.
CABS was used to model five molecular transformations which RXR molecule does or hypothetically may undergo. In all five cases receptor molecule consisted of residues 225–462 (238 residues) and in all simulations but first (where only structure of the receptor was modeled) cofactor molecule consisted of residues 641–651 (11 residues). Secondary structure was assigned to all residues in the following way: DSSP [22] was run on both 1lbd and 1xdk structures, whenever secondary structure assignment for 1lbd agreed with the one for 1xdk it was also assigned to the modeled residue, otherwise secondary structure was set to coil for that residue. All simulations were run with the same force field parameters set, in the same temperature and for the same number of cycles. Brief description of all five runs is presented below.
Run I (reorientation of the receptor in the presence of the ligand)
Starting structure of the receptor was taken from 1lbd. Conformational flexibility of the residues marked as conserved (Fig. 1) was strongly restricted by a network of distance restraints imposed on Ca atoms. Distances were measured in both 1lbd and 1xdk structures and subsequently averaged. Similarly, residues marked as ligand active site were restrained as well, but this time only distances from 1xdk were used. This was done to indirectly reflect the presence of the ligand in the binding pocket, since CABS is so far capable of handling only protein molecules. Rest of the residues was left unrestrained.
Run II (cofactor binding to the receptor-ligand complex)
Starting structure of the receptor was taken from 1xdk. Set of distance restraints imposed on the receptor was the same as in the first run. Initial cofactor conformation was random. Moreover the cofactor molecule was shifted away 15 Å from the receptor surface in respect to the 1xdk structure (all cofactor atoms were shifted by a 15Å -long vector pointing from receptor’s center of gravity (COG) to cofactor’s COG). No structural restraints were imposed on cofactor structure.
Run III (simultaneous reorientation of the receptor in the presence of the ligand and cofactor binding)
Initial structure of the receptor was the same as in the first run and the cofactor structure was the same as in the second run. Identical set of restraints was used as in the first two runs.
Run IV (cofactor binding to the receptor in apo form)
Initial structures of both receptor and cofactor were the same as in the third run, but distance restraints imposed on active site residues were derived from 1lbd.
Run V (holo to apo transformation upon ligand dissociation)
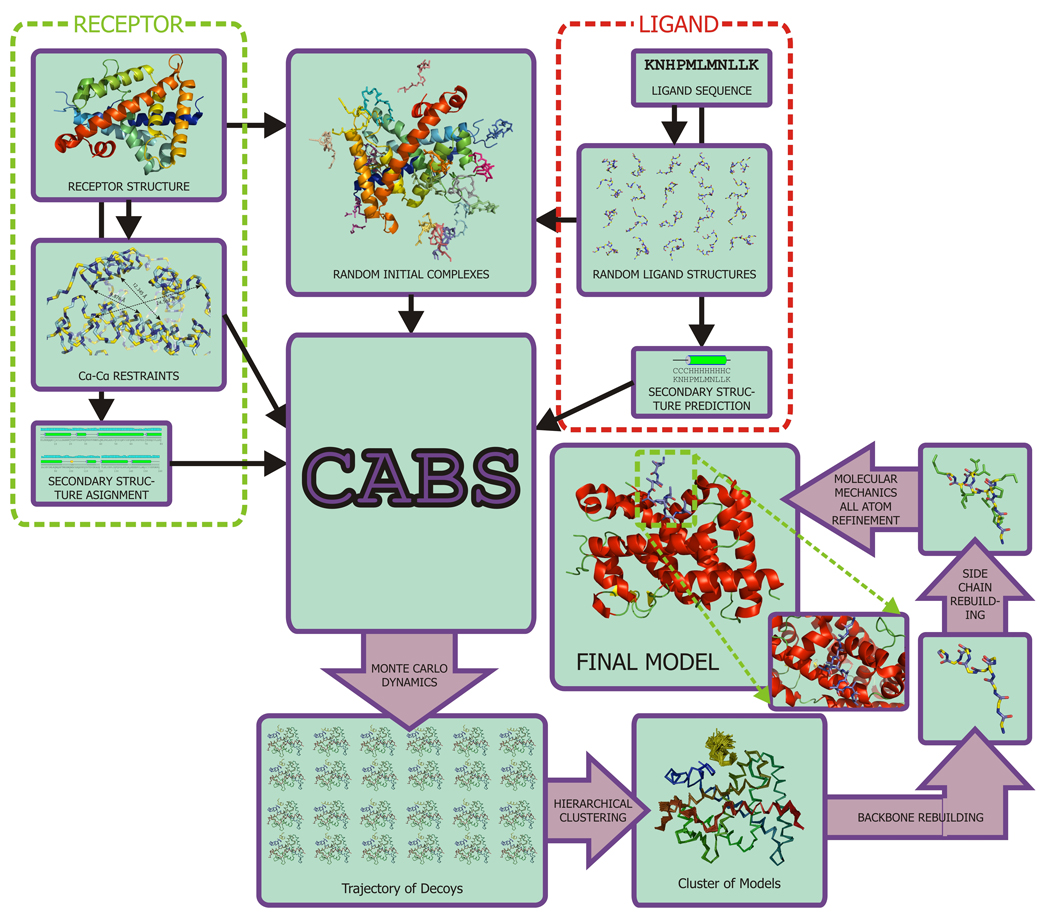
Initial structures of both the receptor and the ligand were taken from 1xdk, but distance restraints imposed on active site residues were derived from 1lbd. Simulation conditions of all five runs are summarized in Table 2. Every run consisted of ten separate iterations differing only by random seed. Every iteration produced 1000 models to sum up to total 10000 models per run. These models were grouped together by their mutual similarity in two-stage hierarchical clustering procedure [23] - first within 1000 models produced in individual iterations, next medoids of the clusters found in the first stage were clustered again. Medoids of top three biggest clusters were further refined: first reconstructed to all-atom representation using BBQ [24] and SCWRL3.0 [25] and energy-minimized in empirical force field Amber99 [26]. Final model for each run was selected according to final energy after the minimization [27]. Scheme of the described methodology is presented in Fig. 2.
Table 2.
Initial conditions in CABS simulations
| RUN | INITIAL STRUCTURE | LIGAND | |
|---|---|---|---|
| COFACTOR | RECEPTOR | ||
| I | apo | - | present |
| II | holo | random | present |
| III | apo | random | present |
| IV | apo | random | absent |
| V | holo | native | absent |
Fig. 2.
Modeling scheme used in current work. Complete process is automated and controlled by user-configured scripts.
3. Results and Discussion
Run 1 (reorientation of the receptor in the presence of the ligand)
The process of receptor reorientation was modeled with high accuracy, as confirmed by rmsd between top-ranked model and 1xdk structure in all categories. Cofactor binding site was modeled with 1.97 Å accuracy in respect to the 1xdk structure, which suggests, that additional to ligand-induced conformational adjustment is required upon binding with the cofactor.
Run 2 (cofactor binding to the receptor-ligand complex)
Rmsd between top-ranked model and 1xdk structure calculated on all residues is below 0.8 Å. Additional adjustment in the cofactor binding site is reflected in rmsd decrease to 0.70 Å in that region, when compared to the model obtained in the first run. This suggests that hormone and cofactor activate the receptor sequentially rather than simultaneously.
Run 3 (simultaneous reorientation of the receptor in the presence of the ligand and cofactor binding)
Although generally low rmsd on the complete structure (2.38 Å), in this run top ranked model had incorrect orientation of the docked cofactor (rmsd on the cofactor after superposition of the receptor with 1xdk structure was over 10 Å). Furthermore, cofactor itself was in wrong conformation (rmsd 2.17 Å in respect to the 1xdk).The shape of the cofactor binding site was deformed as well (rmsd 1.41 Å). This is another premise suggesting sequential binding of the ligand/cofactor molecules.
Run 4 (cofactor binding to the receptor in apo form)
In this case receptor remained in its apo form, which inactivated it for cofactor reception as shown by high rmsd values.
Run 5 (holo to apo transformation upon ligand dissociation)
This case was run to model holo to apo transformation upon ligand dissociation. Indeed shape of both ligand and cofactor active site has changed significantly. Also cofactor structure and location has changed. However, apo-form was not reconstructed completely - final model shows some structural similarities to both apo and holo form (rmsd 5.38 Å and 4.76 Å respectively).
In runs I and II the process of binding the ligand and subsequently the cofactor was modeled with great accuracy. At the same time runs III and IV demonstrate that different sequence of binding leads to wrong conformations of final structures. Rmsd values are presented in Table 3. Initial and final structures from all simulations are shown in Figure 3.
Table 3.
RMSD values calculated on selected models from CABS simulations vs. crystallographic structures of both apo and holo forms of RXR-alpha.
| RUN | REFERENCE STRUCTURE |
1LBD | 1XDK | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RESIDUE SELECTION |
RECEPTOR | CONSERVED | LIGAND ACTIVE SITE |
COFACTOR ACTIVE SITE |
RECEPTOR | CONSERVED | LIGAND ACTIVE SITE |
COFACTOR ACTIVE SITE |
COFACTOR | COFACTOR AFTER RECE- PTOR SUPER- POSITION |
RECEPTOR + COFACTOR |
|
| NUMBER OF RESIDUES |
217 | 71 | 19 | 16 | 217 | 71 | 19 | 16 | 11 | 11 | 228 | |
| 1 | TOP RANKED | 7.23 | 0.50 | 3.00 | 14.40 | 0.79 | 0.36 | 0.32 | 1.97 | - | - | - |
| BEST | 5.34 | 0.43 | 2.83 | 10.57 | 0.58 | 0.31 | 0.32 | 0.96 | - | - | - | |
| LAST | 7.32 | 0.50 | 2.99 | 14.88 | 0.78 | 0.35 | 0.38 | 1.83 | - | - | - | |
| LOW ENERGY | 7.18 | 0.48 | 3.04 | 14.05 | 0.72 | 0.35 | 0.37 | 1.97 | - | - | - | |
| AVERAGE | 7.23 | 0.48 | 3.01 | 14.33 | 0.88 | 0.35 | 0.45 | 2.15 | - | - | - | |
| 2 | TOP RANKED | 7.35 | 0.81 | 2.97 | 14.83 | 0.64 | 0.71 | 0.47 | 0.70 | 0.95 | 1.81 | 0.74 |
| BEST | 4.93 | 0.42 | 2.81 | 10.78 | 0.58 | 0.29 | 0.35 | 0.28 | 0.73 | 1.07 | 0.66 | |
| LAST | 7.38 | 0.80 | 3.04 | 14.83 | 0.64 | 0.70 | 0.42 | 0.63 | 2.17 | 2.96 | 0.91 | |
| LOW ENERGY | 7.34 | 0.43 | 3.00 | 14.65 | 0.88 | 0.31 | 0.45 | 1.86 | 2.75 | 9.84 | 2.31 | |
| AVERAGE | 7.33 | 0.78 | 3.01 | 14.75 | 0.73 | 0.69 | 0.45 | 0.88 | 1.62 | 3.11 | 0.99 | |
| 3 | TOP RANKED | 7.39 | 0.48 | 3.07 | 14.78 | 0.80 | 0.33 | 0.35 | 1.41 | 2.17 | 10.24 | 2.38 |
| BEST | 3.41 | 0.43 | 2.14 | 6.19 | 0.76 | 0.31 | 0.32 | 1.19 | 0.69 | 4.32 | 1.95 | |
| LAST | 7.40 | 0.48 | 3.09 | 14.78 | 0.81 | 0.34 | 0.47 | 1.41 | 2.40 | 10.00 | 2.34 | |
| LOW ENERGY | 7.39 | 0.47 | 3.09 | 14.78 | 0.81 | 0.34 | 0.47 | 1.41 | 2.40 | 10.00 | 2.34 | |
| AVERAGE | 7.30 | 0.48 | 3.02 | 14.65 | 0.95 | 0.35 | 0.44 | 1.78 | 2.57 | 10.28 | 2.46 | |
| 4 | TOP RANKED | 0.78 | 0.33 | 0.23 | 1.20 | 7.25 | 0.42 | 2.99 | 14.57 | 1.67 | 16.39 | 7.80 |
| BEST | 0.69 | 0.33 | 0.23 | 0.96 | 6.22 | 0.40 | 2.59 | 14.24 | 0.75 | 4.17 | 6.20 | |
| LAST | 0.77 | 0.33 | 0.23 | 1.07 | 7.24 | 0.42 | 2.99 | 14.56 | 3.31 | 13.34 | 7.51 | |
| LOW ENERGY | 0.74 | 0.33 | 0.23 | 1.18 | 7.26 | 0.42 | 2.99 | 14.61 | 1.77 | 15.88 | 7.78 | |
| AVERAGE | 0.79 | 0.33 | 0.24 | 1.19 | 7.25 | 0.42 | 2.99 | 14.65 | 2.86 | 15.93 | 7.77 | |
| 5 | TOP RANKED | 5.38 | 0.32 | 0.28 | 10.51 | 4.76 | 0.42 | 2.94 | 7.32 | 2.32 | 14.47 | 5.48 |
| BEST | 4.28 | 0.30 | 0.19 | 6.67 | 3.04 | 0.40 | 2.89 | 1.88 | 0.80 | 2.40 | 3.19 | |
| LAST | 7.72 | 0.30 | 0.19 | 15.11 | 5.20 | 0.42 | 2.97 | 4.16 | 2.47 | 10.51 | 5.61 | |
| LOW ENERGY | 5.91 | 0.30 | 0.19 | 11.82 | 5.30 | 0.42 | 2.97 | 6.58 | 3.60 | 16.25 | 6.23 | |
| AVERAGE | 6.17 | 0.31 | 0.26 | 11.45 | 5.24 | 0.42 | 2.96 | 6.70 | 3.24 | 15.73 | 6.10 | |
TOP RANKED - model selected in clustering/refinement procedure; BEST - lowest values of listed measures found in all models; LAST - last model in CABS trajectory; LOW ENERGY - model with the lowest CABS energy; AVERAGE - mean value of listed measures, averaged over whole trajectory
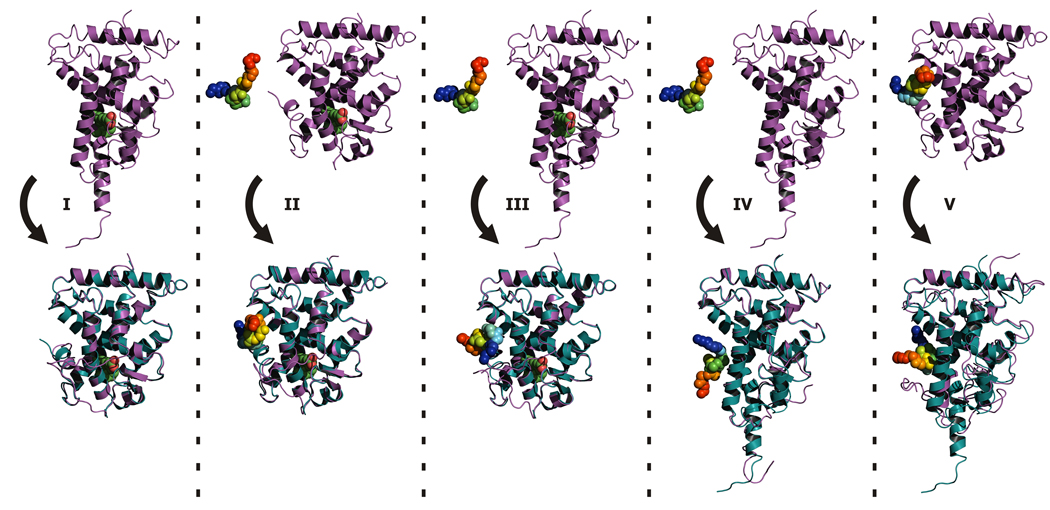
Fig. 3.
Initial and final states of the modeled systems in simulations I–V. In the upper row initial structures of the receptor are shown in purple cartoons. Structure of the cofactor is shown in rainbow spheres. 9-cis retinoic acid - in green. In the bottom row presented are top ranked models (in purple cartoons (receptor) and rainbow spheres (cofactor)) superposed onto crystallographic structures: 1xdk(I–III) and 1lbd (IV–V).
4. Conclusions
We present a methodology for investigation of receptor-cofactor binding mechanisms. Case study included prediction of binding TRAP220 coactivator to RXR-alpha receptor in both holo and apo forms, in simultaneous with the ligand, or sequential order. Also ligand-activated apo-holo transformation was investigated. Obtained results remain in agreement with experimental data (crystallographic structures), therefore similar procedure may be applied to investigation of binding pathways in other NR/hormone/cofactor complexes. We also propose a two-stage sequential mechanism of RXR-alpha activation by 9-cis retinoic acid and TRAP220 coactivator, as this matter still remains unresolved. We hope that methodology presented here may find wide spectrum of applications as it provides fast and inexpensive alternative to experimental techniques.
Acknowledgements
This work was supported by the NIH grant no. 1R01GM081680 and Polish Ministry of Science and Higher Education, grant no. NN301465634. Computational part of this work was done using the computer cluster at the Computing Center of Faculty of Chemistry, University of Warsaw. A commercial version of CABS-based modeling software was used http://www.selvita.com/selvita-protein-modeling-platform.html
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240(4854):889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chawla A, et al. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 4.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 5.Rotkiewicz P, et al. Model of three-dimensional structure of vitamin D receptor and its binding mechanism with 1alpha,25-dihydroxyvitamin D(3) Proteins. 2001;44(3):188–199. doi: 10.1002/prot.1084. [DOI] [PubMed] [Google Scholar]
- 6.Sicinska W, Rotkiewicz P, DeLuca HF. Model of three-dimensional structure of VDR bound with Vitamin D3 analogs substituted at carbon-2. J Steroid Biochem Mol Biol. 2004;89–90(1–5):107–110. doi: 10.1016/j.jsbmb.2004.03.102. [DOI] [PubMed] [Google Scholar]
- 7.Sicinski RR, et al. 2-Ethyl and 2-ethylidene analogues of 1alpha,25-dihydroxy-19-norvitamin D(3): synthesis, conformational analysis, biological activities, and docking to the modeled rVDR ligand binding domain. J Med Chem. 2002;45(16):3366–3380. doi: 10.1021/jm020007m. [DOI] [PubMed] [Google Scholar]
- 8.Bourguet W, et al. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375(6530):377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 9.Pogenberg V, et al. Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J Biol Chem. 2005;280(2):1625–1633. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- 10.Kolinski A, Bujnicki JM. Generalized protein structure prediction based on combination of fold-recognition with de novo folding and evaluation of models. Proteins. 2005;61 Suppl 7:84–90. doi: 10.1002/prot.20723. [DOI] [PubMed] [Google Scholar]
- 11.Kolinski A, Gront D. Comparative modeling without implicit sequence alignments. Bioinformatics. 2007;23(19):2522–2527. doi: 10.1093/bioinformatics/btm380. [DOI] [PubMed] [Google Scholar]
- 12.Boniecki M, et al. Protein fragment reconstruction using various modeling techniques. J Comput Aided Mol Des. 2003;17(11):725–738. doi: 10.1023/b:jcam.0000017486.83645.a0. [DOI] [PubMed] [Google Scholar]
- 13.Kmiecik S, Kolinski A. Characterization of protein-folding pathways by reduced-space modeling. Proc Natl Acad Sci U S A. 2007;104(30):12330–12335. doi: 10.1073/pnas.0702265104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kmiecik S, et al. Denatured proteins and early folding intermediates simulated in a reduced conformational space. Acta Biochim Pol. 2006;53(1):131–144. [PubMed] [Google Scholar]
- 15.Kmiecik S, Kolinski A. Folding pathway of the b1 domain of protein G explored by multiscale modeling. Biophys J. 2008;94(3):726–736. doi: 10.1529/biophysj.107.116095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurcinski M, Kolinski A. Hierarchical modeling of protein interactions. J Mol Model. 2007;13(6–7):691–698. doi: 10.1007/s00894-007-0177-8. [DOI] [PubMed] [Google Scholar]
- 17.Kurcinski M, Kolinski A. Steps towards flexible docking: modeling of three-dimensional structures of the nuclear receptors bound with peptide ligands mimicking co-activators' sequences. J Steroid Biochem Mol Biol. 2007;103(3–5):357–360. doi: 10.1016/j.jsbmb.2006.12.059. [DOI] [PubMed] [Google Scholar]
- 18.Swendsen RH, Wang JS. Replica Monte Carlo simulation of spin glasses. Phys Rev Lett. 1986;57(21):2607–2609. doi: 10.1103/PhysRevLett.57.2607. [DOI] [PubMed] [Google Scholar]
- 19.Gront D, Kolinski A. Utility library for structural bioinformatics. Bioinformatics. 2008;24(4):584–585. doi: 10.1093/bioinformatics/btm627. [DOI] [PubMed] [Google Scholar]
- 20.Gront D, Kolinski A. BioShell--a package of tools for structural biology computations. Bioinformatics. 2006;22(5):621–622. doi: 10.1093/bioinformatics/btk037. [DOI] [PubMed] [Google Scholar]
- 21.Kolinski A. Protein modeling and structure prediction with a reduced representation. Acta Biochim Pol. 2004;51(2):349–371. [PubMed] [Google Scholar]
- 22.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogenbonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 23.Gront D, Kolinski A. HCPM--program for hierarchical clustering of protein models. Bioinformatics. 2005;21(14):3179–3180. doi: 10.1093/bioinformatics/bti450. [DOI] [PubMed] [Google Scholar]
- 24.Gront D, Kmiecik S, Kolinski A. Backbone building from quadrilaterals: a fast and accurate algorithm for protein backbone reconstruction from alpha carbon coordinates. J Comput Chem. 2007;28(9):1593–1597. doi: 10.1002/jcc.20624. [DOI] [PubMed] [Google Scholar]
- 25.Bower MJ, Cohen FE, Dunbrack RL., Jr Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol. 1997;267(5):1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 26.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys J. 2005;88(4):2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kmiecik S, Gront D, Kolinski A. Towards the high-resolution protein structure prediction. Fast refinement of reduced models with all-atom force field. BMC Struct Biol. 2007;7:43. doi: 10.1186/1472-6807-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]