Abstract
The prime objective for every life-form is to deliver its genetic material, intact and unchanged, to the next generation. This must be achieved despite constant assaults by endogenous and environmental agents on the DNA. To counter this threat, life has evolved several systems to detect DNA damage, signal its presence and mediate its repair. Such responses, which impact a wide range of cellular events, are biologically significant because they prevent diverse human diseases. Our improving understanding of DNA-damage responses is providing new avenues for disease management.
Each of the ~1013 cells in the human body receives tens of thousands of DNA lesions per day1. These lesions can block genome replication and transcription, and if they are not repaired or are repaired incorrectly, they lead to mutations or wider-scale genome aberrations that threaten cell or organism viability. Some DNA aberrations arise via physiological processes, such as DNA mismatches occasionally introduced during DNA replication and DNA strand breaks caused by abortive topoisomerase I and topoisomerase II activity. In addition, hydrolytic reactions and non-enzymatic methylations generate thousands of DNA-base lesions per cell per day. DNA damage is also produced by reactive-oxygen compounds arising as byproducts from oxidative respiration or through redox-cycling events involving environmental toxins and Fenton reactions mediated by heavy metals2. Reactive oxygen and nitrogen compounds are also produced by macrophages and neutrophils at sites of inflammation and infections3. Such chemicals can attack DNA, leading to adducts that impair base-pairing and/or block DNA replication and transcription, base loss, or DNA single-strand breaks (SSBs). Furthermore, when two SSBs arise in close proximity, or when the DNA-replication apparatus encounters a SSB or certain other lesions, double-strand breaks (DSBs) are formed. While DSBs do not occur as frequently as the other lesions listed above, they are difficult to repair and extremely toxic4.
The most pervasive environmental DNA-damaging agent is ultraviolet light (UV). While the ozone layer absorbs the most dangerous part of the solar UV spectrum (UV-C), residual UV-A and UV-B in strong sunlight can induce ~100,000 lesions per exposed cell per hour. Ionizing radiation (IR) also generates various forms of DNA damage, the most toxic of these being DSBs5. Some IR results from radioactive decay of naturally-occurring radioactive compounds. For example, uranium decay produces radioactive radon gas that accumulates in some homes and contributes to lung-cancer incidence. Exposure to natural or man-made radioisotopes also occurs during cancer radiotherapy, while the radioactive compounds iodine-131 and technetium-99m are exploited to diagnose and treat benign and malignant thyroid diseases. Lessons about the health consequences of excessive radiation exposure are provided by the aftermaths of the Chernobyl nuclear-reactor disaster and nuclear detonations over Japan in World War II.
Today, probably the most prevalent environmental cancer-causing chemicals are those produced by tobacco products, which trigger various cancers, most notably those of the lung, oral cavity and adjacent tissues6, 7. Cancer-causing DNA-damaging chemicals can also contaminate foods, such as aflatoxins found in contaminated peanuts and heterocyclic amines in over-cooked meats7. DNA-damaging chemicals have also been used in warfare, and on a more positive note, are widely used to treat cancer8 and ailments such as psoriasis9.
Here, we describe how DNA lesions are dealt with at the molecular level. We then explain how such responses affect many cellular processes, their biological significance and their roles in preventing human diseases. Finally, we illustrate how our increasing knowledge of DNA-damage responses is providing opportunities for improving disease detection and management.
An integrated signaling and genome-maintenance network
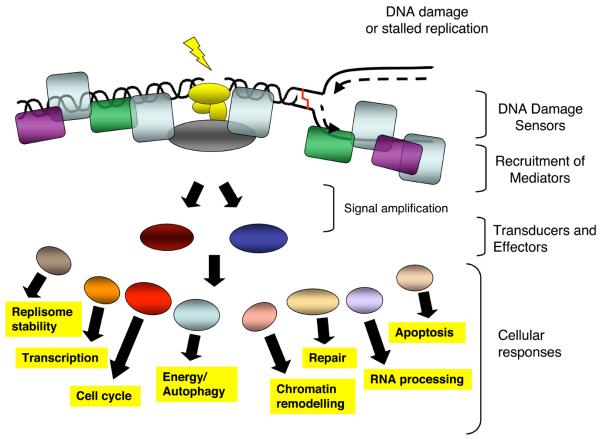
To combat threats posed by DNA damage, cells have evolved mechanisms – collectively termed the DNA-damage response (DDR) – to detect DNA lesions, signal their presence and promote their repair10-12. Cells defective in these mechanisms generally display heightened sensitivity towards DNA-damaging agents and, as described further below, many such defects cause human disease. Although responses differ for different classes of DNA lesions, they usually occur by a common general program (Fig. 1). Although we focus on DNA repair and DNA-damage signaling separately, we stress that these operate collectively and share many components.
Figure 1. Model for the DDR.
The presence of a lesion in the DNA, which can lead to replication stalling, is recognized by various sensor proteins. These sensors initiate signaling pathways that impact a wide variety of cellular processes. See text for details.
DNA-repair pathways
The wide diversity of DNA-lesion types necessitates multiple, largely distinct DNA-repair mechanisms (Table 1). While some lesions are subject to direct protein-mediated reversal, most are repaired by a sequence of catalytic events mediated by multiple proteins. In mismatch repair (MMR), detection of mismatches and insertion/deletion loops triggers a single-strand incision that is then acted upon by nuclease, polymerase and ligase enzymes13. In base-excision repair (BER), a damaged base is often recognized by a DNA glycosylase enzyme that mediates base removal before nuclease, polymerase and ligase proteins complete the repair14 in processes overlapping with those used in single-strand break repair (SSBR)15, 16. The nucleotide excision repair (NER) system, which recognizes helix-distorting base lesions, operates via two sub-pathways that differ in the mechanism of lesion recognition: transcription-coupled NER, which specifically targets lesions that block transcription, and global-genome NER15. A key aspect of NER is that the damage is excised as a 22-30 base oligonucleotide, producing single-stranded DNA (ssDNA) that is acted upon by DNA polymerases and associated factors before ligation ensues15. Notably, some DNA lesions are not repaired but are instead bypassed during DNA replication by polymerases with less stringent base-pairing requirements than replicative polymerases17.
Table 1.
DDR mechanisms and components. See text for details.
| DDR mechanism | Prime lesions acted upon | Key protein components |
|---|---|---|
| Direct DNA-lesion reversal |
UV photo-products O6 alkylguanine |
Photolyase O6-methylguanine methyltransferase (MGMT) |
| Mismatch repair (MMR) | DNA mismatches and insertion/deletion loops arising from DNA replication |
Sensors MSH2-MSH6 and MSH2-MSH3 plus MLH1-PMS2, MLH1-PMS1, PLH1-MLH3, EXO1, polymerases δ and ε, PCNA, RFC, RPA, ligase I |
| Base excision repair (BER) and single-strand break repair (SSBR) |
Abnormal DNA bases, simple base- adducts, SSBs generated as BER intermediates, by oxidative damage or by abortive topoisomerase I activity |
DNA glycosylases (sensors), APE1 endonuclease, DNA polymerases (β, δ, ε) and associated factors, flap endonuclease FEN1, ligase I or ligase III. SSBR can also involve polymerase β lyase activity, XRCC1, PARP-1, PARP-2, polynucleotide kinase (PNK) and aprataxin (APTX) |
| Nucleotide excision repair (NER) |
Lesions that disrupt the DNA double-helix, such as bulky base adducts and UV photo-products |
Sensors elongating RNA polymerase, XPC-HR23B and DDB1/2, plus XPA, XPE, XPF/ERCC1, XPG, CSA, CSB, TFIIH (containing helicases XPB and XPD), DNA polymerases and associated factors, RPA, ligase I |
| Trans-lesion bypass mechanisms |
Base damage blocking replication- fork progression |
“Error-prone” DNA polymerases, including polymerases eta, iota, kappa, REV3 and REV1; plus associated factors |
| Non-homologous end- joining (NHEJ) |
Radiation- or chemically-induced DSBs plus V(D)J and CSR intermediates |
Sensors Ku and DNA-PKcs plus XRCC4, XLF/Cernunnos and ligase IV. Can also employ the MRE11-RAD50-NBS1 complex, Artemis nuclease, PNK, Aprataxin and polymerases μ and λ |
| Homologous recombination (HR) |
DSBs, stalled replication forks, inter-strand DNA cross-links and sites of meiotic recombination and abortive Topoisomerase II action |
RAD51, RAD51-related proteins (XRCC2, XRCC3, RAD51B, RAD51C, RAD51D, DMC1), RAD52, RAD54, BRCA2, RPA, FEN1, DNA polymerase and associated factors. Promoted by MRN, CtIP, BRCA1, and the ATM signalling pathway |
| Fanconi anaemia (FANC) pathway |
Inter-strand DNA cross-links | FA-A, C, D1/BRCA2, D2, E, F, G, I, J, L, M, N plus factors including PALB2 and HR factors |
| ATM-mediated DDR signalling |
DSBs | ATM, MRN and CHK2. Promoted by mediator proteins such as MDC1, 53BP1 MCPH1/BRIT1, and by ubiquitin ligases RNF8, RNF168/RIDDLIN and BRCA1 |
| ATR-mediated DDR signalling |
ssDNA, resected DSBs | Sensors ATR ATRIP and RPA plus the RAD9-RAD1-HUS1 (911) complex, RAD17 (RFC1-like) and CHK1. Promoted by MRN, CtIP and mediator proteins such as TOPBP1, Claspin, MCPH1/BRIT1 and BRCA1 |
For DSB repair, two principal mechanisms are used: non-homologous end-joining (NHEJ)18 and homologous recombination (HR)19. In NHEJ, DSBs are recognized by the Ku protein that then binds and activates the protein kinase DNA-PKcs, leading to recruitment and activation of end-processing enzymes, polymerases and DNA ligase IV. A less-well-characterized Ku-independent NHEJ pathway, called microhomology-mediated end-joining (MMEJ) or alternative end-joining (AEJ), also exists; it always results in sequence deletions20. While both NHEJ and MMEJ are error-prone, they can operate in any phase of the cell cycle. By contrast, HR is generally restricted to S and G2 because it uses sister-chromatid sequences as the template to mediate faithful repair. Although there are several HR sub-pathways19, HR is always initiated by ssDNA generation, which is promoted by various proteins including the MRE11-RAD50-NBS1 (MRN) complex. In events catalyzed by RAD51 and the breast-cancer susceptibility proteins BRCA1 and BRCA2, the ssDNA then invades the undamaged template and, following the actions of polymerases, nucleases, helicases and other components, DNA ligation and substrate resolution occur. HR is also used to restart stalled replication forks and to repair interstrand DNA crosslinks, the repair of which also involves the Fanconi Anaemia (FA) protein complex21.
DNA-damage signaling and cell-cycle checkpoints
Key DDR-signaling components in mammalian cells are the protein kinases ATM and ATR, which are recruited to and activated by DSBs and RPA-coated ssDNA, respectively (Table 1)22-24. Two of the best studied ATM/ATR targets are the protein kinases CHK1 and CHK2 which, together with ATM and ATR, act to reduce cyclin-dependent kinase (CDK) activity by various mechanisms, some of which are mediated by activation of the p53 transcription factor23, 25, 26. Inhibition of CDKs slows down or arrests cell-cycle progression at the G1-S, intra-S and G2-M “cell-cycle checkpoints”, which is thought to increase the time available for DNA repair before replication or mitosis ensues. In parallel, ATM/ATR signaling enhances repair by inducing DNA-repair-proteins transcriptionally or post-transcriptionally; by recruiting repair factors to the damage; and by activating DNA-repair proteins by modulating their phosphorylation, acetylation, ubiquitylation or sumoylation27. Proteomics studies have recently identified a great many as-yet uncharacterized ATM/ATR-mediated phosphorylation sites, suggesting that the DDR modulates additional cellular processes28. If the above events allow effective DNA repair, DDR inactivation ensues, allowing the resumption of normal cell functioning. Alternatively, if the damage cannot be removed, chronic DDR signaling triggers cell death by apoptosis or cellular senescence (i.e. permanent cell-cycle withdrawal), both of which have potential anti-tumour functions29, 30. As discussed in later sections, another important SSB- and DSB-signaling protein is the enzyme poly(ADP-ribose) polymerase (PARP).
It is becoming increasingly clear that chromatin structure impacts the DDR and is modulated in response to DNA damage23, 31. The best characterized example of this is ATM/ATR/DNA-PK-mediated phosphorylation of serine-139 of the histone H2A variant, H2AX, on chromatin flanking DSB sites. This brings about ubiquitin-adduct formation in such regions and the recruitment of DDR factors plus other chromatin-modifying components which, together, are thought to promote DSB repair and amplify DSB signalling27. Notably, ATM activation leads to chromatin relaxation at sites of DSBs32, and H2AX tyrosine-142 phosphorylation was recently shown to function in the DDR33, 34. It therefore seems likely that further DDR-induced chromatin modifications await discovery.
DDR events operate in diverse biological settings
Generating immune-receptor diversity
The only known programmed genome alterations in vertebrates are V(D)J recombination, class-switch recombination (CSR) and somatic hyper-mutation (SHM)35, 36. These occur in developing B- and T-lymphocytes to generate immunoglobulin (Ig) and T-cell receptor (TCR) diversity, thus allowing effective recognition of diverse pathogens and antigens. Ig and TCR proteins comprise variable regions that specify antigen binding, and constant regions that endow specific properties to the TCR or the various Ig classes. Exons encoding the antigen-binding portions of these molecules are composed of V, D and J segments that are combined in various ways to generate mature Ig and TCR genes. Each segment is flanked by recombination-signal sequences that are recognized by the RAG1/RAG2 protein complex, which generates a blunt DSB at the signal sequence and a covalently-closed DNA hairpin at the coding end. These structures are then processed and ligated by the NHEJ apparatus35. Consequently, besides causing IR hypersensitivity, NHEJ defects yield severe-combined immune-deficiency.
A rearranged Ig heavy-chain variable domain is initially expressed fused to an Igμ constant region but, during antigen-stimulated B-cell differentiation, CSR can juxtapose a V region to any of several constant regions that bestow distinct properties on the encoded Ig. B-cells undergoing antigen stimulation also activate SHM to increase mutation rates in the heavy- and light-chain V regions, thus expanding the repertoire of variable segments and allowing selection of B-cells expressing Ig molecules with heightened antigen affinity. Unlike V(D)J recombination, CSR and SHM require activation-induced deaminase (AID). AID targeting to variable-region exons and IgH switch regions is believed to trigger deamination of cytosine to uracil, resulting in U:G mismatches that are processed by MMR and/or BER to yield SSBs. In SHM, error-prone repair of these SSBs is thought to yield mutations within the variable exon; while in CSR, SSBs are converted into DSBs that are acted upon by NHEJ to juxtapose the Ig variable exon to a constant-region exon35.
Production of gametes for sexual reproduction
The DDR also plays a key role in generating genetic diversity via sexual reproduction, a stage in which is meiosis, the cell-division pathway that generates haploid gametes. Following DNA replication, meiosis proceeds by two successive cell divisions: MI that reductionally-segregates the two copies of individual chromosomes; and MII that separates resulting sister-chromatid pairs. Prior to MI, homologous chromosomes align and exchange genetic information by HR37. In species ranging from yeast to man, meiotic HR is triggered by the topoisomerase II-related enzyme, Spo11, which generates Spo11-bound DSBs. Spo11 removal and DSB resection then ensue by mechanisms requiring the MRN complex, resulting in ssDNA that promotes HR with the homologous chromosome. These events require all mitotic HR components together with the meiosis-specific RAD51-like protein DMC137. Consequently, mice deficient in Spo11 or Dmc1 are healthy but infertile. In addition, DDR factors such as ATM, MRN and H2AX monitor and coordinate meiotic HR progression.
Telomere homeostasis and ageing
In most organisms, the ends of chromosomes are organized into telomeres that comprise stretches of short-tandem-DNA repeats terminating in a 3′ protruding ssDNA overhang. These repeats are normally generated by the ribonucleoprotein complex, telomerase but in some cancer cells they are maintained by HR-based “alternative lengthening of telomeres” (ALT) mechanisms38. Although telomeres possess DNA ends, their sequestration into a complex termed Shelterin prevents them from engaging in NHEJ-mediated fusions or activating ATM/ATR signaling39. Nevertheless, various DDR proteins play important roles at normal telomeres (Table 2); and consequently, their defects cause telomere shortening and/or telomere dysfunction that trigger chromosome fusions and ensuing chromosomal instability38, 40. Furthermore, mammalian telomeres are recognized by MRN and ATM during G2, possibly to trigger a localized DDR that promotes telomere end-processing and Shelterin-complex formation. HR proteins such as RAD51D are also required for telomere integrity, which might reflect their involvement in establishing T-loop structures wherein the telomeric 3′ overhang invades the DNA duplex of internal telomeric sequences38-41.
Table 2.
DDR proteins acting at normal telomeres. See text for details and Table 1 for protein functions.
| DDR protein | Role at normal telomeres |
|---|---|
| MRN | Telomere length regulator, role in end-processing |
| ATM, Chk2 and ATR | Maintain telomere length, phosphorylate Shelterin- complex components, possible roles in telomerase activation and recruitment |
| Ku and DNA-PKcs | Telomere components and telomere-length regulators, possible telomere-capping functions |
| 9-1-1 | Telomere component and telomere-length regulator; aids telomerase recruitment/activation |
| Nucleases EXO1, FEN1, XPF/ERCC1 and Apollo |
Processing of telomeric termini to promote telomerase action; regulate telomere integrity |
| PARP1 | Potential telomere-length regulator |
| BRCA1 | Telomere maintenance |
| RPA | Telomere component, role in telomerase recruitment |
| WRN | Maintains telomere structure and functions in telomere replication |
| RAD51D and other HR proteins | Regulate telomeric integrity |
Excepting specialized cells such as stem cells, human cells generally do not express sufficient telomerase to counteract telomere shortening caused by the inability of the DNA replication machinery to fully replicate chromosomal ends. Thus, human telomeres generally shorten with each cell division until some retain hardly any terminal telomeric repeats. These then fail to act as telomeres and are instead recognized as DSBs, triggering chromosomal fusions and ensuing breakage-fusion-breakage cycles. Under such situations of chronic DDR activation, cells enter into apoptosis or senescence40, 41. There is evidence that such processes occur during natural ageing and under circumstances of high cell turnover, such as chronic inflammation or persistent infections. Consistent with ageing in part reflecting the accumulation of shortened telomeres and/or DNA damage, markers of unrepaired DSBs accumulate with age in human and mouse cells, and certain DDR-defective mice strains display hallmarks of accelerated ageing42-44. Furthermore, senescent cells occur at sites of age-related pathologies in man, including atherosclerotic lesions, skin ulcers and arthritic joints29, 44.
Physiological control of the DDR
The differing physiologies of various cells presumably impose different DDR requirements. Indeed, some DNA-repair pathways are down-regulated upon cell differentiation, possibly reflecting rapid DNA repair being less imperative for non-dividing cells. For example, work on terminally-differentiated neurons and macrophages has indicated the existence of a new type of NER, termed differentiation-associated repair, in which both transcribed and non-transcribed DNA strands are repaired effectively but non-transcribed loci are repaired poorly or not at all45. DSB-repair requirements also change during mammalian nervous-system development, with HR being crucial during neuron proliferation, while NHEJ becomes critical as neurons terminally differentiate46. This could reflect NHEJ being the only DSB mechanism available to post-mitotic neurons, while dividing neural precursors also use HR. Because of their importance for tissue homeostasis and renewal, it has been speculated that stem cells will rely heavily on the DDR. Indeed, defects in BER, NER, MMR, HR or the FA complex impair stem-cell function47, and NER or NHEJ defects trigger age-related haematopoietic stem-cell failure in mice48, 49.
Many organisms regulate physiological processes in synchrony with the light-dark cycle via the circadian rhythm/biological clock that is controlled by light stimuli. Recent work has established molecular linkages between the biological clock and DDR events50. For instance, the Caenorhabditis elegans biological-clock gene clk-2 affects radiation sensitivity, and CLK-2 and its mammalian counterpart control the S-phase checkpoint in response to replication stress51. Also, it was recently reported that NER is regulated by the circadian clock52. Perhaps such linkages allow cells to enhance DDR proficiency at times when physiological or environmentally-induced DNA lesions are most prevalent.
Life cycles of pathogens
The cells of pathogens also possess DDR proteins to mitigate the effects of DNA damage. Furthermore, mutational repair and recombination occasionally occur in viruses, thus fueling evolution of pathogens such as avian influenza (H5N1) and swine-flu (H1N1) viruses53. In addition, certain pathogens use DDR mechanisms to promote virulence. For example, African trypanosomes – unicellular eukaryotic parasites that infect mammals, including humans – evade immune-surveillance by using HR to periodically alter their protective variant-surface-glycoprotein coat54. Furthermore, acquisition of drug or pesticide resistance in certain bacteria, plants and unicellular eukaryotic pathogens often involves integration of resistance genes into the organism's genome via DSB-repair mechanisms. DDR activation is also triggered when cells are infected by viruses, including retroviruses (such as HIV-1), adenoviruses, Herpes simplex viruses 1 and 2, Hepatitis B virus, Epstein-Barr virus, cytomegalovirus, Kaposi's sarcoma virus, Simian virus 40 and polyomavirus55. Indeed, DDR factors often provide a line of defense against these pathogens, and in many cases viruses have evolved ways to evade such responses. For example, the E6 protein of human papilloma virus types 16 and 18 targets the p53 tumor suppressor for proteolytic degradation to prevent apoptosis of infected cells56. Furthermore, the MRN complex and NHEJ components curtail adenovirus infectivity by concatamerizing the viral genome; the virus circumvents this by impairing DNA-PK activity, disrupting complexes containing MRN, and targeting MRN for degradation. Conversely, host cell DDR activities sometimes facilitate viral infectivity. For instance, NHEJ-mediated conversion of linear viral double-stranded (ds) DNA into circles seems important for Herpes simplex virus replication57. Furthermore, CHK2 deficiency or ATM inhibition impair HSV-1 growth58. Retroviruses have dsRNA genomes that are converted into dsDNA, which must then integrate into the host genome to produce new retroviruses; notably, ATM, MRN and NHEJ proteins are required for efficient retrovirus infection, probably by promoting repair of viral integration intermediates55, 59.
The DDR and human disease
Cancer and DNA damage: an intimate relationship
A fundamental feature of cancer is genome instability60. For example, genomic instability in lymphoid tumours frequently corresponds to chromosomal translocations, wherein proto-oncogene loci are fused to those of antigen receptors, apparently by aberrant antigen-receptor recombination35, 36. In addition, MMR defects cause microsatellite instability (MIN) that predisposes to colorectal and endometrial carcinomas13. Furthermore, chromosomal instability (CIN) is seen in most sporadic solid tumours61. It is likely that transient CIN arises when telomeres in a nascent tumour become critically short and prone to chromosomal fusions62, while activated oncogenes and ensuing DNA-replication stress with DSB formation fuel CIN continuously30. At later stages of cancer progression, chronic hypoxia and/or cycles of hypoxia and re-oxygenation might also contribute to genomic instability and deregulate DDR pathways63.
Most carcinogens operate by generating DNA damage and causing mutations15, 26. Furthermore, inherited DDR defects commonly predispose to cancer, contribute to the “mutator phenotype” of many malignancies, and may allow tumour-cell survival and proliferation despite enhanced mutation rates and genome instability (Table 3). Notably, aberrant cell proliferation, caused by oncogene activation or inactivation of certain tumour-suppressors, elicits DNA-replication stress and ongoing DNA-damage formation. Such damage activates ATR/ATM-mediated signaling, causing cell death or senescence in cell-culture models and during tumorigenesis in vivo29, 30, 64, 65. Indeed, the DDR is commonly activated in early neoplastic lesions and likely protects against malignancy64, 65. It has been suggested that breaches to this barrier, arising through mutational or epigenetic inactivation of DDR components, are subsequently selected for during tumor development, thus allowing malignant progression. This model for the DDR as an anti-cancer barrier helps explain the high frequency of DDR defects in human cancers30.
Table 3.
Inherited DDR defects cause cancers and neurodegenerative diseases. See text for more details and Table 1 for protein functions
| Syndrome | Phenotypes | Mutated Genes |
|---|---|---|
| GGR/NER deficiency | ||
| Xeroderma pigmentosum | Neurodegeneration and microcephaly, Photosensitivity, skin cancer |
XPA-XPG Pol η |
| Cockayne syndrome | Microcephaly and neuron demyelination | CSA, CSB, XPB, XPD, XPG |
| Trichothiodystrophy | Neurodevelopmental defects and dysmyelination (abnormal myelin) |
XPD, XPB, TTD-A |
| Cerebro-oculo-facio- skeletal (COFS) syndrome |
Demyelination (loss of myelin), dysmyelination, brain calcification, microcephaly |
XPD, XPG, CSB, ERCC1 |
| DNA helicase deficiency | ||
| Bloom's syndrome | Microcephaly, short stature, dysmorphic features, mild/moderate mental retardation, susceptible to infections, Elevated predisposition to all cancers |
BLM |
| Werner's syndrome | Premature ageing, cancer predisposition | WRN |
| Rothmund–Thompson syndrome |
Stunted growth, skeletal abnormalities, early cataracts, accelerated ageing, chromosomal instability and cancer predisposition |
RECQL4 |
| Ataxia with oculomotor apraxia 2 |
Ataxia, neurodegeneration and oculomotor apraxia |
SETX |
| Exonuclease deficiency | ||
| Aicardi-Goutières syndrome (AGS) |
De/dysmyelination, brain calcification, microcephaly, elevated CSF (cerebrospinal fluid) IFN-α, CSF lymphocytosis (CSF lymphocytes increased) |
TREX1, RNASEH2 |
| NHEJ/V(D)J recombination deficiency | ||
| Inactivation of Ku70 or Ku80 in mouse models |
Premature aging, cancer predisposition, lymphomas |
Ku70 Ku80 |
| Lig4 syndrome/Human immunodeficiency with microcephaly |
Microcephaly, leukemia, immunodeficiency, and developmental and growth delay |
DNA Ligase IV, XLF/Cerunnos |
| RS-SCID (radiosensitive- SCID) |
Severe-combined immunodeficiency, lymphomas and hypersensitivity to ionizing radiation |
Artemis |
| HR deficiency | ||
| Breast cancer 1, early onset | Breast and ovarian cancer | BRCA1 |
| Breast cancer 2, early onset | Breast and ovarian cancer; predisposition to pancreatic, prostate and gastric cancer and melanoma |
BRCA2 |
| DNA SSB repair deficiency | ||
| Spinocerebellar ataxia with axonal neuropathy (SCAN1) |
Ataxia, neurodegeneration, peripheral axonal motor and sensory neuropathy, muscle weakness |
TDP1 |
| Ataxia with oculomotor apraxia 1 (AOA1) |
Ataxia, neurodegeneration, oculomotor apraxia and peripheral neuropathy |
APTX |
| DNA cross-link repair deficiency | ||
| Fanconi anaemia | Congenital abnormalities, progressive bone marrow failure, prone to AML, squamous carcinomas of head, neck or gynaecological system. |
FANCA-FANCL, BRCA2 (FANCD1) |
| Mismatch repair deficiency | ||
| Hereditary non-polyposis colorectal cancer (HNPCC) |
Colon and gynaecologic cancers | MSH2, MSH3, MSH6, MLH1, PMS2, APC |
| DNA DSB-repair and signal-transduction deficiency | ||
| Li-Fraumeni syndrome | Soft tissue sarcomas, breast cancer, brain tumours | p53 |
| Familial breast cancer (non BRCA1/2) |
Predisposition to medium/late-onset breast cancer | Chk2, MRN, ATM BRIP1, PALB2, |
| Ataxia telangiectasia | Cerebellar ataxia, telangiectases, immune defects, predisposition to malignancy (mainly lymphomas but also breast cancer) |
ATM |
| Ataxia telangiectasia-like disorder |
Mild A-T like features, possibly cancer predisposed |
MRE11 |
| Nijmegen breakage syndrome (NBS) |
Microcephaly, growth retardation, mental retardation, immunodeficiency, cancer predisposition |
NBS1 |
| NBS-like syndrome | NBS-like phenotype | RAD50 |
| RIDDLE syndrome | Radiosensitivity, immunodeficiency, dysmorphic features and learning difficulties |
RNF168 (RIDDLIN) |
| Seckel syndrome | Marked microcephaly, primordial dwarfism, dysmorphic facial features and mental retardation, possibly AML |
ATR, SCKL2, SCKL3 |
| Primary microcephaly 1 | Microcephaly, mental retardation | MCPH1/BRIT1 |
| Hutchinson-Gilford progeria syndrome (HGPS) and restrictive dermopathy(RD) |
Accelerated ageing (HGPS); neonatal lethality (RD) |
Lamin-A |
| DNA-damage-response impairment and defective DNA repair | ||
| Down syndrome | Mental retardation, progeria | Trisomy of chromosome 21 |
| Alzheimer's disease | Progressive neurodegeneration leading to dementia, memory loss and cognitive decline |
Increased oxidative stress and damage |
| Parkinson's disease | Tremor, bradykinesia, posture rigidity and postural instability, degeneration of dopaminergic neurons in substantia nigra area |
Mutations in α- Synuclein and Parkin variants |
| Huntington's disease | Progressive chorea and dementia, severe neuronal loss in the striatum and cerebral cortex |
CAG repeat expansion in huntingtin (HD) |
| Several spinocerebellar ataxias |
Problems with bodily movements (similar to Huntington's disease), progressive neuron loss |
Expanded CAG repeats in various genes |
| Friedreich's ataxia | Limb ataxia, cerebellar dysarthria, sensory loss, skeletal deformities |
GAA expanded repeats in frataxin (FXN) |
| Myotonic dystrophy types 1 and 2 |
Muscle weakness and wasting, cataracts, testicular atrophy, cognitive decline |
CTG expansion (type 1), CCTG expansion (type 2) |
| Triple-A syndrome | Adrenal insufficiency, achalasia, alacrima, neurodegeneration, autonomic dysfunction |
Mutation in AAAS gene |
| Amyotrophic lateral sclerosis |
Progressive degeneration of motor neurons, muscle weakness and atrophy, leading to fatality |
Defective Cu-Zn superoxide dismutase (SODC, SOD1); mitochondrial DNA |
Additional human diseases with DDR defects and nuclear or mitochondrial DNA instability can be found at: http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=gnd; http://www.mitomap.org; or http://www.cepearsonlab.com (under ‘Repeat Disease Database’).
Neurodegenerative disorders
Accumulation of DNA lesions in neurons is associated with neurodegenerative disorders, including ataxias together with Alzheimer's, Huntington's and Parkinson's diseases (Table 3)66, 67. One reason for this may be that neurons generally exhibit high mitochondrial respiration and associated reactive-oxygen-species production that can damage mitochondrial and nuclear DNA68. Consistent with a role for BER and SSBR in repairing such lesions, defects in these pathways trigger neuronal dysfunction and degeneration66, 69. Another reason why the nervous system is particularly vulnerable to DNA damage is its limited capacity for cell replacement in adulthood, potentially leading to accumulation of damaged but irreplaceable terminally-differentiated neurons. Furthermore, being in G0, such cells do not repair DSBs by HR but must employ error-prone NHEJ66. It is also noteworthy that neurons rely heavily on transcription and that oxidative DNA damage can block this. Thus, accumulation of DNA lesions in repair-defective patients - and possibly in ageing normal individuals – might progressively deprive neurons of vital transcripts, leading to cell dysfunction or apoptosis70. Such processes presumably contribute to the neurodegeneration observed in ataxias and in Cockayne syndrome, which are caused by defects in DNA strand-break repair and transcription-coupled NER, respectively66, 67.
Genome instability in other heritable human diseases
DNA-repeat instability causes some forty known diseases that result from expansions or contractions of genetically-unstable DNA repeat sequences, usually a trinucleotide motif, within a specific locus for each disease. This instability is thought to arise through the repetitive nature of these regions allowing aberrant DNA-secondary-structure formation during DNA replication or DNA-repair processes71, 72. These neuromuscular and neurodegenerative diseases include Fragile X syndromes, Friedrich's ataxia, spinocerebellar ataxias, diabetes mellitus type 2, Creutzfeldt-Jakob disease, myotonic dystrophy and Huntington's disease (see Table 3 for examples). Analogously, mutations or rearrangements of mitochondrial DNA can lead to impaired mitochondrial function as found in amyotrophic lateral sclerosis, mitochondrial encephalomyopathy, Leigh syndrome, myoclonic epilepsy, Leber's hereditary optic neuropathy, and additional neuro- and myopathies73.
Immune deficiencies and infertility
Genome rearrangements involving DDR factors occur during immune-system development, meaning that DDR defects can cause immune deficiency. For instance, mutations in NHEJ factors yield B- and T-cell immune deficiency, while AT and NBS patients (defective in ATM and NBS1, respectively) are prone to sometimes-fatal infections, partly due to impaired immunity (CSR is particularly affected in AT patients). Furthermore, many cancers arising in such conditions are lymphomas and leukaemias of B- and T-cell origin that can result from impaired V(D)J recombination. Human infertility is a significant issue, with ~20% of males in western countries being affected74. As meiotic recombination involves DSB generation, it seems likely that certain DDR defects would cause human infertility. Indeed, DDR signaling is readily detectable during human spermatogenesis75, and various inherited DDR deficiencies are characterized by infertility or sub-fertility74. A significant proportion of human infertility might therefore be caused by DDR deficiencies.
Ageing, stem cell dysfunction, cardiovascular disease and metabolic syndrome
There is evidence that ageing is in part caused by accumulated DNA damage76. First, various endogenously-arising DNA lesions accumulate with age in the nuclear and mitochondrial genomes of healthy mammals, including humans42, 76, 77. This may reflect not only ongoing DNA-damage induction but also declining DNA-repair capacity over time68, 73. Second, patients with inherited DDR defects often display features of premature ageing (Table 3). Third, work in various organisms has implicated growth hormone and insulin-like growth factor 1 signaling in regulating longevity, and notably, such signaling is down-regulated in response to DNA damage76. The evolutionary honing of longevity-regulating pathways may serve as an example of antagonistic pleiotropy, where processes selected as advantageous early in life (such as active DDR signaling) because they promote reproduction and prevent tumourigenesis are detrimental later in life because they lead to stem-cell depletion, hence contributing to ageing.
Cell senescence and apoptosis are suspected causes of ageing under conditions where attempted tissue regeneration causes stem-cell exhaustion44, 78. Suggesting that DNA damage contributes to such episodes, impaired stem-cell function is exhibited in mice with defects in the FA, NER, MMR or NHEJ pathways48, 49, 79, 80. Furthermore, whereas p53-induced cell death protects against tumorigenesis, pro-apoptotic p53 activity is harmful in settings such as stroke or heart attack81, 82. Induction of p53 by oxidative stress and other sources of DNA damage can also affect the development of atherosclerosis, thus providing a link between the DDR and cardiovascular disease83. Indeed, growing evidence points to human atherosclerosis being characterized by enhanced DNA damage and DDR signaling, leading to senescence of vascular smooth muscle cells and death of other cells to yield atherosclerotic lesions. Modulating ROS production and the DDR therefore represent potential therapeutic opportunities for atherosclerosis.
Metabolic syndrome is a relatively common condition characterized by aberrant glucose metabolism, insulin resistance and atherosclerosis. Interestingly, ATM-defective patients commonly exhibit insulin resistance and glucose intolerance, while mice heterozygous or homozygous for ATM mutations display features associated with metabolic syndrome and atherosclerosis84, 85. Furthermore, DDR-regulated kinases target multiple substrates involved in glucose metabolism and the insulin-AKT kinase signaling network28, 84, 85. Thus, although some linkages between the DDR and metabolic syndrome might be indirect, it is possible that the DDR directly modulates certain aspects of energy metabolism and vascular physiology of relevance to metabolic syndrome.
Harnessing DDR knowledge for treating disease
Cancer
Other than surgery, the most prevalent cancer treatments are radiotherapy and chemotherapies that function by generating DNA damage (Table 4). Although such therapies generate dose-limiting toxicities in normal tissues, they are often efficacious. In part, this reflects most cancer cells being DDR-impaired and them proliferating more rapidly than most normal cells (S phase is a particularly vulnerable time for DNA-damage exposure). Nevertheless, DNA repair provides a common mechanism for cancer-therapy resistance. For instance, it has been reported that glioma stem cells display a heightened DDR and are refractory to radiation treatment86, thus potentially helping to explain why glioblastoma is difficult to cure (radiation- and chemotherapy-resistance of cancer stem cells might more generally reflect unique properties of their DDR machinery. It has therefore been speculated that DDR inhibition might enhance the effectiveness of radiotherapy and DNA-damaging chemotherapies; and indeed, various DDR-inhibitory drugs are in pre-clinical and clinical development to test this premise (Table 4)87, 88. Another possible application for DDR inhibitors is to block apoptotic events, such as those mediated by CHK2 and p53, thus alleviating toxicities to normal tissues.
Table 4.
Examples of DNA-damaging drugs used to treat cancer. See text for details (modifed from Helleday et al98).
| Cancer Treatment | Types of DNA lesions induced |
|---|---|
| Radiotherapy and radiomimetics | |
| Ionizing radiation Bleomycin |
Single-strand breaks, double-strand breaks, base damage |
| Monofunctional alkylators | |
| Alkylsulphonates Nitrosourea compounds Temozolomide |
Base damage, replication lesions, bulky DNA adducts |
| Bifunctional alkylators | |
| Nitrogen mustard Mitomycin C Cisplatin |
Double-strand breaks, DNA cross-links, replication lesions, bulky DNA adducts |
| Antimetabolites | |
| 5-Fluorouracil (5FU) Thiopurines Folate analogs |
Cytotoxic metabolite, inhibits base pairing leading to base damage and replication lesions |
| Topoisomerase inhibitors | |
| Camptothecins (Topo I); Etoposide (Topo II), anthracyclines (Doxorubicin, Epirubicin, Daunorubicin (Topo II) |
Double-strand breaks, single-strand breaks, replication lesions; anthracyclines also generate free radicals |
| Replication inhibitors | |
| Aphidicolin Hydroxyurea |
Double-strand breaks, replication lesions |
Many, and possibly all, cancer cells lack one or other aspects of the DDR due to selective pressures operating during tumour evolution (see above). Indeed, reduced or absence of DDR factors correlates, usually positively, with therapeutic outcome (exceptions are defects in p53 and other pro-apoptotic proteins, which commonly yield therapy resistance82, 89). Because different DNA-repair pathways can overlap in function, and as one pathway can sometimes “back-up” for defects in another, inhibition of pathways present in a cancer cell should in some cases have a greater impact on the cancer than on normal tissues (Fig. 2a). A paradigm for this is provided by drugs targeting the enzyme PARP-1, which binds SSBs and BER intermediates to facilitate these repair processes. Notably, PARP inhibitors are relatively nontoxic to normal cells but striking cytotoxic towards HR-defective cells, particularly those impaired in BRCA1 or BRCA2 (Fig. 2b)90, 91. Based on promising Phase 1 data, Phase 2 trials are currently underway to test PARP-1 inhibitors in BRCA-defective breast cancer and ovarian cancer patients (http://www.cancer.gov/search/ResultsClinicalTrialsAdvanced.aspx?protocolsearchid=5678174). Significantly, some sporadic breast, ovarian, prostate, pancreatic and other tumours also possess HR defects due to mutation or epigenetic inactivation of HR components, suggesting that PARP inhibitors might be more broadly applicable. Furthermore, as other DDR pathways are frequently impaired in cancers, there may be additional situations where DDR inhibitors would display selective anti-tumour effects. Consistent with this idea, CHK1 inhibition reportedly sensitizes p53-deficient cells to DNA-damaging agents more than p53-proficient cells92. The development of diagnostic procedures to identify DDR differences between cancer and normal cells therefore holds great promise for intelligent tailoring of DNA-damaging therapies and DDR-inhibitor therapies for the individual patient. Furthermore, as DDR activation is prevalent during oncogenesis, screening for DDR-markers could enhance the reliability and sensitivity of cancer detection, and might allow effective detection of pre-malignant disease. In the longer term, it might be possible to develop drugs that enhance DDR events, thus reducing cancer incidence. In this regard, it is noteworthy that mice engineered to exhibit enhanced p53-dependent DNA-damage responses are less tumour-prone than wild-type mice93.
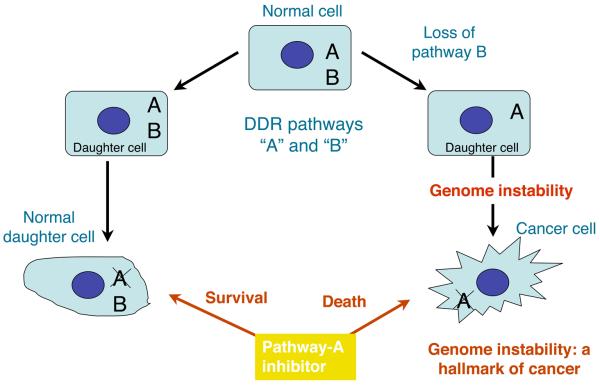
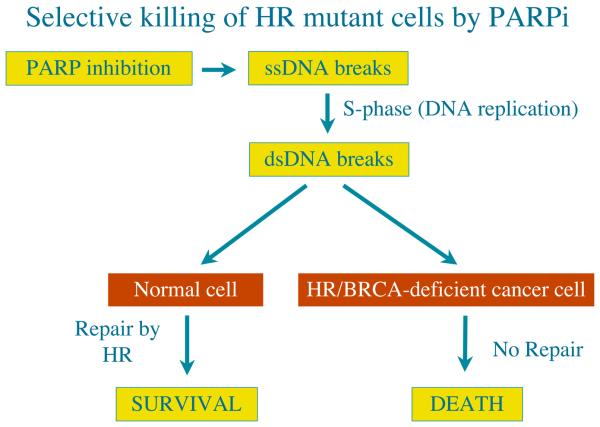
Figure 2. Exploitation of the DDR pathways to enhance therapeutic responses.
a, Normal cells have two DDR pathways, A and B. If one pathway (B) is eliminated, genome instability results, which can foster the evolution of a cancer cell. Addition of a inhibitor targeting the second pathway (A) leads to cell death. Normal cells that still retain an active B pathway, however, survive. b, Treatment with a PARP inhibitor selectively kills HR/BRCA-deficient cells. PARP inhibition impairs the repair of SSBs, which are converted to DSBs in S-phase. Such DSBs are effectively repaired by HR in non-cancerous cells but not in BRCA-deficient cells.
Ischaemia-reperfusion injury, inflammatory diseases and ageing
While DDR mechanisms generally protect against disease, their hyper-activation can contribute to pathology. A prime example of this is in ischaemia-reperfusion episodes associated with stroke and myocardial infarction, where PARP-1 can become hyper-activated through DNA damage caused by re-oxygenation and nitric-oxide production. This depletes intracellular pools of nicotinamide adenine dinucleotide, resulting in impaired ATP production and cell death, often by necrosis. Strikingly, genetic inactivation or pharmacological inhibition of PARP-1 in rodents provides considerable protection towards such cell death, consequently diminishing ensuing organ dysfunction. Similarly, animal models have shown PARP-1 inhibition to protect against traumatic brain injury, endotoxic shock, tissue damage caused by chronic inflammation and drug-induced diabetes (for example see94, 95). Thus, PARP-1 inhibitors might find utility in treating such conditions in people. It is also noteworthy that p53 dysfunction is associated with inflammatory diseases and atherosclerosis96, 97, suggesting that pharmacological modulation of p53, and its upstream activator ATM, could ameliorate such pathologies85. With our growing realization that DNA damage and sub-optimal DDR-events are associated with neurodegenerative disease and various other age-related degenerative pathologies, it is also tempting to speculate that DDR-modulatory drugs will one day be used to slow down or prevent such condition…and perhaps even certain aspects of the normal ageing process.
Viral, parasitic and other diseases
DDR proteins function in the life-cycles of human parasites and pathogens, suggesting that DDR inhibitors could be used to treat their associated pathologies. For instance, the reliance of HIV on host-cell DDR factors suggests a potential for DDR inhibitors in AIDS therapy98, 99. Although such treatments would need to be evaluated for potential side-effects, a possible advantage over conventional treatments that target the pathogen itself is that they would not be easily subject to evolution of resistance. Furthermore, antibacterial agents could be developed that target aspects of bacterial DDR mechanisms that are distinct from those of host cells.
Gene therapy
Correcting gene dysfunction is a long sought-after treatment for many human maladies, including immune-deficiencies, cystic fibrosis, muscular dystrophy and hereditary blindness. While some success has been achieved, such approaches have been plagued by safety issues, largely arising through unwanted NHEJ-mediated integration of the introduced gene into tumour-suppressor loci. While it is difficult to imagine such obstacles being surmounted in the near future, the development of methods to interfere with NHEJ or promote gene integration into desired loci (for example100) offers exciting prospects for gene-therapy optimization.
Future challenges
Great progress has been made towards understanding the DDR but much remains to be learned. One major future challenge is to understand in more detail how the activities of DDR proteins are controlled. Other challenges are to determine precisely how and why the DDR impacts on myriad cellular functions and how such complex programmes are orchestrated. Additional important issues to be addressed are how the DDR can be shaped and fine-tuned by other pathways and events, and how the same DDR stimulus can yield markedly different responses in different cells and tissues, including cancer cells and stem cells. Such knowledge will not only enhance our appreciation of DDR functions but will undoubtedly present exciting opportunities for better understanding and managing human health and disease.
Acknowledgements
We thank Sophie Polo and Pablo Huertas for advice, and Kate Dry for expert help with the text and figures. The SPJ laboratory is supported by grants from Cancer Research UK, the European Commission (projects GENICA and DNA Repair), the Wellcome Trust and the Biotechnology and Biological Sciences Research Council. The JB laboratory is supported by grants from the Danish Cancer Society, the Danish National Research Foundation and the European Commission (projects GENICA, Active p53, TRIREME and DNA Repair).
References
- 1.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. An excellent overview of the extent of endogenous DNA damage, the types of DNA lesions arising from cell autonomous sources, and the pathways that repair such lesions. [DOI] [PubMed] [Google Scholar]
- 2.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Kawanishi S, Hiraku Y, Pinlaor S, Ma N. Oxidative and nitrative DNA damage in animals and patients with inflammatory diseases in relation to inflammation-related carcinogenesis. Biol Chem. 2006;387:365–372. doi: 10.1515/BC.2006.049. [DOI] [PubMed] [Google Scholar]
- 4.Khanna KK, Jackson SP. DNA double-trand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 5.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 6.Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst. 1981;66:1191–1308. Classical overview of epidemiological evidence for DNA-damaging environmental insults implicated as cancinogens, and suggestions for measures to prevent such tumors. [PubMed] [Google Scholar]
- 7.Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14:473–486. doi: 10.1016/j.semcancer.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Espinosa E, Zamora P, Feliu J, Gonzalez Baron M. Classification of anticancer drugs--a new system based on therapeutic targets. Cancer Treat Rev. 2003;29:515–523. doi: 10.1016/s0305-7372(03)00116-6. [DOI] [PubMed] [Google Scholar]
- 9.Lebwohl M, Ting PT, Koo JY. Psoriasis treatment: traditional therapy. Annals of the rheumatic diseases. 2005;64 Suppl 2:ii83–86. doi: 10.1136/ard.2004.030791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 12.Harrison JC, Haber JE. Surviving the Breakup: The DNA Damage Checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 13.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 14.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. A highly informative review of the links between DNA damage, DNA repair pathways and their defects contributing to tumorigenesis. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg EC, et al. DNA Repair and Mutagenesis. 2nd Edition. ASM Press; Washington, DC: 2006. An excellent, comprehensive multi-author book covering essentially the entire field of DNA repair, from basic mechanisms in diverse organisms, to human diseases associated with defective DNA repair. [Google Scholar]
- 17.Loeb LA, Monnat RJ., Jr. DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 18.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 19.San Filippo J, Sung P, Klein H. Mechanism of Eukaryotic Homologous Recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 20.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 22.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature Reviews Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. Describes the key DDR kinases ATM, ATR and DNA-PK, provides an overview of their substrates, and outlines the cellular pathways affected by DNA-damage signaling. [DOI] [PubMed] [Google Scholar]
- 25.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53- regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 26.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 27.Huen MS, Chen J. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 2008;18:8–16. doi: 10.1038/cr.2007.109. [DOI] [PubMed] [Google Scholar]
- 28.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. Milestone report on the proteomic identification of ATM/ATR substrates and their assignment to various cellular functions, including RNA processing and other protein networks not previously recognized as DDR targets. [DOI] [PubMed] [Google Scholar]
- 29.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 30.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 31.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 33.Xiao A, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassing CH, Alt FW. The cellular response to general and programmed DNA double-strand breaks. DNA Repair. 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Schlissel MS, Kaffer CR, Curry JD. Leukemia and lymphoma: a cost of doing business for adaptive immunity. Genes Dev. 2006;20:1539–1544. doi: 10.1101/gad.1446506. [DOI] [PubMed] [Google Scholar]
- 37.Richardson C, Horikoshi N, Pandita TK. The role of the DNA double-strand break response network in meiosis. DNA Repair. 2004;3:1149–1164. doi: 10.1016/j.dnarep.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 38.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 39.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 40.d'Adda di Fagagna F, Teo SH, Jackson SP. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. References 39 and 40 illustrate the intimate links between the telomere maintenance and DDR machineries. [DOI] [PubMed] [Google Scholar]
- 41.Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22:125–140. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedelnikova OA, et al. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 43.Niedernhofer LJ, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. Reports a powerful mouse model of multifaceted premature ageing, based on engineered deficiency in the Xpf gene involved in transcription-coupled NER. [DOI] [PubMed] [Google Scholar]
- 44.Jeyapalan JC, Sedivy JM. Cellular senescence and organismal aging. Mechanisms of Ageing and Development. 2008;129:467–474. doi: 10.1016/j.mad.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26:8722–8730. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci U S A. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park Y, Gerson SL. DNA repair defects in stem cell function and aging. Annual review of medicine. 2005;56:495–508. doi: 10.1146/annurev.med.56.082103.104546. [DOI] [PubMed] [Google Scholar]
- 48.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 49.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 50.Collis SJ, Boulton SJ. Emerging links between the biological clock and the DNA damage response. Chromosoma. 2007;116:331–339. doi: 10.1007/s00412-007-0108-6. [DOI] [PubMed] [Google Scholar]
- 51.Collis SJ, et al. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol. 2007;9:391–401. doi: 10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- 52.Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci U S A. 2009;106:2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdel-Ghafar AN, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 54.McCulloch R, Barry JD. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15:119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Narisawa-Saito M, Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505–1511. doi: 10.1111/j.1349-7006.2007.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muylaert I, Elias P. Knock-down of DNA ligase IV/ XRCC4 by RNAi inhibits herpes simplex virus type I DNA replication. J Biol Chem. 2007;282:10865–10872. doi: 10.1074/jbc.M611834200. [DOI] [PubMed] [Google Scholar]
- 58.Li H, et al. Chk2 is required for HSV-1 ICP0-mediated G2/M arrest and enhancement of virus growth. Virology. 2008;375:13–23. doi: 10.1016/j.virol.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith J, Daniel R. Following the Path of the Virus: The Exploitation of Host DNA Repair Mechanisms by Retroviruses. ACS Chem. Biol. 2006;1:217–226. doi: 10.1021/cb600131q. [DOI] [PubMed] [Google Scholar]
- 60.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. A comprehensive overview of cancer-predisposing mutations and advances in cancer genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 62.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 63.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 64.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 65.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. References 64 and 65 provide evidence for activation of the DDR machinery in early human oncogenic lesions and models of oncogenic transformation, and propose that the DNA-damage checkpoint activated by oncogene-evoked replication stress and DNA breakage provides an inducible barrier against tumor progression. [DOI] [PubMed] [Google Scholar]
- 66.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 67.Kulkarni A, Wilson DM., 3rd The involvement of DNA-damage and -repair defects in neurological dysfunction. Am J Hum Genet. 2008;82:539–566. doi: 10.1016/j.ajhg.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007;145:1318–1329. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 69.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 70.Ljungman M, Lane DP. Transcription - guarding the genome by sensing DNA damage. Nat Rev Cancer. 2004;4:727–737. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- 71.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 72.Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- 73.Yang JL, Weissman L, Bohr VA, Mattson MP. Mitochondrial DNA damage and repair in neurodegenerative disorders. DNA Repair (Amst) 2008;7:1110–1120. doi: 10.1016/j.dnarep.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bartkova J, Rajpert-De Meyts E, Skakkebaek NE, Lukas J, Bartek J. DNA damage response in human testes and testicular germ cell tumours: biology and implications for therapy. Intl J Andrology. 2007;30:282–291. doi: 10.1111/j.1365-2605.2007.00772.x. [DOI] [PubMed] [Google Scholar]
- 76.Schumacher B, Garinis GA, Hoeijmakers JH. Age to survive: DNA damage and aging. Trends Genet. 2008;24:77–85. doi: 10.1016/j.tig.2007.11.004. A thought-provoking review of the evidence for causative links between DNA-damage accumulation and organismal ageing, which proposes the concept of a survival response that allows the organism's resources to be shifted from emphasis on growth, to survival of DNA damage and other stresses. [DOI] [PubMed] [Google Scholar]
- 77.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 78.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 79.Navarro S, et al. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol Ther. 2006;14:525–535. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 80.Reese JS, Liu L, Gerson SL. Repopulating defect of mismatch repair-deficient hematopoietic stem cells. Blood. 2003;102:1626–1633. doi: 10.1182/blood-2002-10-3035. [DOI] [PubMed] [Google Scholar]
- 81.Mocanu MM, Yellon DM. p53 down-regulation: a new molecular mechanism involved in ischaemic preconditioning. FEBS Lett. 2003;555:302–306. doi: 10.1016/s0014-5793(03)01260-2. [DOI] [PubMed] [Google Scholar]
- 82.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 83.Mercer J, Mahmoudi M, Bennett M. DNA damage, p53, apoptosis and vascular disease. Mutat Res. 2007;621:75–86. doi: 10.1016/j.mrfmmm.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 84.Schneider JG, et al. ATM-dependent suppression of stress signaling reduces vascular disease in metabolic syndrome. Cell Metab. 2006;4:377–389. doi: 10.1016/j.cmet.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 85.Kastan MB. DNA damage responses: mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial Award Lecture. Mol Cancer Res. 2008;6:517–524. doi: 10.1158/1541-7786.MCR-08-0020. [DOI] [PubMed] [Google Scholar]
- 86.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 87.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 88.Martin SA, Lord CJ, Ashworth A. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev. 2008;18:80–86. doi: 10.1016/j.gde.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 89.Jiang H, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes & Development. 2009 doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 91.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. References 90 and 91 document the potential of personalized cancer treatment, based on the exceptional sensitivity of tumour cells defective in BRCA1/BRCA2-dependent HR towards small molecule inhibitors of PARP1; these studies support the principle of synthetic-lethal relationships between complementary DDR pathways. [DOI] [PubMed] [Google Scholar]
- 92.Chen Z, et al. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int J Cancer. 2006;119:2784–2794. doi: 10.1002/ijc.22198. [DOI] [PubMed] [Google Scholar]
- 93.García-Cao I, et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pacher P, Szabo C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: the therapeutic potential of PARP inhibitors. Cardiovasc Drug Rev. 2007;25:235–260. doi: 10.1111/j.1527-3466.2007.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moroni F. Poly(ADP-ribose)polymerase 1 (PARP-1) and postischemic brain damage. Current Opinion Pharmacol. 2008;8:96–103. doi: 10.1016/j.coph.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 96.Guevara NV, Kim HS, Antonova EI, Chan L. The absence of p53 accelerates atherosclerosis by increasing cell proliferation in vivo. Nat Med. 1999;5:335–339. doi: 10.1038/6585. [DOI] [PubMed] [Google Scholar]
- 97.Andreassi MG. DNA damage, vascular senescence and atherosclerosis. J Mol Med. 2008;86:1033–1043. doi: 10.1007/s00109-008-0358-7. [DOI] [PubMed] [Google Scholar]
- 98.Lau A, et al. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat Cell Biol. 2005;7:493–500. doi: 10.1038/ncb1250. [DOI] [PubMed] [Google Scholar]
- 99.Smith JA, et al. Evidence that the Nijmegen breakage syndrome protein, an early sensor of double-strand DNA breaks (DSB), is involved in HIV-1 post-integration repair by recruiting the ataxia telangiectasia-mutated kinase in a process similar to, but distinct from, cellular DSB repair. Virol J. 2008;5:11. doi: 10.1186/1743-422X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moehle EA, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]