Abstract
Spermatogenesis in man starts with spermatogonial stem cells (SSCs), and leads to the production of sperm in ∼64 days, common to old and young men. Sperm from elderly men are functional and able to fertilize eggs and produce offspring, even though daily sperm production is more than 50% lower and damage to sperm DNA is significantly higher in older men than in those who are younger. Our hypothesis is that the SSC/spermatogonial progenitors themselves age. To test this hypothesis, we studied the gene expression profile of mouse SSC/progenitor cells at several ages using microarrays. After sequential enzyme dispersion, we purified the SSC/progenitors with immunomagnetic cell sorting using an antibody to GFRA1, a known SSC/progenitor cell marker. RNA was isolated and used for the in vitro synthesis of amplified and labeled cRNAs that were hybridized to the Affymetrix mouse genome microarrays. The experiments were repeated twice with different cell preparations, and statistically significant results are presented. Quantitative RT-PCR analysis was used to confirm the microarray results. Comparison of four age groups (6 days, 21 days, 60 days, and 8 months old) showed a number of genes that were expressed specifically in the older mice. Two of them (i.e. Icam1 and Selp) have also been shown to mark aging hematopoietic stem cells. On the other hand, the expression levels of the genes encoding the SSC markers Gfra1 and Plzf did not seem to be significantly altered by age, indicating that age affects only certain SSC/progenitor properties.
Introduction
Age has a deleterious effect on most tissues and organs, eventually leading to system failure. However, the mechanisms of aging in the various systems have been difficult to investigate and fully comprehend. Although men appear to be fertile until very late in life, there are definitely genetic defects in the offspring due to DNA damage and increased mutation rates that are associated with advanced paternal age (Tsitouras 1987). Quantitative studies of sperm parameters in men younger than 35 years or older than 55 years have shown that sperm motility and semen volume are inversely related to age, while sperm concentration is unaffected (Eskenazi et al. 2003, Levitas et al. 2007). Furthermore, histological studies have shown that age is associated with a decline in the number of Leydig and Sertoli cells, a thickening of the basement of the seminiferous tubules, and an increase in arrested divisions of germ cells (Tsitouras 1987). In addition, studies using the Brown Norway rat as a model for male reproductive aging have shown that advanced paternal age (24 months) has a significant effect on the frequency of neonatal death in the litter, although there seems to be no significant effect on fertility (Tsitouras 1987).
The relation between age and sperm DNA damage has been studied in men by Singh et al. (Tsitouras 1987). Sperm with damaged DNA were found at a significantly higher frequency in men aged 36–57 years than in those aged 20–35 years. In addition, a number of reports have suggested an association between advanced paternal age and a variety of genetic syndromes in the offspring, including schizophrenia, achondroplasia, Apert syndrome, autism, Down syndrome, and Marfan syndrome (Glaser et al. 2000, 2003, Cantor et al. 2007, Croen et al. 2007, Kolevzon et al. 2007). However, a common confounder in these reports is that the age of the mother is also advanced (>40 years), and thus, it is difficult to isolate the paternal influence. The best documented studies suggesting a link between paternal age and adverse outcomes in the offspring come from two Israeli groups (Malaspina et al. 2001, Reichenberg et al. 2006). In both these studies, there is a significant association between paternal age and schizophrenia or autism, even after controlling for maternal age and other confounding factors, i.e., gender of offspring, education, and socioeconomic status. Taken together, an increase of threefold for schizophrenia and fivefold for autism has been demonstrated for the offspring of fathers aged 50 years or older. Similarly, Wilkin et al. (1998) reported 154 new mutations of three different genes – FGFR3, FGFR2, and RET – causing six genetic disorders – Apert, Crouzon, and Pfeiffer syndromes, multiple endocrine neoplasias 2A and 2B, and achondroplasia, which all have a paternal origin.
Recently, Desai et al. (2009) reviewed the advances toward the understanding of the mechanism behind decreasing sperm quality with age in men, suggesting that reactive oxygen species (ROS) production is a possible cause (Cocuzza et al. 2008). Oxidative stress may result in unfavorable, physiological changes in the reproductive organs, including the epididymis and accessory glands (Sloter et al. 2006). Damage to the epididymis may affect normal sperm maturation processes. Reduced semen volume caused by damaged accessory glands is another physical manifestation of oxidative stress (Kidd et al. 2001, Levitas et al. 2007). Finally, it was reported that oxidative stress compromised normal chromatin packaging and integrity in the spermatozoa of Brown Norway aged rats, suggesting a direct effect of ROS production on male fertility (Zubkova & Robaire 2006).
Stem cells are present in most adult tissues, and in the testes, the spermatogonial stem cells (SSCs) are located on the basement membrane, interacting with surrounding Sertoli cells that produce the stem cell niche. To achieve optimal tissue maintenance, the niche has to be strictly regulated (Kostereva & Hofmann 2008, de Rooij 2009). Aberrant regulation of the stem cell/niche compartments could be the result of aging, leading to compromised stem cell renewal capability and, therefore, tissue failure. The effect of aging on SSCs and their niche has been previously addressed by two groups that used similar experimental designs but reached somewhat contradicting results (Ryu et al. 2006, Zhang et al. 2006). Ryu et al. (2006) who used the SSC transplantation technique to assay stem cell number and function in young (2–12 months), aging (14–18 months), and old (20–22 months) mice observed a decline in fertility after mating them with females and in testis weight of the mice aged 14 months or older, as well as a high percentage of seminiferous tubules devoid of sperm in aging and old males (16–24 months). Interestingly, expression of Gfra1 could be only detected in young and aging fertile animals and not in the aging infertile animals. In addition, the number of functional stem cells, as assayed by transplantation into busulphan-treated recipient testes, was 3–4 times lower in mice aged 14–18 months than in those aged 2–12 months, while in mice older than 20 months, the SSC activity was nearly absent. However, no significant difference was observed between the different age groups of the donors, when the size of the established colonies was compared in the recipient testis, or when serial transplantations were performed in young busulphan-treated testes. Taken together, the results of this study supported the notion that stem cells are long-lived and potentially immortal, and that declining function of supporting cells that constitute the stem cell niche might be a major factor in age-related male infertility. This conclusion, however, is not supported by the results reported by Zhang et al. (2006), who demonstrated that a decline in SSC function due to aging is independent of the niche, as they observed a decline in colony number and length when SSCs from 2-year-old atrophied testis were transplanted into young, busulphan-treated recipients. They also noted that SSCs from young, fertile males were unable to establish colonies of spermatogenesis when transplanted into the testes of old, infertile males. The difference between the two studies remains to be resolved. However, in both the above-mentioned studies, there was a decline in SSC function in the aged mice.
Similarly, Leydig cells have been shown to be affected by age, by both intrinsic and extrinsic factors, leading to reduced testosterone production, as reviewed recently by Chen et al. (2009). Although it is not clear whether the Leydig stem cells are also affected by age, experiments using Brown Norway rats demonstrated that the steroidogenic capacity of the Leydig cells is reduced by about 50% in aged rats, and provided evidence that ROS, derived from the mitochondrial electron transport chain, steroidogenesis, and/or macrophages, might cause damage to Leydig cell membrane lipids and proteins by altering the redox environment of the aging Leydig cells, and that this, in turn, might result in the reduced LH signaling that characterizes aged Leydig cells. Reduced LH signaling results in decreased cAMP production, less efficient steroidogenesis, and reduced cholesterol transport, all of which are observed in aged Leydig cells. Although it is not clear whether the Leydig cells are part of the SSC niche, the selective position of the SSC niches in proximity to the interstitial tissue that has been observed in mammalian testicular sections (Yoshida et al. 2007) has led to the hypothesis that factors coming from outside the seminiferous tubules must determine either directly or indirectly via the Sertoli cells whether self-renewal or differentiation of an SSC will be favored at a particular site of the tubule (de Rooij 2009). In support of this model, colony-stimulating factor 1 (CSF1) produced by the Leydig cells has been shown to promote self-renewal or in vitro proliferation of SSCs (Ryan et al. 2001, Kokkinaki et al. 2009, Oatley et al. 2009).
In the present study, our aim was to investigate whether age differences among fertile male mice reflect differences in gene expression profiles of their spermatogonial stem/progenitor cells. The A-single spermatogonia are the SSCs, and they give rise to the A-paired (Apr) and A-aligned (Aal) spermatogonia. The Apr and the Aal are the progenitor spermatogonia giving rise to more differentiated cell types. The SSCs as well as some Apr and Aal possess the GFRA1. Therefore, we compared the gene expression of SSC/progenitors isolated from mice belonging to four different age groups: 6 days old (immature), 21 days old (young), 60 days old (young adult), and 8 months old (older fertile adult). The 6-day-old mice contain type A spermatogonia, but lack more differentiated germ cell types. SSC/progenitors were purified from the germ cell population with immunomagnetic cell sorting with the antibody to GFRA1, an established SSC/progenitor marker (de Rooij & Grootegoed 1998, Hofmann et al. 2005).
Results
Decline in spermatogonial stem/progenitor cell number in the testes of the 8-month-old mice
We used immunomagnetic cell sorting (magnetic activated cell sorting; MACS) with anti-GFRA1 to separate the spermatogonial stem/progenitor cells from the testes of mice from four different age groups (6 days, 21 days, 60 days, and 8 months old). We used eighty 6-day-old mice, forty 21-day-old mice, twenty 60-day-old mice, and twenty 8-month-old mice in each isolation, and we repeated the isolations twice. No cell freezing was involved. The dissection of the testes from the mice, the enzymatic digestion of the testes, and the immunomagnetic cell sorting were performed on the same day. Following the overnight antibody incubation, RNA was isolated from the sorted cells the next day. RNA samples from each age group were frozen at −80 °C until they were used for the microarray experiment. The number of cells used in each preparation and the number of cells in the positive fraction after immunomagnetic cell sorting were measured on a hematocytometer, and are shown in Fig. 1. In addition, immunostaining with anti-GFRA1 on a sample (∼1/10) of the positive fraction was performed (Fig. 1), and the positively staining cells were also counted. Specifically, we counted a total number of 500 DAPI-stained cells from the GFRA1-positive fraction of each preparation, and calculated the percentage of those that were also positive for anti-GFRA1 after immunostaining. The s.d. from the two different cell isolations was also calculated (Fig. 1). We then estimated the percentage of SSC/progenitor cells in the tissue using the following formula: (Number of GFRA1-positive cells after MACS/Cell number before MACS) × Percentage of cells staining with anti-GFRA1 × 100 (%). Although the calculated percentage of SSC/progenitor cells was approximately the same (0.6–0.7% of the total number of cells) between the 6-, 21-, and 60-day-old mice, we observed a significant decrease (∼10×) in the 8-month-old age group (Fig. 1).
Figure 1.
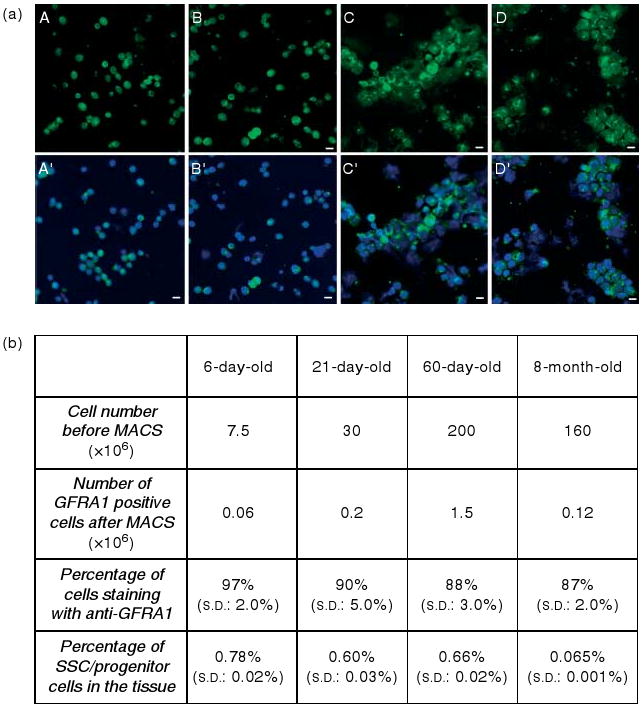
(a) GFRA1-positive cells isolated by immunomagnetic cell sorting from 6-day- (A, A′), 21-day- (B, B′), 60-day- (C, C′) and 8-month-old mouse testes, and stained with goat anti-GFRA1 and anti-goat-FITC. Nuclei were stained by DAPI. GFRA1-positive cells are shown in A–D, and merged with DAPI in A′, B′, C′, and D′ respectively. Scale bars = 10 μm. (b) Cell numbers before MACS in each age group, number of GFRA1-positive cells after MACS, percentage of cells stained with anti-GFRA1, and the estimated percentage of SSC/progenitor cells in the tissue. For example, the percentage of SSC/progenitors for the 6-day-old testis is (0.06/7.5) × 0.97 × 100 = 0.78%. s.d. from the two different cell isolations are also shown.
Comparison of the gene expression profiles of GFRA1-positive and GFRA1-negative cells in the different age groups by microarray analysis
Evaluation of the microarray experiments
The fidelity of microarray experiments relies on their reproducibility. Therefore, principal component analysis (PCA) mapping was performed on the microarray data obtained from two independent experiments, with samples representing different cell isolations, to test the similarity of the results derived from each of them. For each age group, this analysis showed that the duplicate GFRA1-positive experiments map closer to each other than to the GFRA1-negative experiments. Therefore, the gene expression profile that we identified for the GFRA1-positive cells in each of the four age groups is reliable. Finally, the GFRA1-positive cells of the 8-month-old age group map closer to the GFRA1-positive cells of the 60-day-old age group than to those of the 6-day-old or 21-day-old age group, which may reflect the gene expression changes during development and aging. The PCA mapping is shown in Fig. 2.
Figure 2.
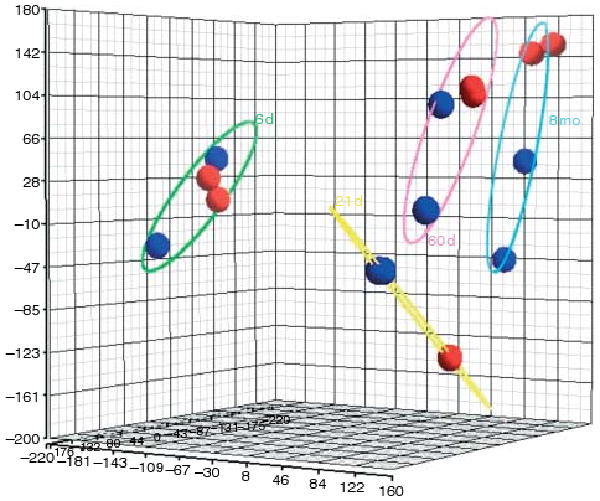
PCA mapping for the microarray data. The gene expression data for each cell type are represented by different colors. GFRA1-positive cell gene expression is illustrated in red, and GFRA1-negative cell gene expression is illustrated in blue. The elliptical lines represent the four age groups, 6 days old (6 d), 21 days old (21 d), 60 days old (60 d), and 8 months old (8 mo). Duplicate red and blue circles represent the two different microarray experiments, representing different RNA isolations. In the 21-day-old age group, the duplicate experiments were almost identical. In all the groups, the gene expression profile of the GFRA1-positive cells appears distinct from that of the GFRA1-negative cells.
Differentially expressed genes between the different age groups
As ‘differentially expressed’, we defined the genes that were 1) over-expressed in the GFRA1-positive versus the GFRA1-negative cells, with at least a twofold increase, within the same age group, and 2) present in the GFRA1-positive cells of one, two, and three but not four age groups. Although we have optimized the isolation of GFRA1-positive cells from adult mice with immunomagnetic cell sorting, we could not absolutely exclude contamination from other cell types in the adult and old age groups. Therefore, we chose to compare the gene expression levels between the GFRA1-positive and GFRA1-negative cells within each age group and not between age groups to minimize the risk of contamination from genes expressed in the contaminating cell types that would appear as ‘over-expressed’ in the adult and old age groups versus the immature group, where they are normally not expressed. Based on the above-mentioned criteria, 3321 Affymetrix probes corresponding to 2819 different genes were shown to be differentially expressed among the four age groups by the microarray analysis (Supplementary Table 2, see section on supplementary data given at the end of this article).
Confirmation of microarray results by quantitative RT-PCR
Our goal for the microarray analysis is to determine age-related changes in gene expression of spermatogonial stem/progenitor cells. To confirm the microarray results, we chose a number of genes that based on the microarray were over-expressed specifically in the GFRA1-positive cells of the 8-month-old age group (Gene set 1), or over-expressed in the GFRA1-positive cells of the immature testis and not in the 8-month-old age group (Gene set 2). Gene set 1 included the genes of G protein-coupled receptor-116, -146, and -56 (Gpr116, Gpr146, and Gpr56), as well as growth factor receptor-bound protein 2 (Grb2), intercellular adhesion molecule 1 (Icam1), LIM and senescent cell antigen-like domains containing 1 (Lims1), Lin-52 Caenorhabditis elegans homolog, selectin P (Selp), and triple functional domain (Trio). Gene set 2 included the genes of Gpr107, protein tyrosine kinase-binding protein (Tyrobp), similar to MAD homolog 4 in Drosophila (Smad4), membrane-spanning 4-domains, subfamily A, member 7 (Ms4a7), and mannose receptor C type 1 (Mrc1). Notably, Gpr107 and Gpr146 were selected based on their fold change between GFRA1-positive and GFRA1-negative cell populations in the 6-day-old and 8-month-old mice respectively (fourfold for Gpr107 and threefold for Gpr146). Both these genes had a P value higher than the threshold of 0.05 (0.08 for Gpr107 and 0.07 for Gpr146), and thus, are not included in Supplementary Table 2. In addition, three of the above-mentioned genes (Icam1, Selp, and Smad4) were selected because they have been reported as affected by age in hematopoietic stem cells (HSCs; see Discussion). The same pattern of differential expression that was observed using microarray for all the 14 genes that we tested was also confirmed by quantitative RT-PCR (qRT-PCR; Table 1), using RNA from three independent cell isolations of the GFRA1-positive cells from the testes of each of the four age groups. Detailed qRT-PCR results are shown in Supplementary Figure 1, see section on supplementary data given at the end of this article.
Table 1.
Genes showing differential expression of spermatogonial stem cell/progenitor cells among the four mouse age groups, confirmed by real-time quantitative PCR.
| Gene name | Gene symbol | 6 days | 21 days | 60 days | 8 months |
|---|---|---|---|---|---|
| G protein-coupled receptor 107 | Gpr107 | + | − | − | − |
| G protein-coupled receptor 116 | Gpr116 | − | − | − | + |
| G protein-coupled receptor 146 | Gpr146 | − | − | − | + |
| G protein-coupled receptor 56 | Gpr56 | − | − | − | + |
| Growth factor receptor-bound protein 2 | Grb2 | − | − | − | + |
| Intercellular adhesion molecule 1 | Icam1a | − | − | − | + |
| LIM and senescent cell antigen-like domains 1 | Lims1 | − | − | − | + |
| Lin-52 homolog (C. elegans) | Lin52 | − | − | − | + |
| Selectin, platelet | Selpa | − | − | − | + |
| Triple functional domain (PTPRF interacting) | Trio | − | − | − | + |
| TYRO protein tyrosine kinase-binding protein | Tyrobp | + | + | − | − |
| Similar to MAD homolog 4 (Drosophila) | Smad4a | + | − | − | − |
| Membrane-spanning 4-domains, subfamily A, member 7 | Ms4a7 | + | + | + | − |
| Mannose receptor, C type 1 | Mrc1 | + | + | − | − |
The genes that were also found to mark aging hematopoietic stem cells (see Discussion).
We also tested the expression of known SSC/progenitor cell markers (i.e. Gfra1, Plzf, and Pou5f1) by qRT-PCR. Gfra1 and Plzf levels of expression did not show significant differences between the four age groups, which is in agreement with the microarray results, while the expression of Pou5f1 was higher in the 6-day-old SSC/progenitors than in the other three age groups. However, Pou5f1 was not shown as differentially expressed by the microarray analysis (Table 2 and Supplementary Figure 1).
Table 2.
The expression of mouse spermatogonial stem cell/progenitor cell marker genes tested by real-time quantitative PCR.
| Gene name | Gene symbol | 6 days | 21 days | 60 days | 8 months |
|---|---|---|---|---|---|
| GDNF family receptor α1 | Gfra1 | +++ | +++ | +++ | +++ |
| Promyelocytic leukemia zinc finger | Plzf | +++ | +++ | +++ | +++ |
| POU domain, class 5, transcription factor 1 | Pou5f1 | ++ | + | + | + |
Discussion
A significant number of recent studies support the notion that stem cells age. Most recently, Choi & Artandi (2009) reported that melanocyte stem cells undergo DNA damage during aging, which results in cell death due to premature differentiation. In addition, Geiger & Rudolph (2009) reviewed the effect of aging on HSCs in mice, and concluded that aging is associated with a reduction in both HSC frequency and function. Aged HSCs demonstrate reduced differentiation toward the lymphoid lineage. The effect of aging in spermatogenesis has been recently reported for Drosophila. Boyle et al. (2007) showed that the number of male germline stem cells (GSCs) decrease in older flies due to a decline in self-renewal factors. Furthermore, Cheng et al. (2008) reported that the decline in spermatogenesis observed in older male flies can be attributed to the inability of GSCs to maintain the correct centrosome orientation during asymmetric divisions, which significantly reduces the number of functional GSCs. Although the molecular mechanisms underlying the aging of stem and progenitor cells are still unclear, it seems that DNA replication defects and loss of transcriptional and epigenetic control are common aging mechanisms.
Gene expression studies of spermatogonial stem/progenitor cells have been performed in both immature and young adult mice, and have led to the characterization of their molecular phenotype. The GDNF receptor GFRA1 has been shown to mark mouse spermatogonial stem/progenitor cells by our group and others, and also to be required for the self-renewal of SSCs (Meng et al. 2000, Hofmann et al. 2005, Naughton et al. 2006, He et al. 2007). In order to reveal the downstream molecules affected by GDNF, Oatley et al. (2006) performed a gene expression study of cultured SSCs in the presence or absence of GDNF and identified six genes affected by GDNF, and thus important for the self-renewal and survival of SSCs, i.e. B-cell CLL/lymphoma 6, member B (Bcl6b), early growth response 2 (Egr2), early growth response 3 (Egr3), Ets variant 5, ERM (Etv5), LIM homeobox 1 (Lhx1), and tetraspanin 8 (Tspan8). POU5F1 (OCT-4), a marker of pluripotent stem cells (Nichols et al. 1998), was also shown to be a nuclear marker for spermatogonial stem and progenitor cells (Ohbo et al. 2003, Ohmura et al. 2004, Braydich-Stolle et al. 2005), and it is required for the self-renewal of mouse SSCs (Dann et al. 2008). PLZF is another nuclear transcription factor that has been shown to mark mouse SSCs and progenitors (Buaas et al. 2004, Costoya et al. 2004), while CD49f, also known as integrin α6 (ITGA6), THY1 (CD90), and GPR125 are surface markers for mouse SSCs and progenitors (Shinohara et al. 1999, Kubota et al. 2003). Various high-throughput gene expression studies have also identified molecules important for SSCs and their progenitors, such as procollagen type I α2 (COLA2) and matrix metallopeptidase 2, which were identified by our group both by serial analysis of gene expression (Wu et al. 2004) and by microarray analysis of GFRA1-expressing cells in the immature mouse testis (Kokkinaki et al. 2009). In the latter study, we also identified the receptor of the CSF1, CSF1R, to be highly over-expressed in the SSC/progenitor cells of the immature mouse testis than in the nonstem cells, and also showed that CSF1 could stimulate spermatogonial cell proliferation in vitro. Interestingly, the positive effect of CSF1 on mouse SSC self-renewal was recently confirmed by Oatley et al. (2009). Finally, the most complete developmental gene expression study of mouse spermatogenesis at ages ranging from day 0 to day 56 post partum was performed by Shima et al. (2004). They provided a thorough resource of gene expression data by analyzing specific patterns of gene expression and identifying clusters of transcripts enriched or specific for each testicular cell type, describing both somatic and germ cells. However, none of the above-mentioned gene expression studies has used animals older than 2 months.
Therefore, the current study focuses on the identification of genes that are expressed in spermatogonial stem/progenitor cells and show an age-related pattern of expression. The major difficulty for this study was to exclude contamination of the GFRA1-positive fraction from spermatocytes and/or spermatids that could create false differentially expressed genes between the adult or old and the immature testes, due to the absence of those cell types in the latter. To minimize this problem, we optimized our cell sorting technique of the adult and old mice testicular cells, with additional washing steps and an additional passage through the column. However, we recognize that even a higher than 85% purity does not necessarily exclude the possibility of contamination from spermatocyte and spermatid genes in our results, and understand the need to confirm our results by other methods, as we did for a number of the identified genes. Moreover, we were able to identify genes that have also been shown to be affected by age in other types of stem cells. We identified genes that were down-regulated (e.g. Gpr107, Tyrobp, Smad4, Ms4a7, and Mrc1) and genes that were up-regulated (e.g. Gpr116, Gpr146, Gpr56, Grb2, Icam1, Lims1, Lin52, Selp, and Trio) in the 8-month-old mice. Several of these have been identified in other age-related gene expression studies of stem cells. Specifically, Selp was also noted to be up-regulated with age by Chambers et al. (2007) in a comparative gene expression study of HSCs isolated from young and old mice. SELP is a cell surface adhesion molecule that serves as a marker for physiological stress, such as inflammation, aging, and cardiovascular disease (Chambers et al. 2007). Similarly, ICAM1, a transmembrane glycoprotein that regulates the key signaling pathways associated with cell-to-cell interactions (Rahman et al. 2000), is up-regulated with aging, common in our study and that of Chambers et al. (2007). Notably, ICAM1 protein levels have also been shown to increase in cultured human pulmonary artery endothelial cells by aging (late passage) and induction of inflammation response by tumour necrosis factor α, resulting in an age-dependent transmembrane signaling disorder (Zhou et al. 2006). In addition, LIN52, which is up-regulated in the spermatogonial stem/progenitor cells of the old mice, has also been found as a component of a multisubunit protein complex that acts as a transcriptional repressor of cell-cycle-dependent genes in quiescence (Litovchick et al. 2007). On the other hand, membrane-spanning 4-domains, subfamily A, member 7 (MS4A7) which was found to be down-regulated in the aging spermatogonial/progenitor cells, is also known to be involved in the proliferation of human blastocyst inner cell mass cells, progenitors of human embryonic stem cells (Brink et al. 2008), suggesting an age-induced decline in stem cell proliferation. It has also been realized that the molecular mechanism of aging involves DNA damage due to the accumulation of ROS and also a stimulation of the DNA repair mechanisms in response to the damage (Johnson et al. 1999). In our study, we found molecules that are known to be involved in DNA repair (e.g. ERCC1, ERCC5, and RAD18) or induced by DNA damage (e.g. GADD45B) or oxidative stress (OSGIN2) to be specifically up-regulated in the 8-month-old SSC/progenitor cells (Table 3), probably indicative of the first signs of aging in the fertile mice. It would be interesting to monitor the expression of these genes at more advanced ages, and perhaps to correlate infertility with reduced expression of DNA repair genes. In conclusion, the genes that have been identified in this study may be a useful resource to aid in the understanding of the effect of aging on mammalian spermatogenesis and probably enhance regenerative efforts of aged testis in the future.
Table 3.
DNA damage-, oxidative stress-induced and DNA repair genes over-expressed in the 8-month-old mouse spermatogonial stem cell/progenitor cells.
| Probe ID | Gene symbol | Description | Fold changea |
|---|---|---|---|
| 1450971_at | Gadd45b | Growth arrest and DNA-damage-inducible 45β | 11.8 |
| 1444184_at | Alkbh3 | alkB, alkylation repair homolog 3 (Escherichia coli) | 7.7 |
| 1441568_at | Osgin2 | Oxidative stress-induced growth inhibitor family member 2 | 5.0 |
| 1453539_at | Alkbh8 | alkB, alkylation repair homolog 8 (E. coli) | 3.7 |
| 1428306_at | Ddit4 | DNA-damage-inducible transcript 4 | 3.3 |
| 1451751_at | Ddit4l | DNA-damage-inducible transcript 4-like | 3.0 |
| 1450935_at | Ercc5 | Excision repair cross-complementing rodent repair deficiency 5 | 2.6 |
| 1437447_s_at | Ercc1 | Excision repair cross-complementing rodent repair deficiency 1 | 2.5 |
| 1429850_x_at | Alkbh4 | alkB, alkylation repair homolog 4 (E. coli) | 2.5 |
| 1451928_a_at | Rad18 | RAD18 homolog (S. cerevisiae) | 2.4 |
Fold change represents the change in transcript levels between GFRA1-positive and GFRA1-negative cells in the 8-month-old age group. No significant change has been observed by microarray analysis for these genes in the other younger age groups.
Materials and Methods
Isolation of germ cells from mouse testes
Male Balb/c mice that were 6 days old, 21 days old, 60 days old, and 8 months old (Charles River Breeding Laboratories, Wilmington, MA, USA) were killed by carbon dioxide inhalation in accordance with the protocols approved by the Georgetown University Animal Care and Use Committee. Briefly, the testes were decapsulated and minced in ice-cold DMEM–F12. Leydig cells, other interstitial cells, and peritubular myoid cells were eliminated by a first enzymatic digestion with collagenase IV (1 mg/ml) and DNase I (2 μg/ml) at 34 °C for 10 min and extensive washes with DMEM to obtain only the seminiferous tubules devoid of myoid cells. Germ cells and Sertoli cells were isolated by a second enzymatic digestion using collagenase IV (1 mg/ml), DNase I (2 μg/ml), hyaluronidase (1.5 mg/ml), and trypsin (1 mg/ml) under the same conditions. The resulting cell suspension, containing mainly germ cells and Sertoli cells, was cultured for 2 h on fetal bovine serum-coated dishes to further eliminate contamination of Sertoli cells by differential plating. For the differential plating, 3 × 106 cells were plated on each petri dish of 10-mm diameter.
Separation of SSCs by immunomagnetic cell sorting
Germ cells isolated by enzymatic dispersion of the testes were resuspended in 1 ml of DMEM containing 10% Nu-serum (BD Biosciences, San Jose, CA, USA) and incubated with 25 μl (200 ng/μl) of rabbit anti-GFRA1 antibody (H-70, Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 16 h at 4 °C with continuous rotation. The cells were washed three times with ice-cold PBS containing 0.5% BSA and 2 mM EDTA (wash buffer), and were then resuspended in 80 μl of the wash buffer and incubated with 20 μl of goat anti-rabbit IgG MicroBeads (Miltenyi Biotech, Auburn, CA, USA) for 20 min at 4 °C with rotation. The labeled cells were first filtered through a 30-μm pore size mesh (Miltenyi Biotech) to remove cell aggregates, and were then immediately sorted through a separation MS column attached to a MiniMACS separator (Miltenyi Biotech). GFRA1-positive cells, the magnetically labeled cells, were retained within the column, whereas nonlabeled cells passed through the column and were collected as the GFRA1-negative cell fraction. To increase the purity of the separation, the column was washed three times with 500 μl of wash buffer. In order to collect the GFRA1-positive cells, the column was removed from the magnetic separator, and 1 ml of wash buffer was added to the reservoir. The positive cell fraction was separated once more through a second MS column to achieve high purity. GFRA1-positive and GFRA1-negative cells were counted on a hematocytometer and used immediately for RNA isolation. Two independent cell isolation experiments were performed for each age group.
RNA isolation
Total RNA was extracted from the cells using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. The RNA samples were treated with RNase-free DNase I (Qiagen) to remove traces of genomic DNA. The concentration and quality of total RNA were determined using the NanoDrop-1000 Spectrophotometer.
cDNA synthesis and target preparation
cDNA was synthesized from 30 ng total RNA and amplified using the Ovation Biotin RNA Amplification and Labeling System, version 1.0 (NuGEN Technologies, San Carlos, CA, USA, Cat no: 2300-12). The amplified cDNAs were purified using the Zymo Research (Orange, CA, USA) Clean and Concentrator-25 columns and, subsequently, fragmented and biotin labeled using the reagents of the Ovation system. The biotin-labeled and fragmented cDNAs were purified using the DyeEx 2.0 Spin kit (Qiagen, Cat no: 63204). Finally, their yield and purity were measured by absorbance at 260, 280, and 320 nm. Only cDNA samples showing an OD260–OD320/OD280–OD320 ratio higher than 1.8 were used for microarray hybridization.
Hybridization on mouse genome 430 v.2 cDNA arrays (Affymetrix, Santa Clara, CA, USA)
Each of the arrays used contains over 39 000 mouse transcripts. Each of the transcripts is represented on the array by multiple probes to ensure maximum accuracy and reproducibility of the hybridization results. The sequences from which these probe sets were derived were selected from GenBank, dbEST, and RefSeq. The sequence clusters were created from the UniGene database (Build 107, June 2002), and were then refined by analysis and comparison with the publicly available draft assembly of the mouse genome from the Whitehead Institute for Genome Research (MGSC, April 2002). The Affymetrix protocol for eukaryotic sample and array processing (http://www.affymetrix.com/support/technical/manual/expression_manual.affx) was followed by staining, washing, and scanning of the arrays. Briefly, the fragmented cDNA probe (0.011 μg/μl) was mixed with 50 pM of the control oligonucleotide B2 and 1.5, 5, 25, and 100 pM of each of the eukaryotic hybridization controls (bioB, bioC, bioD, and cre respectively). Herring sperm DNA (0.1 mg/ml), BSA (0.5 mg/ml), 10% DMSO, and 1× hybridization buffer were also added at a total volume of 200 μl (49 format array), and the mix was heated to 99 °C for 5 min before it was applied to the arrays which had been pre-hybridized with 1× hybridization buffer for 10 min at 45 °C. The hybridization was performed for 16 h at 45 °C with 60-r.p.m. rotation. Arrays were washed on Affymetrix's GeneChip Fluidics Station 450 using a primary streptavidin phycoerythrin (SAPE) stain, subsequent biotinylated antibody stain, and secondary SAPE stain, following the EukGE-WS2v5 protocol. Arrays were scanned on Affymetrix's GeneChip Scanner 3000 7G with AutoLoader. Scanned images obtained by the Affymetrix GeneChip Operating Software (GCOS) v1.4 were used to extract raw signal intensity values per probe set on the array and calculate detection calls (absent, marginal, or present). All raw chip data were scaled in GCOS to normalize signal intensities for interarray comparisons.
Statistical analysis
Raw signal intensity and detection call data obtained per array from the GCOS were uploaded into the Partek Genomics Suite software (Partek Inc., St Louis, MO, USA) for statistical analysis. The t-tests were applied to locate genes whose expression levels are statistically significant in differentiating expression between GFRA1-positive and GFRA1-negative cells in each of the four age groups (6 days, 21 days, 60 days, and 8-months old). To check the overall general trends of expression over the two cell populations and compare the results of the two different experiments for each cell population, PCA was performed. For each cell population, genes that had a maximum P value <0.05 and false discovery rate <0.01 were selected. From these gene lists, those genes that showed consistent fold changes above a minimum threshold (>2-fold increase or decrease) were identified as ‘differentially expressed’ between the GFRA1-positive and GFRA1-negative cell populations. Finally, we compared the genes that were over-expressed in GFRA1-positive cells between the four age groups applying the above-mentioned criteria.
RT, qPCR and data analysis
For the qRT-PCR analyses, the SuperScript III First-Strand Synthesis System (Invitrogen) was used to perform RT with 500 ng of the oligo-dT primer and 25 ng of total RNA from GFRA1-positive and GFRA1-negative germ cells isolated from the four different age groups of mouse testes. The RNA was treated with RNAse-free Dnase I (Qiagen) to remove traces of genomic DNA. Reactions were performed in duplicates; RT enzyme was added in one of the two, with the other serving as a negative control for the presence of genomic DNA in the samples. Primers were designed for real-time qPCR using the PrimerQuest tool (www.idtdna.com/Scitools/Applications/Primerquest/, Integrated DNA Technologies, San Diego, CA, USA). All primers had similar length (22–24 nucleotides) and ∼50% GC content, and melting temperatures (Tm) were 58–60 °C (Supplementary Table 1, see section on supplementary data given at the end of this article). To avoid false positive results due to contamination by genomic DNA, we chose to design the primers spanning at least one intron in the genomic sequence, which was possible in most cases, except for genes Gpr146, Actb, and Pou5f1 for which the primers were designed on the same exon. The expected PCR product sizes for all the primer sets that were designed were 100–120 bp. Real-time qPCR was carried out in triplicates on an ABI7900HT (Applied Biosystems, Foster City, CA, USA). Reactions were performed in 96-well plates at a final volume of 25 μl. PCR mixture contained 1× QuantiTect SYBR Green PCR master mix (Qiagen), 6 pmol of each primer, and 1 μl of the RT reaction product. Cycling parameters shared for all genes were 95 °C for 15 min, followed by 40 cycles at 94 °C for 15 s, 58 °C for 30 s, and 72 °C for 30 s. Finally, a temperature-determining dissociation step was performed at 95 °C for 15 s, 60 °C for 15 s, and 95 °C for 15 s at the end of the amplification phase. For all primer combinations, PCR product authenticity was assessed by gel electrophoresis. To normalize the data obtained from different RNA samples, the mouse β-actin gene (Actb) was used as an internal control. For the construction of a standard curve, a quantified cDNA (mouse testis-derived Biobank cDNA purchased from PrimerDesign Ltd, Southampton, UK) was used as a template for all the primer sets that were assayed and for β-actin at an initial concentration of 4 ng/μl and three serial dilutions (1/10, 1/100, and 1/1000).
QPCR data were collected using SDS 2.2 software (Applied Biosystems), and used to estimate the cycle threshold (Ct) for each reaction replicate. Once Ct for all replicates was obtained, analyses were conducted using Qgene software (Muller et al. 2002). Briefly, for each primer combination, the amplification efficiency (E) was determined as E = 1(−1/slope), where the slope was estimated by plotting the Ct in a serial dilution of cDNA. To compare the mRNA levels among different age groups, the normalized expression (NE) of each target gene (assayed in triplicates) was related to Actb reference gene expression levels by the formula: NE = (Etarget)Ct target/(Eref)Ct ref. For each gene, differences in expression levels among the different age groups were analyzed by one-way ANOVA. Differences were considered statistically significant at P < 0.05.
Immunostaining
Small fraction of cells (500–1000 cells/slide) were attached on microscope slides using the Cell Adherence Solution (Crystalgene Inc., Commack, NY, USA), fixed with 4% paraformaldehyde, and stored at 4 °C until they were used for immunostaining. The cells were then blocked with 10% normal donkey serum in 1 × PBS and 0.1% Tween-20 for 1 h. The primary antibody, goat anti-GFRA1 (Santa Cruz Biotechnology Inc.), was diluted at 1:100 in 1 × PBS containing 1% BSA and 0.1% Tween-20, and was incubated at 4 °C for 12 h. The fluorescence-conjugated secondary antibody (donkey anti-goat FITC, Jackson Immunoresearch, West Grove, PA, USA) was diluted 1:250 in 1 × PBS containing 1% BSA and 0.1% Tween-20, and was incubated at room temperature for 1 h. Nuclei were stained fluorescently with DAPI.
Supplementary Material
Acknowledgments
Funding: This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), Eunice Kennedy Shriver National Institute of Child Health and Human Development (W-Y Chan), and NIH grants HD044543 (M-C Hofmann) and HD033728 (M Dym).
Footnotes
Supplementary data: This is linked to the online version of the paper at http://dx.doi.org/10.1530/REP-09-0566.
Declaration of interest: The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1:470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Braydich-Stolle L, Nolan C, Dym M, Hofmann MC. Role of glial cell line-derived neurotrophic factor in germ-line stem cell fate. Annals of the New York Academy of Sciences. 2005;1061:94–99. doi: 10.1196/annals.1336.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink TC, Sudheer S, Janke D, Jagodzinska J, Jung M, Adjaye J. The origins of human embryonic stem cells: a biological conundrum. Cells, Tissues, Organs. 2008;188:9–22. doi: 10.1159/000112843. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nature Genetics. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Cantor RM, Yoon JL, Furr J, Lajonchere CM. Paternal age and autism are associated in a family-based sample. Molecular Psychiatry. 2007;12:419–421. doi: 10.1038/sj.mp.4001966. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biology. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ge RS, Zirkin BR. Leydig cells: from stem cells to aging. Molecular and Cellular Endocrinology. 2009;306:9–16. doi: 10.1016/j.mce.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Artandi S. Stem cell aging and aberrant differentiation within the niche. Cell Stem Cell. 2009;5:6–8. doi: 10.1016/j.stem.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Cocuzza M, Athayde KS, Agarwal A, Sharma R, Pagani R, Lucon AM, Srougi M, Hallak J. Age-related increase of reactive oxygen species in neat semen in healthy fertile men. Urology. 2008;71:490–494. doi: 10.1016/j.urology.2007.11.041. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nature Genetics. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Croen LA, Najjar DV, Fireman B, Grether JK. Maternal and paternal age and risk of autism spectrum disorders. Archives of Pediatrics & Adolescent Medicine. 2007;161:334–340. doi: 10.1001/archpedi.161.4.334. [DOI] [PubMed] [Google Scholar]
- Dann CT, Alvarado AL, Molyneux LA, Denard BS, Garbers DL, Porteus MH. Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated during retinoic acid-induced differentiation. Stem Cells. 2008;26:2928–2937. doi: 10.1634/stemcells.2008-0134. [DOI] [PubMed] [Google Scholar]
- Desai N, Sabanegh E, Jr, Kim T, Agarwal A. Free radical theory of aging: implications in male infertility. Urology. 2009;75:14–19. doi: 10.1016/j.urology.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Eskenazi B, Wyrobek AJ, Sloter E, Kidd SA, Moore L, Young S, Moore D. The association of age and semen quality in healthy men. Human Reproduction. 2003;18:447–454. doi: 10.1093/humrep/deg107. [DOI] [PubMed] [Google Scholar]
- Geiger H, Rudolph KL. Aging in the lympho-hematopoietic stem cell compartment. Trends in Immunology. 2009;30:360–365. doi: 10.1016/j.it.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Glaser RL, Jiang W, Boyadjiev SA, Tran AK, Zachary AA, Van Maldergem L, Johnson D, Walsh S, Oldridge M, Wall SA, et al. Paternal origin of FGFR2 mutations in sporadic cases of Crouzon syndrome and Pfeiffer syndrome. American Journal of Human Genetics. 2000;66:768–777. doi: 10.1086/302831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser RL, Broman KW, Schulman RL, Eskenazi B, Wyrobek AJ, Jabs EW. The paternal-age effect in Apert syndrome is due, in part, to the increased frequency of mutations in sperm. American Journal of Human Genetics. 2003;73:939–947. doi: 10.1086/378419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Jiang J, Hofmann MC, Dym M. Gfra1 silencing in mouse spermatogonial stem cells results in their differentiation via the inactivation of RET tyrosine kinase. Biology of Reproduction. 2007;77:723–733. doi: 10.1095/biolreprod.107.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann MC, Braydich-Stolle L, Dym M. Isolation of male germ-line stem cells; influence of GDNF. Developmental Biology. 2005;279:114–124. doi: 10.1016/j.ydbio.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB, Sinclair DA, Guarente L. Molecular biology of aging. Cell. 1999;96:291–302. doi: 10.1016/s0092-8674(00)80567-x. [DOI] [PubMed] [Google Scholar]
- Kidd SA, Eskenazi B, Wyrobek AJ. Effects of male age on semen quality and fertility: a review of the literature. Fertility and Sterility. 2001;75:237–248. doi: 10.1016/s0015-0282(00)01679-4. [DOI] [PubMed] [Google Scholar]
- Kokkinaki M, Lee TL, He Z, Jiang J, Golestaneh N, Hofmann MC, Chan WY, Dym M. The molecular signature of spermatogonial stem cells in the 6-day-old mouse testis. Biology of Reproduction. 2009;80:707–717. doi: 10.1095/biolreprod.108.073809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolevzon A, Gross R, Reichenberg A. Prenatal and perinatal risk factors for autism: a review and integration of findings. Archives of Pediatrics & Adolescent Medicine. 2007;161:326–333. doi: 10.1001/archpedi.161.4.326. [DOI] [PubMed] [Google Scholar]
- Kostereva N, Hofmann MC. Regulation of the spermatogonial stem cell niche. Reproduction in Domestic Animals. 2008;43(Supplement 2):386–392. doi: 10.1111/j.1439-0531.2008.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. PNAS. 2003;100:6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitas E, Lunenfeld E, Weisz N, Friger M, Potashnik G. Relationship between age and semen parameters in men with normal sperm concentration: analysis of 6022 semen samples. Andrologia. 2007;39:45–50. doi: 10.1111/j.1439-0272.2007.00761.x. [DOI] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Molecular Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Malaspina D, Harlap S, Fennig S, Heiman D, Nahon D, Feldman D, Susser ES. Advancing paternal age and the risk of schizophrenia. Archives of General Psychiatry. 2001;58:361–367. doi: 10.1001/archpsyc.58.4.361. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. 1376, 1378–1379. [PubMed] [Google Scholar]
- Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biology of Reproduction. 2006;74:314–321. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. PNAS. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Oatley MJ, Avarbock MR, Tobias JW, Brinster RL. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development. 2009;136:1191–1199. doi: 10.1242/dev.032243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbo K, Yoshida S, Ohmura M, Ohneda O, Ogawa T, Tsuchiya H, Kuwana T, Kehler J, Abe K, Scholer HR, et al. Identification and characterization of stem cells in prepubertal spermatogenesis in mice small star, filled. Developmental Biology. 2003;258:209–225. doi: 10.1016/s0012-1606(03)00111-8. [DOI] [PubMed] [Google Scholar]
- Ohmura M, Yoshida S, Ide Y, Nagamatsu G, Suda T, Ohbo K. Spatial analysis of germ stem cell development in Oct-4/EGFP transgenic mice. Archives of Histology and Cytology. 2004;67:285–296. doi: 10.1679/aohc.67.285. [DOI] [PubMed] [Google Scholar]
- Rahman A, Anwar KN, Malik AB. Protein kinase C-zeta mediates TNF-α-induced ICAM-1 gene transcription in endothelial cells. American Journal of Physiology Cell Physiology. 2000;279:C906–C914. doi: 10.1152/ajpcell.2000.279.4.C906. [DOI] [PubMed] [Google Scholar]
- Reichenberg A, Gross R, Weiser M, Bresnahan M, Silverman J, Harlap S, Rabinowitz J, Shulman C, Malaspina D, Lubin G, et al. Advancing paternal age and autism. Archives of General Psychiatry. 2006;63:1026–1032. doi: 10.1001/archpsyc.63.9.1026. [DOI] [PubMed] [Google Scholar]
- de Rooij DG. The spermatogonial stem cell niche. Microscopy Research and Technique. 2009;72:580–585. doi: 10.1002/jemt.20699. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Grootegoed JA. Spermatogonial stem cells. Current Opinion in Cell Biology. 1998;10:694–701. doi: 10.1016/s0955-0674(98)80109-9. [DOI] [PubMed] [Google Scholar]
- Ryan GR, Dai XM, Dominguez MG, Tong W, Chuan F, Chisholm O, Russell RG, Pollard JW, Stanley ER. Rescue of the colony-stimulating factor 1 (CSF-1)-nullizygous mouse (Csf1(op)/Csf1(op)) phenotype with a CSF-1 transgene and identification of sites of local CSF-1 synthesis. Blood. 2001;98:74–84. doi: 10.1182/blood.v98.1.74. [DOI] [PubMed] [Google Scholar]
- Ryu BY, Orwig KE, Oatley JM, Avarbock MR, Brinster RL. Effects of aging and niche microenvironment on spermatogonial stem cell self-renewal. Stem Cells. 2006;24:1505–1511. doi: 10.1634/stemcells.2005-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biology of Reproduction. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Shinohara T, Avarbock MR, Brinster RL. β1- and α6-integrin are surface markers on mouse spermatogonial stem cells. PNAS. 1999;96:5504–5509. doi: 10.1073/pnas.96.10.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloter E, Schmid TE, Marchetti F, Eskenazi B, Nath J, Wyrobek AJ. Quantitative effects of male age on sperm motion. Human Reproduction. 2006;21:2868–2875. doi: 10.1093/humrep/del250. [DOI] [PubMed] [Google Scholar]
- Tsitouras PD. Effects of age on testicular function. Endocrinology and Metabolism Clinics of North America. 1987;16:1045–1059. [PubMed] [Google Scholar]
- Wilkin DJ, Szabo JK, Cameron R, Henderson S, Bellus GA, Mack ML, Kaitila I, Loughlin J, Munnich A, Sykes B, et al. Mutations in fibroblast growth-factor receptor 3 in sporadic cases of achondroplasia occur exclusively on the paternally derived chromosome. American Journal of Human Genetics. 1998;63:711–716. doi: 10.1086/302000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM, Baxendale V, Chen Y, Pang AL, Stitely T, Munson PJ, Leung MY, Ravindranath N, Dym M, Rennert OM, et al. Analysis of mouse germ-cell transcriptome at different stages of spermatogenesis by SAGE: biological significance. Genomics. 2004;84:971–981. doi: 10.1016/j.ygeno.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nabeshima Y. A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science. 2007;317:1722–1726. doi: 10.1126/science.1144885. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ebata KT, Robaire B, Nagano MC. Aging of male germ line stem cells in mice. Biology of Reproduction. 2006;74:119–124. doi: 10.1095/biolreprod.105.045591. [DOI] [PubMed] [Google Scholar]
- Zhou X, Perez F, Han K, Jurivich DA. Clonal senescence alters endothelial ICAM-1 function. Mechanisms of Ageing and Development. 2006;127:779–785. doi: 10.1016/j.mad.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Zubkova EV, Robaire B. Effects of ageing on spermatozoal chromatin and its sensitivity to in vivo and in vitro oxidative challenge in the Brown Norway rat. Human Reproduction. 2006;21:2901–2910. doi: 10.1093/humrep/del193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


